Background: Cell surface-associated enolase-1 regulates plasmin formation and thus pericellular proteolysis.
Results: STIM1/ORAI1-mediated Ca2+ influx controls enolase-1 exteriorization in cancer cells.
Conclusion: Extracellular enolase-1 regulates migratory and invasive properties of cancer cells.
Significance: Enhanced exteriorization of enolase-1 may aggravate the malignant behavior of cancer cells and thus contribute to metastasis formation.
Keywords: calcium, cancer, inflammation, membrane trafficking, stromal interaction molecule 1 (STIM1), enolase-1
Abstract
Tumor cells use broad spectrum proteolytic activity of plasmin to invade tissue and form metastatic foci. Cell surface-associated enolase-1 (ENO-1) enhances plasmin formation and thus participates in the regulation of pericellular proteolysis. Although increased levels of cell surface bound ENO-1 have been described in different types of cancer, the molecular mechanism responsible for ENO-1 exteriorization remains elusive. In the present study, increased ENO-1 protein levels were found in ductal breast carcinoma and on the cell surface of highly metastatic breast cancer cell line MDA-MB-231. Elevated cell surface-associated ENO-1 expression correlated with augmented MDA-MB-231 cell migratory and invasive properties. Exposure of MDA-MB-231 cells to LPS potentiated translocation of ENO-1 to the cell surface and its release into the extracellular space in the form of exosomes. These effects were independent of de novo protein synthesis and did not require the classical endoplasmic reticulum/Golgi pathway. LPS-triggered ENO-1 exteriorization was suppressed by pretreatment of MDA-MB-231 cells with the Ca2+ chelator BAPTA or an inhibitor of endoplasmic reticulum Ca2+-ATPase pump, cyclopiazonic acid. In line with these observations, the stromal interaction molecule (STIM) 1 and the calcium release-activated calcium modulator (ORAI) 1-mediated store-operated Ca2+ entry were found to regulate LPS-induced ENO-1 exteriorization. Pharmacological blockage or knockdown of STIM1 or ORAI1 reduced ENO-1-dependent migration of MDA-MB-231 cells. Collectively, our results demonstrate the pivotal role of store-operated Ca2+ channel-mediated Ca2+ influx in the regulation of ENO-1 exteriorization and thus in the modulation of cancer cell migratory and invasive properties.
Introduction
Proteolytic systems are required for many physiological processes, including fibrin clot formation and dissolution, angiogenesis, and bone remodeling (1, 2). The same enzyme systems are used by cancer cells to grow and spread. Among diverse proteases known to be involved in cancer invasion and metastasis, plasmin is the most extensively studied (3–5). Plasmin is generated from plasminogen (PLG)3 via limited proteolysis by either a tissue-type plasminogen activator (t-PA) or a urokinase-type plasminogen activator (6). Accumulating evidence suggests that immobilization of PLG and its activators on the cell surface accelerates the conversion of PLG to plasmin (7, 8), enhances the catalytic activity of plasmin itself (9), and protects bound plasmin from inactivation by means of inhibitors (7, 10). Plasminogen binding to the cell surface is mediated by a heterogenous group of PLG receptors, which are widely distributed on eukaryotic as well as prokaryotic cells (11). Cell surface bound enolase-1 (ENO-1) is one of them. It primarily acts as a glycolytic enzyme localized in the cytoplasm (12). However, under stimulatory conditions, it can be translocated to the cell surface where it interacts with PLG and thus participates in the regulation of pericellular proteolysis, allowing cells to invade tissue (13–15). This fact can explain the positive correlation between high levels of ENO-1 present on the cell surface of cancer cells (16–19) and their increased invasive potential (20, 21). In agreement with this notion, patients with non-small cell lung cancer whose tumors expressed high levels of cell surface bound ENO-1 were found to have a poor survival outcome (22).
Inflammation has long been associated with tumor progression. LPS was shown to increase the invasion of hepatic cancer cells in mice (23) and to potentiate lung metastasis upon intravenous injection of mouse mammary carcinoma cells into animals (24). LPS may contribute to tumor progression by activation of TLR-4 signaling pathway in cancer cells (25) and by the recruitment of inflammatory cells into the tumor microenvironment (26). LPS-induced migration and invasion of inflammatory cells is accompanied by increased production and release of proinflammatory cytokines (27), which display protumorigenic activities (26, 28). Given the fact that LPS up-regulates cell surface expression of ENO-1 on monocytoic cells, thereby increasing their invasive potential (14), LPS-triggered regulation of ENO-1-dependent pericellular proteolytic activity could represent another mechanism that contributes to cancer cell spreading and thus to metastasis formation.
Despite strong evidence demonstrating the importance of cell surface-associated ENO-1 in cancer cell invasion and metastasis formation, the driving factors governing ENO-1 translocation to the cell surface have not yet been identified. In the present study, we investigated the mechanism and the functional consequence of LPS-triggered ENO-1 transport to the cell surface and to the extracellular milieu.
EXPERIMENTAL PROCEDURES
Cell Culture
Human MDA-MB-435 highly metastatic breast carcinoma, human MCF-7 breast adenocarcinoma (both from ATCC, Manassas, VA), and human MDA-MB-231 metastatic breast carcinoma (LGC Standards GmbH, Wesel, Germany) cell lines were maintained in RPMI 1640 medium (Invitrogen Life Technologies) supplemented with 10% heat-inactivated FCS (Hyclone, Cramlington, UK), 2 mm GlutaMAX, and 1% penicillin/streptomycin (both from Invitrogen Life Technologies). Human mammary epithelial cells (Invitrogen Life Technologies) were cultured in DMEM (Invitrogen Life Technologies) supplemented with FCS and 1% penicillin/streptomycin. Cell cultures were maintained at 37 °C in a humidified incubator with 5% CO2.
Immunohistochemistry
Formalin-fixed tissues were obtained from patients with ductal breast carcinoma (n = 6), squamous cell lung carcinoma (n = 5), colon adenocarcinoma (n = 11), bronchoalveolar carcinoma (n = 5), and lung adenocarcinoma (n = 12) who underwent surgical resection. The investigations have been conducted according to the Declaration of Helsinki principles and were approved by the local institutional review board and ethics committee. 5-μm tissue sections were deparaffinized in xylene and rehydrated through graded ethanol washes. Antigen retrieval was performed by the treatment of tissue sections with Fast Enzyme (Zymed Laboratories Inc.) for 10 min at room temperature. Immunohistochemistry was performed using a ZytoChem-Plus AP Polymer-Kit according to the manufacturer's instructions (Zymed Laboratories Inc.). A rabbit anti-ENO-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was applied overnight at 4 °C. Negative control was performed by replacing the primary antibody with a species-matched isotype control. The slides were scanned with a Mirax desk digital slide scanner (Zeiss) and analyzed using a Mirax viewer.
Western Blotting
100 μg of biotinylated proteins or 20 μl of exosomal fraction were separated on a 10% SDS-PAGE under reducing conditions, followed by electrotransfer to a PVDF membrane (GE Healthcare). After blocking the membrane with 5% nonfat milk (Sigma-Aldrich) in TBS-T (5 mm Tris-Cl, 150 mm NaCl, 0.1% Tween 20, pH 7.5), the membrane was probed with one of the following antibodies: rabbit anti-ENO-1, mouse anti-GFP (both from Santa Cruz Biotechnology), mouse anti-26S proteasome subunit (P26S; Abcam, Berlin, Germany), mouse anti-β1-integrin, mouse anti-CD63 (both from Millipore, Schwalbach, Germany), mouse anti-heat shock protein 70 (Hsp70; generous gift from Dr. M. Korfei, Department of Internal Medicine, University of Giessen Lung Centre, Giessen, Germany). Afterward, the membrane was incubated with peroxidase-labeled secondary antibody (all from Dako, Gostrup, Denmark). Final detection of proteins was performed using an ECL Plus kit (Amersham Biosciences). To determine the amounts of protein loaded on the gel, blots were stripped and reprobed using a mouse anti-β-actin antibody (Sigma-Aldrich).
Cell Surface Biotinylation Assay
MDA-MB-231, MCF-7, and MDA-MB-435 cells were treated for 2, 4, and 6 h with 10 μg/ml LPS serotype O111:B4 (Calbiochem, Darmstadt, Germany), 50 ng/ml TNF-α, 20 ng/ml TGF-β1, or 100 ng/ml chemokine (C-C motif) ligand 2 (CCL2; all from R&D, Wiesbaden, Germany). In other experiments MDA-MB-231 cells were pretreated for 1 h with brefeldin A (BD Biosciences, Heidelberg, Germany), glyburide, methylamine, ouabain, ionophore A23187, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′ tetraacetic acid (BAPTA), cyclopiazonic acid (CPA), or YM58483 (all from Sigma-Aldrich) and then stimulated with 10 μg/ml LPS for 2 h. Afterward, the cells were labeled with 1 mg/ml EZ-link NHS-SS-biotin (Thermo Scientific, Schwerte, Germany) for 1 h at 4 °C, rinsed three times with PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4) containing 100 mm glycine, and solubilized in cell lysis buffer (50 mm Tris, pH 7.4, 100 mm NaCl, 50 mm NaF, 5 mm β-glycerophosphate, 2 mm EDTA, 2 mm EGTA, 1 mm sodium orthovanadate, 0.1% Triton X-100) containing protease inhibitor mixture (Roche Diagnostics). Protein concentration was determined using a Pierce BCA protein assay kit (Thermo Scientific) according to the manufacturer's instructions. 100 μg of proteins were incubated overnight at 4 °C with end over end shaking with NeutrAvidin-agarose resin beads (Thermo Scientific). Finally, beads were washed and resuspended in 25 μl of 2× Laemmli sample buffer (10% SDS, 20% glycerol, 0.2 m Tris-HCl, 0.05% bromphenol blue, 10% β mercaptoethanol). The samples were analyzed by Western blotting as described above.
Generation of MDA-MB-231 Cells Stably Expressing GFP-tagged ENO-1
MDA-MB-231 cells stably transfected with pEGFP-C1 (GFP-EV) and pEGFP-C1-ENO-1WT (GFP-ENO-1) were generated as previously described (21).
Cell Proliferation Assay
Proliferation of MDA-MB-231 cells and stable transfectants was determined by a DNA synthesis assay based on the uptake of [3H]thymidine (PerkinElmer Life Sciences). Cells were cultured in 48-well plates, growth-arrested in serum-free RPMI medium and left unstimulated or stimulated with 10 μg/ml LPS for 8 h. Subsequently, cells were pulsed with 0.2 μCi/ml [3H]thymidine for 16 h. Afterward, cells were solubilized in 0.5 m NaOH, and [3H]thymidine incorporation was determined by liquid scintillation spectrometry.
Trichloroacetic Acid Precipitation of Proteins
Proteins present in conditioned cell culture media were precipitated with TCA (Sigma-Aldrich). Briefly, MDA-MB-231 cells were stimulated with 10 μg/ml LPS for 2, 4, or 6 h. After the indicated time points, supernatants were collected, mixed with TCA (final concentration, 10%), vortexed, and incubated for 10 min at 4 °C. The precipitated proteins were collected by centrifugation at 20,000 × g for 45 min at 4 °C. The pellets were washed twice with 70% ice-cold ethanol, air-dried, and resuspended in 5× Laemmli sample buffer.
Exosome Isolation
Exosomes were isolated either from unstimulated GFP-EV and GFP-ENO-1 cells or stimulated MDA-MB-231, MCF-7, and MDA-MB-435 cells. Briefly, MDA-MB-231, MCF-7, and MDA-MB-435 cells were treated for 24 h with 1 μg/ml LPS, 50 ng/ml TNF-α, 20 ng/ml TGF-β1, or 100 ng/ml CCL2. In other experiments MDA-MB-231 cells were preincubated with A23187, BAPTA, or YM58483 for 1 h and then stimulated with 1 μg/ml LPS for 24 h. Exosomes were isolated from 10 ml of conditioned culture media that were first centrifuged at 800 × g for 10 min at room temperature to sediment cells and then centrifuged at 10,000 × g for 10 min at 4 °C (Optima LE-80K ultracentrifuge; Beckman, Ramsey, MN) to remove the cellular debris. Exosomes were pelleted by centrifugation at 100,000 × g for 3 h at 4 °C. Finally, the exosome pellet was washed once with PBS and resuspended in 100 μl of PBS. A 20-μl exosomal fraction was mixed with 5× Laemmli sample buffer and analyzed by Western blotting. The viability of the treated cells was assessed in each experiment using a cytotoxicity detection kit (Roche Diagnostics).
Exosome Uptake
Exosomes were purified from cell culture supernatants of GFP-EV and GFP-ENO-1 stably transfected cells according to the above mentioned protocol. The purified exosomes were resuspended in 100 μl of PBS. MDA-MB-231 cells were cultured in complete RPMI medium on microscope coverslips in 6-well plates. Cells were serum-starved overnight and then incubated with purified exosomes for 30 min at 37 °C. Subsequently, the cells were washed three times with cold PBS, fixed with 4% paraformaldehyde for 10 min at 4 °C, incubated with rhodamine-conjugated phalloidin (Invitrogen Life Technologies) for 10 min at room temperature, and mounted with Vectashield mounting medium (Vector). The nuclei were visualized by DAPI staining. The images were captured by Leica DMR microscope (Leica, Heidelberg, Germany). Post processing and image analyses were done with a MetaMorphTM (Leica Microsystems, Wetzlar, Germany).
Electron Microscopy
Exosomes were fixed with 2% paraformaldehyde and deposited onto Butvar carbon-coated grids. The vesicle-coated grids were washed twice with PBS, twice with PBS containing 50 mm glycine, and finally with PBS containing 0.5% BSA (PBS, 0.5% BSA), stained with 2% uranyl acetate, and then viewed with a transmission electron microscope (Zeiss EM900; Zeiss, Jena, Germany). For the immunogold labeling, exosome samples were absorbed onto a carbon coated Butvar film on 300 mesh nickel grids, washed in PBS containing 10 mm glycine for 5 min, and then washed in PBS only. Samples were placed onto 25-μl drops of 0.4% skim milk in water for 5 min, blotted dry on filter paper, and then placed on 25-μl drops of the rabbit anti-ENO-1 antibodies (1:25 dilution; Santa Cruz Biotechnology) and incubated for 1 h at 30 °C. After washing with PBS, samples were incubated with 0.4% skim milk for 5 min, blotted dry, then placed onto 25-μl drops of a mixture of Protein A/G gold and goat anti-rabbit gold nanoparticles (1:75 dilution), and incubated for 30 min at room temperature. Samples were then washed with PBS and TE buffer (20 mm Tris, 2 mm EDTA, pH 7.0) before air-drying. Samples were examined in a transmission electron microscope 910 Zeiss at an acceleration voltage of 80 kV.
Live Cell Ca2+ Imaging
The cells were cultured on 25-mm glass coverslips that were loaded with 2 μm fura-2/AM in dark for 45 min followed by a washing step in Ringer solution (5.8 mm KCl, 141 mm NaCl, 0.5 mm KH2PO4, 0.4 mm NaH2PO4, 11.1 mm glucose, 10 mm Hepes, 1.8 mm CaCl2, 1 mm MgCl2, pH 7.4) as previously described (29). After 15 min, the single glass coverslip was mounted on the stage of a Zeiss 200M inverted epifluorescence microscope coupled to a PolyChrome V monochromator (Till Photonics, Munich, Germany) light source in a sealed temperature-controlled RC-21B imaging chamber (Warner Instruments, Hamden, CT) and perfused with prewarmed solution (32 °C). Fluorescence images were obtained with alternate excitation at 340 and 380 nm. The emitted light was collected at 510 nm by an air-cooled Andor Ixon camera (Andor Technology, Belfast, Ireland). Measurements were made every 3 s. Background fluorescence was recorded from each coverslip and subtracted before calculation. The acquired images were stored and processed offline with TillVision software (Till Photonics). [Ca2+]i was calculated as described by Grynkiewicz et al. (30). Maximal and minimal ratio values were determined at the end of each experiment by first treating the cells with 10 μm ionomycin (maximal ratio) and then chelating all free Ca2+ with 20 mm EGTA (minimal ratio). Cells that did not respond to ionomycin were discarded. After 3 min of baseline measurement, the cells were stimulated with 10 μg/ml LPS for 10 min in the absence or presence of extracellular Ca2+. In the next set of experiments, after baseline measurement, cells were pretreated with 20 μm BAPTA or 50 μm CPA followed by 10 min of 10 μg/ml LPS treatment. For data analysis, the basal level of Ca2+ was determined as an average value of the first 50 s of the curve. Then, after subtracting the baseline, the LPS-induced Ca2+ peak height was calculated and is presented as Δ[Ca2+]i. All chemicals were dissolved and diluted to the desired concentrations in Ringer solution. All the solutions were freshly prepared on the day of the experiment and stored at 4 °C until they were used.
Antisense Oligonucleotides
Commercially available siRNA sequence directed against human stromal interaction molecule (STIM) 1 (Thermo Scientific), human calcium release-activated calcium modulator (ORAI) 1 (Life Technologies), and a universal negative-control siRNA (Thermo Scientific) were employed. Cells were starved overnight and then treated with 100 nm siRNA using the siLentFectTM lipid transfection reagent (Bio-Rad) according to the manufacturer's instructions. After 72 h, cells were split and seeded onto 6-well tissue cultures plates, serum-starved overnight, and then treated one more time with 100 nm siRNA for 48 h. The efficacy of STIM1 and ORAI1 knockdown was assessed by real time RT-PCR.
RNA Isolation and Real Time RT-PCR
Isolation of RNA from formalin-fixed, paraffin-embedded tumor tissue and adjacent nontumorous tissue was performed as previously described (31). Isolation of RNA from MDA-MB-231 cells was performed using a peqGOLD total RNA kit (Peqlab, Erlangen, Germany) according to the manufacturer's instructions. Real time RT-PCR (qRT-PCR) was performed as described previously (14) to amplify transcripts of human ENO-1, human STIM1, human ORAI1, subunit of human L-type calcium channel (LTCC Cav1.2), human β-actin (β-ACT), and human porphobilinogen deaminase (PBGD) (Table 1). PBGD and β-ACT were used as reference genes. Cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Melting curve analysis and gel electrophoresis were performed to confirm the exclusive amplification of the expected PCR product. All changes in the target gene mRNA levels are presented as ΔCt, which was calculated by subtracting the Ct value of the target gene from the Ct value of the reference gene. The higher values of ΔCt correspond to higher relative expression of the gene of interest.
TABLE 1.
List of primers used for qRT-PCR
F, forward; R, reverse; Tm, melting temperature.
| Gene | Accession number | Nucleotide sequences (5′ → 3′) | Tm | Amplicon size |
|---|---|---|---|---|
| °C | nt | |||
| ENO-1 | NM_001428.3 | F: GAA TAA AGA AGG CCT GGA GC | 60 | 217 |
| R: TAG ACA CCA CTG GGT AGT CC | ||||
| STIM1 | NM_001277961.1 | F: AGT GAG AAG GCG ACA GGA | 60 | 130 |
| R: ATG TTA CGG ACT GCC TCG | ||||
| ORAI1 | NM_032790.3 | F: ACC TCG GCT CTG CTC TCC | 60 | 86 |
| R: CAG GCA CTG AAG GCG ATG | ||||
| Cav1.2 | NM_001167625.1 | F: TGG TCC ATG GTC AAT GAG | 60 | 107 |
| R: CGC ATT GGC ATT CAT GTT | ||||
| PBGD | NM_000190.3 | F: CCC ACG CGA ATC ACT CTC AT | 60 | 69 |
| R: TGT CTG GTA ACG GCA ATG CG | ||||
| β-ACT | NM_001101.3 | F: ATTGCCGACAGGATGCAGGAA | 60 | 149 |
| R: GCTGATCCACATCTGCTGGAA |
Lactate Dehydrogenase Release or Cytotoxicity Assay
MDA-MB-231 cells were stimulated with 10 μg/ml LPS for 2, 4, or 6 h. After the indicated time points, lactate dehydrogenase release was assessed using a cytotoxicity detection kit (Roche Diagnostics) according to the manufacturer's instructions. MDA-MB-231 cells treated with 1% Triton X-100 for 5 min were used as a positive control.
Wound Healing Assay
MDA-MB-231 cells or cells stably transfected with GFP-EV and GFP-ENO-1 were seeded onto 6-well tissue culture plates and serum-starved overnight. MDA-MB-231 cells were stimulated either with 10 μg/ml LPS alone or in combination with a peptide blocking binding of PLG to ENO-1 (KFAGRNFRNPLAK; kindly provided by Dr. S. Bergmann, Institute of Microbiology, Technical University Braunschweig, Braunschweig, Germany) or a scramble peptide (KFAGRNFRNPLA; Thermo Scientific). Cells were washed with PBS, and wounds were incised by scratching the cell monolayers using a pipette tip. Images of the scratch were captured with a microscope immediately after incision (0 h) and 8 h after scratching to assess the rate of gap closure.
Transwell Invasion Assay
Invasion assay was performed either with unstimulated GFP-EV and GFP-ENO-1 cells or stimulated MDA-MB-231 cells. Briefly, MDA-MB-231 cells were starved overnight and stimulated either with 1 μg/ml LPS or with exosomes isolated from GFP-EV and GFP-ENO-1 cells. Cells (5 × 104) were added into the upper chamber containing the fibronectin-coated polycarbonate membrane (8-μm pore size; BD Biosciences). 500 μl of RPMI medium containing 2% FCS was added into the lower chamber of the Transwell. Cells were then cultured for 16 h at 37 °C. Afterward, cells on the upper surface of the polycarbonate membrane of the Transwell were removed with a cotton swab, and the cells that migrated onto the underside of the membrane were fixed with acetone/methanol (1:1) solution, washed with PBS, and stained with 0.5% crystal violet for 30 min. Cells that migrated to the lower surface of the filter were counted.
Statistics
The statistical analysis was performed using a GraphPad Prism version 5.02 for Windows (GraphPad Software, La Jolla, CA). The data are presented as mean values ± S.E. unless otherwise stated. Differences between two groups were tested using a Student's t test. Comparison of multiple groups was performed by analysis of variance followed by Tukey's post hoc test or using a Bonferroni's multiple comparison test for the live cell Ca2+ imaging data. All tests were performed with an undirected hypothesis. A p value less than 0.05 was considered as statistically significant.
RESULTS
Expression of ENO-1 Is Elevated on the Cell Surface of Cancer Cells and Contributes to Cancer Cell Invasion
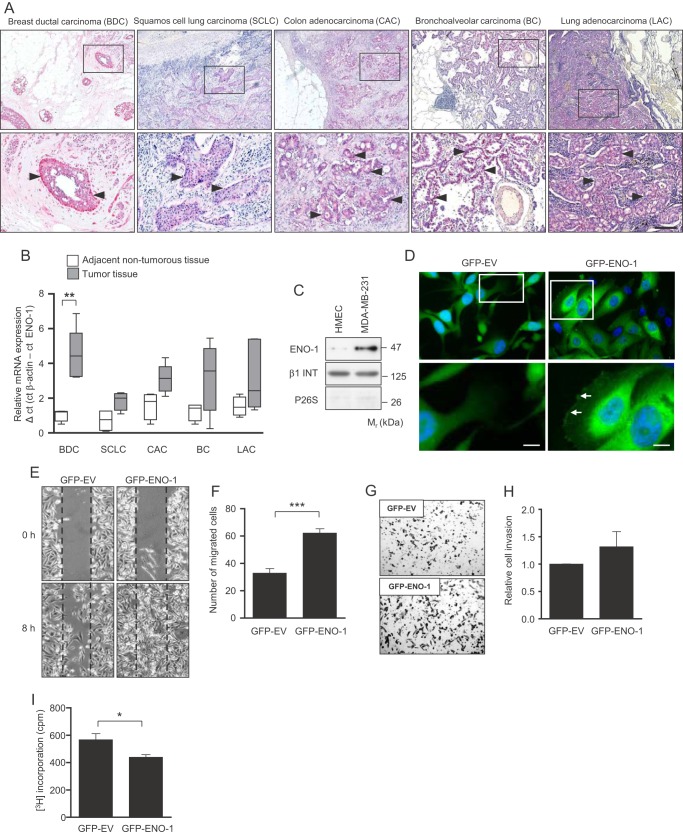
To investigate the role of cell surface-bound ENO-1 for cancer cell invasion, we first determined the expression levels of this protein in different tumor types. Immunohistochemical staining of breast ductal carcinoma, squamous cell lung carcinoma, colon adenocarcinoma, bronchoalveolar carcinoma, and lung adenocarcinoma revealed high ENO-1 protein expression in cancer cells (Fig. 1A, indicated by arrowheads). qRT-PCR confirmed high ENO-1 mRNA expression in tumor tissue as compared with adjacent nontumorous tissue (Fig. 1B). Because the highest ENO-1 expression was detected in ductal carcinoma of the breast, in further studies we focused on the role of ENO-1 in breast cancer cell motility. Cell fractionation revealed markedly increased levels of cell surface-associated ENO-1 in metastatic breast cancer cell line MDA-MB-231 as compared with primary human mammary epithelial cells (Fig. 1C). To verify ENO-1 distribution in cancer cells, MDA-MB-231 cells were transfected either with GFP alone (GFP-EV) or GFP-tagged ENO-1 (GFP-ENO-1), and the localization of ENO-1 was examined by fluorescence microscopy. Although GFP was exclusively expressed in the cytoplasm (Fig. 1D, left panel), GFP-ENO-1 was present in the cytoplasm, as well as on the cell surface (Fig. 1D, right panel, indicated by arrows). To explore the functional consequence of increased ENO-1 expression in cancer cells, wound healing and Transwell invasion assays with MDA-MB-231 cells that overexpress GFP-ENO-1 were performed. Transfection of MDA-MB-231 cells with GFP-ENO-1 enhanced their migratory (Fig. 1, E and F) and invasive (Fig. 1, G and H) properties. These effects were dependent on the impact of ENO-1 on cancer cell motility because overexpression of GFP-ENO-1 did not increase the number of cells (Fig. 1I).
FIGURE 1.
ENO-1 is expressed on the cell surface of cancer cells. A, representative breast ductal carcinoma (BDC; n = 6), squamous cell lung carcinoma (SCLC; n = 5), colon adenocarcinoma (CAC; n = 11), bronchoalveolar carcinoma (BC; n = 5), and lung adenocarcinoma (LAC; n = 12) tissue sections stained for ENO-1. ENO-1-positive staining is indicated by arrowheads. Bar, 100 μm. B, qRT-PCR analysis of ENO-1 mRNA expression in tumor and adjacent nontumorous tissue. qRT-PCR data are expressed as ΔCt using β-actin as a reference gene (n = 5 per group). C, cell surface expression of ENO-1 in primary human mammary epithelial cells (HMEC) and in MDA-MB-231 cells. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1-integrin (β1 INT) and P26S, respectively (n = 3). Representative Western blots are shown. D, distribution of ENO-1 in MDA-MB-231 cells stably transfected with GFP-EV or GFP-ENO-1. Arrows indicate the cell surface-associated GFP-tagged ENO-1. Bar, 5 μm. E, confluent monolayers of stable transfectants expressing either GFP (GFP-EV) or GFP-tagged ENO-1 (GFP-ENO-1) were scratched and incubated for 8 h at 37 °C in serum-free RPMI medium. Representative pictures from the wound healing assay at times 0 and 8 h are shown. F, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 3). ***, p < 0.001 G, stable transfectants expressing either GFP (GFP-EV) or GFP-tagged ENO-1 (GFP-ENO-1) were seeded onto a fibronectin-coated membrane and allowed to invade for 16 h. Representative images of the cells that invaded the underside of the membrane are shown. H, cells that invaded the underside of the membrane were counted. The data represent mean values ± S.E. (n = 3). I, proliferation of GFP-EV- and GFP-ENO-1-overexpressing cells as assessed by [3H]thymidine incorporation. The data represent mean values ± S.E. (n = 3). *, p < 0.05.
LPS Up-regulates Cell Surface Expression and Release of ENO-1 into the Extracellular Space
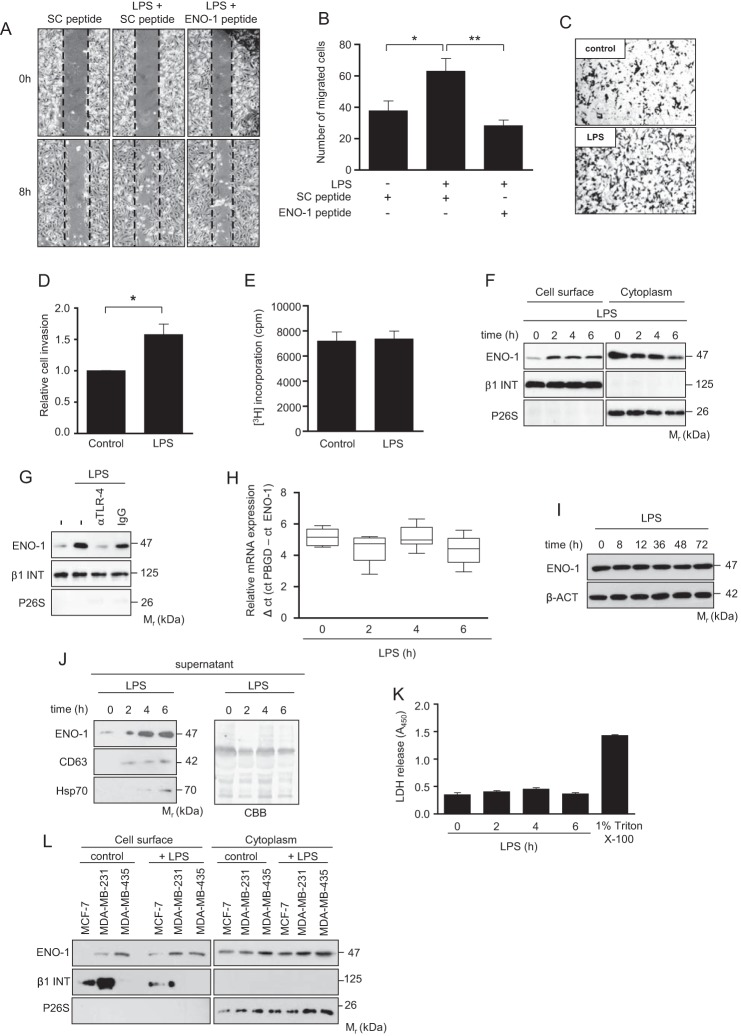
Inflammation has long been thought to contribute to tumor progression, for example, by influencing the invasive potential of cancer cells (24). In line with this notion, treatment of MDA-MB-231 cells with LPS markedly augmented their migratory and invasive properties (Fig. 2, A–D). The increased migration of MDA-MB-231 cells observed in the presence of LPS was abolished by a peptide directed against the C-terminal part of ENO-1 involved in the binding of PLG (Fig. 2, A and B) (15). Stimulation of cells with LPS did not affect their growth (Fig. 2E). LPS-mediated increase in cell migration was accompanied by elevated cell surface expression of ENO-1. Alterations in cell membrane-bound ENO-1 levels were visible 2 h after stimulation and remained unchanged thereafter (Fig. 2F). Moreover, LPS-induced mobilization of ENO-1 on the cell surface was TLR-4-dependent because preincubation of cells with an anti-TLR-4 blocking antibody inhibited ENO-1 translocation (Fig. 2G). Augmented levels of cell surface bound ENO-1 occurred in the absence of any detectable changes in total cellular expression of ENO-1 mRNA and protein (Fig. 2, H and I), suggesting that alterations in cell surface ENO-1 abundance are independent of new protein synthesis. Interestingly, there was not only an increased ENO-1 translocation to the cell surface but also an increased release into the extracellular space (Fig. 2J). Elevated extracellular levels of ENO-1 were positively correlated with the markers of exosomes, CD63 and Hsp70, implying that ENO-1 is secreted from LPS-treated MDA-MB-231 cells in the form of vesicles (Fig. 2J). To exclude that the release of ENO-1 is a result of cell damage, a lactate dehydrogenase assay was performed. This experimental procedure revealed no impact of LPS on cell viability (Fig. 2K). To examine whether the level of cell surface ENO-1 associates with the metastatic potential of cancer cells, ENO-1 cell surface expression was explored in a less metastatic breast cancer cell line, MCF-7, and a highly metastatic breast cancer cell line, MDA-MB-435. In contrast to MDA-MB-435 cells, which exhibited high cell surface-bound ENO-1 levels under basal conditions, cell membrane-associated ENO-1 was not detectable on MCF-7 cells (Fig. 2L). Following LPS exposure, levels of cell surface-bound ENO-1 increased on MCF-7 cells but remained unchanged on MDA-MB-435 cells (Fig. 2L), suggesting that the level of ENO-1 on the cell surface of MDA-MB-435 cells is already saturated under basal conditions. Altogether, these results imply that there is a positive correlation between the level of cell surface ENO-1 and the invasive potential of cancer cells.
FIGURE 2.
LPS up-regulates ENO-1 cell surface expression and its release into the extracellular space. A, impact of ENO-1 peptide on LPS-driven MDA-MB-231 cell migration. Representative pictures from the wound healing assay at times 0 and 8 h are shown. B, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 4). *, p < 0.05; **, p < 0.01. SC, scramble. C, LPS stimulated MDA-MB-231 cells were seeded onto a fibronectin-coated membrane and allowed to invade for 16 h. Representative images of cells that invade the underside of the membrane are demonstrated. D, cells that invaded the underside of the membrane were counted. The data represent mean values ± S.E. (n = 3). *, p < 0.05. E, proliferation of MDA-MB-231 cells stimulated with LPS for 8 h as assessed by [3H]thymidine incorporation. The data represent mean values ± S.E. (n = 3). F, cell surface expression of ENO-1 in MDA-MB-231 cells exposed to LPS for the indicated time points. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are shown. G, cell surface expression of ENO-1 in MDA-MB-231 cells stimulated with LPS in the absence or presence of an anti-TLR-4 antibody. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are demonstrated. IgG, isotype control. H and I, time course of ENO-1 mRNA and protein expression after treatment of MDA-MB-231 cells with LPS for indicated time points as assessed by qRT-PCR (H) and Western blotting (I). qRT-PCR data are expressed as ΔCt using PBGD as a reference gene. Representative Western blots are shown. J, levels of ENO-1 in conditioned culture media collected after stimulation of MDA-MB-231 cells with LPS for indicated time points. Hsp70 and CD63 served as exosome markers. Coomassie Brilliant Blue (CBB) staining served as a loading control (n = 3). Representative Western blots are shown. K, release of lactate dehydrogenase to cell culture media collected from the cells exposed to LPS for indicated time points. Triton X-100 was used as a positive control. The data represent mean values ± S.E. (n = 3). LDH, lactate dehydrogenase. L, cell surface expression of ENO-1 in MCF-7, MDA-MB-231, and MDA-MB-435 cells exposed to LPS for 2 h. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are shown.
ENO-1 Exteriorization Is Triggered by Stimuli Promoting Tumor Progression
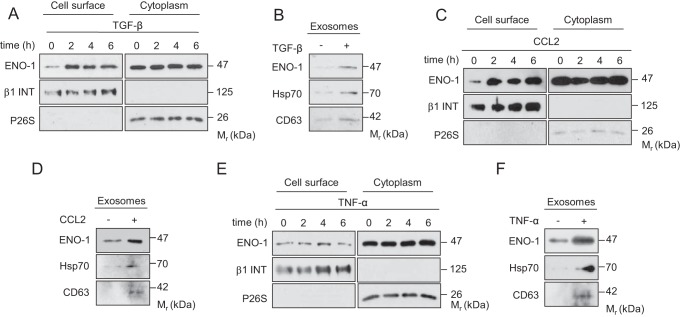
To study whether ENO-1 exteriorization is stimulus-dependent, MDA-MB-231 cells were treated with TGF-β1, CCL2, or TNF-α and cell surface ENO-1 mobilization, as well as exosome release were analyzed. All aforementioned stimuli were shown to promote cancer progression by enhancing cancer cell proliferation, migration, and invasion (32–34). Although TGF-β1 and CCL2 increased cell surface expression of ENO-1 (Fig. 3, A and C), TNF-α did not promote ENO-1 translocation to the outer leaflet of the plasma membrane (Fig. 3E). TGF-β1, CCL2, and TNF-α augmented ENO-1 release into the extracellular space, which was accompanied by a concomitant increase in the levels of Hsp70 and CD63 (Fig. 3, B, D, and F). Collectively, these results demonstrate that ENO-1 exteriorization is promoted by factors associated with tumor progression.
FIGURE 3.
ENO-1 exteriorization is triggered by stimuli promoting tumor progression. A, C, and E, cell surface expression of ENO-1 in MDA-MB-231 cells exposed to 20 ng/ml TGF-β1 (A), 100 ng/ml CCL2 (C), or 50 ng/ml TNF-α (E) for the indicated time points. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are shown. B, D, and F, abundance of ENO-1 in exosomes isolated from MDA-MB-231 cells exposed to 20 ng/ml TGF-β1 (B), 100 ng/ml CCL2 (D), or 50 ng/ml TNF-α (F) as assessed by Western blotting. Hsp70 and CD63 served as exosome markers (n = 3). Representative Western blots are demonstrated.
ENO-1 Released from MDA-MB-231 Cells in the Form of Exosomes Enhances Tumor Cell Migration
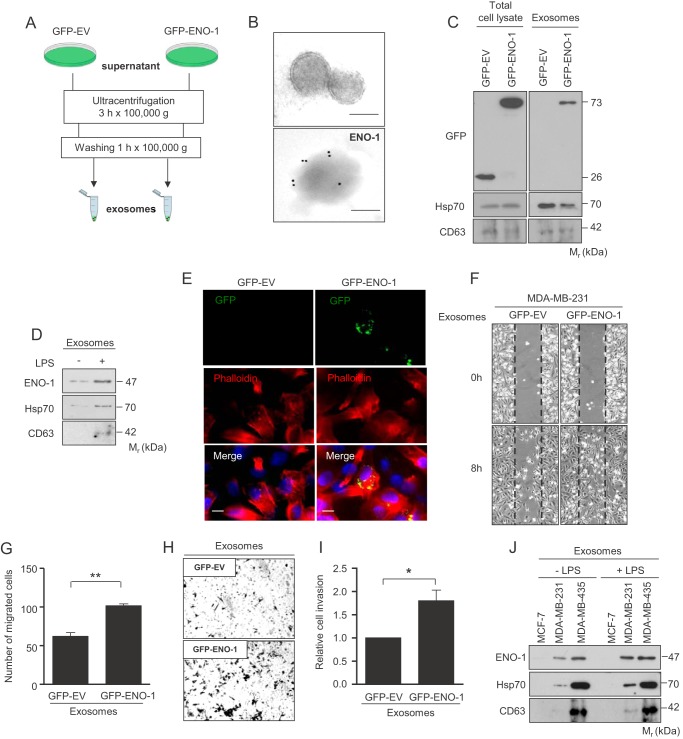
Accumulating evidence suggests that cancer cells secrete abundant levels of exosomes, small (30–150 nm in diameter) membranous vesicles (35). Thus, we examined whether ENO-1 detected in the cell culture media of MDA-MB-231 cells is associated with exosomes. Exosomes were isolated by serial ultracentrifugation steps from cell culture supernatants of MDA-MB-231 cells overexpressing GFP-EV or GFP-ENO-1 (Fig. 4A). Electron microscopy confirmed the presence of double membrane vesicles with a size range between 100 and 150 nm in the recovered high speed pellets (Fig. 4B, upper panel). Enolase-1, detected by an anti-ENO-1 antibody coupled to gold particles, was observed on the surface of exosomes (Fig. 4B, lower panel). Purified exosomes were further analyzed by Western blotting. Despite similar expression levels of GFP and GFP-tagged ENO-1 in MDA-MB-231 total cell lysates, only GFP-ENO-1 was found in exosomes (Fig. 4C). In addition to GFP-ENO-1, isolated exosomes were positive for CD63 and Hsp70 (Fig. 4C). To assess whether the LPS-triggered increase in the amount of extracellular ENO-1 is a result of augmented exosome secretion, exosomes were isolated from LPS-treated MDA-MB-231 cells and subjected to Western blotting with an anti-ENO-1 antibody. Stimulation of cells with LPS resulted in increased levels of ENO-1, which was accompanied by a concomitant increase in the levels of Hsp70 and CD63 (Fig. 4D). Because LPS did not potentiate cell proliferation, changes in the cell number could not account for the observed effect. To assess a functional role of exosomal ENO-1 in cancer cells, an exosome uptake assay was performed. Immunofluorescence analysis demonstrated uptake of GFP-ENO-1-loaded exosomes by parental MDA-MB-231 cells and their redistribution in the perinuclear region (Fig. 4E). To determine whether exosomal ENO-1 is able to influence migratory and invasive properties of recipient cells, wound healing and Transwell invasion assays were performed in the presence of exosomes isolated from cells overexpressing either GFP alone or GFP-tagged ENO-1. The rate of wound closure was markedly increased when GFP-ENO-1 loaded vesicles were applied (Fig. 4, F and G). Similarly, invasion of MDA-MB-231 cells was enhanced when exosomes isolated from cells overexpressing GFP-ENO-1 were used (Fig. 4, H and I). These results support the functional role of exosomal ENO-1 in the regulation of cancer cell motility. To assess whether LPS potentiates release of ENO-1 into the extracellular space from other breast cancer cell lines, MCF-7 and MDA-MB-435 cells were analyzed for exosome production. ENO-1, Hsp70, and CD63 were not detected under basal conditions and upon LPS stimulation when MCF-7 cells were employed (Fig. 4J). MDA-MB-435 already displayed high extracellular levels of ENO-1, Hsp70, and CD63 under basal condition. LPS stimulation did not further increase exosome production in these cells (Fig. 4J). The observed differences in exosome secretion cannot arise from dissimilarities in cell growth because exosomes were always isolated from the same number of cells, irrespective of the conditions applied. Together, our results suggest that not only the level of cell surface-bound ENO-1 but also the amount of ENO-1 released into the extracellular space in the form of exosomes correlate with the invasive/metastatic potential of cancer cells.
FIGURE 4.
ENO-1 released from MDA-MB-231 cells in the form of exosomes enhances tumor cell migration. A, schematic representation of exosome purification from the cells overexpressing GFP-EV or GFP-ENO-1. B, transmission electron micrographs of exosomes isolated from MBA-MB-231 cells. Exosomes negatively stained with 2% uranyl acetate (upper panel). Exosomal ENO-1 visualized by a gold particle-coupled anti-ENO-1 antibody (lower panel). Bar, 100 nm. C, levels of ENO-1 in total protein extracts (left panel) and exosomes (right panel) isolated from GFP-EV and GFP-ENO-1-overexpressing MDA-MB-231 cells as assessed by Western blotting. Hsp70 and CD63 served as exosome markers (n = 3). Representative Western blots are shown. D, abundance of ENO-1 in exosomes isolated from MDA-MB-231 cells exposed to LPS as assessed by Western blotting. Hsp70 and CD63 served as exosome markers (n = 3). Representative Western blots are demonstrated. E, representative pictures demonstrating uptake of exosomes isolated from GFP-EV- and GFP-ENO-1-overexpressing cells by MDA-MB-231 cells. Bar, 5 μm (n = 3). F, impact of exosomes isolated from GFP-EV or GFP-ENO-1 cells on MDA-MB-231 cell migration. Representative pictures from the wound healing assay at times 0 and 8 h are shown. G, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 3). **, p < 0.01. H, MDA-MB-231 cells stimulated with exosomes isolated from cells overexpressing either GFP-EV or GFP-ENO-1 were seeded onto a fibronectin-coated membrane and allowed to invade for 16 h. Representative images of cells that invaded the underside of the membrane are shown. I, cells that invaded the underside of the membrane were counted. The data represent mean values ± S.E. (n = 3). *, p < 0.05. J, levels of ENO-1 in exosomes isolated from MCF-7, MDA-MB-231, and MDA-MB-435 cells exposed to LPS as assessed by Western blotting. Hsp70 and CD63 served as exosome markers (n = 3). Representative Western blots are demonstrated.
Translocation of ENO-1 to the Cell Surface of MDA-MB-231 Cells Occurs via a Nonclassical Secretory Pathway
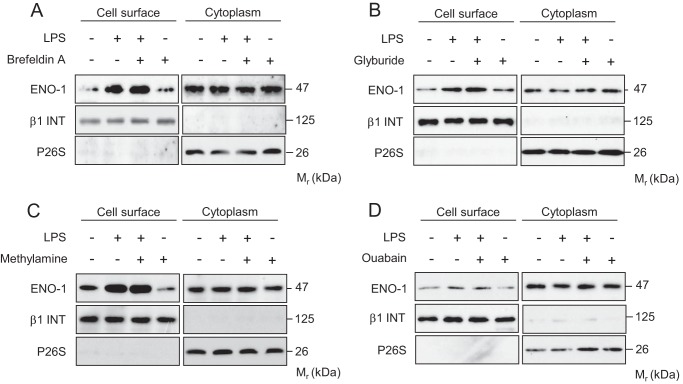
Because ENO-1 lacks an N-terminal signal peptide motif, which is required for endoplasmic reticulum/Golgi targeting (36), a nonconventional protein secretion pathway has been suggested to explain the transport of ENO-1 to the cell surface. To verify this notion, several biochemical approaches were applied. Preincubation of MDA-MB-231 cells with brefeldin A, a known blocker of endoplasmic reticulum/Golgi transport, did not inhibit translocation of ENO-1 to the cell surface upon exposure of cells to LPS, implying that a nonclassical secretion pathway is involved (Fig. 5A). Several mediators of nonconventional cell surface protein expression have been described, including ABC transporter, endosomal recycling, and Na+/K+ ATPase. However, glyburide, an ABC transporter inhibitor; methylamine, an endosomal recycling blocker; and ouabain, a Na+/K+ ATPase antagonist had no effect on LPS driven transport of ENO-1 to the cell surface (Fig. 5, B–D).
FIGURE 5.
Translocation of ENO-1 to the cell surface of MDA-MB-231 cells occurs via a nonclassical secretory pathway. A–D, cell surface expression of ENO-1 in MDA-MB-231 cells stimulated with LPS in the absence or presence of 10 μg/ml brefeldin A (A), 5 mm glyburide (B), 5 mm methylamine (C), or 100 nm ouabain (D). The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are demonstrated.
LPS-driven ENO-1 Exteriorization Is Mediated by Ca2+
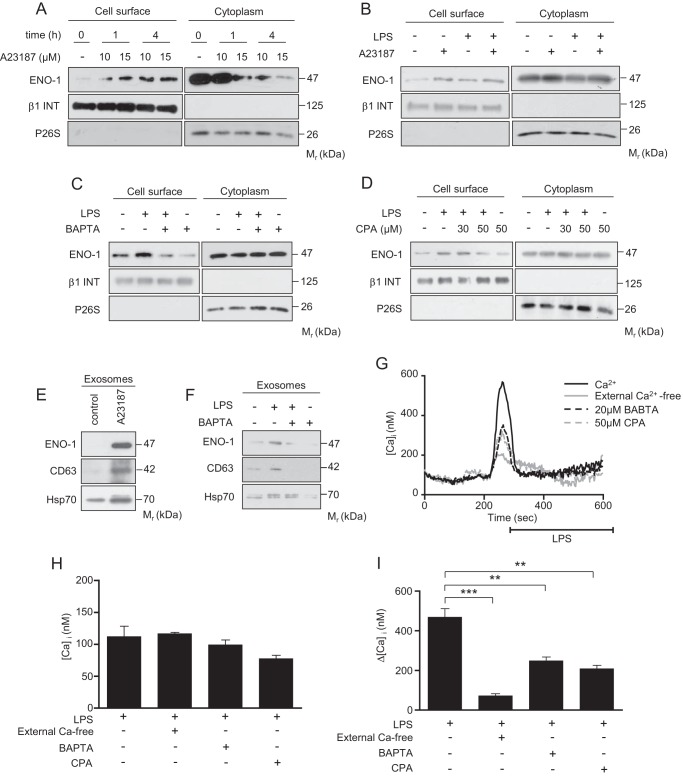
Because LTCCs have been implicated in the exteriorization of another PLG receptor, histone 2B (37), we next investigated the role of Ca2+ in the transport of ENO-1 to the cell surface. First, preincubation of MDA-MB-231 cells with the Ca2+ ionophore A23187 induced a time and concentration-dependent translocation of ENO-1 to the cell surface (Fig. 6A). To elucidate whether LPS and A23187 share a common pathway to regulate ENO-1 exteriorization, MDA-MB-231 cells were simultaneously treated with these two reagents. Although A23187 and LPS increased the transport of ENO-1 to the cell surface, this effect was not additive when both stimuli were applied at the same time (Fig. 6B), indicating that A23187 and LPS use a similar mechanism to regulate ENO-1 cell surface abundance. To further explore the involvement of Ca2+ in LPS-triggered transport of ENO-1 to the cell surface, the levels of intracellular Ca2+ were depleted by pretreatment of MDA-MB-231 cells with the Ca2+ chelator BAPTA. As depicted in Fig. 6C, BAPTA blocked LPS-driven ENO-1 translocation. Similar results were obtained when CPA, an inhibitor of endoplasmic reticulum Ca2+ATPase pump, was used (Fig. 6D). To investigate whether the release of ENO-1 into the extracellular space also depends on Ca2+ levels, MDA-MB-231 cells were first preincubated with the Ca2+ ionophore A23187. A23187 augmented exosomal levels of ENO-1, which positively correlated with elevated levels of CD63 and Hsp70 (Fig. 6E). Similar results were obtained when MDA-MB-231 cells were exposed to LPS. Moreover, LPS-driven release of exosomal ENO-1 was blocked by BAPTA. Notably, BAPTA alone completely abolished the release of exosomes into the extracellular space (Fig. 6F). To demonstrate the direct effect of LPS on Ca2+ levels, cells were treated either with LPS alone or in combination with BAPTA or CPA, and intracellular Ca2+ was measured by live cell imaging. There was no difference in the basal intracellular calcium level of the cells, regardless of the treatment or the presence of extracellular calcium (Fig. 6, G and H). The LPS-induced intracellular calcium increase was significantly diminished by removal of the extracellular calcium or 10 min of BAPTA or CPA treatment (Fig. 6, G and I). These data indicate that LPS-induced ENO-1 exteriorization is calcium-dependent.
FIGURE 6.
LPS-driven translocation of ENO-1 to the cell surface is mediated by Ca2+. A–D, cell surface expression of ENO-1 in MDA-MB-231 cells stimulated with LPS in the absence or presence of 10 or 15 μm A23187 (A and B) or 20 μm BAPTA (C) or CPA (D). The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are demonstrated. E and F, levels of ENO-1 in exosomes isolated from MDA-MB-231 cells stimulated with 1 μm A23187 (E) or with combination of LPS and 20 μm BAPTA (F) as assessed by Western blotting. Hsp70 and CD63 served as exosome markers (n = 3). Representative Western blots are shown. G, representative traces of live cell calcium measurements upon LPS stimulus in the absence or presence of extracellular calcium, 20 μm BAPTA, and 50 μm CPA. H and I, quantitative data illustrating basal calcium levels (H) and LPS-induced acute intracellular calcium change (I) in MDA-MB-231 cells. The data represent mean values ± S.E. (n experiments = 4; n cells = 26–52 in each group). **, p < 0.01; ***, p < 0.001.
Blockage of STIM1/ORAI1 Inhibits LPS-induced ENO-1 Exteriorization
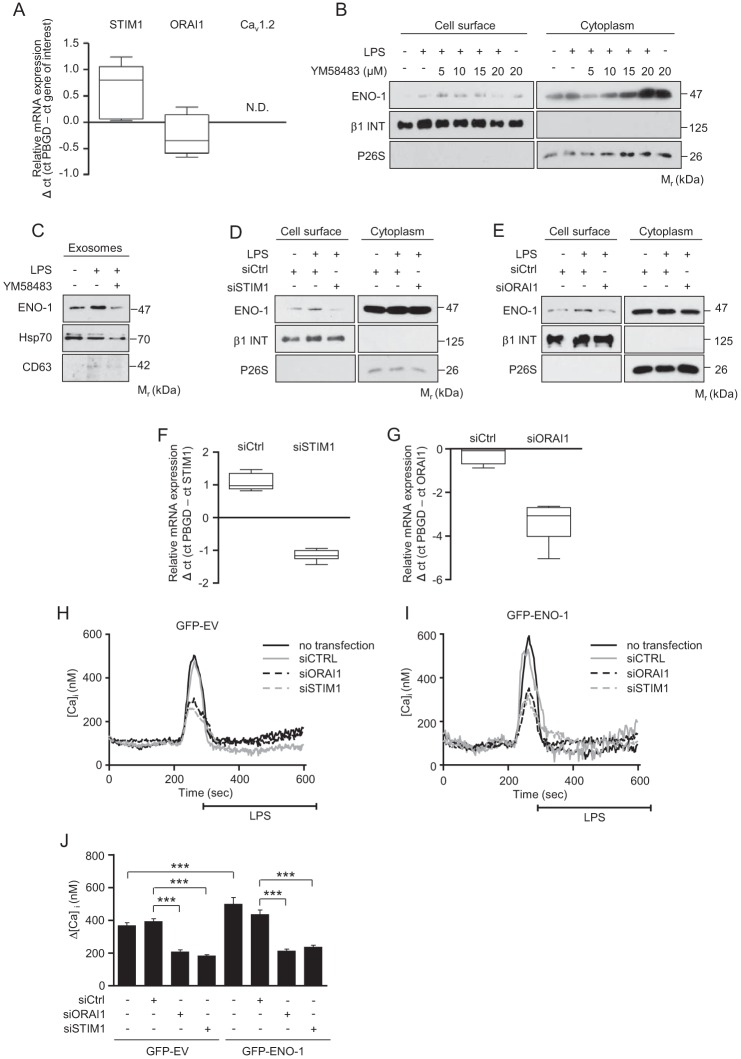
To further investigate the role of Ca2+ in the regulation of ENO-1 translocation, the expression STIM1 and its interacting partner ORAI1, a member of store-operated calcium (SOC) channels, as well as the Cav1.2 subunit of LTCC, was assessed. We focused on STIM1, ORAI1, and Cav1.2 LTCC because these Ca2+ channels have been found to control translocation of other PLG receptors to the cell surface and to regulate LPS-induced inflammatory responses (37–39). Whereas STIM1 and ORAI1 mRNA were detected in MDA-MB-231 cells, Cav1.2 was not expressed in this cell line (Fig. 7A). To determine the potential role of STIM1 and ORAI1 in LPS triggered transport of ENO-1 to the cell surface and to the extracellular space, a selective SOC channel inhibitor YM58483 was employed. Pretreatment of MDA-MB-231 cells with YM58483 suppressed, in a dose-dependent manner, LPS-induced translocation of ENO-1 to the cell surface (Fig. 7B) and to the extracellular space (Fig. 7C), suggesting an essential role of SOC channels in LPS-mediated ENO-1 exteriorization. To exclude nontarget effects of the YM58483 inhibitor, siRNA directed against STIM1 and ORAI1 was applied. Depletion of STIM1 markedly decreased LPS-driven translocation of ENO-1 to the cell surface (Fig. 7D). Similar results were obtained when ORAI1 siRNA was employed (Fig. 7E). Efficiency of STIM1 and ORAI1 knockdown in MDA-MB-231 cells is demonstrated in Fig. 7 (F and G). To prove the direct involvement of STIM1/ORAI1 in LPS-mediated Ca2+ influx in GFP-EV and GFP-ENO-1 cells, the cells were transfected with STIM1 or ORAI1 siRNA, and the changes in intracellular Ca2+ were measured by live cell imaging. As depicted in Fig. 7 (H–J), GFP-ENO-1 cells displayed increased intracellular Ca2+ levels upon LPS stimulation as compared with LPS-treated GFP-EV cells. Depletion of STIM1 or ORAI1 significantly reduced LPS-triggered Ca2+ influx as opposed to the cells treated with control siRNA in both cell types (Fig. 7J). These results suggest that overexpression of ENO-1 alters intracellular Ca2+ levels in MDA-MB-231 cells.
FIGURE 7.
Blockage of STIM1/ORAI1 inhibits LPS-induced ENO-1 exteriorization. A, qRT-PCR analysis of STIM1, ORAI1, and Cav1.2 mRNA expression in MDA-MB-231 cells. The data are expressed as ΔCt using PBGD as a reference gene (n = 3). N.D., not detectable. B, cell surface expression of ENO-1 in MDA-MB-231 cells exposed to LPS in the absence or presence of YM58483. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are shown. C, levels of ENO-1 in exosomes isolated from MDA-MB-231 cells exposed to LPS in the absence or presence of 5 μm YM58483 as assessed by Western blotting. Hsp70 and CD63 served as exosome markers (n = 3). Representative Western blots are shown. D and E, effect of STIM1 (D) and ORAI1 (E) depletion on ENO-1 cell surface levels in MDA-MB-231 stimulated with LPS. The purity of cytosolic and cell membrane fractions was assessed by probing the samples for β1 INT and P26S, respectively (n = 3). Representative Western blots are demonstrated. siSTIM1, siRNA directed against STIM1; siORAI1, siRNA directed against ORAI1; siCtrl, control siRNA. F and G, efficacy of STIM1 (F) and ORAI1 (G) knockdown in MDA-MB-231 cells as assessed by qRT-PCR. The data are expressed as ΔCt using PBGD as a reference gene (n = 3). H and I, representative traces of live cell calcium measurements demonstrating LPS-induced changes in intracellular calcium level in GFP-EV (H) and GFP-ENO-1 (I) cells upon STIM1 or ORAI1 knockdown. J, quantitative data illustrating LPS-induced changes in intracellular calcium level in GFP-EV and GFP-ENO-1 cells upon STIM1 or ORAI1 depletion. The data represent mean values ± S.E. (n experiments = 3; n cells = 44–119 in each group). ***, p < 0.001.
Depletion of STIM1/ORAI1 Reduces ENO-1-dependent MDA-MB-231 Cell Motility
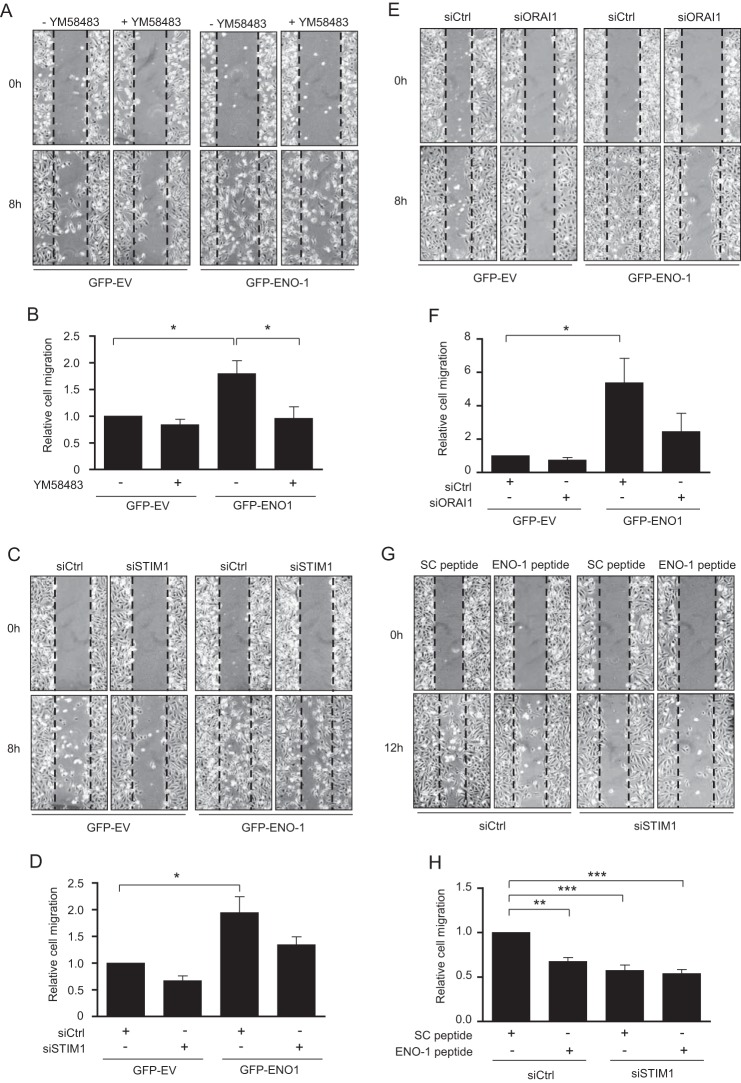
To elucidate the potential role of STIM1 and ORAI1 in LPS-driven ENO-1 exteriorization and thus in the regulation of cell motility, a wound healing assay employing MDA-MB-231 cells overexpressing GFP alone or GFP-ENO-1 was performed. Overexpression of GFP-ENO-1 markedly increased the rate of wound closure as compared with the cells transfected with GFP-EV. YM58483 inhibited the effect of GFP-ENO-1 overexpression, decreasing cell migration to the level observed in YM58483-treated GFP-EV-overexpressing MDA-MB-231 cells (Fig. 8, A and B). To confirm these results, cells transfected either with GFP-EV or GFP-ENO-1 were treated with siRNA directed against STIM1 or control siRNA. A clear tendency toward reduced cell migration was observed in ENO-1-overexpressing cells treated with STIM1 siRNA as compared with the control siRNA transfected cells (Fig. 8, C and D). Similar results were obtained when ORAI1 siRNA was employed (Fig. 8, E and F). To further confirm that suppressed migration of MDA-MB-231 cells is a result of decreased ENO-1 cell surface expression, STIM-1 depleted nonoverexpressing MDA-MB-231 cells were treated with a peptide directed against the C-terminal part of ENO-1 involved in the binding of PLG, and a wound healing assay was performed. Treatment of cells with the ENO-1 peptide or STIM1 siRNA considerably reduced their migratory properties; however, this effect was not potentiated when two reagents were applied at the same time (Fig. 8, G and H). These results support the crucial role of STIM1-mediated Ca2+ influx in ENO-1-dependent cell migration.
FIGURE 8.
Suppression of STIM1 or ORAI1 reduces ENO-1-mediated migration of MDA-MB-231 cells. A, effect of 5 μm YM58483 on nondirectional migration of MDA-MB-231 cells stably transfected with GFP-EV or GFP-ENO-1. Cells were scratched and incubated for 8 h at 37 °C in serum-free RPMI medium. Representative pictures from the wound healing assay at times 0 and 8 h are shown. B, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 5). *, p < 0.05 C, impact of STIM1 knockdown on nondirectional migration of MDA-MB-231 cells stably transfected with GFP-EV or GFP-ENO-1. Cells were scratched and incubated for 8 h at 37 °C in serum-free RPMI medium. Representative pictures from the wound healing assay at times 0 and 8 h are shown. D, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 3). *, p < 0.05. E, effect of ORAI1 knockdown on nondirectional migration of MDA-MB-231 cells stably transfected with GFP-EV or GFP-ENO-1. Cells were scratched and incubated for 8 h at 37 °C in serum-free RPMI medium. Representative pictures from the wound healing assay at times 0 and 8 h are shown. F, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 3). *, p < 0.05. G, effect of STIM1 knockdown and/or ENO-1 peptide on nondirectional migration of MDA-MB-231 cells. Cells were scratched and incubated for 12 h at 37 °C in serum-free RPMI medium. Representative pictures from the wound healing assay at times 0 and 12 h are shown. H, the rate of wound closure was assessed by counting the cells that migrated into the same-sized square fields. The data represent mean values ± S.E. (n = 3). **, p < 0.01; ***, p < 0.001.
DISCUSSION
Enolase-1 is a glycolytic enzyme primarily localized in the cytoplasm of prokaryotic and eukaryotic cells (12). However, under stimulatory conditions, ENO-1 can be translocated to the cell surface, where it acts as a PLG receptor (13). Accumulation of PLG on the cell surface and its subsequent conversion to plasmin contribute to the modulation of pericellular proteolytic activity and thus to the regulation of migratory properties of numerous cell types including cancer cells (14, 40). Although increased abundance of ENO-1 was described on the cell surface of cancer cells, the contribution of cell surface-bound ENO-1 to the increased migratory and the invasive properties of tumor cells, as well as the mechanism underlying ENO-1 exteriorization, have not yet been reported. In the present study, we demonstrate that LPS potentiates ENO-1 exteriorization in cancer cells, thereby increasing their motility. This phenomenon is mediated by a nonclassical protein secretion pathway and depends on Ca2+ levels regulated by STIM1 and ORAI1.
Enolase-1 expression is altered in many cancer types, and several mechanisms seem to account for the indicated changes in ENO-1 production. First, ENO-1 is located in the chromosomal region 1p36 (41), which is frequently rearranged in human malignancies. Second, hypoxia drives transcription of the ENO-1 gene through a hypoxia-inducible factor 1 binding element (42). Third, the expression of ENO-1 is elevated in cells overexpressing prooncogenic c-Myc (43). In line with these considerations, high levels of ENO-1 were found in many types of cancer, including head, neck, breast, and lung cancer (44). High levels of ENO-1 in cancer cells correlate with cancer progression and a poor clinical outcome of the affected patients (12). Ectopic overexpression of ENO-1 promotes cell proliferation, migration, invasion, and colony formation, thereby contributing to metastasis formation (19, 21, 45). The ability of ENO-1 to regulate so many processes related to cancer cell biology results from its participation in glucose metabolism and therefore in energy production, as well as its capability to modulate expression of genes involved in cell growth and inflammation (45). Our data indicate that altered cell surface expression of ENO-1 and thus augmented pericellular proteolytic activity may also contribute to its prooncogenic functions. Consistent with this notion, a direct link between the cell surface amount of PLG receptors and invasiveness of cancer cells has been demonstrated (46).
Although high cell surface expression of ENO-1 has been detected on cancer cells, our results indicate that inflammatory conditions may further increase cell surface levels of this glycolytic enzyme. LPS-induced inflammatory responses have been shown to regulate cancer development/progression in several ways: (i) they may potentiate expression of proteins involved in the breakdown of the extracellular matrix (47); (ii) increase adhesive properties of cancer cells that are essential for the metastatic colonization of a normal tissue (25); (iii) induce recruitment of inflammatory cells into the tumor microenvironment, which in turn can contribute to extracellular matrix turnover (48); (iv) regulate angiogenesis (49); and (v) modulate, as indicated by our study, the cell surface-associated proteolysis by triggering PLG receptor exteriorization. LPS-triggered ENO-1 translocation was found to be independent of the classical endoplasmic reticulum-Golgi pathway and de novo protein synthesis, and it was associated with increased levels of Ca2+. The elevated levels of cell surface bound ENO-1 contributed to increased cancer cell motility, highlighting the functional importance of this phenomenon.
Strikingly, our results imply that ENO-1 is not only translocated to the cell surface of tumor cells but also released into the extracellular space in the form of exosomes. A growing body of evidence suggests that exosomes may act as regulators of cell to cell communication. This concept is based on the finding that exosomes released from a given cell type may interact with other cells, leading to target cell stimulation (50). Exosomes may influence the behavior of target cells in multiple ways. For example, they can activate signaling complexes by direct stimulation of target cells or deliver genetic information/proteins to recipient cells (50). Thus, exosomes were found to regulate a variety of processes, including cell proliferation, differentiation, migration, and invasion (50). Tumor cells were found to release large amounts of exosomes (35). Exosomes derived from the tumor may transfer oncogenes and proangiogenic signals to stromal cells and thus promote tumor vascularization. Additionally, they can contribute to stromal remodeling and tumor cell invasion by carrying active matrix metalloproteinases (50). In view of these results, it is tempting to speculate that exosomal ENO-1 could contribute to tumor progression either by concentrating proteolytic activity on the cancer cell surface or by enlarging the cytoplasmic pool of ENO-1 and, by doing this, regulating expression of the genes involved in cell growth, migration, and inflammation. However, it is unclear whether exosomal ENO-1 increases the migratory and invasive properties of target cells by being reattached to the cell membrane or by being endocytosed and consequently translocated to the cell surface. The fact that the mechanism of ENO-1 release into the extracellular space in response to LPS is similar to the mechanism responsible for translocation of this glycolytic enzyme to the cell surface implies that these two processes may occur simultaneously or successively. Namely, cell surface localization of ENO-1 may assure its association with the intralumenal vesicles of the multivesicular endosomes and its further release into the extracellular space in the form of exosomes. Rebinding of exteriorized ENO-1 to the cell surface closes the cycle of ENO-1 extracellular transport. A similar mechanism has been proposed to explain extracellular localization of annexin A2 (51). Nonetheless, it still remains to be elucidated which part of ENO-1 stays on the cell surface and which one is packed into exosomes and where the decision about ENO-1 sorting is made.
Our previous study points toward the role of caveolin-1 (cav-1), the main scaffolding protein of caveolae, in the transport of ENO-1 to the outer leaflet of the plasma membrane (21). A growing body of evidence suggests that elevated expression of cav-1 in advanced stages of cancer promotes tumor cell proliferation and spreading (52, 53). This phenomenon can be partially explained by cav-1-dependent stimulation of store operated Ca2+ entry (SOCE) and thus activation of multiple signaling pathways (54), which in turn may initiate cav-1 expression (55), thereby creating a positive feedback loop. In view of these results, it is tempting to speculate that altered levels of intracellular Ca2+ not only affect expression of cav-1 but also facilitate its binding to ENO-1 and thus the transport of this glycolytic enzyme to the cell surface, finally leading to increased pericellular proteolytic activity and thus cell spreading. LPS, as a modulator of SOCE activity (38) and cav-1 expression (56), may perpetuate all events.
Although molecular details of the pathway(s) governing Ca2+-mediated ENO-1 extracellular localization in tumor cells remain to be deciphered, we show here a critical role for STIM1 and ORAI1 in this process. STIM1 molecules in the endoplasmic reticulum and ORAI1 proteins in the plasma membrane are two main components of SOCE that can couple into a pore-forming complex to regulate levels of intracellular Ca2+. Stimulation of STIM1 can activate SOCE, leading to sustained extracellular calcium influx (57). Several studies demonstrated a critical role of STIM1 in cancer cell migration and metastasis formation (58, 59). Concomitantly, overexpression of STIM1 has been observed in various types of human cancer, and it was found to have a diagnostic as well as prognostic value (60). Given the fact that STIM1- and ORAI1-dependent ENO-1 exteriorization markedly contributes to increased breast cancer cell motility, pharmacological blockers of either STIM1 or ORAI1 could represent one possible approach in anti-cancer therapy. Supporting this concept, suppression of STIM1 in the animal model of human glioblastoma significantly inhibited tumor growth and metastasis formation (58).
Collectively, our data provide new insights into the mechanism responsible for translocation of ENO-1 to the surface of cancer cells and its release into the extracellular space. The pivotal role of STIM1/ORAI1-mediated Ca2+ influx in the aforementioned processes may, in part, explain the beneficial effect of STIM1 inhibition in the experimental models of cancer.
Acknowledgments
We thank Horst Thiele and Yvonne Horn for excellent technical assistance.
This work was supported by the University Medical Center Giessen and Marburg (to M. W.), Deutsche Forschungsgemeinschaft Grant WY119/1-1 (to M. W. and K. T. P.), Sonderforschungsbereich-Transregio 84 Project A2 (to M. W. and K. T. P.), the Excellence Cluster Cardiopulmonary System (to M. W. and D. Z.), and the Deutsches Zentrum für Lungenforschung.
- PLG
- plasminogen
- ENO-1
- enolase-1
- TLR-4
- Toll-like receptor-4
- CCL2
- chemokine (C-C) motif ligand 2
- Hsp70
- heat shock protein 70
- β1 INT
- β1 integrin
- P26S
- 26S proteasome
- BAPTA
- 1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- CPA
- cyclopiazonic acid
- qRT-PCR
- quantitative real time RT-PCR
- LTCC
- L-type Ca2+ channel
- STIM1
- stromal interaction molecule 1
- ORAI1
- calcium release-activated calcium modulator 1
- SOCE
- store-operated calcium entry
- cav-1
- caveolin-1.
REFERENCES
- 1. Werb Z., Vu T. H., Rinkenberger J. L., Coussens L. M. (1999) Matrix-degrading proteases and angiogenesis during development and tumor formation. APMIS 107, 11–18 [DOI] [PubMed] [Google Scholar]
- 2. Lund L. R., Romer J., Bugge T. H., Nielsen B. S., Frandsen T. L., Degen J. L., Stephens R. W., Danø K. (1999) Functional overlap between two classes of matrix-degrading proteases in wound healing. EMBO J. 18, 4645–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnsen M., Lund L. R., Rømer J., Almholt K., Danø K. (1998) Cancer invasion and tissue remodeling: common themes in proteolytic matrix degradation. Curr. Opin. Cell Biol. 10, 667–671 [DOI] [PubMed] [Google Scholar]
- 4. Matrisian L. M. (1999) Cancer biology: extracellular proteinases in malignancy. Curr. Biol. 9, R776–R778 [DOI] [PubMed] [Google Scholar]
- 5. Stamenkovic I. (2000) Matrix metalloproteinases in tumor invasion and metastasis. Semin. Cancer Biol. 10, 415–433 [DOI] [PubMed] [Google Scholar]
- 6. Robbins K. C., Summaria L., Hsieh B., Shah R. J. (1967) The peptide chains of human plasmin: mechanism of activation of human plasminogen to plasmin. J. Biol. Chem. 242, 2333–2342 [PubMed] [Google Scholar]
- 7. Plow E. F., Freaney D. E., Plescia J., Miles L. A. (1986) The plasminogen system and cell surfaces: evidence for plasminogen and urokinase receptors on the same cell type. J. Cell Biol. 103, 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellis V., Behrendt N., Danø K. (1991) Plasminogen activation by receptor-bound urokinase: a kinetic study with both cell-associated and isolated receptor. J. Biol. Chem. 266, 12752–12758 [PubMed] [Google Scholar]
- 9. Gonzalez-Gronow M., Stack S., Pizzo S. V. (1991) Plasmin binding to the plasminogen receptor enhances catalytic efficiency and activates the receptor for subsequent ligand binding. Arch. Biochem. Biophys. 286, 625–628 [DOI] [PubMed] [Google Scholar]
- 10. Hall S. W., Humphries J. E., Gonias S. L. (1991) Inhibition of cell surface receptor-bound plasmin by α 2-antiplasmin and α 2-macroglobulin. J. Biol. Chem. 266, 12329–12336 [PubMed] [Google Scholar]
- 11. Plow E. F., Doeuvre L., Das R. (2012) So many plasminogen receptors: why? J. Biomed. Biotechnol. 2012, 141806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capello M., Ferri-Borgogno S., Cappello P., Novelli F. (2011) α-Enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 278, 1064–1074 [DOI] [PubMed] [Google Scholar]
- 13. Miles L. A., Dahlberg C. M., Plescia J., Felez J., Kato K., Plow E. F. (1991) Role of cell-surface lysines in plasminogen binding to cells: identification of α-enolase as a candidate plasminogen receptor. Biochemistry 30, 1682–1691 [DOI] [PubMed] [Google Scholar]
- 14. Wygrecka M., Marsh L. M., Morty R. E., Henneke I., Guenther A., Lohmeyer J., Markart P., Preissner K. T. (2009) Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood 113, 5588–5598 [DOI] [PubMed] [Google Scholar]
- 15. Redlitz A., Fowler B. J., Plow E. F., Miles L. A. (1995) The role of an enolase-related molecule in plasminogen binding to cells. Eur. J. Biochem. 227, 407–415 [DOI] [PubMed] [Google Scholar]
- 16. Cappello P., Tomaino B., Chiarle R., Ceruti P., Novarino A., Castagnoli C., Migliorini P., Perconti G., Giallongo A., Milella M., Monsurrò V., Barbi S., Scarpa A., Nisticò P., Giovarelli M., Novelli F. (2009) An integrated humoral and cellular response is elicited in pancreatic cancer by α-enolase, a novel pancreatic ductal adenocarcinoma-associated antigen. Int. J. Cancer 125, 639–648 [DOI] [PubMed] [Google Scholar]
- 17. He P., Naka T., Serada S., Fujimoto M., Tanaka T., Hashimoto S., Shima Y., Yamadori T., Suzuki H., Hirashima T., Matsui K., Shiono H., Okumura M., Nishida T., Tachibana I., Norioka N., Norioka S., Kawase I. (2007) Proteomics-based identification of α-enolase as a tumor antigen in non-small lung cancer. Cancer Sci. 98, 1234–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seweryn E., Pietkiewicz J., Bednarz-Misa I. S., Ceremuga I., Saczko J., Kulbacka J., Gamian A. (2009) Localization of enolase in the subfractions of a breast cancer cell line. Z. Naturforsch. C. 64, 754–758 [DOI] [PubMed] [Google Scholar]
- 19. Didiasova M., Wujak L., Wygrecka M., Zakrzewicz D. (2014) From plasminogen to plasmin: role of plasminogen receptors in human cancer. Int. J. Mol. Sci. 15, 21229–21252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsiao K. C., Shih N. Y., Fang H. L., Huang T. S., Kuo C. C., Chu P. Y., Hung Y. M., Chou S. W., Yang Y. Y., Chang G. C., Liu K. J. (2013) Surface α-enolase promotes extracellular matrix degradation and tumor metastasis and represents a new therapeutic target. PLoS One 8, e69354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zakrzewicz D., Didiasova M., Zakrzewicz A., Hocke A. C., Uhle F., Markart P., Preissner K. T., Wygrecka M. (2014) The interaction of enolase-1 with caveolae-associated proteins regulates its subcellular localization. Biochem. J. 460, 295–307 [DOI] [PubMed] [Google Scholar]
- 22. Chang G. C., Liu K. J., Hsieh C. L., Hu T. S., Charoenfuprasert S., Liu H. K., Luh K. T., Hsu L. H., Wu C. W., Ting C. C., Chen C. Y., Chen K. C., Yang T. Y., Chou T. Y., Wang W. H., Whang-Peng J., Shih N. Y. (2006) Identification of α-enolase as an autoantigen in lung cancer: its overexpression is associated with clinical outcomes. Clin. Cancer Res. 12, 5746–5754 [DOI] [PubMed] [Google Scholar]
- 23. McDonald B., Spicer J., Giannais B., Fallavollita L., Brodt P., Ferri L. E. (2009) Systemic inflammation increases cancer cell adhesion to hepatic sinusoids by neutrophil mediated mechanisms. Int. J. Cancer 125, 1298–1305 [DOI] [PubMed] [Google Scholar]
- 24. Pidgeon G. P., Harmey J. H., Kay E., Da Costa M., Redmond H. P., Bouchier-Hayes D. J. (1999) The role of endotoxin/lipopolysaccharide in surgically induced tumour growth in a murine model of metastatic disease. Br. J. Cancer 81, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu R. Y., Chan C. H., Spicer J. D., Rousseau M. C., Giannias B., Rousseau S., Ferri L. E. (2011) LPS-induced TLR4 signaling in human colorectal cancer cells increases β1 integrin-mediated cell adhesion and liver metastasis. Cancer Res. 71, 1989–1998 [DOI] [PubMed] [Google Scholar]
- 26. Grivennikov S. I., Karin M. (2011) Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Ann. Rheum. Dis. 70, i104–i108 [DOI] [PubMed] [Google Scholar]
- 27. Perera P. Y., Mayadas T. N., Takeuchi O., Akira S., Zaks-Zilberman M., Goyert S. M., Vogel S. N. (2001) CD11b/CD18 acts in concert with CD14 and Toll-like receptor (TLR) 4 to elicit full lipopolysaccharide and taxol-inducible gene expression. J. Immunol. 166, 574–581 [DOI] [PubMed] [Google Scholar]
- 28. Szkaradkiewicz A., Marciniak R., Chudzicka-Strugala I., Wasilewska A., Drews M., Majewski P., Karpinski T., Zwozdziak B. (2009) Proinflammatory cytokines and IL-10 in inflammatory bowel disease and colorectal cancer patients. Arch. Immunol. Ther. Exp. (Warsz.) 57, 291–294 [DOI] [PubMed] [Google Scholar]
- 29. Bálint Z., Zabini D., Konya V., Nagaraj C., Végh A. G., Váró G., Wilhelm I., Fazakas C., Krizbai I. A., Heinemann A., Olschewski H., Olschewski A. (2014) Double-stranded RNA attenuates the barrier function of human pulmonary artery endothelial cells. PLoS One 8, e63776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grynkiewicz G., Poenie M., Tsien R. Y. (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 31. Zhang F., Wang Z. M., Liu H. Y., Bai Y., Wei S., Li Y., Wang M., Chen J., Zhou Q. H. (2010) Application of RT-PCR in formalin-fixed and paraffin-embedded lung cancer tissues. Acta Pharmacol. Sin. 31, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jakowlew S. B. (2006) Transforming growth factor-β in cancer and metastasis. Cancer Metastasis Rev. 25, 435–457 [DOI] [PubMed] [Google Scholar]
- 33. Bonapace L., Coissieux M. M., Wyckoff J., Mertz K. D., Varga Z., Junt T., Bentires-Alj M. (2014) Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature 515, 130–133 [DOI] [PubMed] [Google Scholar]
- 34. Fang W. B., Jokar I., Zou A., Lambert D., Dendukuri P., Cheng N. (2012) CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J. Biol. Chem. 287, 36593–36608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peinado H., Lavotshkin S., Lyden D. (2011) The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin. Cancer Biol. 21, 139–146 [DOI] [PubMed] [Google Scholar]
- 36. Plow E. F., Das R. (2009) Enolase-1 as a plasminogen receptor. Blood 113, 5371–5372 [DOI] [PubMed] [Google Scholar]
- 37. Das R., Burke T., Van Wagoner D. R., Plow E. F. (2009) L-type calcium channel blockers exert an antiinflammatory effect by suppressing expression of plasminogen receptors on macrophages. Circ. Res. 105, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun R., Zhu Z., Su Q., Li T., Song Q. (2012) Toll-like receptor 4 is involved in bacterial endotoxin-induced endothelial cell injury and SOC-mediated calcium regulation. Cell Biol. Int. 36, 475–481 [DOI] [PubMed] [Google Scholar]
- 39. Gandhirajan R. K., Meng S., Chandramoorthy H. C., Mallilankaraman K., Mancarella S., Gao H., Razmpour R., Yang X. F., Houser S. R., Chen J., Koch W. J., Wang H., Soboloff J., Gill D. L., Madesh M. (2013) Blockade of NOX2 and STIM1 signaling limits lipopolysaccharide-induced vascular inflammation. J. Clin. Invest. 123, 887–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andronicos N. M., Ranson M. (2001) The topology of plasminogen binding and activation on the surface of human breast cancer cells. Br. J. Cancer 85, 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. D'Ancona G. G., Chern C. J., Benn P., Croce C. M. (1977) Assignment of the human gene for enolase 1 to region pter in equilibrium p36 of chromosome 1. Cytogenet. Cell Genet. 18, 327–332 [DOI] [PubMed] [Google Scholar]
- 42. Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 43. Ray R. B., Steele R., Seftor E., Hendrix M. (1995) Human breast carcinoma cells transfected with the gene encoding a c-myc promoter-binding protein (MBP-1) inhibits tumors in nude mice. Cancer Res. 55, 3747–3751 [PubMed] [Google Scholar]
- 44. Altenberg B., Greulich K. O. (2004) Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84, 1014–1020 [DOI] [PubMed] [Google Scholar]
- 45. Díaz-Ramos A., Roig-Borrellas A., García-Melero A., López-Alemany R. (2012) α-Enolase, a multifunctional protein: its role on pathophysiological situations. J. Biomed. Biotechnol. 2012, 156795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andreasen P. A., Egelund R., Petersen H. H. (2000) The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol. Life Sci. 57, 25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Del Pozo J. L. (2010) Primers on molecular pathways: lipopolysaccharide signaling-potential role in pancreatitis and pancreatic cancer. Pancreatology 10, 114–118 [DOI] [PubMed] [Google Scholar]
- 48. Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Cancer-related inflammation. Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 49. Pei Z., Lin D., Song X., Li H., Yao H. (2008) TLR4 signaling promotes the expression of VEGF and TGFβ1 in human prostate epithelial PC3 cells induced by lipopolysaccharide. Cell Immunol. 254, 20–27 [DOI] [PubMed] [Google Scholar]
- 50. Urbanelli L., Magini A., Buratta S., Brozzi A., Sagini K., Polchi A., Tancini B., Emiliani C. (2013) Signaling pathways in exosomes biogenesis, secretion and fate. Genes 4, 152–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Valapala M., Vishwanatha J. K. (2011) Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. J. Biol. Chem. 286, 30911–30925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Senetta R., Stella G., Pozzi E., Sturli N., Massi D., Cassoni P. (2013) Caveolin-1 as a promoter of tumour spreading: when, how, where and why. J. Cell Mol. Med. 17, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goetz J. G., Lajoie P., Wiseman S. M., Nabi I. R. (2008) Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Rev. 27, 715–735 [DOI] [PubMed] [Google Scholar]
- 54. Zhu H., Weisleder N., Wu P., Cai C., Chen J. W. (2008) Caveolae/caveolin-1 are important modulators of store-operated calcium entry in Hs578/T breast cancer cells. J. Pharmacol. Sci. 106, 287–294 [DOI] [PubMed] [Google Scholar]
- 55. Yang X. Y., Huang C. C., Kan Q. M., Li Y., Liu D., Zhang X. C., Sato T., Yamagata S., Yamagata T. (2012) Calcium regulates caveolin-1 expression at the transcriptional level. Biochem. Biophys. Res. Commun. 426, 334–341 [DOI] [PubMed] [Google Scholar]
- 56. Tiruppathi C., Shimizu J., Miyawaki-Shimizu K., Vogel S. M., Bair A. M., Minshall R. D., Predescu D., Malik A. B. (2008) Role of NF-κB-dependent caveolin-1 expression in the mechanism of increased endothelial permeability induced by lipopolysaccharide. J. Biol. Chem. 283, 4210–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li G., Zhang Z., Wang R., Ma W., Yang Y., Wei J., Wei Y. (2013) Suppression of STIM1 inhibits human glioblastoma cell proliferation and induces G0/G1 phase arrest. J. Exp. Clin. Cancer Res. 32, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang S., Zhang J. J., Huang X. Y. (2009) Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 15, 124–134 [DOI] [PubMed] [Google Scholar]
- 60. Chen Y. F., Chen Y. T., Chiu W. T., Shen M. R. (2013) Remodeling of calcium signaling in tumor progression. J. Biomed. Sci. 20, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]