Background: MTDH is associated with poor prognosis in cancer, yet its biologic function is unclear.
Results: Mtdh-null male mice are infertile, with a lack of mature sperm in testes and altered expression of small non-coding RNAs.
Conclusion: Mtdh deficiency results in male infertility due to impaired spermatogenesis.
Significance: The oncogene MTDH plays an indispensable role in male fertility.
Keywords: Development, DNA Damage, Meiosis, MicroRNA (miRNA), Reproduction, Spermatogenesis, MTDH, Male Infertility, Pachytene, piRNA, AEG-1, Mili, Rad18
Abstract
Increased expression of metadherin (MTDH, also known as AEG-1 and 3D3/LYRIC) has been associated with drug resistance, metastasis, and angiogenesis in a variety of cancers. However, the specific mechanisms through which MTDH is involved in these processes remain unclear. To uncover these mechanisms, we generated Mtdh knock-out mice via a targeted disruption of exon 3. Homozygous Mtdh knock-out mice are viable, but males are infertile. The homozygous male mice present with massive loss of spermatozoa as a consequence of meiotic failure. Accumulation of γ-H2AX in spermatocytes of homozygous Mtdh knock-out mice confirms an increase in unrepaired DNA breaks. We also examined expression of the DNA repair protein Rad18, which is regulated by MTDH at the post-transcriptional level. In testes from Mtdh exon 3-deficient mice, Rad18 foci were increased in the lumina of the seminiferous tubules. The Piwi-interacting RNA (piRNA)-interacting protein Mili was expressed at high levels in testes from Mtdh knock-out mice. Accordingly, genome-wide small RNA deep sequencing demonstrated altered expression of piRNAs in the testes of Mtdh knock-out mice as compared with wild type mice. In addition, we observed significantly reduced expression of microRNAs (miRNAs) including miR-16 and miR-19b, which are known to be significantly reduced in the semen of infertile men. In sum, our observations indicate a crucial role for MTDH in male fertility and the DNA repair mechanisms required for normal spermatogenesis.
Introduction
Metadherin (MTDH2; also known as AEG-1 and 3D3/LYRIC) has been implicated in many cancer-related processes, including cellular proliferation, survival, invasion, chemoresistance, and metastasis (1–3). MTDH is located on chromosome 8q22, a region that is frequently amplified in cancers and is correlated with poor survival in patients with breast cancer and hepatocellular carcinoma (4). MTDH is overexpressed in a wide variety of cancers and in 90% of cases of hepatocellular carcinoma (5). Other mechanisms that are known to cause the up-regulation of MTDH include reduced post-transcriptional regulation by miR-375, increased stabilization by monoubiquitination, and interaction with cytoplasmic polyadenylation element-binding protein 1 (CEBP1) (6, 7).
MTDH is a poorly understood protein with few conserved domains that indicate its biological function. There are, however, three lysine-rich nuclear localization signals and several regions required to interact with other proteins such as PLZF (promyelocytic leukemia zinc finger) (8), BCCIPα (BRCA2 and CDKN1A-interacting protein α isoform) (9), SND1 (staphylococcal nuclease and Tudor domain-containing 1) (10, 11), and NFκB subunit p65 (12). Thus, although the underlying biological functions of MTDH have not been completely elucidated, others have suggested that it acts in the nucleus as a co-factor to promote the transcription of downstream genes involved in metastasis and drug resistance (1, 2, 12). Alternatively, several studies have identified roles for MTDH in regulation of protein translation. The interaction between MTDH and SND1 facilitates RNAi-mediated gene silencing in the cytoplasm. MTDH also increases the translation of multidrug resistance protein 1 (MDR1) and coagulation factor XII (FXII) proteins by promoting loading of their respective mRNAs onto the polysome (13–15). We recently reported that MTDH contains several putative RNA binding sites and that it interacts with a large number of mRNAs and ribosomal proteins in the cytoplasm (16).
As the vast majority of studies have been performed in malignant cells, the primary function(s) of MTDH in non-cancerous cells remains to be determined. MTDH was originally reported to be induced by HIV infection in astrocytes (accounting for its original name, AEG-1, astrocyte elevated gene 1) and as a lysine-rich CEACAM1 co-isolated (LYRIC) protein associated with tight junctions in prostate epithelial cells (17). Unlike cancer cells where the predominant location of MTDH is in the cytoplasm, MTDH is also present in the nucleus and nucleolus of normal cells (15). Although MTDH has been implicated in diverse physiological and pathological processes (18, 19), the precise mechanisms remain undefined. Using Mtdh exon 3-deficient mice, we herein report a novel functional role for MTDH in male fertility through the regulation of small non-coding RNAs and spermatogenesis.
EXPERIMENTAL PROCEDURES
Generation of Mtdh Knock-out Mice
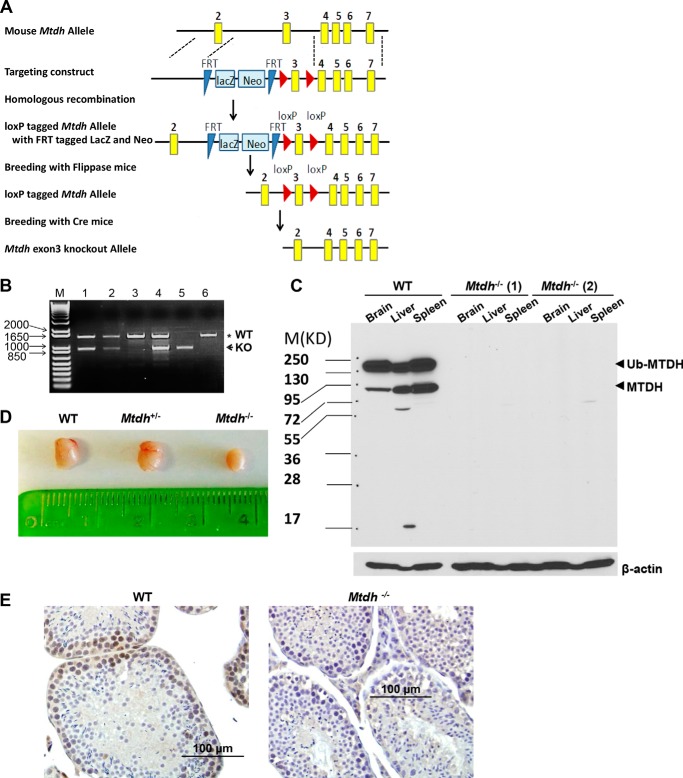
Chimeric mice were generated by injecting the mouse embryonic stem cell (ESC) line generated by the International Knock-out Mouse Consortium (IKMC) (Mtdh, project CSD 48311) into the blastocysts of mouse embryos. High-contribution chimeras were mated with C57BL/6 females to confirm germline transmission of the ESCs. Mtdh heterozygous mice were bred with FLPe (The Jackson Laboratory, Stock number: 003800) and Cre (The Jackson Laboratory, Stock number: 003724) mice to delete the reporter genes and exon 3 of Mtdh, respectively (see Fig. 1A). Mtdh heterozygous knock-out mice were genotyped by PCR using Mtdh-specific genotype primers (5′-TGGAAAATGATGGTGGATTG-3′; 5′-CACGTTTACGCTGTTGTCGT-3′) and were interbred to obtain Mtdh homozygous knock-out mice. The Mtdh heterozygous knock-out mice were maintained by intercrosses on the C57BL/6 background. Animal protocol 4071085 has been approved by Institutional Animal Care and Use Committee Office of Animal Resources (through the Animal Care and Use Review Form (ACURF)).
FIGURE 1.
Generation of Mtdh knock-out mice by targeted deletion of exon 3. A, promoterless targeting cassettes for the generation of a “KO first allele” in C57BL/6N embryonic stem cells were used to target Mtdh exon 3, an exon common to all transcript variants that, when deleted, creates a frameshift mutation. The KO first allele produced reporter-null alleles following subsequent exposure to site-specific recombinases Flp and Cre. B, representative genotyping of litters obtained by crossing Mtdh+/− mice. Mtdh+/+: lanes 3 and 6; Mtdh+/−: lanes 1, 2, and 4; Mtdh−/−: lane 5. M, molecular weight markers. C, Mtdh expression in liver, brain, and spleen lysates from Mtdh+/+ and two different Mtdh−/− mice, denoted as 1 and 2. Arrowheads indicate full-length Mtdh and ubiquitinated (Ub) Mtdh. D, representative images of testes from WT, heterozygous, and homozygous Mtdh exon 3-deficient adult male mice. E, representative images of Mtdh expression by immunohistochemistry in testes from Mtdh+/+ and Mtdh−/− mice.
Western Blotting
The following antibodies were used: anti-MTDH (40-6500, Invitrogen); anti-β-actin (A1978, Sigma); anti-Rad51 (PC130, EMD Millipore); and anti-Mili and anti-Miwi (5940 and 6915, Cell Signaling Technology). Whole-cell protein lysates were prepared and analyzed by Western blotting as described previously (16).
Histology
Formalin-fixed, paraffin-embedded (FFPE) testes sections from WT or homozygous Mtdh exon 3 knock-out mice were stained using H&E, dehydrated using three washes of ethanol and xylene, mounted with coverslips, and analyzed by a pathologist blinded to the genotype.
Immunohistochemistry
Expression of Mtdh was analyzed in FFPE sections of testes or ovary from WT or homozygous Mtdh exon 3 knock-out mice using anti-MTDH antibody as described previously (20).
Quantitative RT-PCR
mRNA levels of Rad18 were examined in testes and liver tissue isolated from WT or homozygous Mtdh exon 3 knock-out mice by quantitative RT-PCR (RT-qPCR) using the SYBR Green-based real time RT-qPCR assay (Applied Biosystems). Primers specific for Rad18 were from Qiagen (Mm_Rad18_1_SG). Data were normalized to β-actin mRNA levels. Levels of miR-101, -16, -182, -19, and -340 were analyzed in liver lysates from WT or homozygous Mtdh exon 3 knock-out mice by TaqMan microRNA RT-qPCR kit (Applied Biosystems) per the manufacturer's instructions. Data were normalized to U6.
FISH
An X/Y-specific centromeric FISH probe (Vysis Inc.) was used to hybridize FFPE testes sections according to the manufacturer's protocol.
Electron Microscopy
Buffered glutaraldehyde (2.5%)-fixed, paraffin-embedded testes sections from WT or homozygous Mtdh exon 3 knock-out mice were analyzed by transmission electron microscopy for ultrastructure of organelles.
Immunostaining
FFPE testes sections from WT or homozygous Mtdh exon 3 knock-out mice were hydrated, deparaffinized, and stained with specific primary antibodies Mili, Miwi, SYCP3 (ab97672, Abcam), SYCP1 (ab15090, Abcam), Rad18 (ab188235, Abcam), and γ-H2AX (2577, Cell Signaling Technology), followed by staining with Alexa Fluor 546-conjugated anti-rabbit or Alexa Fluor 488-conjugated anti-mouse secondary antibodies and visualization by confocal microscopy. Nuclei were counterstained with DAPI (Vector Laboratories) or TO-PRO 3 iodide (Invitrogen).
miRNA Sequencing and Data Analysis by Illumina HiSeq 2000
Total RNA from individual testis of two WT mice and three homozygous Mtdh exon 3-depleted mice was used to prepare the miRNA sequencing library by the following steps: 1) 3′-adapter ligation with T4 RNA ligase 2 (truncated); 2) 5′-adapter ligation with T4 RNA ligase; 3) cDNA synthesis with RT primer; 4) PCR amplification; and 5) separation by PAGE, extraction, and purification of ∼135–155-bp PCR-amplified fragments (corresponding to ∼15–35-nt small RNAs). After the completed libraries were quantified with an Agilent 2100 bioanalyzer, the DNA fragments in the libraries were denatured with 0.1 m NaOH to generate single-stranded DNA molecules, captured on Illumina flow cells, amplified in situ, and sequenced for 36 cycles on an Illumina HiSeq 2000 according to the manufacturer's instructions. Image analysis and base calling were performed using Off-Line Basecaller software (OLB V1.8.0). Subsequently, 3′-adapter sequences were trimmed from clean reads (reads that passed a Solexa CHASTITY quality filter), and any reads shorter than 15 nt were discarded. Next the 3′-adapter-trimmed-reads (≥15 nt) were aligned to the latest known human reference miRNA precursor set (Sanger miRBase 19) using NovoAlign (v2.07.11). Reads (counts < 2) were discarded when calculating miRNA expression. To characterize the miRNA isoform variability, any sequences that matched the miRNA precursors in the mature miRNA region ±4 nt (no more than one mismatch) were accepted as mature miRNA isoforms, which were grouped according to the 5-prime (5p) or 3-prime (3p) arm of the precursor hairpin. The full dataset has been deposited at GEO (accession number GSE62330).
piRNA Sequencing and Data Analysis by Illumina HiSeq 2000
Total testicular RNA from two WT mice and three homozygous Mtdh exon 3-deficient mice was sequenced individually by the same procedures used for miRNAs. Specifically, the 3′-adapter-trimmed reads (length ≥15 nt) were aligned to the latest piRNA set in piRNABank using NovoAlign software (v2.07.11). Expression of each piRNA was defined as the mapped tag counts. The full dataset has been deposited at GEO (accession number GSE62330).
Statistical Analysis
Differentially expressed miRNAs or piRNAs between WT mice and homozygous Mtdh exon 3-depleted mice were identified through t test filtering (-fold change ≥1.5, p value ≤ 0.05). For RT-qPCR experiments, statistical significance was assessed using Student's t test, and a p value <0.05 was considered statistically significant.
RESULTS
Mtdh Exon 3-deficient Male Mice Are Sterile
To generate Mtdh-knock-out mice, we used the mouse ESC line generated by IKMC (Mtdh, project CSD 48311), which contains loxP sites flanking exon 3 of Mtdh (Fig. 1A). Injection of CSD 48311 ES cells into blastocysts generated chimeric mice, and germline transmission was confirmed (Fig. 1A). Mtdh heterozygous knock-out (Mtdh+/−) mice were bred with FLPe mice to delete the reporter genes and with Cre mice to delete exon 3 of Mtdh (Fig. 1B). Mtdh+/− mice were interbred to obtain Mtdh homozygous knock-out (Mtdh−/−) mice. In Mtdh+/+ adult mice, Mtdh protein was detected in a variety of tissues, including liver, brain, and spleen, whereas it was undetectable in Mtdh−/− tissues (Fig. 1C). These data indicate that this deletion strategy results in a truncated Mtdh protein that either is not expressed or is unstable.
Unexpectedly, breeding Mtdh+/− male and female mice produced Mtdh−/− offspring at Mendelian ratios significantly below the expected ratio (Mtdh+/+, 52.7%; Mtdh+/−, 41.3%; Mtdh−/−, 6%; supplemental Table S1A). The surviving male Mtdh−/− mice were found to be infertile (supplemental Table S1B) and significantly smaller as compared with their WT or heterozygous littermates (data not shown). In addition, Mtdh−/− males had smaller testes than WT and heterozygotes (Fig. 1D). Female Mtdh−/− mice were also smaller in weight than other littermates (data not shown). Crossing Mtdh−/− female mice with Mtdh+/− male mice resulted in only 9.5% Mtdh−/− offspring (supplemental Table S1C), indicating that the female Mtdh−/− mice are subfertile.
Data available through the Human Protein Atlas demonstrate expression of MTDH in seminiferous ducts, Leydig cells, follicle cells, and ovarian stroma cells of human origin. We confirmed that testes from WT mice retain Mtdh expression by immunohistochemistry, whereas testes from Mtdh−/− male mice are devoid of Mtdh (Fig. 1E). Similar results were observed with ovaries from WT and Mtdh−/− female mice (supplemental Fig. S1).
Morphological Alterations of the Testes from Mtdh Exon 3-deficient Mice
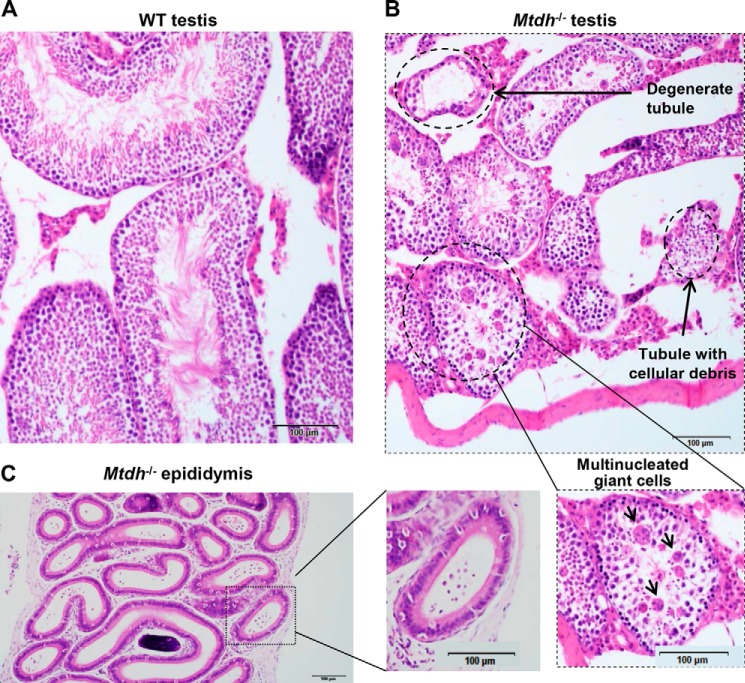
To determine the mechanism of infertility in Mtdh−/− male mice, H&E staining of testes from WT, Mtdh+/−, and Mtdh−/− mice was performed. No significant morphological differences were observed between WT and heterozygous Mtdh exon 3-deficient mice. However, all four of the homozygous Mtdh exon 3-deficient mice had moderate to severe multifocal to coalescing testicular degeneration characterized by the accumulation of multinucleated giant cells within the seminiferous tubules and a lack of spermatozoa within the lumina of the seminiferous tubules (Fig. 2, A and B). The epithelial lining of the seminiferous tubules in Mtdh−/− testes was intact and composed of the appropriate germ cell types along with scattered Sertoli cells. By contrast, there was a complete lack of spermatozoa and rare spermatids within the majority of Mtdh−/−seminiferous tubules and epididymis (Fig. 2, B and C).
FIGURE 2.
Testes from homozygous Mtdh exon 3-deficient male mice display testicular degeneration and a lack of mature spermatozoa. A and B, morphology of testes from WT mice (A) and homozygous Mtdh exon 3-deficient mice (B) by H&E staining. Multinucleated giant cells are indicated by arrows. C, H&E staining of a representative section of the epididymis from an Mtdh exon 3-deficient mouse.
To further examine morphological changes in testes associated with Mtdh deficiency, we performed electron microscopy. Both chromatoid bodies (Fig. 3A) and inter-mitochondrial cement (also called inter-mitochondrial material/bar/cloud, Fig. 3B) were observed in WT and homozygous Mtdh exon 3-deficient testicles. However, multinuclear cells were only observed in homozygous Mtdh exon 3-deficient testicles (Fig. 3C). Prominent clusters of mitochondria were also observed in the multinuclear giant cells in homozygous Mtdh exon 3-deficient testicles as opposed to the random distribution of mitochondria in WT testes (Fig. 3D). Vacuolated vesicles were observed in homozygous Mtdh exon 3-deficient testes but not in WT testes (Fig. 3, A and D).
FIGURE 3.
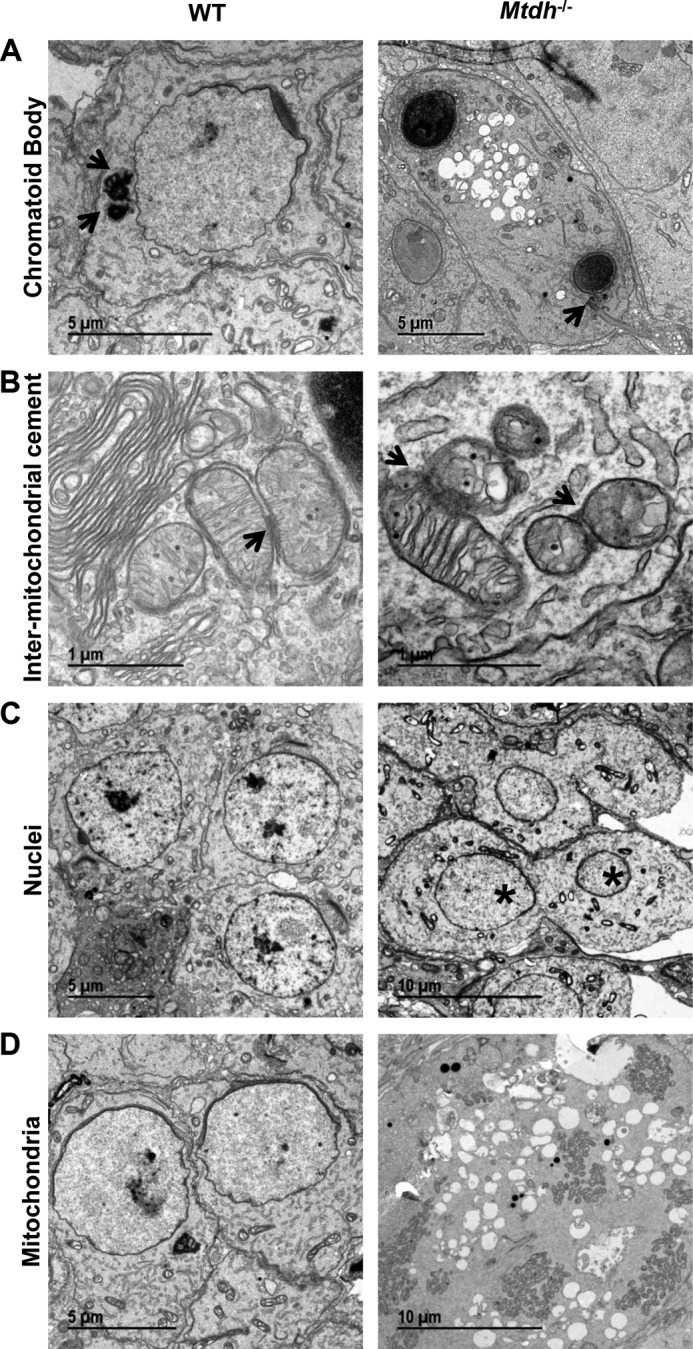
Deficiency of Mtdh in testes is characterized by vacuolated vesicles, multinuclear cells, and mitochondrial clusters. Testes sections from WT or Mtdh−/− mice were subjected to electron microscopy for visualization of ultrastructure. A, chromatoid bodies as indicated by arrows. Note two condensed nuclei and vacuolated bulbs in testes from Mtdh exon 3-deficient mice. B, inter-mitochondria cement as indicated by arrows. C, multinuclear cells were detected only in Mtdh−/− testes. * indicates two nuclei within a single cell. D, mitochondrial clusters were observed in Mtdh knock-out giant cells, whereas WT cells displayed random mitochondrial distribution.
Synapsis Is Impaired in Homozygous Mtdh Exon 3-deficient Spermatocytes
SYCP3 is a synaptonemal complex marker, highly expressed in zygotene spermatocytes and confined to the axes of paired chromosomes during pachytene (21). To determine whether Mtdh−/− spermatocytes have impaired progression through prophase I during meiosis, the expression and localization patterns of SYCP3 were determined. SYCP3 was mainly present in the epithelial lining of the seminiferous tubules in WT mice (Fig. 4A). However, SYCP3 expression was dramatically higher both in the epithelial lining and near the lumina of the seminiferous tubules of Mtdh exon 3-deficient mice, indicating that many spermatocytes accumulated at pachytene of meiosis prophase I (Fig. 4B). We also analyzed expression of SYCP1, another component of the synaptonemal complex, in Mtdh exon 3-deficient testes. As shown in Fig. 4C, the staining pattern of SYCP1 is similar to that observed with SYCP3. These data support that deletion of Mtdh results in accumulation of spermatocytes in pachytene.
FIGURE 4.
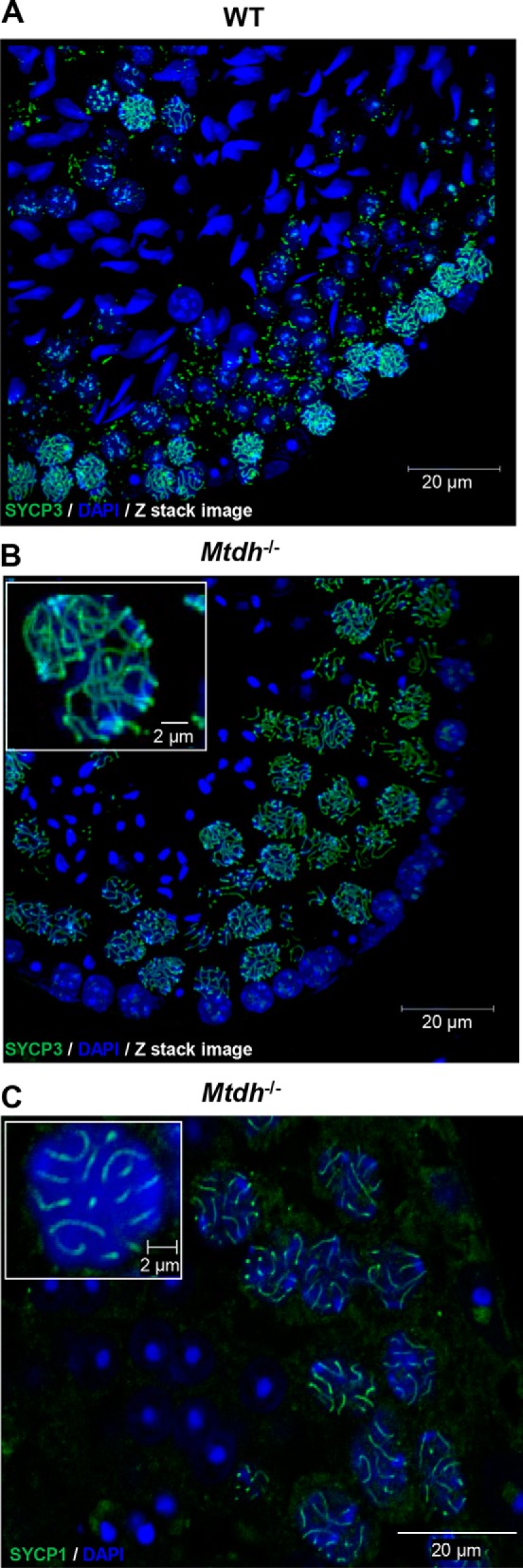
Mtdh exon 3-deficient spermatocytes accumulate at pachytene of meiosis prophase I. A and B, immunofluorescence detecting the synaptonemal complex marker, SYCP3, in WT (A) and Mtdh−/− (B) seminiferous tubules. Green, SYCP3; blue, nuclei (DAPI). Abnormal accumulation of SYCP3-positive spermatocytes was seen in Mtdh−/− seminiferous tubules. C, immunofluorescence detecting another synaptonemal complex marker, SYCP1, in Mtdh−/− seminiferous tubules. Green, SYCP1; blue, nuclei (DAPI). In B and C, insets are enlarged images to depict SYCP3 or SYCP1 staining in single cells from Mtdh−/− seminiferous tubules.
piRNA-interacting Protein Mili Is Overexpressed in the Testes of Mtdh Exon 3-deficient Mice
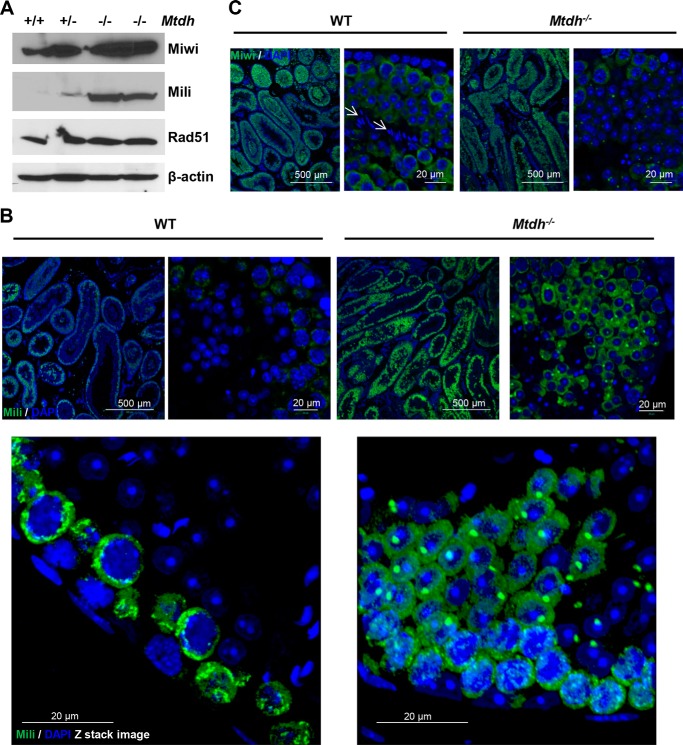
Previous studies have demonstrated that MTDH associates with SND1 to regulate the activity of the RNA-induced silencing complex (RISC) in the cytoplasm (11). The MTDH/SND1 interaction promotes oncogenic miRNA-mediated degradation of some tumor suppressor mRNAs during carcinogenesis (11). SND1 belongs to the Tudor family and shares a common Tudor domain. Multiple members of the Tudor family, including Tdrd1, Tdrd5, and Tdrd9, are required for spermatocyte development (22). Similar to MTDH and SND1, the Piwi family of genes plays important roles in RNA silencing and translational regulation during the meiotic differentiation of spermatocytes (23). Mili is one of three mouse homologs of Piwi (23). Previous observations from Mili-null mice indicate that Mili plays an essential role in meiotic differentiation of spermatocytes (24). Others have confirmed that spermatogenesis is blocked completely at zygotene of meiosis I in Mili-null mice, causing male infertility (24–26). Because Mtdh exon 3-deficient mice also display defective spermatogenesis, we examined Mili expression in WT and Mtdh exon 3-deficient testes. Interestingly, Mili expression was dramatically increased in Mtdh exon 3-deficient testes (Fig. 5A) as compared with WT and heterozygotes, indicating a correlation with accumulation of spermatocytes at pachytene when Mili is normally expressed. Similar to SYCP3 staining, increased expression of Mili was observed in multiple layers both in the epithelial lining and near the lumina of the seminiferous tubules in homozygous Mtdh exon 3-deficient mice as compared with WT (Fig. 5B). In contrast, no significant alteration of Miwi expression was observed in homozygous Mtdh exon 3-deficient testes as compared with WT (Fig. 5C).
FIGURE 5.
Expression of piRNA-associated protein Mili is increased in testes of Mtdh−/− mice. A, expression of Miwi, Mili, and Rad51 in testes lysates from WT (+/+), heterozygous (+/−), and homozygous Mtdh knock-out mice (−/−) by Western blotting. B, Mili expression in testes from WT and Mtdh−/− mice by immunofluorescent staining. Green, Mili; blue, nuclei (DAPI). C, Miwi expression in testes from WT and Mtdh−/− mice by immunofluorescent staining. Arrows indicate nuclei of sperm in WT testes, which are devoid of Miwi staining. Green, Miwi; blue, nuclei (DAPI), blue.
Expression of miRNAs and piRNAs Is Altered in the Testes of Mtdh Exon 3-deficient Mice
Both MTDH and Mili play critical roles in the function of non-coding small RNAs (23). To identify the effect of Mtdh exon 3 depletion on the expression levels of miRNAs and piRNAs, these were assayed from homozygous Mtdh exon 3-deficient testicles and WT adult testicles. Reduced expression of miRNAs was detected in homozygous Mtdh exon 3-deficient testes (Table 1). By contrast, miRNA levels were not reduced in the liver from Mtdh exon 3-deficient as compared with WT mice (supplemental Table S2), indicating that the observed miRNA changes in testes are germ cell-specific. As shown in Table 2, piRNA expression levels were also dysregulated, with some being reduced and others increased in homozygous Mtdh exon 3-deficient testes as compared with WT mice.
TABLE 1.
Deficiency of Mtdh results in miRNA down-regulation in testes
The full dataset has been deposited at GEO (accession number GSE62330).
| miRNA | Average copy number |
Mtdh−/−
vs. WT |
||
|---|---|---|---|---|
| Mtdh−/− | WT | -Fold change | p value | |
| mmu-miR-101b-3p | 309 | 485 | 0.64 | 0.012 |
| mmu-miR-106b-3p | 148 | 257 | 0.57 | 0.034 |
| mmu-miR-16–2–3pa | 11 | 20 | 0.55 | 0.006 |
| mmu-miR-182–5p | 637 | 1022 | 0.62 | 0.040 |
| mmu-miR-19b-3pa | 473 | 1250 | 0.38 | 0.016 |
| mmu-miR-28a-3p | 35 | 60 | 0.59 | 0.008 |
| mmu-miR-293–5p | 12 | 24 | 0.49 | 0.0105 |
| mmu-miR-340–5p | 905 | 1561 | 0.58 | 0.0009 |
a miRNAs that are also down-regulated in infertile men (34).
TABLE 2.
Deficiency in Mtdh results in altered expression of piRNAs in testes
The full dataset has been deposited at GEO (accession number GSE62330).
| piRNA ID | piRNA Sequence | Average copy number |
Mtdh−/−
vs. WT |
||
|---|---|---|---|---|---|
| Mtdh−/− | WT | -Fold change | p value | ||
| Up-regulated piRNAs in Mtdh−/− testes | |||||
| mmu_piR_000219 | ACGGGAAACCTCACCCGGCCCGGACAC | 118.63 | 24 | 4.94 | 0.026 |
| mmu_piR_006207 | TTAAAAAGATTTTAAAAAGTGCTATTGAGG | 224.57 | 80.2 | 2.80 | 0.042 |
| mmu_piR_033479 | TTAAAAATTTTAATACTGGGGACTGAAAA | 68.57 | 27.05 | 2.531 | 0.028 |
| mmu_piR_031363 | TAAAATGTCCAGGACATTTCACTGAGGGAC | 558.43 | 230.2 | 2.43 | 0.011 |
| mmu_piR_023012 | TAGGACAAAATGAAGATCCCAGAAGGCTCT | 1584.7 | 653.85 | 2.42 | 0.030 |
| mmu_piR_038202 | TAGGCTAAGTGACAACAAGTGGAGAAAGGGC | 368.57 | 152.45 | 2.42 | 0.007 |
| mmu_piR_022877 | CTGAAATGAAGAGAATACTCTTGCTGATC | 188.2 | 78.9 | 2.38 | 0.024 |
| mmu_piR_002024 | TGAAAAGTGTACAGAGTAGAGAAAAAGGG | 68.1 | 28.7 | 2.37 | 0.034 |
| mmu_piR_019994 | TGAGAAATGATCATAAAAGTGAAGGGCTAG | 370.8 | 157.25 | 2.36 | 0.011 |
| mmu_piR_009031 | TCACAGTAGCCTGGTGACCGTTACCAACTC | 55.23 | 23.85 | 2.32 | 0.033 |
| mmu_piR_021054 | TGGGAAGTGGGATAGGCTTGCTGAGCTG | 35.67 | 15.7 | 2.27 | 0.021 |
| mmu_piR_038322 | AAGAACGAAAGTCGGAGGTTCGAAGACGATC | 35.57 | 15.75 | 2.26 | 0.009 |
| mmu_piR_022855 | TGGGTTGTCTGTAATGGCCTTTTTCTTGGGC | 61.87 | 28.95 | 2.13 | 0.019 |
| mmu_piR_008329 | TAGGGTCTGTGTTTGTTGCTTTAGTCTTT | 39.03 | 18.65 | 2.09 | 0.039 |
| Down-regulated piRNAs in Mtdh−/− testes | |||||
| mmu_piR_009188 | TCAGTCCTTGACTGAGAGCCTCGTTCTGCCC | 188.93 | 451.1 | −2.39 | 0.017 |
| mmu_piR_029292 | TGTAAATCTGTATGGTTATTAAGGTTGGC | 14.7 | 30 | −2.04 | 0.0357 |
| mmu_piR_031889 | TAAACATGAGTTGTACTCTGTAGAGAGCCT | 129.33 | 250.6 | −1.94 | 0.0437 |
| mmu_piR_027538 | TATGTAGATTGTCATGGCGCTCTGCTCAG | 16 | 30.5 | −1.91 | 0.0202 |
| mmu_piR_005912 | TGTGCTTCATTCTGTGGATCTGACATGCCTT | 404.4 | 750 | −1.85 | 0.0115 |
| mmu_piR_014481 | TGTGATTAAAGGCGTGCACTACCATGCTGG | 23.23 | 42.9 | −1.85 | 0.005 |
| mmu_piR_002469 | TGACCTTCAGGCTAAACTCCTCCCAGTTA | 16 | 29.2 | −1.83 | 0.010 |
| mmu_piR_034273 | TGATCTAGTCAAGTGGCTCTGAGCAGAC | 12.47 | 22.7 | −1.82 | 0.0007 |
| mmu_piR_008406 | TAGGTTCTGATGGAGCATTGTTAGCAGGC | 18.1 | 32.75 | −1.81 | 0.0067 |
| mmu_piR_039588 | TCATCTTCTCTGACGACTCTGGGCATTGG | 17.6 | 31.45 | −1.79 | 0.0087 |
| mmu_piR_037472 | TCATGTTTTCTGATACATGGATCTGTACTC | 13.23 | 23.3 | −1.76 | 0.0053 |
| mmu_piR_012706 | TGATTGATTGGCTATAGTCCTTGAATGGC | 13.83 | 24.35 | −1.76 | 0.0071 |
| mmu_piR_016098 | TGATTGTGGATGTGATGTGACCCGCACCA | 37.7 | 65.8 | −1.75 | 0.0019 |
| mmu_piR_004659 | TGGCAATAAAGGGTTGAAAGCACACAGC | 11.7 | 20.05 | −1.71 | 0.0204 |
| mmu_piR_014665 | TGGGAATTTGATAGGATTTCACAGCTGTTT | 13.77 | 23.45 | −1.70 | 0.0049 |
| mmu_piR_019387 | TGCCAAAGATCTTTGGAAAGTCCAAGTGGC | 16.67 | 28.3 | −1.70 | 0.0265 |
| mmu_piR_025712 | TTCACTTTGCTATTATTAAAATGGATGACAA | 13.17 | 22.35 | −1.697 | 0.0347 |
| mmu_piR_011190 | TGCCTATGGATCCCAAGCAGGGATGACTGGA | 11.17 | 18.65 | −1.67 | 0.0236 |
| mmu_piR_015224 | TCTTGGAAATATCGAACTCTGAGCCTTGTT | 48.5 | 80.7 | −1.66 | 0.0008 |
| mmu_piR_025912 | TTGTTTTTGTTGACTTTGGATGTGTCCTA | 25.43 | 42.25 | −1.66 | 0.0186 |
| mmu_piR_021121 | TGTTCTAGAATGGAGCCTGAAATAGAAGTC | 52.37 | 85.05 | −1.62 | 0.0296 |
| mmu_piR_001140 | TCCCAAGCTTCTGGTGATCTCTCAATGCC | 15.73 | 25.45 | −1.62 | 0.0152 |
| mmu_piR_035733 | CAGTAAATATGAAAGGATGAAACGCAACAT | 14.03 | 22.65 | −1.61 | 0.0468 |
| mmu_piR_005510 | TGGTTTCTGCAATGCATTGCCATGGAAGG | 10.2 | 16.45 | −1.61 | 0.0428 |
| mmu_piR_000553 | CCCATCTGTTGCACTGTTGATCTTGGCTGT | 41.8 | 67.2 | −1.61 | 0.0302 |
| mmu_piR_019939 | TGAAAGGAACTTTCCTAGAGTTATGCACTGT | 47.4 | 76.2 | −1.61 | 0.0297 |
| mmu_piR_022992 | TAGTCGAGATTTTGTCATCTCAGGGAACC | 19.9 | 31.85 | −1.60 | 0.0391 |
γ-H2AX Is Increased in Homozygous Mtdh Exon 3-deficient Spermatocytes
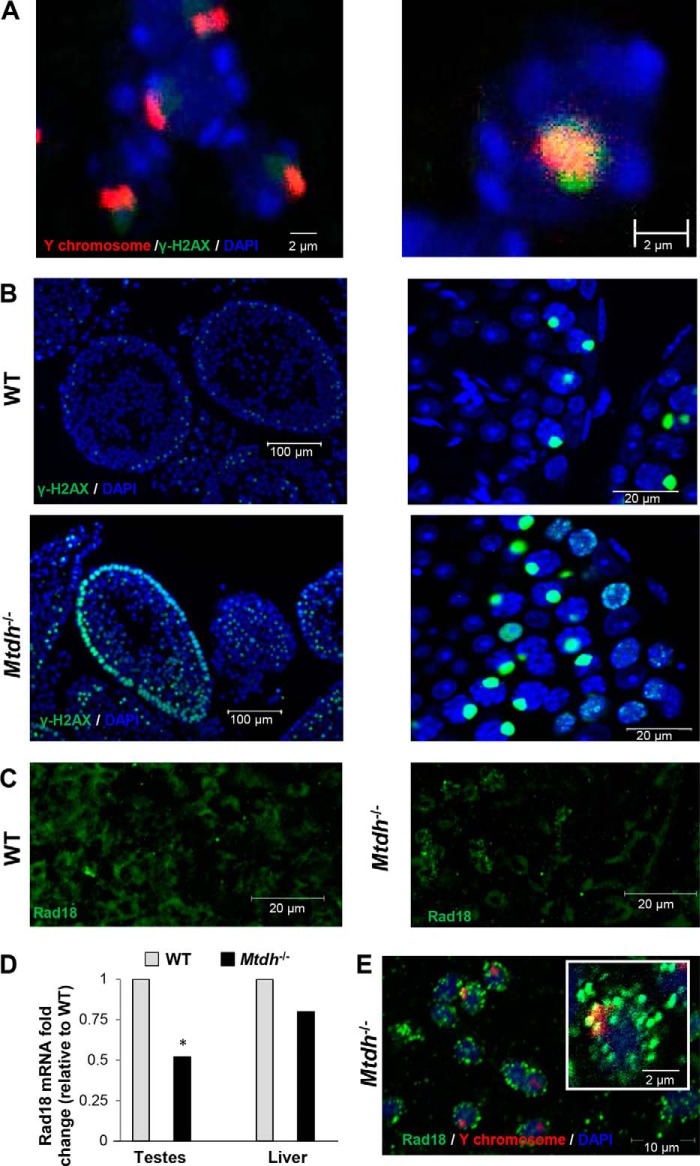
Abnormally high γ-H2AX levels indicate DNA damage in cells. Expression normally increases during leptotene, but then diminishes as DNA damage is repaired during zygotene, eventually becoming confined mainly to the XY body, which is prominent during pachytene (27). The XY body forms as a consequence of chromosomal alignment, particularly the more difficult pairing of the X and Y chromosomes. As a result, the X and Y chromosomes only partially synapse through their pseudo-autosomal regions during normal pachytene (27, 28). The co-localization of γ-H2AX with a Y chromosome FISH probe confirms the accumulation of γ-H2AX at the XY body in testis from a WT mouse (Fig. 6A). In Mtdh−/− testes, abnormally high expression of γ-H2AX was observed in the epithelial lining and near the lumina of the seminiferous tubules as compared with WT testes (Fig. 6B). This is an indicator of ongoing and persistent DNA damage in Mtdh−/− testes.
FIGURE 6.
Alterations in the DNA damage marker γ-H2AX and DNA repair protein Rad18 in testes of Mtdh−/− mice. A, co-localization of γ-H2AX with Y chromosome in seminiferous tubule WT mice. Green, γ-H2AX; red, Y chromosome-specific FISH probe; blue, nuclei (DAPI). B, expression of γ-H2AX is increased in the seminiferous tubule of Mtdh exon 3-deficient mice as compared with WT mice. Green, γ-H2AX; blue, nuclei (DAPI). C, effect of Mtdh deficiency on Rad18 protein levels as detected by immunofluorescent staining in seminiferous tubules. Green, Rad18. D, effect of Mtdh deficiency on Rad18 mRNA levels as detected by RT-qPCR in testes or liver. Data were normalized to β-actin and reported as the -fold change related to WT. p < 0.01 versus WT. E, co-localization of Rad18 with Y chromosome in seminiferous tubule from Mtdh exon 3-deficient mice. Green, Rad18; red, Y chromosome-specific FISH probe; blue, nuclei (DAPI). Inset is an enlarged image to depict punctate staining of Rad18 that does not co-localize with the Y chromosome in Mtdh−/− seminiferous tubules.
Deficiency of Mtdh Modifies DNA Damage Repair Protein Rad18 in Testes
We previously reported that MTDH associates with multiple mRNAs, including the DNA damage repair protein RAD18 (16). Because RAD18 has been implicated in spermatogenesis (29), we examined a possible mechanistic link between MTDH and RAD18 in infertility in Mtdh exon 3-deficient male mice. We found that mRNA and protein levels of Rad18 were reduced in homozygous Mtdh exon 3-deficient testes (Fig. 6, C and D). However, testes from Mtdh exon 3-depleted mice had a pronounced increase in Rad18 foci, specifically near the lumina of the seminiferous tubules. In addition, the majority of the Rad18 foci did not co-localize with Y chromosome probe (Fig. 6E), suggestive of a role for MTDH in DNA damage repair in somatic chromosomes as well as in sex chromosomes.
DISCUSSION
To date, extensive studies of MTDH have identified it as a key driver of cellular behavior, including promoting metastasis and chemoresistance in cancer. However, the function of MTDH in normal physiology is still unknown. Others have reported that in WT mice, Mtdh co-localizes with Ki-67, a biomarker of cell proliferation (19). Mtdh is expressed in mid-to-hindbrain, fronto-nasal processes, limbs, and pharyngeal arches in the early developmental period from E8.5 to E9.5. At stages E12.5–E18.5, Mtdh is localized in the brain, liver, olfactory, and skeletal systems as well as in skin, including hair follicles (19). Herein we identify a role for Mtdh in the regulation of spermatogenesis, male fertility, and the expression of small non-coding RNAs during spermatogenesis. Specifically, we found that Mtdh deletion mediated by exon 3 disruption results in the altered expression profile of small non-coding RNAs, increased expression of Mili, and accumulation of spermatocytes in pachytene uniquely in male mice. As a result, spermatogenesis is disrupted, and depleted mice demonstrate significantly abnormal testicular histology with accumulation of multinucleated giant cells demonstrating abnormal mitochondrial distribution. In addition, Mtdh-null mice largely fail to develop spermatozoa and are therefore infertile. There also appears to be an enhanced loss of Mtdh−/− fetuses in utero as birth Mendelian ratios are skewed, suggesting that Mtdh deficiency is sublethal.
Interestingly, our model differs from the Mtdh knock-out mice generated by Wan et al. (30), who recently reported normal Mendelian ratios and no significant phenotypic alterations. To create the knock-out animals, these investigators used the ESC line XB780 from the Bay Genomics gene trap database, which contains an insertion in the second intron of Mtdh resulting in premature termination of transcription at exon 2. Only a transient delay in ductal outgrowth of mammary glands from 3- and 5-week-old knock-out mice and no significant differences in branching morphogenesis at later time points or during pregnancy and lactation were observed as compared with WT littermates (30). The mice were reported to be fertile. In contrast, our Mtdh knock-out mice, generated through a different targeting strategy, clearly show abnormal Mendelian ratios and have a pronounced sterile phenotype in males. Robertson et al. (5) generated another Mtdh (also called AEG-1) knock-out mouse model with deletion of the promoter region, exon 1, and part of intron 1 of the Mtdh gene using a Cre-loxP system. These mice were viable; however, the litter sizes generated from homozygous Mtdh knock-out mice were very small (1–2 pups per litter). Litters generated by crossing heterozygous Mtdh+/− breeding pairs were also reported to be very small (2–3 pups per litter), indicating that there may be some reproductive and/or sublethal consequences of Mtdh depletion in this model as well.
There are several possible explanations for the differences between the three models. First, our model utilizes the ESC line of EPD0291_7_A08 from IKMC Project 48311 with the deletion of exon 3, which results in an unstable peptide fragment that is degraded. We were unable to detect any Mtdh species (even truncated molecules) in homozygous Mtdh knock-out mice reported herein. The Robertson et al. model (5) targeted the N terminus, and again, no expression was found. In addition, the Wan et al. model (30) utilized an insertion in the first intron to cause premature termination at exon 2, but theoretically, these animals may retain expression of exon 1. If this region is present in the Wan et al. model (30), there are functional implications that could contribute to the differing impact of the deletion strategies. For example, the 162 amino acid residues corresponding to exons 1 and 2, if expressed, contain the transmembrane domain and the first nuclear localization signal motif. This region is also required for the protein-protein interaction between MTDH and BCCIP, PLZF, CBP (CREB-binding protein), and YY1 and is involved in the association of MTDH with mRNA targets (1–3). Recently, the N-terminal region encoded by exon 1 was shown to interact with retinoid X receptor (RXR) and profoundly inhibit retinoid X receptor/retinoic acid receptor (RAR)-mediated transcriptional activation (31). In our model, where no fragments of Mtdh were detected by Western blotting, all of the functionality as described above is lost. Therefore, we believe that our findings, unlike those of Wan et al. (30), clearly point to Mtdh as an important factor in spermatogenesis. When Mtdh is lost, normal DNA processes required for meiosis and male fertility are greatly impacted. We hypothesize that the stable expression of the putative Mtdh N terminus peptide encoded by exons 1 and 2 may be adequate to compensate for the loss of full-length Mtdh in reproduction. Another possible explanation for the difference in fertility among models is the contribution of different backgrounds. Our model and the Robertson et al. model (5) are on the C57BL/6 background, whereas the mice generated by Wan et al. (30) were backcrossed to FVB background.
Our observations of increased expression of Mili and altered expression of piRNAs in the testes of Mtdh exon 3-deficient mice identify an essential role for MTDH in the regulation of small non-coding RNAs in spermatogenesis. Previous experiments have demonstrated that MTDH associates with SND1 and is involved in functional regulation of the RNA-induced silencing complex to inhibit tumor suppressor genes in cancer cells (10, 11). SND1 belongs to the Tudor domain-containing protein family. Our data support the hypothesis that MTDH is involved in the functional regulation of piRNAs via a mechanism similar to the Tudor family proteins. Argonaute proteins are crucial components in diverse gene-silencing processes. The Piwi family is a subgroup of Argonaute proteins that are required for germ and stem cell development. Mili and Miwi are Piwi family homologs essential for spermatogenesis in the mouse, and Mili expression is highest during the early stages of spermatogenesis (25). We observed Mili overexpression in our mouse model, which could simply be the result of severely impaired spermatogenesis at an early stage as Mili is expressed uniquely early in spermatogenesis (23, 25).
There are two populations of piRNAs that are expressed at the pre-pachytene and pachytene stages of meiosis in the male germ cells of mammals (32). Pre-pachytene piRNAs are mostly encoded by retrotransposons and silence the respective retrotransposons. Pachytene piRNAs are derived from ∼3,000 genomic clusters and function in maintaining post-meiotic genome integrity by regulating other key aspects of translational control during spermatogenesis. Accordingly, we found that Mtdh exon 3 deletion alters the expression profile of piRNAs in male testes. Although the role of piRNAs in spermatogenesis is well defined, few studies have addressed the role of miRNAs in this process (33). We found a significant decrease in nine miRNAs, including miR-16, miR-19b, miR-28a, miR-101b, miR-106b, miR-144, miR-182, miR-293, and miR-340, in testes but not liver in response to loss of Mtdh expression. Importantly, low miR-19b and miR-16 levels are also present in the semen of infertile men (34).
What is the role of Mtdh in pachytene of male mice? The pachytene checkpoint is necessary to effect alignment of the X and Y chromosomes and to exchange genetic information during meiosis in male mice (35). The XY body, which is prominently seen during this period, is required to recruit DNA repair proteins to silence the sex chromosomes and to accomplish the unique sex chromosome partial synapsis at meiotic prophase in males. Accumulation of high levels of γ-H2AX during pachytene indicates significant DNA damage as a result of Mtdh loss. Our studies also suggest that deficiency of Mtdh induces the pachytene checkpoint because the majority of spermatocytes fail to progress further in meiosis.
A key step in meiosis is the generation and repair of DSBs. Because we observed a pronounced accumulation of cells in pachytene, along with increased expression of γ-H2AX, we speculate that MTDH is required for DSB repair, perhaps specifically in male germ cells. Furthermore, several lines of additional evidence support the role of MTDH in DNA repair in general. First, MTDH has been reported to interact with BCCIP, a BRCA2- and CDKN1-interacting protein involved in DNA damage repair and cell cycle regulation (36). Second, as with previously reported knock-out mouse models targeting other classic, general DNA repair molecules, Mtdh deficiency using our strategy results in male sterility. For example, deficiency of Atm, Brca2, and Rad18 similarly results in defects in spermatogenesis (29, 37–39). ATM, the gene mutated in the human autosomal recessive disorder ataxia telangiectasia, plays a crucial role in the detection of DSBs and is a signal transduction protein in the DNA damage surveillance network. Male Atm-null mice are sterile, and spermatogenesis is arrested at mid-pachytene, consistent with our observations in our Mtdh knock-out model (37). BRCA2 and BRCA1 play critical roles in homologous recombination repair to maintain genomic stability. Brca2-deficient spermatocytes can undergo the early steps of recombination (DNA DSB formation), but fail to complete this process and to progress beyond prophase I of meiosis (38, 39). In contrast to spermatocytes, some Brca2-deficient oocytes can progress through meiotic prophase I and can be fertilized to produce embryos. Deficiency of Rad18 is also associated with impaired spermatogenesis (29). Further confirming a role for MTDH in DNA damage repair, we observed increased Rad18 foci in the lumina of the seminiferous tubules in testes from Mtdh exon 3-depleted mice, and some punctate staining that did not co-localize with the Y chromosome. In addition, the decrease in Rad18 expression under Mtdh deficiency is consistent with our previous report that MTDH associates with multiple mRNAs, including Rad18, to regulate their translation (16). These data suggest a novel mechanism whereby MTDH regulates DNA damage repair of both somatic and sex chromosomes in spermatogenesis.
In conclusion, we report that complete deletion of Mtdh in mice, based upon the targeting strategy described herein, results in male infertility characterized by accumulation of spermatocytes in pachytene and the absence of spermatozoa. Mtdh deficiency has significant effects on the expression of multiple non-coding RNAs critical for spermatogenesis and repair of DSBs. These findings further define the normal function of MTDH, which was heretofore best known as an amplified, pro-survival gene in cancer that controls gene expression and protein translation.
Supplementary Material
Acknowledgments
We thank Katherine Gibson-Corley (Department of Pathology) for histology analysis, Sarit Smolikove (Department of Biology) for commenting on the meiosis pachytene checkpoint, Eric J. Devor (Department of Obstetrics and Gynecology) for assistance in manuscript editing, and the Central Microscopy Research Facility at the University of Iowa for image acquisition. We also thank Drs. Jac Nickoloff (Colorado State University) and Amy Sparks (Department of Obstetrics and Gynecology, University of Iowa) for critically reviewing the manuscript.
This work was supported by National Institutes of Health Grants R01CA184101 (to X. M. and K. K. L.) and R01CA99908 (to K. K. L.) and by the Department of Obstetrics and Gynecology Research Development Fund (to K. K. L.).

This article contains supplemental Fig. S1 and Tables S1, A–C, and S2.
- MTDH
- metadherin
- ESC
- embryonic stem cell
- FFPE
- formalin-fixed, paraffin-embedded
- RT-qPCR
- quantitative RT-PCR
- E
- embryonic day
- miRNA
- microRNA
- piRNA
- Piwi-interacting RNA
- miR
- miRNA
- DSB
- double-strand break
- nt
- nucleotide(s)
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1. Sarkar D., Fisher P. B. (2013) AEG-1/MTDH/LYRIC: clinical significance. Adv. Cancer Res. 120, 39–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu G., Wei Y., Kang Y. (2009) The multifaceted role of MTDH/AEG-1 in cancer progression. Clin. Cancer Res. 15, 5615–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meng X., Thiel K. W., Leslie K. K. (2013) Drug resistance mediated by AEG-1/MTDH/LYRIC. Adv. Cancer Res. 120, 135–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu G., Chong R. A., Yang Q., Wei Y., Blanco M. A., Li F., Reiss M., Au J. L., Haffty B. G., Kang Y. (2009) MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robertson C. L., Srivastava J., Siddiq A., Gredler R., Emdad L., Rajasekaran D., Akiel M., Shen X. N., Guo C., Giashuddin S., Wang X. Y., Ghosh S., Subler M. A., Windle J. J., Fisher P. B., Sarkar D. (2014) Genetic deletion of AEG-1 prevents hepatocarcinogenesis. Cancer Res. 74, 6184–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He X. X., Chang Y., Meng F. Y., Wang M. Y., Xie Q. H., Tang F., Li P. Y., Song Y. H., Lin J. S. (2012) MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 31, 3357–3369 [DOI] [PubMed] [Google Scholar]
- 7. Zhao J., Wang W., Huang Y., Wu J., Chen M., Cui P., Zhang W., Zhang Y. (2014) HBx elevates oncoprotein AEG-1 expression to promote cell migration by downregulating miR-375 and miR-136 in malignant hepatocytes. DNA Cell Biol. 33, 715–722 [DOI] [PubMed] [Google Scholar]
- 8. Thirkettle H. J., Mills I. G., Whitaker H. C., Neal D. E. (2009) Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene 28, 3663–3670 [DOI] [PubMed] [Google Scholar]
- 9. Ash S. C., Yang D. Q., Britt D. E. (2008) LYRIC/AEG-1 overexpression modulates BCCIPα protein levels in prostate tumor cells. Biochem. Biophys. Res. Commun. 371, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanco M. A., Alečkovič M., Hua Y., Li T., Wei Y., Xu Z., Cristea I. M., Kang Y. (2011) Identification of staphylococcal nuclease domain-containing 1 (SND1) as a metadherin-interacting protein with metastasis-promoting functions. J. Biol. Chem. 286, 19982–19992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoo B. K., Santhekadur P. K., Gredler R., Chen D., Emdad L., Bhutia S., Pannell L., Fisher P. B., Sarkar D. (2011) Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 53, 1538–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sarkar D., Park E. S., Emdad L., Lee S. G., Su Z. Z., Fisher P. B. (2008) Molecular basis of nuclear factor-κB activation by astrocyte elevated gene-1. Cancer Res. 68, 1478–1484 [DOI] [PubMed] [Google Scholar]
- 13. Sarkar D. (2013) AEG-1/MTDH/LYRIC in liver cancer. Adv. Cancer Res. 120, 193–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoo B. K., Chen D., Su Z. Z., Gredler R., Yoo J., Shah K., Fisher P. B., Sarkar D. (2010) Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res. 70, 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Srivastava J., Siddiq A., Emdad L., Santhekadur P. K., Chen D., Gredler R., Shen X. N., Robertson C. L., Dumur C. I., Hylemon P. B., Mukhopadhyay N. D., Bhere D., Shah K., Ahmad R., Giashuddin S., Stafflinger J., Subler M. A., Windle J. J., Fisher P. B., Sarkar D. (2012) Astrocyte elevated gene-1 promotes hepatocarcinogenesis: novel insights from a mouse model. Hepatology 56, 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meng X., Zhu D., Yang S., Wang X., Xiong Z., Zhang Y., Brachova P., Leslie K. K. (2012) Cytoplasmic Metadherin (MTDH) provides survival advantage under conditions of stress by acting as RNA-binding protein. J. Biol. Chem. 287, 4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarkar D., Fisher P. B. (2013) Advances in Cancer Research. AEG-1/MTDH/LYRIC implicated in multiple human cancers. Preface. Adv. Cancer Res. 120, xi–xiv [DOI] [PubMed] [Google Scholar]
- 18. Lee S. G., Kang D. C., DeSalle R., Sarkar D., Fisher P. B. (2013) AEG-1/MTDH/LYRIC, the beginning: initial cloning, structure, expression profile, and regulation of expression. Adv. Cancer Res. 120, 1–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeon H. Y., Choi M., Howlett E. L., Vozhilla N., Yoo B. K., Lloyd J. A., Sarkar D., Lee S. G., Fisher P. B. (2010) Expression patterns of astrocyte elevated gene-1 (AEG-1) during development of the mouse embryo. Gene Expr. Patterns 10, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meng X., Brachova P., Yang S., Xiong Z., Zhang Y., Thiel K. W., Leslie K. K. (2011) Knockdown of MTDH sensitizes endometrial cancer cells to cell death induction by death receptor ligand TRAIL and HDAC inhibitor LBH589 co-treatment. PLoS One 6, e20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Syrjänen J. L., Pellegrini L., Davies O. R. (2014) A molecular model for the role of SYCP3 in meiotic chromosome organisation. Elife 10.7554/eLife.02963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hosokawa M., Shoji M., Kitamura K., Tanaka T., Noce T., Chuma S., Nakatsuji N. (2007) Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 301, 38–52 [DOI] [PubMed] [Google Scholar]
- 23. Kuramochi-Miyagawa S., Kimura T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T. (2001) Two mouse piwi-related genes: miwi and mili. Mech. Dev. 108, 121–133 [DOI] [PubMed] [Google Scholar]
- 24. Unhavaithaya Y., Hao Y., Beyret E., Yin H., Kuramochi-Miyagawa S., Nakano T., Lin H. (2009) MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284, 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuramochi-Miyagawa S., Kimura T., Ijiri T. W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Lin H., Matsuda Y., Nakano T. (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839–849 [DOI] [PubMed] [Google Scholar]
- 26. Wang J., Saxe J. P., Tanaka T., Chuma S., Lin H. (2009) Mili interacts with Tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 19, 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blanco-Rodríguez J. (2012) Programmed phosphorylation of histone H2AX precedes a phase of DNA double-strand break-independent synapsis in mouse meiosis. Reproduction 144, 699–712 [DOI] [PubMed] [Google Scholar]
- 28. Fernandez-Capetillo O., Mahadevaiah S. K., Celeste A., Romanienko P. J., Camerini-Otero R. D., Bonner W. M., Manova K., Burgoyne P., Nussenzweig A. (2003) H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4, 497–508 [DOI] [PubMed] [Google Scholar]
- 29. Sun J., Yomogida K., Sakao S., Yamamoto H., Yoshida K., Watanabe K., Morita T., Araki K., Yamamura K., Tateishi S. (2009) Rad18 is required for long-term maintenance of spermatogenesis in mouse testes. Mech. Dev. 126, 173–183 [DOI] [PubMed] [Google Scholar]
- 30. Wan L., Lu X., Yuan S., Wei Y., Guo F., Shen M., Yuan M., Chakrabarti R., Hua Y., Smith H. A., Blanco M. A., Chekmareva M., Wu H., Bronson R. T., Haffty B. G., Xing Y., Kang Y. (2014) MTDH-SND1 interaction is crucial for expansion and activity of tumor-initiating cells in diverse oncogene- and carcinogen-induced mammary tumors. Cancer Cell 26, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Srivastava J., Robertson C. L., Rajasekaran D., Gredler R., Siddiq A., Emdad L., Mukhopadhyay N. D., Ghosh S., Hylemon P. B., Gil G., Shah K., Bhere D., Subler M. A., Windle J. J., Fisher P. B., Sarkar D. (2014) AEG-1 regulates retinoid X receptor and inhibits retinoid signaling. Cancer Res. 74, 4364–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chuma S., Nakano T. (2013) piRNA and spermatogenesis in mice. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 368, 20110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran M., Chen B., Yin J., Yang A., Jiang M. (2014) Advances in miRNA research related to testis development and spermatogenesis. Yi. Chuan 36, 646–654 [DOI] [PubMed] [Google Scholar]
- 34. Liu T., Cheng W., Gao Y., Wang H., Liu Z. (2012) Microarray analysis of microRNA expression patterns in the semen of infertile men with semen abnormalities. Mol. Med. Rep. 6, 535–542 [DOI] [PubMed] [Google Scholar]
- 35. Sciurano R., Rahn M., Rey-Valzacchi G., Solari A. J. (2007) The asynaptic chromatin in spermatocytes of translocation carriers contains the histone variant γ-H2AX and associates with the XY body. Hum. Reprod. 22, 142–150 [DOI] [PubMed] [Google Scholar]
- 36. Emdad L., Das S. K., Dasgupta S., Hu B., Sarkar D., Fisher P. B. (2013) AEG-1/MTDH/LYRIC: signaling pathways, downstream genes, interacting proteins, and regulation of tumor angiogenesis. Adv. Cancer Res. 120, 75–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takubo K., Hirao A., Ohmura M., Azuma M., Arai F., Nagamatsu G., Suda T. (2006) Premeiotic germ cell defect in seminiferous tubules of Atm-null testis. Biochem. Biophys. Res. Commun. 351, 993–998 [DOI] [PubMed] [Google Scholar]
- 38. Gudmundsdottir K., Ashworth A. (2004) BRCA2 in meiosis: turning over a new leaf. Trends Cell Biol. 14, 401–404 [DOI] [PubMed] [Google Scholar]
- 39. Thorslund T., Esashi F., West S. C. (2007) Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 26, 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.