Background: Scorpion venoms are an ample source of toxins targeting potassium channels.
Results: A comprehensive search for new toxins was performed by combining transcriptomics and peptidomics with a fluorescent test system.
Conclusion: We identified five new high affinity potassium channel blockers in the venom of Mesobuthus eupeus.
Significance: The proposed integrated approach is of general utility for potassium channel pharmacology.
Keywords: Drug Screening, Fluorescence, Molecular Pharmacology, Neurotoxin, Potassium Channel, Protein Chimera, Protein Drug Interaction, Proteomics, Toxin, Transcriptomics
Abstract
The lesser Asian scorpion Mesobuthus eupeus (Buthidae) is one of the most widely spread and dispersed species of the Mesobuthus genus, and its venom is actively studied. Nevertheless, a considerable amount of active compounds is still under-investigated due to the high complexity of this venom. Here, we report a comprehensive analysis of putative potassium channel toxins (KTxs) from the cDNA library of M. eupeus venom glands, and we compare the deduced KTx structures with peptides purified from the venom. For the transcriptome analysis, we used conventional tools as well as a search for structural motifs characteristic of scorpion venom components in the form of regular expressions. We found 59 candidate KTxs distributed in 30 subfamilies and presenting the cysteine-stabilized α/β and inhibitor cystine knot types of fold. M. eupeus venom was then separated to individual components by multistage chromatography. A facile fluorescent system based on the expression of the KcsA-Kv1.1 hybrid channels in Escherichia coli and utilization of a labeled scorpion toxin was elaborated and applied to follow Kv1.1 pore binding activity during venom separation. As a result, eight high affinity Kv1.1 channel blockers were identified, including five novel peptides, which extend the panel of potential pharmacologically important Kv1 ligands. Activity of the new peptides against rat Kv1.1 channel was confirmed (IC50 in the range of 1–780 nm) by the two-electrode voltage clamp technique using a standard Xenopus oocyte system. Our integrated approach is of general utility and efficiency to mine natural venoms for KTxs.
Introduction
Found throughout the tree of life, potassium channels are probably one of the most fundamental components of cells. Voltage-gated potassium channels (Kv)3 control membrane potential in both electro-excitable and nonexcitable cells, and are the key players in shaping the action potential in neurons and muscle cells (1). It is therefore not surprising that these channels are targets for a variety of toxins from venomous animals (2) and are involved in many pathological processes (3).
In particular, the voltage-gated Kv1.1 channel is widely expressed in the nervous system and plays an important role in controlling neuronal excitability (4). In some disorders (multiple sclerosis and spinal cord injury), Kv1.1 channels become exposed at juxtaparanodal sites and thus largely prevent electrical conduction along neurons. In these cases, blocking of Kv1.1 channels alleviates disease symptoms (5, 6). Recently, involvement of Kv1.1 channels in proliferation of tumor cells was revealed in vitro (7) and confirmed in vivo (8). An anti-tumor effect was achieved with dendrotoxin-κ, a selective blocker of Kv1.1 channels, suggesting that Kv1.1 can be considered as an emerging target in cancer therapy (9).
A search for new specific Kv1.1 channel ligands will contribute to the study of structure and function of these channels and will provide new pharmacological tools. Moreover, potent and specific blockers need to be discovered to avoid possible toxic side effects in clinical practice.
Some venomous creatures, including sea anemones, scorpions, spiders, and snakes, produce powerful blockers of potassium channels. Usually, these molecules are peptides containing several disulfide bridges (2). Two modes of Kv blockage may be delineated, namely physical occlusion of the ion-conducting pore (in the case of pore blockers) and inhibition of the channel activation by voltage sensor trapping (in gating modifiers) (10). Importantly, some of the toxins display remarkable selectivity with respect to different potassium channel isoforms (11–15), a property that justifies their potential as drug leads (16). Alternatively, high selectivity may be achieved by means of chemical modification and targeted or random mutagenesis of natural peptide molecules (17–20).
Scorpion venoms are a particularly ample source of toxins that specifically affect potassium channels (21). Potassium channel pore blockers from scorpions described thus far are known to belong to at least five types of fold as follows: (i) cysteine-stabilized α/β motif (CSα/β) (22); (ii) inhibitor cystine knot (ICK) (23); (iii) Kunitz bovine pancreatic trypsin inhibitor-type fold (14); (iv) α-helical hairpin with two disulfide bridges (CSα/α 2(S-S)) (24); and (v) α-helical hairpin with three disulfide bridges (CSα/α 3(S-S)) (25). The most populated class is CSα/β, in turn consisting of the following three families: α-, β-, and γ-KTx (26). The α-KTx family is the most numerous and currently includes at least two dozen subfamilies that are numbered consecutively: α-KTx 1, α-KTx 2, etc. Many of α-KTxs are potent blockers of Kv1 channels with nano- and sub-nanomolar affinity.
Traditional approach to screening and study of Kv blockers from scorpion venom necessitates bioactivity-guided purification of the active compounds by the means of multistage chromatography as well as utilization of electrophysiology to evaluate their blocking activity. The purified individual compounds are then subjected to protein sequencing. More recently, venom gland transcriptomics was introduced to aid high throughput sequencing of scorpion toxins (27–29). The recovered sequence information from a transcriptome database is then analyzed based on identified similarities with the already known toxins.
Here, we present a comprehensive study on the variability of Kv1.1 blockers in the venom of the scorpion Mesobuthus eupeus. We combined venom gland transcriptomics and computational analysis with multidimensional chromatography and mass spectrometry. To streamline bioactivity tests, we developed a fluorescent system based on bacterial expression of hybrid KcsA-Kv1.1 channels and operating in the facile “mix and read” mode. The KcsA-Kv1.1 system was utilized by us to search for Kv1.1 ligands in M. eupeus venom. Proving that our integrated approach is a powerful method to discover KTxs in scorpion venoms, we report on 59 candidate KTxs encoded in M. eupeus cDNA and five novel Kv1.1 blockers found in the venom.
EXPERIMENTAL PROCEDURES
cDNA Library Construction and Analysis
All stages of cDNA library construction were completed in collaboration with DuPont Agriculture and Nutrition as described earlier (30). Briefly, venom glands of M. eupeus were harvested, homogenized, and lysed by TRIzol (Life Technologies). Poly(A)+ RNA was purified with the mRNA purification kit (Amersham Biosciences). First- and second-strand cDNA synthesis, linker addition, and cloning into pBlueScript SK+ were all carried out according to the Stratagene cDNA kit. cDNA was purified using a cDNA column (Life Technologies). Sequencing was performed using the M13 forward primer by the ABI PRISM BigDye Terminator Cycle Sequencing Ready reaction kit with AmpliTaq DNA polymerase, FS (PerkinElmer Life Sciences), on a model 373 DNA sequencer (Applied Biosystems).
Common structural motifs of peptides with specific cysteine spacing can be presented through appropriate regular expressions and sequences of characters and metacharacters that form a search pattern. cDNA sequences were translated with an on-line tool from The Sequence Manipulation Suite. Notepad++ version 5.9 (Notepad++ team) was used to search peptides with specific cysteine spacing. Searches were performed using the following regular expressions: MX∞CX3CX∞Cy1CX∞* for CSα/β and CSα/α 3(S-S); MX∞CX6CX5CCX∞* for ICK; MX∞CX24CX20CX3CX∞* for Kunitz bovine pancreatic trypsin inhibitor-type fold; and MX∞Cy3Cy∞Cy3Cy∞* for CSα/α 2(S-S), where M is methionine and C is cysteine; X is any amino acid; y is any amino acid excluding cysteine; and * indicates the stop codon. Subscripts show the number of amino acid residues, and ∞ indicates any number of amino acids.
The obtained sequences were aligned and grouped using ClustalW2. BLAST (blast.ncbi.nlm.nih.gov) was used for search and comparison with known related amino acid sequences. SignalP 4.1 program was used not only for identification of putative typical N-terminal signal peptides but also for detection of nonsense and incomplete sequences without signal peptides. Phylogenetic trees were constructed in MEGA6 (31) using the neighbor-joining method (32).
Venom Separation
All venom separation procedures, toxin purification, modification, and sequencing were performed according to our detailed protocols that can be found elsewhere (33). Crude venom of M. eupeus (4 mg of freeze-dried powder) was dissolved in 300 μl of deionized water and centrifuged in Ultrafree-MC centrifugal filter units (with a 30-kDa nominal molecular mass cutoff, Millipore). The flow-through was fractionated using size-exclusion chromatography on a TSK 2000SW column (7.5 × 600 mm, 12.5-nm pore size, 10-μm particle size; Toyo Soda Manufacturing). The eluent contained 10% acetonitrile in 0.1% aqueous trifluoroacetic acid (TFA). Chromatography was performed using a flow rate of 0.5 ml/min, and effluent absorbance was monitored at 210 nm. Active fractions were separated by reverse phase-HPLC (RP-HPLC) on a Vydac 218TP54 C18 column (4.6 × 250 mm, 30-nm pore size, 5-μm particle size; Separations Group) using a 60-min linear gradient of acetonitrile concentration (0–60%) in 0.1% TFA at a flow rate of 1 ml/min. In this case absorbance was monitored at 210 and 280 nm. At the final step all active components were purified on the same Vydac column using a 90-min linear gradient of solvent containing 50% acetonitrile and 20% isopropyl alcohol in 0.1% TFA or 50% acetonitrile in 10 mm Tris-HCl (pH 7.0).
Mass Spectrometry
For molecular mass measurements an Ultraflex TOF-TOF (Bruker Daltonik GmbH) spectrometer was used as described (34).
Reduction of Disulfide Bonds and Modification of Thiol Groups
Purified peptides were reduced-alkylated following all steps as recommended in Ref. 33. Modified peptides were subjected to separation by RP-HPLC on a Jupiter C5 column (4.6 × 250 mm, 30-nm pore size, 5-μm particle size; Phenomenex) using a 60-min linear gradient of acetonitrile concentration (5–60%) in 0.1% TFA at a flow rate of 1 ml/min.
Cleavage of N-terminal Pyroglutamic Acid Residue
Pyroglutamate residues were removed from N-terminally blocked peptides by pyroglutamate aminopeptidase following the published guidelines (33). The products were isolated by RP-HPLC on a Vydac C18 column using a 60-min linear gradient of acetonitrile concentration (0–60%) in 0.1% TFA at a flow rate of 1 ml/min.
Peptide Sequencing
The N-terminal amino acid sequences of modified peptides were determined by automatic Edman degradation using a Procise Model 492 protein sequencer (Applied Biosystems) according to the protocol recommended by the manufacturer.
Concentration Measurements
Peptide concentrations were determined by UV spectrophotometry. Absorbance was measured at 280, 274.5, and 205 nm for peptides that contain tryptophan, tyrosine but no tryptophan, and do not contain either, respectively. For tryptophan-containing peptides, molar extinction coefficients at 280 nm were calculated using the GPMAW software (Lighthouse Data, Denmark). For peptides containing tyrosine but no tryptophan, molar extinction coefficients at 274.5 nm were calculated assuming the value of 1340 m−1cm−1 for each tyrosine residue. Finally, for peptides containing neither tyrosine nor tryptophan, molar extinction coefficients at 205 nm were calculated as described (35).
KcsA-Kv1.1 Binding Studies
Recombinant kaliotoxin-1 (KTX), tetraethylammonium chloride (TEA), 4-aminopyridine (4AP), and lysozyme from chicken egg white were from Sigma. Recombinant agitoxin-2 (AgTx2) and AgTx2 labeled using 5(6)-carboxytetramethylrhodamine N-succinimidyl ester (R-AgTx2) were produced and purified as described earlier (36). Recombinant OSK1 was produced as described elsewhere (36).
E. coli BL21(DE3) cells that express KcsA-Kv1.1 proteins in the inner membrane were obtained as described earlier (37, 38). Cell cultivation and preparation of spheroplasts were performed as described elsewhere (36).
For binding experiments, KcsA-Kv1.1-presenting spheroplasts were diluted with buffer containing 50 mm Tris-HCl (pH 7.5), 0.25 m sucrose, 0.3 mm EDTA, 4 mm KCl, 50 mm NaCl, 10 mm MgCl2, and 0.1% BSA up to a concentration of 1000 cells/μl. The spheroplasts were incubated with 7.3 nm fluorescently labeled R-AgTx2 in the presence of venom samples (crude venom, venom fractions, and subfractions). Nonlabeled AgTx2 (10 nm) or pure buffer was added to spheroplasts instead of venom samples as a positive and negative control, respectively. For competitive binding, KcsA-Kv1.1-presenting spheroplasts were incubated with R-AgTx2 (9 nm) and increasing concentrations of known pore blockers (AgTx2, OSK1, KTX, TEA, or 4AP) or purified MeKTx13-2. Reaction mixtures were incubated for 90 min with continuous stirring at room temperature, then placed in a 12-well flexiPERM silicon chamber (Perbio) attached to a coverslip, and centrifuged for 6 min at 200 × g.
Fluorescence imaging was performed using an inverted laser scanning confocal microscope LSM 710 (Carl Zeiss) with an α Plan-Apochromat oil immersion objective (63×, NA 1.46). Fluorescence of R-AgTx2 was excited by a He-Ne laser (543.5 nm, 12 microwatts on the sample), and emission was registered within the range of 550–685 nm. Quantitative analysis of R-AgTx2 binding to spheroplasts was performed at ∼0.25 μm lateral and 1.5 μm axial resolution, and all parameters of the measurements that could affect the intensity of the detected signal were fixed. The ImageJ software (National Institutes of Health) was used for spheroplast identification and estimation of average fluorescence signal intensity per cell (If). For each data point, If values of 150–200 spheroplasts were averaged to get the Iav value, and standard deviation was calculated. All experiments were repeated two or three times.
Expression of Voltage-gated Ion Channels in Xenopus laevis Oocytes
For the expression of the rat channel rKv1.1 in Xenopus oocytes, its gene was transcribed using the mMESSAGE mMACHINE T7 transcription kit (Ambion). The harvesting of stage V to VI oocytes from an anesthetized female X. laevis frog was described previously (39). Oocytes were injected with 50 nl of cRNA at a concentration of 1 ng/nl using a micro-injector (Drummond Scientific). The oocytes were incubated in ND96 solution containing the following: 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES (pH 7.4), supplemented with 50 mg/liter gentamycin sulfate.
Electrophysiological Recordings
Two-electrode voltage clamp recordings were performed at room temperature (18–22 °C) using a Geneclamp 500 amplifier (Molecular Devices) controlled by a pClamp data acquisition system (Axon Instruments) as described (13). Whole-cell currents from oocytes were recorded 1–4 days after injection. Oocytes were kept in ND96. Voltage and current electrodes were filled with 3 m KCl. Resistances of both electrodes were kept in the range of 0.7–1.5 megohms. The elicited currents were filtered at 500 Hz and sampled at 2 kHz using a four-pole low-pass Bessel filter. Leak subtraction was performed using a –P/4 protocol. Kv1.1 currents were evoked by 250-ms depolarization to 0 mV followed by a 250-ms pulse to −50 mV from a holding potential of −90 mV.
To investigate the current-voltage relationship, currents were evoked by 10-mV depolarization steps from a holding potential of −90 mV. The values of IK were plotted as a function of voltage and fitted using the Boltzmann Equation 1,
where Imax represents maximal IK; V½ is the voltage corresponding to half-maximal current; and k is the slope factor.
To assess the concentration dependence of the toxin-induced inhibitory effects, a concentration-response curve was constructed, in which the percentage of current inhibition was plotted as a function of toxin concentration. Data were fitted with the Hill Equation 2,
where y is the amplitude of the toxin-induced effect; IC50 is the toxin concentration at half-maximal inhibition; [C] is the toxin concentration; and h is the Hill coefficient. Comparison of two sample means was made using a paired Student's t test (p < 0.05). All data represent at least three independent experiments (n ≥ 3) and are presented as means ± S.E.
RESULTS
Assembling and Analysis of cDNA Database from M. eupeus Venom Glands
A raw cDNA library from the lesser Asian scorpion Mesobuthus eupeus (Buthidae) venom glands was obtained using a conventional approach (30). The total number of cDNA sequences in the database was 1911, and the number of corresponding putative peptide sequences was 11,466 (translation was performed in three frames, both direct and reverse).
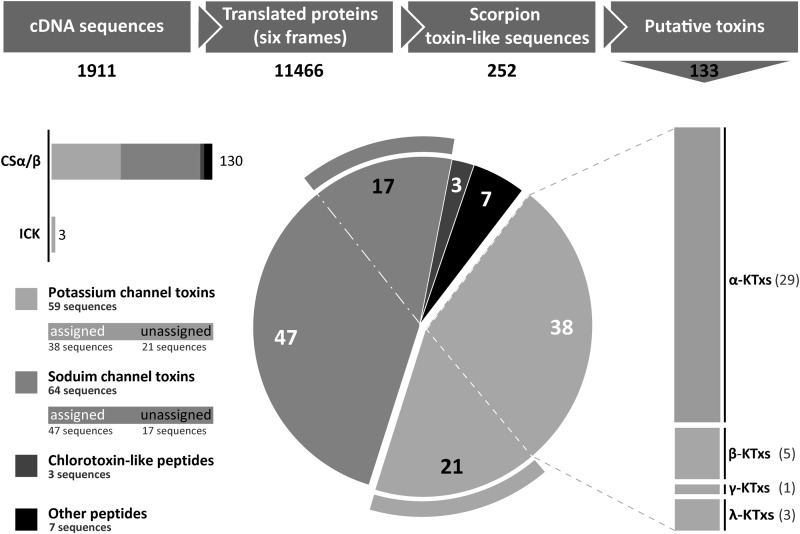
To search for putative scorpion toxin-like peptides, all these sequences were transferred to Notepad++ software and analyzed using the Search tool through regular expressions. As a result, we obtained 244 sequences of putative CSα/β peptides, 4 of ICK, and 4 of CSα/α 2(S-S) toxins satisfying the search conditions (252 sequences in total). All these sequences were analyzed by the SignalP 4.1 program and BLAST. Those sequences that did not contain a putative signal peptide were rejected. The remaining 155 sequences were analyzed using BLAST to remove identical entries, and finally we identified 133 unique sequences. Among these, three sequences show a typical ICK signature, and 130 sequences conform to the CSα/β structural fold. We distinguish the following groups of CSα/β peptides: 56 KTxs; 64 sodium channel toxins; 3 chlorotoxin-like peptides; and 7 sequences with no significant similarity with known toxins. Proper sequences for CSα/α 2(S-S) and Kunitz-type toxins were not found.
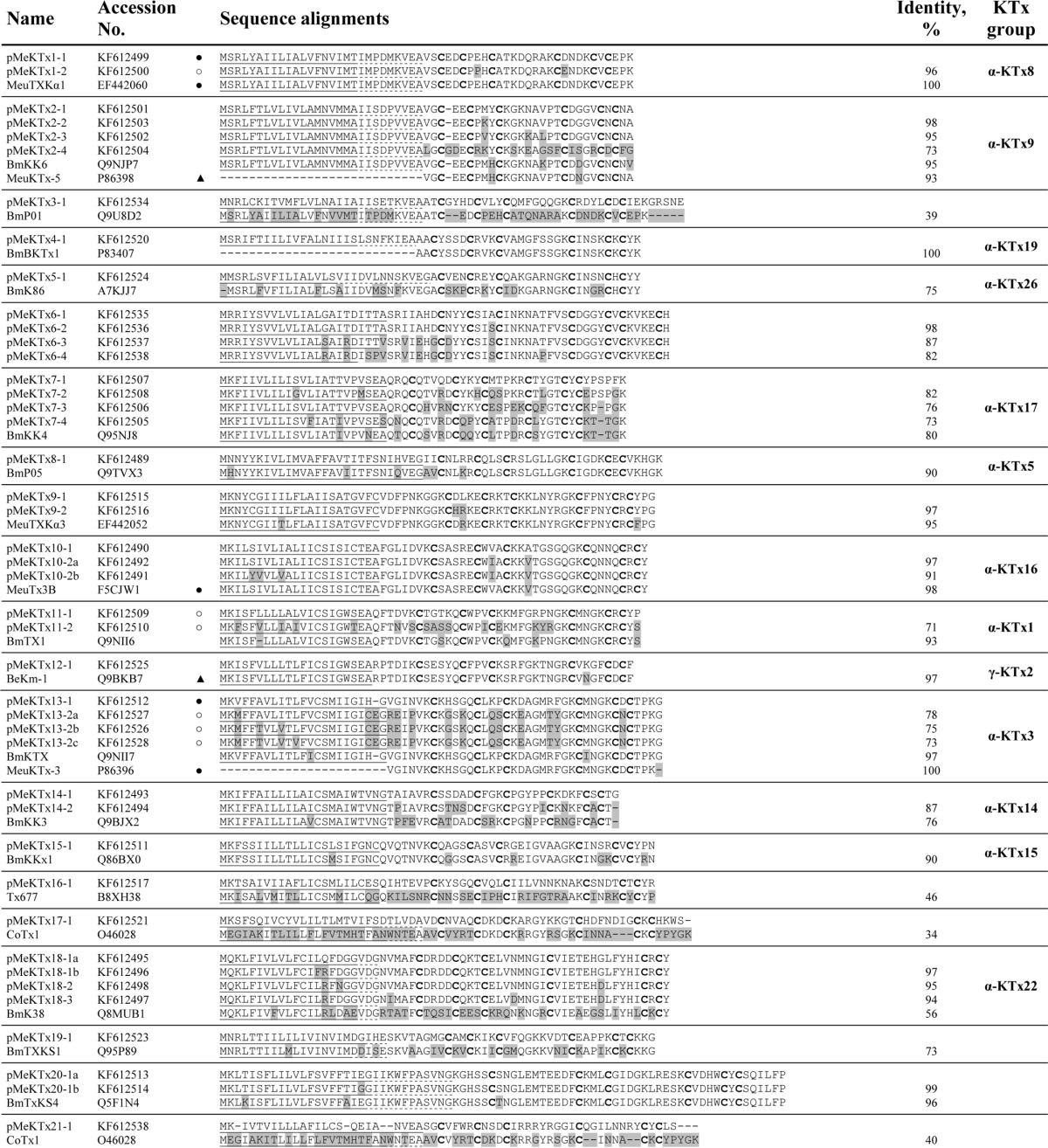
The main stages of the search process and the distribution of putative toxins among different taxonomic and structural groups are presented in Fig. 1. All cDNA sequences coding for putative KTxs were submitted to GenBankTM (translated sequences are shown in Table 1).
FIGURE 1.
Stages of identification of putative toxins in the M. eupeus cDNA database, and their distribution among different structural and functional groups. Raw cDNA sequences were translated and analyzed using regular expressions. Then toxin-like sequences were identified by the presence of a signal peptide, and iterative sequences were rejected. Putative toxins were divided in four groups (potassium and sodium channel toxins, chlorotoxin-like peptides, and others). The different families of assigned KTXs are indicated at right. For all families and groups, the total numbers of identified sequences are shown.
TABLE 1.
Putative KTx precursors identified in M. eupeus transcriptome
Sequences were classified into 30 subfamilies according to their amino acid sequence similarity. The most similar sequences of known KTxs from this or other scorpions are included in each group for comparison. Predicted signal peptides are underlined. Dotted lines indicate alternative signal peptide processing. Prosequences are in italics. Differences are shaded. ○, peptides identified in this work; ▴, previously; and ●, previously and in this work.

Classification of Putative Toxins and Comparison with Reviewed Potassium Channel Blockers
All deduced KTx precursors were aligned using ClustalW2 and divided into 30 groups (from one to four members in each group) according to their amino acid sequences similarity. Putative toxin precursors were named pMeKTx, where p is precursor; Me is M. eupeus; and KTx is potassium channel toxin. Using SignalP 4.1, typical signal peptides were predicted. For some precursors, it was impossible to identify signal peptides definitely, so we considered several most probable alternatives of signal peptide cleavage. For example, in the case of pMeKTx1-1, SignalP indicates that the signal peptide ends with Thr-19 or Ala-28, and although according to the program the most probable variant is cleavage after Thr-19, a mature peptide starting from Val-29 was actually purified from the venom (see below). For two precursors pMeKTx8-1 and pMeKTx20-1b, signal peptides were not detected by SignalP (in both cases no putative cleavage site was found). Nevertheless, in both cases signal peptides were inferred from homology. Moreover, these sequences showed all attributes of toxin-like peptides and were not rejected from the analysis. All new sequences were analyzed by BLAST to search for homologs, and in Table 1 the most closely related peptides found in public databases are shown for each group. Most of the obtained sequences are members of different well described groups such as α-KTx-1, β-KTx, γ-KTx-2, etc. Some pMeKTxs are similar to sequences that were found in transcriptomes of scorpions belonging to the Buthidae family but were not purified from venom. A small fraction of putative toxins demonstrates low homology with reviewed sequences in UniProt or even absence of similarity to the known sequences.
Development and Characterization of Analytical KcsA-Kv1.1 System
KcsA-Kv1.1 chimeric protein is able to bind peptide pore blockers of human Kv1.1 channels due to the transfer of the toxin-binding site from Kv1.1 into the corresponding region of the bacterial potassium channel KscA (36–38, 40). KcsA-Kv1.1 was obtained by substituting a part of the KcsA P-loop (52RGAPGAQLITYPR64) with a homologous region from Kv1.1 (349AEEAESHFSSIPD361). KcsA-Kv1.1 is readily expressed in E. coli and forms tetramers in the plasma membrane with the outer pore vestibule of the channel oriented extracellularly. Spheroplasts prepared from E. coli cells with membrane-embedded KcsA-Kv1.1 bind specifically fluorescently labeled agitoxin-2 (R-AgTx2) (38). Therefore, R-AgTx2 and spheroplasts bearing KcsA-Kv1.1 can be used as components of a fluorescent system for investigation of Kv1.1 ligands just as it was realized recently for the KcsA-Kv1.3 system (36).
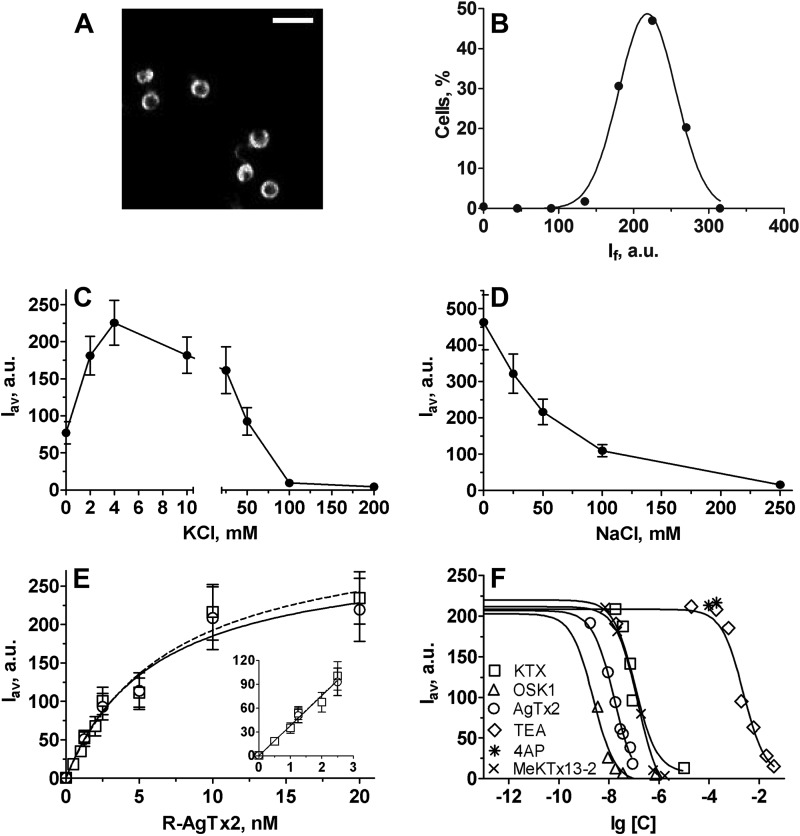
Specific binding of R-AgTx2 to spheroplasts bearing KcsA-Kv1.1 is detected with laser scanning confocal microscopy (Fig. 2A) and characterized with (i) an average fluorescence signal intensity per cell (If) for each measured spheroplast and (ii) parameter Iav for a particular spheroplast sampling (Iav is equal to If averaged over this sampling). Dependence of Iav on the concentration of added R-AgTx2 (Fig. 2E) presents a shape that is typical for saturation binding curves observed in fluorescent and radioligand analyses of ligand-receptor interactions. Nonspecific binding of R-AgTx2 to native E. coli spheroplasts or to spheroplasts presenting KcsA was not observed. All spheroplasts bearing KcsA-Kv1.1 were stained with R-AgTx2 (Fig. 2A), and frequency distribution of If is described by a normal (Gaussian) distribution (Fig. 2B). This means that the spheroplast population is uniform in terms of the amount of membrane-embedded KcsA-Kv1.1, and the parameter Iav is valid to estimate the average quantity of complexes between R-AgTx2 and KcsA-Kv1.1.
FIGURE 2.
Characteristics of a cellular system for the search and study of Kv1.1 pore ligands that is based on KcsA-Kv1.1-bearing spheroplasts. A, distribution of complexes between R-AgTx2 and KcsA-Kv1.1 at the membrane of spheroplasts measured with laser scanning confocal microscopy. The bar corresponds to 4 μm. B, frequency distribution of parameter If measured for KcsA-Kv1.1-bearing spheroplasts preincubated with R-AgTx2 (10 nm). Data were fitted with a normal (Gaussian) distribution (R2 = 0.98). C, effect of KCl concentration on the binding of R-AgTx2 (19 nm) to KcsA-Kv1.1 in buffer A supplemented with 50 mm NaCl. D, effect of NaCl concentration on the binding of R-AgTx2 (6 nm) to KcsA-Kv1.1 in buffer A supplemented with 4 mm KCl. E, saturation curves of R-AgTx2 binding to KcsA-Kv1.1 in buffer A supplemented with 50 mm NaCl and 4 mm KCl at different concentrations of spheroplasts. The inset shows a zoom on the saturation curves in the range of low R-AgTx2 concentrations. Circles refer to 3000 cells/μl, and squares refer to 30,000 cells/μl. F, competition between R-AgTx2 (9 nm) and different ligands for binding to KcsA-Kv1.1-bearing spheroplasts in buffer A supplemented with 50 mm NaCl and 4 mm KCl. Content of buffer A is as follows: 50 mm Tris-HCl, 0.25 m sucrose, 0.3 mm EDTA, 10 mm MgCl2, 0.1% BSA (pH 7.5).
Influence of particular buffer components on ligand binding to KcsA-Kv1.1 was characterized. We found that the binding of R-AgTx2 to spheroplasts bearing KcsA-Kv1.1 depends on the concentration of KCl and NaCl (Fig. 2, C and D). A monotonic decrease in R-AgTx2 binding with the increase in NaCl concentration can be explained by a negative influence of ionic strength. In contrast, R-AgTx2 binding to KcsA-Kv1.1 has a maximum at ∼4 mm KCl. Low concentrations of potassium ions enhance ligand binding to both natural and artificial Kv1 channels (36, 41, 42), whereas high KCl concentrations produce a trivial effect of ionic strength. Our experiments revealed that sucrose (up to 0.25 m) and BSA (0.1%) do not affect interactions between R-AgTx2 and KcsA-Kv1.1 (data not shown).
To simplify the quantitative analysis of ligand-receptor interactions, the titration of receptors with a ligand was performed at the condition of [L] ≫ [R], where [L] and [R] are concentrations of a ligand and receptor, respectively. The saturation binding curves did not depend on the concentration of cells (i.e. KcsA-Kv1.1 receptors, Fig. 2E) in the range from 3000 to 30,000 cells/μl and therefore met the condition of [L] ≫ [R]. In this case, Equation 3 can be applied,
where [L] is R-AgTx2 concentration, and Isat is equal to the Iav value at the plateau. The Kd value for the complexes between R-AgTx2 and KcsA-Kv1.1 was estimated to be 3.4 ± 0.9 nm.
Further characterization of the new system revealed that Kv1.1 ligands such as AgTx2, OSK1, KTX, and TEA compete specifically with R-AgTx2 for binding to KcsA-Kv1.1 (Fig. 2F). The competitive binding curves of R-AgTx2 displacement by Kv1.1 ligands were measured when the concentration of free R-AgTx2 was much higher than that of the bound ligand and analyzed with Equation 4,
where [C] is the concentration of the added competitive ligand; Im is Iav at [C] = 0, and InC50 is the ligand concentration that displaces 50% of R-AgTx2 from the complex with KcsA-Kv1.1. The estimated InC50 values were used to calculate the apparent dissociation constants (Kap) of competitive ligands as shown in Equation 5,
Binding of high affinity OSK1 and KTX, and low affinity TEA to KcsA-Kv1.1 is characterized with Kap values that are close to the corresponding Kd values obtained for these ligands in patch clamp measurements on Kv1.1 channels (Table 2). Binding of TEA to KcsA-Kv1.1 was observed because it has a binding site in the outer vestibule of Kv (43) in addition to another site situated in the inner cavity of the pore (44, 45). As expected, the internal pore blocker 4AP did not compete with R-AgTx2 for binding to KcsA-Kv1.1 channel up to the concentration of 5 mm.
TABLE 2.
Affinities of ligands to KcsA-Kv1.1 and native Kv1.1 channels measured with different methods
We therefore conclude that the new analytical system (i) recognizes both peptide and low molecular weight ligands that bind to the extracellular vestibule of Kv1.1, (ii) discerns high and low affinity pore blockers, (iii) provides correct estimation of ligand affinity to Kv1.1, and (iv) supplements the previously developed analytical system targeted to Kv1.3 ligands.
Venom Fractionation
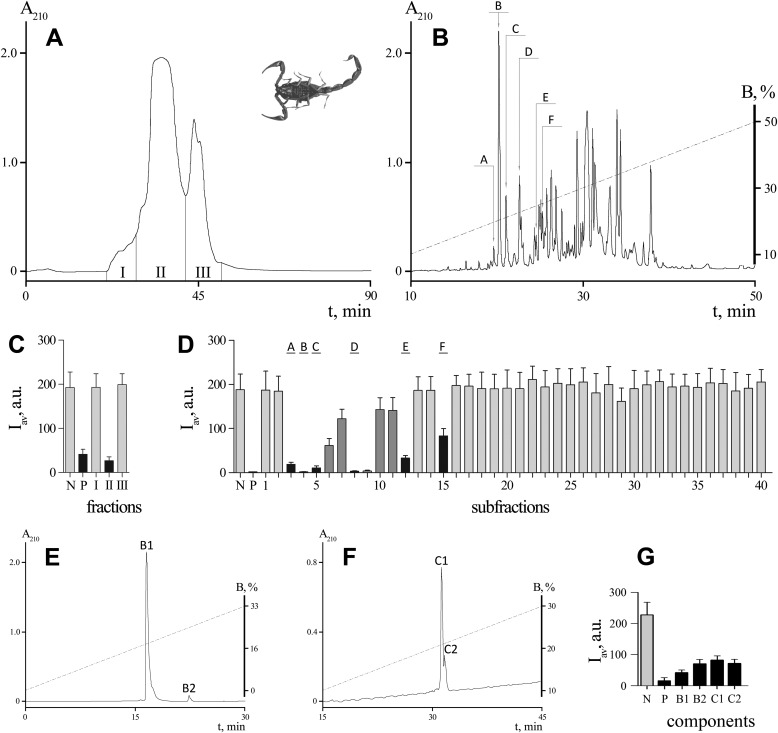
Crude venom of M. eupeus was fractionated according to our common protocol (33). We used the KcsA-Kv1.1 setup for screening M. eupeus venom components to detect novel Kv1.1-specific toxins. On the first step, venom components were separated to three broad fractions using size-exclusion chromatography (I–III, see Fig. 3A). These fractions were tested for the ability to bind to KcsA-Kv1.1 hybrids, and activity was observed only in fraction II (Fig. 3C). Negative results, which were obtained for fractions I and III, indicated that these fractions contained no high affinity Kv1.1 ligands.
FIGURE 3.
M. eupeus venom fractionation and analysis with the spheroplast-based fluorescent system. A, separation of the crude venom into three fractions with size-exclusion chromatography on a TSK 2000SW column. Fractions are indicated by roman numerals. B, separation of fraction II into subfractions using RP-HPLC in a linear gradient of acetonitrile. Active subfractions are indicated by capital letters. C, D, and G, fluorescence intensity distribution of KcsA-Kv1.1 presenting spheroplasts. Pure buffer (N, negative control), AgTx2 (10 nm; P, positive control), or venom fractions/subfractions/components were added. Black bars indicate active fractions/subfractions/components; dark gray bars, subfractions that were contaminated with minor impurities of the active components, and light gray bars represent inactive fractions/subfractions/components. E and F, additional round of RP-HPLC for subfractions B and C containing several active components that are indicated as B1 and B2 and C1 and C2.
To isolate active components, fraction II was subjected to separation by RP-HPLC (Fig. 3B). As a result, 40 subfractions were obtained and tested against KcsA-Kv1.1 (Fig. 3D). Six subfractions (named A–F) competed specifically with R-AgTx2 for binding to the hybrid channels and were enriched in active components. Small inhibition effects were observed in some neighboring subfractions due to the contamination with active components. Subfractions 28 and 29 were characterized by a weak and unstable ability to reduce signal intensity, but they did not compete with R-AgTx2 in a concentration-dependent manner.
The second round of RP-HPLC led to the purification of active components in individual state. For this, a 90-min linear gradient of solvent containing 50% acetonitrile and 20% isopropyl alcohol in 0.1% TFA was used. Each of the subfractions A, D, E, and F contained a single active component. In subfraction C, two active components (C1 and C2) were identified (Fig. 3, F and G). Subfraction B also contained two active components (B1 and B2), but they could not be separated even after the second round of RP-HPLC. We therefore modified the purification strategy by using a different eluent (50% acetonitrile in 10 mm Tris-HCl (pH 7.0)), and we successfully separated B1 and B2 (Fig. 3, E and G).
In total, eight Kv1.1 blockers were detected and purified from the venom of M. eupeus. Homogeneity of the active compounds A, B1, B2, C1, C2, D, E, and F was confirmed by MALDI-MS.
Analysis of M. eupeus Venom Profile and Sequencing of the Novel Peptides
The following KTxs were previously found in the M. eupeus venom: three α- (MeuKTx-1 (MeuTXKα1), UniProt accession number P86400; MeuKTx-3, P86396; and MeuTx3B, D3JXM1) (46–48); one β- (MeuTXK-β-1, A9XE60) (49), one γ- (BeKm-1, Q9BKB7) (50), and one ICK motif-containing toxin (λ-MeuTx-1, P86399) (23). Whereas β- and λ-KTxs are known as very weak inhibitors of potassium channels, and γ-KTxs are blockers of the ether-a-go-go-related gene (ERG) family of potassium channels, our research was focused on α-KTxs, the most representative and high affinity ligands.
In total, we identified five novel and three already described active peptides in M. eupeus venom by a combination of RP-HPLC, MS, and fluorescent assay (Table 3). Three known α-KTxs (MeuKTx-1, MeuKTx-3, and MeuTx3B) were detected in subfractions B, C, and F, respectively (corresponding to components B1, C2, and F). Their identity was confirmed by partial N-terminal Edman degradation, MS, and comparison of their RP-HPLC retention times with published data.
TABLE 3.
Kv1.1 ligands purified from the venom of M. eupeus
Components were identified exclusively in this work (○) or in this work as well as previously (●). Z, pyroglutamic acid residue.

Besides the known toxins, two additional active components were observed in subfractions B and C. Component B2 (MeKTx1-2) is a very close homolog of MeuKTx-1 with two replacements (E8P and D20E), and component C1 (MeKTx13-3) shows an identical sequence with a toxin from a related species Mesobuthus martensii (BmKTX, Q9NII7) (51). The difference in one amino acid residue between BmKTX (also from M. martensii) and MeuKTx-3 was identified in position 28 (I28M).
The new peptides were sequenced by Edman degradation and named as shown in Table 3; sequences were submitted to UniProt. For toxins MeKTx11-3 and MeKTx11-1 (components D and E), initial Edman sequencing failed due to N-terminal blockage by a pyroglutamic acid residue (<Glu) originating from glutamine (Gln). The blockage was removed using pyroglutamate aminopeptidase. MeKTx11-3 and MeKTx11-1 (components D and E) are close homologs with two amino acid replacements (V9G and S37P). For toxins, MeKTx13-2, MeKTx13-3, and MeuKTx-3 from subfractions A and C, the C-terminal amidation was inferred by comparison of the experimentally measured molecular masses with the calculated values based on amino acid sequences.
Activity of Novel Toxins
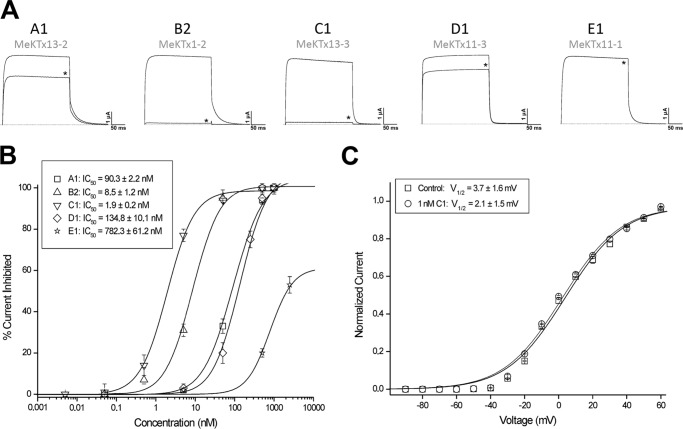
To confirm the pore blocking activity of five newly purified toxins the peptides were investigated on rKv1.1 using the two-electrode voltage clamp technique. Fig. 4A shows the observed inhibition of rKv1.1 channels after application of toxins MeKTx1-2, MeKTx11-1, MeKTx11-3, MeKTx13-2, and MeKTx13-3. At a concentration of 50 nm, MeKTx1-2 and MeKTx13-3 could inhibit completely the ion current through rKv1.1 channels. The same concentration of MeKTx13-2 and MeKTx11-3 reduced the current by ∼30 and 20%, respectively, although no activity was observed for MeKTx11-1 (it should be noted that at higher concentrations MeKTx11-1 did inhibit rKv1.1 channels).
FIGURE 4.
Toxin activity studied by electrophysiology. A, representative traces of currents through Kv1.1 in control and after application of 50 nm toxin (indicated with asterisks). B, concentration-response curves. IC50 values are listed. C, I-V relationship for MeuKTx13-3 indicates no modulation of the channel. V½ values are listed.
Concentration-response curves were constructed for the five tested toxins (Fig. 4B) and yielded the IC50 values of 1.9 ± 0.2 nm for MeKTx13-3, 8.5 ± 1.2 nm (MeKTx1-2), 90 ± 2 nm (MeKTx13-2), 135 ± 10 nm (MeKTx11-3), and 780 ± 60 nm (MeKTx11-1).
The most potent toxin, MeKTx13-3, was used to further investigate the characteristics of channel inhibition. To find out whether the observed current inhibition is due to pore block or rather to altered channel gating, the I-V curve was constructed in control and in the presence of 1 nm toxin (Fig. 4C). The V½ value did not change, i.e. 3.7 ± 1.6 mV in control and 2.1 ± 1.5 mV in toxin conditions, respectively (n = 4), testifying in favor of direct pore block. rKv1.1 block occurred rapidly and was reversible, because the current recovered quickly and completely upon washout (data not shown).
Finally, using KcsA-Kv1.1-presenting spheroplasts, MeKTx13-2 was measured to have a Kap of 44 ± 12 nm (Fig. 2F). This value is similar to the IC50 value reported for rKv1.1-expressing oocytes.
DISCUSSION
Utilization of Regular Expression-based Searches for Transcriptome Analysis
Current research of scorpion venoms more often includes transcriptomic data provided by the fast development of DNA sequencing technologies (52, 53). Transcriptome analysis can be easily performed, on the one hand, and provides a high information content, on the other hand (27). For several scorpion species such studies were carried out and a large variety of peptides and proteins were thus predicted (54–56). This set includes not only active venom components (such as ion channel toxins, cytolytic peptides, and enzymes) but also housekeeping proteins, some of which are responsible for cellular metabolism (29). Of course, the main target of such studies is different bioactive molecules and, in particular, toxins interacting with potassium and sodium channels.
In this work, we investigated the transcriptomic data on the diversity of potassium channel blockers and estimated the variability of predicted peptides in venom glands of M. eupeus. Raw transcriptome data can be analyzed using different strategies as follows: (i) matching transcriptomic and proteomic data obtained by MS or Edman sequencing; (ii) manually, i.e. when every sequence from the database is examined separately; (iii) by BLAST verification, i.e. when all sequences are compared with the current databases; and (iv) through usage of regular expressions, wherein specific motifs are located in primary structures of translated proteins.
Manual analysis is of universal importance and can help to annotate all target sequences from a transcriptome database. In this approach, sequences are checked step by step using all possible tools; one obvious drawback is high time consumption. BLAST verification enables fast comparison of an investigated database with registered sequences from different global databases. The limitation of this approach is a possible absence of homologs for the new sequences in reference databases, which is often the case for new venom components. Analysis through regular expressions permits us to find sequences with different patterns in primary structure. The disadvantage of this method is that it does not allow identification of sequences without certain specific motifs, even for the known polypeptides (for example, linear peptides without any patterns). Moreover, primary structure motif does not necessarily correspond to a particular protein fold. For example, recently a potassium channel-blocking peptide was described from Tityus trivittatus showing typical CSα/β cysteine spacing with an unusual disulfide pattern. This toxin adopts a novel fold named CSα/α (25). But as opposed to BLAST, analysis of regular expressions allows one to detect sequences sharing no homology but showing common motifs. All potassium channel blockers from scorpion venoms are disulfide-containing peptides, and relevant regular expressions can be constructed for each type of motif. Therefore, this strategy is suitable for determination of the Kv blocker sequences.
Usage of KcsA-Kv1 Hybrid Channels in Bioassay-guided Analysis of Scorpion Venoms
cDNA libraries constructed on the basis of transcriptomic data afford significant gain in sequence information, but the deduced components still require confirmation of their presence in the venom, a major problem in most “venomic” studies. Scorpion venoms are complex mixtures, and deep investigation of the content of even one species can take more than 30 years (57, 58). Different bioassay-guided approaches are needed for targeted investigations of the active compounds. Traditionally, ligand interactions with ion channels are examined using electrophysiology (47) (voltage clamp and/or patch clamp) and the radioligand binding assay (59). Electrophysiology is considered as the “gold standard” in ion channel research. Several high throughput screening systems were created on the basis of voltage clamp and patch clamp techniques, but even now they are expensive and have some critical limitations for use. The radioligand binding assay is a robust method for a quantitative analysis of ligand-receptor interactions, but currently it is rarely used because of strict regulations for work with radioactive substances. A modern safe alternative for the radioligand binding assay is the fluorescence detection analysis. Kv1 channels were studied using flow cytometry (60), single molecule fluorescent microscopy (61), and the analysis of membrane potential changes using fluorescent dyes and fluorescence readers (62). Recently, a convenient fluorescent screening system based on KcsA-Kv1.3 hybrid receptors was developed and applied successfully to screen Orthochirus scrobiculosus venom for peptide toxins, which specifically target the pore region of Kv1.3 (36, 38). This system, which represents an efficient, robust, and rapid binding assay, is a powerful alternative to radioligand and patch clamp techniques. Here, we report an analogous system designed to screen for Kv1.1 pore blockers.
We performed all the relevant experiments to control the stability and specificity of the toxin-receptor interaction. We paid particular attention to the ability of the novel test system to bind Kv1 family-specific ligands. KTX, AgTx2, OSK1, and TEA compete with R-AgTx2 for binding to KcsA-Kv1.1 channels, and the apparent dissociation constants (Kap) of these ligands are in accordance with the data provided by other methods (Table 2). Comparing these data, it is important to note that the Kd value obtained for a ligand depends on measurement conditions such as buffer composition, channel state, and type of assay. Among tested ligands, only 4AP did not compete with R-AgTx2 for binding to the pore region of KcsA-Kv1.1, because 4AP binds to the site located on the inner side of the channel pore (44, 45). Another small organic molecule TEA can bind to potassium channels on either side (43). These results provide confirmatory evidence that the hybrid KcsA-Kv1.1 channel possesses, in general, a Kv1.1 ligand affinity profile, and that the novel test system based on KcsA-Kv1.1-expressing spheroplasts can be used for toxin screening.
Our screening approach is limited to ligands that bind to the outer vestibule of the channel pore. Neither gating modifiers nor ligands of cytoplasmic domains can be detected.
The sensitivity of the test system should also be considered. A hypothetical 4-kDa ligand (an average mass of known peptide pore blockers) with Kap = 1 nm displaces >50% of R-AgTx2 (7.3 nm) from complexes with KcsA-Kv1.1 when its concentration is >13 μg/liter. Fractions I and III were tested at concentrations of ∼1 g/liter. Negative results (Fig. 3C) indicate that these fractions contained none or apparently less than 1.3 × 10−6%, 0.0013%, and 1.3% of KcsA-Kv1.1 ligands by mass, having Kap of 1 pm, 1 nm, and 1 μm, respectively. According to our estimations, negative results obtained for the tested subfractions (Fig. 3D) mean that these subfractions were either unable to bind to KcsA-Kv1.1 or had Kap >1 μm. All peptides detected by the novel test system were confirmed to be pore blockers of Kv1.1. No false-positive results were obtained due to careful re-examination of subfractions with marginal activity.
The KcsA-Kv1.1 test system was applied by us in combination with classical chromatographic methods and MS analyses for screening new blockers in M. eupeus venom. We tracked the channel binding activity of venom components throughout all the steps of separation (Fig. 3). Using this approach, we significantly reduced the amount of material needed to be tested and the time of measurement as compared with the more conventional methods. Screening of M. eupeus venom showed the presence of several components having affinity to KcsA-Kv1.1 channels. Most of these components were predicted by transcriptome analysis that simplified considerably the peptide sequencing. Further voltage clamp experiments (Fig. 4) and analysis of published data confirmed that all ligands found in M. eupeus venom using KcsA-Kv1.1 hybrid channels are indeed Kv1.1 pore blockers showing high affinity (with IC50 values below 1 μm).
Variability of KTxs in M. eupeus
At present, more than 200 potassium channel blockers from scorpion venom have been described and submitted to the UniProt database. Most of these peptides were purified from crude venom directly and then sequenced (63, 64), but several dozens of blockers were predicted from scorpion venom gland mRNA (53, 56, 65). Among the five known structural motifs of potassium channel blockers from scorpion venom (66), the dominating position belongs to CSα/β. About 90% of all scorpion KTxs contain this motif, and only less than about 10% of peptides show other fold types. α-KTxs are the major family of CSα/β toxins comprising about 65% of all scorpion KTxs. The prime source of α-KTxs is scorpions from the Buthidae family (about 80% of all known α-KTxs), and all other scorpion families contribute only about 20% of α-KTxs. All known α-KTxs (about 170 peptides) are divided into more than 25 subfamilies.
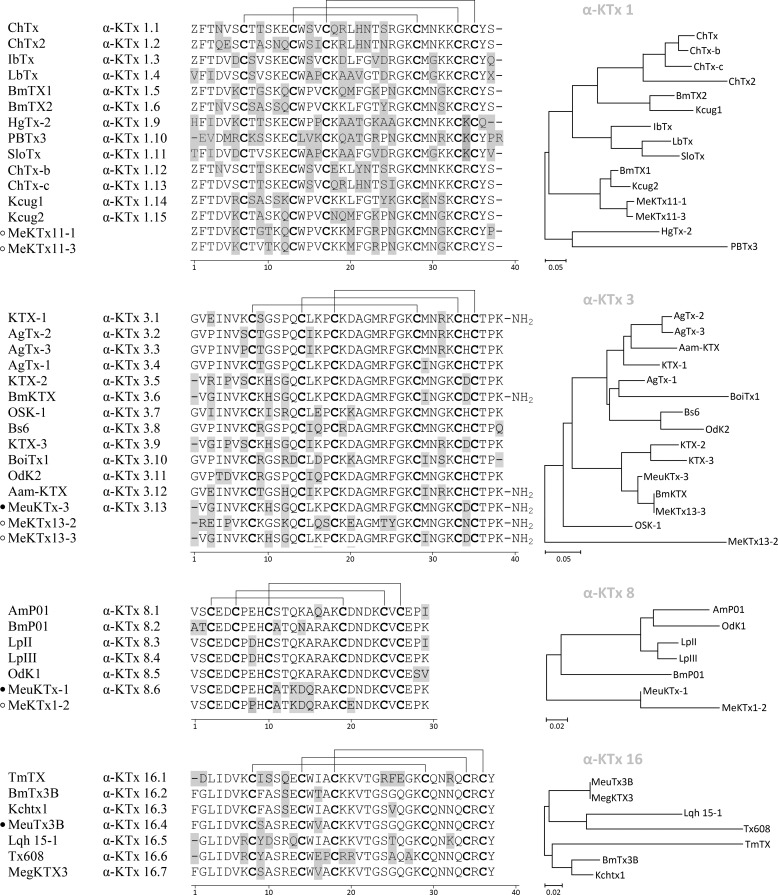
Our transcriptomic data showed that a wide diversity of potential KTxs is present in the M. eupeus venom. For a facile representation of the relative similarity between KTx precursors, an unrooted phylogenetic tree was constructed (Fig. 5). According to amino acid sequence similarity, all predicted toxins were included either in those subfamilies that were physiologically characterized or unassigned subfamilies (peptides from unassigned subfamilies were found only in transcriptomes). Analysis of current data in the field of KTxs from scorpion venom shows that almost all assigned subfamilies contain KTxs targeting Kvs. Exceptions are subfamilies α-KTx 5,α-KTx 9, α-KTx 14, α-KTx 15, α-KTx 17, α-KTx 19, α-KTx 22, α-KTx 24, and α-KTx 25 that contain toxins either demonstrating activity only on Ca2+-activated K+ channels (KCa1 to KCa3), or their targets are still uninvestigated. Kv1 blockers generally belong to α-KTx 1–4, α-KTx 6–8, α-KTx 10–13, α-KTx 16, α-KTx 18, α-KTx 20, α-KTx 21, and α-KTx 23 subfamilies (21, 67).
FIGURE 5.
Phylogenetic tree of mature KTxs obtained from the M. eupeus cDNA database. Assigned groups are shaded.
We assigned 12 α-KTx subfamilies in the M. eupeus transcriptome, and only four of those subfamilies (α-KTx 1, α-KTx 3, α-KTx 8, and α-KTx 16) are known to contain potent Kv1 blockers (Fig. 6). Indeed, further investigation of the venom, purification, and sequencing of the peptides active on Kv1.1 demonstrated that all novel KTxs belong to those four subfamilies. These toxins are short-chain peptides (from 29 to 38 amino acid residues long) and contain six cysteines forming three intramolecular disulfide bridges. Several members of α-KTx 3 bear C-terminal amidation that is the result of C-terminal glycine processing. All sequences of the four subfamilies present a key lysine residue at a conserved location (68). The number of this lysine residue from the N terminus may be different and depends on sequence length, but in all cases it is the first amino acid before the fourth cysteine.
FIGURE 6.
Amino acid sequence alignment of several purified M. eupeus toxins with KTxs from other scorpions. Peptides are allocated into groups according to sequence homology. Differences are shaded gray, and cysteine residues are in bold. Z, pyroglutamic acid residue. Putative disulfide bridges are represented by lines. ○, components described in this work; ●, components described previously. Toxin phylogenetic trees constructed by neighbor-joining algorithm are represented at right. A scale bar is shown below each tree.
Interestingly, among active KTxs in M. eupeus venom, we found MeKTx13-3, which is identical to BmKTX, a peptide purified earlier from M. martensii (69). It seems that the presence of identical peptides in the venom of different species is common for scorpions; MeuTx3B from M. eupeus shows the same amino acid sequence as MegKTx3 purified from Mesobuthus gibbosus (48, 70); three peptides with the same name butantoxin were identified in the venom of Tityus serrulatus, Tityus trivittatus, and Tityus stigmurus (71).
Conclusion
The combination of venom gland transcriptomics and computational analyses with multidimensional chromatography and mass spectrometry is proved to be a state-of-the-art approach to comprehensive investigation of scorpion venoms, especially when it is supplemented with a convenient tool for recognition of target activity, such as our ligand-binding assay. The five novel peptides presented in this communication are high affinity Kv1.1 channel blockers, which extend the panel of potential pharmacologically important Kv1 ligands. Our integrated approach is of general utility and efficiency to mine natural venoms for KTxs.
Acknowledgments
We thank the DuPont Agriculture and Nutrition staff and especially Maureen Dolan, Will Krespan, Bill McCutchen, and Rafi Herrmann for cDNA library construction and sequencing. We also thank Alexey M. Nesterenko from the A. N. Belozersky Institute of Physico-Chemical Biology (Moscow State University) for assistance in construction of regular expressions for database analysis. We also thank Olaf Pongs from the Institute of Physiology (Saarland University Medical Center) for sharing the rKv1.1 clone.
This work was supported in part by Russian Foundation for Basic Research Grants 14-04-32091 and 13-04-01857, Program for Fundamental Research of the Presidium of Russian Academy of Sciences No. 24, and by the Moscow State University Program of Development.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KF612484–KF612542.
The protein sequences of KTx have been submitted to the UniProt Knowledgebase (UniProtKB) under the accession numbers C0HJQ4–C0HJQ8.
- Kv
- voltage-gated potassium channel
- 4AP
- 4-aminopyridine
- AgTx2
- agitoxin-2
- CSα/α
- cysteine-stabilized α/α motif
- CSα/β
- cysteine-stabilized α/β motif
- ICK
- inhibitor cystine knot
- KTX
- kaliotoxin-1
- KTx
- potassium channel toxin
- RP
- reverse-phase
- R-AgTx2
- agitoxin-2 labeled with 5(6)-carboxytetramethylrhodamine N-succinimidyl ester
- TEA
- tetraethylammonium chloride
- TFA
- trifluoroacetic acid.
REFERENCES
- 1. Hille B. (2001) Ion Channels of Excitable Membranes, 3rd Ed., pp. 131–160, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 2. Mouhat S., Andreotti N., Jouirou B., Sabatier J.-M. (2008) Animal toxins acting on voltage-gated potassium channels. Curr. Pharm. Des. 14, 2503–2518 [DOI] [PubMed] [Google Scholar]
- 3. Wulff H., Castle N. A., Pardo L. A. (2009) Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 8, 982–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Camerino D. C., Tricarico D., Desaphy J.-F. (2007) Ion channel pharmacology. Neurotherapeutics 4, 184–198 [DOI] [PubMed] [Google Scholar]
- 5. Beraud E., Viola A., Regaya I., Confort-Gouny S., Siaud P., Ibarrola D., Le Fur Y., Barbaria J., Pellissier J.-F., Sabatier J.-M., Medina I., Cozzone P. J. (2006) Block of neural Kv1.1 potassium channels for neuroinflammatory disease therapy. Ann. Neurol. 60, 586–596 [DOI] [PubMed] [Google Scholar]
- 6. Hayes K. C. (2004) The use of 4-aminopyridine (fampridine) in demyelinating disorders. CNS Drug Rev. 10, 295–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouadid-Ahidouch H., Chaussade F., Roudbaraki M., Slomianny C., Dewailly E., Delcourt P., Prevarskaya N. (2000) KV1.1 K+ channels identification in human breast carcinoma cells: involvement in cell proliferation. Biochem. Biophys. Res. Commun. 278, 272–277 [DOI] [PubMed] [Google Scholar]
- 8. Jeon W. I., Ryu P. D., Lee S. Y. (2012) Effects of voltage-gated K+ channel blockers in gefitinib-resistant H460 non-small cell lung cancer cells. Anticancer Res. 32, 5279–5284 [PubMed] [Google Scholar]
- 9. Jang S. H., Ryu P. D., Lee S. Y. (2011) Dendrotoxin-κ suppresses tumor growth induced by human lung adenocarcinoma A549 cells in nude mice. J. Vet. Sci. 12, 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swartz K. J. (2007) Tarantula toxins interacting with voltage sensors in potassium channels. Toxicon 49, 213–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korolkova Y. V., Kozlov S. A., Lipkin A. V., Pluzhnikov K. A., Hadley J. K., Filippov A. K., Brown D. A., Angelo K., Strøbaek D., Jespersen T., Olesen S. P., Jensen B. S., Grishin E. V. (2001) An ERG channel inhibitor from the scorpion Buthus eupeus. J. Biol. Chem. 276, 9868–9876 [DOI] [PubMed] [Google Scholar]
- 12. Abdel-Mottaleb Y., Vandendriessche T., Clynen E., Landuyt B., Jalali A., Vatanpour H., Schoofs L., Tytgat J. (2008) OdK2, a Kv1.3 channel-selective toxin from the venom of the Iranian scorpion Odonthobuthus doriae. Toxicon 51, 1424–1430 [DOI] [PubMed] [Google Scholar]
- 13. Peigneur S., Billen B., Derua R., Waelkens E., Debaveye S., Béress L., Tytgat J. (2011) A bifunctional sea anemone peptide with Kunitz type protease and potassium channel inhibiting properties. Biochem. Pharmacol. 82, 81–90 [DOI] [PubMed] [Google Scholar]
- 14. Chen Z.-Y., Hu Y.-T., Yang W.-S., He Y.-W., Feng J., Wang B., Zhao R.-M., Ding J.-P., Cao Z.-J., Li W.-X., Wu Y.-L. (2012) Hg1, novel peptide inhibitor specific for Kv1.3 channels from first scorpion Kunitz-type potassium channel toxin family. J. Biol. Chem. 287, 13813–13821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Varga Z., Gurrola-Briones G., Papp F., Rodríguez de la Vega R. C., Pedraza-Alva G., Tajhya R. B., Gaspar R., Cardenas L., Rosenstein Y., Beeton C., Possani L. D., Panyi G. (2012) Vm24, a natural immunosuppressive peptide, potently and selectively blocks Kv1.3 potassium channels of human T cells. Mol. Pharmacol. 82, 372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Panyi G., Possani L. D., Rodríguez de la Vega R. C., Gáspár R., Varga Z. (2006) K+ channel blockers: novel tools to inhibit T cell activation leading to specific immunosuppression. Curr. Pharm. Des. 12, 2199–2220 [DOI] [PubMed] [Google Scholar]
- 17. Kalman K., Pennington M. W., Lanigan M. D., Nguyen A., Rauer H., Mahnir V., Paschetto K., Kem W. R., Grissmer S., Gutman G. A., Christian E. P., Cahalan M. D., Norton R. S., Chandy K. G. (1998) ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide. J. Biol. Chem. 273, 32697–32707 [DOI] [PubMed] [Google Scholar]
- 18. Han S., Yi H., Yin S.-J., Chen Z.-Y., Liu H., Cao Z.-J., Wu Y.-L., Li W.-X. (2008) Structural basis of a potent peptide inhibitor designed for Kv1.3 channel, a therapeutic target of autoimmune disease. J. Biol. Chem. 283, 19058–19065 [DOI] [PubMed] [Google Scholar]
- 19. Takacs Z., Toups M., Kollewe A., Johnson E., Cuello L. G., Driessens G., Biancalana M., Koide A., Ponte C. G., Perozo E., Gajewski T. F., Suarez-Kurtz G., Koide S., Goldstein S. A. (2009) A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library. Proc. Natl. Acad. Sci. U.S.A. 106, 22211–22216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen R., Chung S.-H. (2012) Engineering a potent and specific blocker of voltage-gated potassium channel Kv1.3, a target for autoimmune diseases. Biochemistry 51, 1976–1982 [DOI] [PubMed] [Google Scholar]
- 21. Rodríguez de la Vega R. C., Possani L. D. (2004) Current views on scorpion toxins specific for K+ channels. Toxicon 43, 865–875 [DOI] [PubMed] [Google Scholar]
- 22. Martins J. C., Zhang W. G., Tartar A., Lazdunski M., Borremans F. A. (1990) Solution conformation of leiurotoxin I (scyllatoxin) by 1H nuclear magnetic resonance. Resonance assignment and secondary structure. FEBS Lett. 260, 249–253 [DOI] [PubMed] [Google Scholar]
- 23. Gao B., Harvey P. J., Craik D. J., Ronjat M., De Waard M., Zhu S. (2013) Functional evolution of scorpion venom peptides with an inhibitor cystine knot fold. Biosci. Rep. 33, e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Srinivasan K. N., Sivaraja V., Huys I., Sasaki T., Cheng B., Kumar T. K., Sato K., Tytgat J., Yu C., San BC, Ranganathan S., Bowie H. J., Kini R. M., Gopalakrishnakone P. (2002) κ-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function. Importance of the functional diad in potassium channel selectivity. J. Biol. Chem. 277, 30040–30047 [DOI] [PubMed] [Google Scholar]
- 25. Saucedo A. L., Flores-Solis D., Rodríguez de la Vega R. C., Ramírez-Cordero B., Hernández-López R., Cano-Sánchez P., Noriega Navarro R., García-Valdés J., Coronas-Valderrama F., de Roodt A., Brieba L. G., Domingos Possani L., del Río-Portilla F. (2012) New tricks of an old pattern: structural versatility of scorpion toxins with common cysteine spacing. J. Biol. Chem. 287, 12321–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tytgat J., Chandy K. G., Garcia M. L., Gutman G. A., Martin-Eauclaire M. F., van der Walt J. J., Possani L. D. (1999) A unified nomenclature for short-chain peptides isolated from scorpion venoms: α-KTx molecular subfamilies. Trends Pharmacol. Sci. 20, 444–447 [DOI] [PubMed] [Google Scholar]
- 27. Rodríguez de la Vega R. C., Schwartz E. F., Possani L. D. (2010) Mining on scorpion venom biodiversity. Toxicon 56, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 28. Quintero-Hernández V., Ortiz E., Rendón-Anaya M., Schwartz E. F., Becerril B., Corzo G., Possani L. D. (2011) Scorpion and spider venom peptides: gene cloning and peptide expression. Toxicon 58, 644–663 [DOI] [PubMed] [Google Scholar]
- 29. Ma Y., He Y., Zhao R., Wu Y., Li W., Cao Z. (2012) Extreme diversity of scorpion venom peptides and proteins revealed by transcriptomic analysis: implication for proteome evolution of scorpion venom arsenal. J. Proteomics 75, 1563–1576 [DOI] [PubMed] [Google Scholar]
- 30. Kozlov S., Malyavka A., McCutchen B., Lu A., Schepers E., Herrmann R., Grishin E. (2005) A novel strategy for the identification of toxinlike structures in spider venom. Proteins 59, 131–140 [DOI] [PubMed] [Google Scholar]
- 31. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saitou N., Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 33. Vassilevski A. A., Kozlov S. A., Egorov T. A., Grishin E. V. (2010) Purification and characterization of biologically active peptides from spider venoms. Methods Mol. Biol. 615, 87–100 [DOI] [PubMed] [Google Scholar]
- 34. Vassilevski A. A., Kozlov S. A., Samsonova O. V., Egorova N. S., Karpunin D. V., Pluzhnikov K. A., Feofanov A. V., Grishin E. V. (2008) Cyto-insectotoxins, a novel class of cytolytic and insecticidal peptides from spider venom. Biochem. J. 411, 687–696 [DOI] [PubMed] [Google Scholar]
- 35. Anthis N. J., Clore G. M. (2013) Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 22, 851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kudryashova K. S., Nekrasova O. V., Kuzmenkov A. I., Vassilevski A. A., Ignatova A. A., Korolkova Y. V., Grishin E. V., Kirpichnikov M. P., Feofanov A. V. (2013) Fluorescent system based on bacterial expression of hybrid KcsA channels designed for Kv1.3 ligand screening and study. Anal. Bioanal. Chem. 405, 2379–2389 [DOI] [PubMed] [Google Scholar]
- 37. Nekrasova O., Tagway A., Ignatova A., Feofanov A., Kirpichnikov M. (2009) Studying of membrane localization of recombinant potassium channels in E.coli. Acta Naturae 1, 91–95 [PMC free article] [PubMed] [Google Scholar]
- 38. Nekrasova O. V., Ignatova A. A., Nazarova A. I., Feofanov A. V., Korolkova Y. V., Boldyreva E. F., Tagvei A. I., Grishin E. V., Arseniev A. S., Kirpichnikov M. P. (2009) Recombinant Kv channels at the membrane of Escherichia coli bind specifically agitoxin2. J. Neuroimmune Pharmacol. 4, 83–91 [DOI] [PubMed] [Google Scholar]
- 39. Liman E. R., Tytgat J., Hess P. (1992) Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9, 861–871 [DOI] [PubMed] [Google Scholar]
- 40. Legros C., Schulze C., Garcia M. L., Bougis P. E., Martin-Eauclaire M.-F., Pongs O. (2002) Engineering-specific pharmacological binding sites for peptidyl inhibitors of potassium channels into KcsA. Biochemistry 41, 15369–15375 [DOI] [PubMed] [Google Scholar]
- 41. Helms L. M., Felix J. P., Bugianesi R. M., Garcia M. L., Stevens S., Leonard R. J., Knaus H. G., Koch R., Wanner S. G., Kaczorowski G. J., Slaughter R. S. (1997) Margatoxin binds to a homomultimer of K(V)1.3 channels in Jurkat cells. Comparison with K(V)1.3 expressed in CHO cells. Biochemistry 36, 3737–3744 [DOI] [PubMed] [Google Scholar]
- 42. Legros C., Pollmann V., Knaus H. G., Farrell A. M., Darbon H., Bougis P. E., Martin-Eauclaire M. F., Pongs O. (2000) Generating a high affinity scorpion toxin receptor in KcsA-Kv1.3 chimeric potassium channels. J. Biol. Chem. 275, 16918–16924 [DOI] [PubMed] [Google Scholar]
- 43. Andalib P., Consiglio J. F., Trapani J. G., Korn S. J. (2004) The external TEA-binding site and C-type inactivation in voltage-gated potassium channels. Biophys. J. 87, 3148–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kirsch G. E., Shieh C. C., Drewe J. A., Vener D. F., Brown A. M. (1993) Segmental exchanges define 4-aminopyridine binding and the inner mouth of K+ pores. Neuron 11, 503–512 [DOI] [PubMed] [Google Scholar]
- 45. Shieh C. C., Kirsch G. E. (1994) Mutational analysis of ion conduction and drug-binding sites in the inner mouth of voltage-gated K+ channels. Biophys. J. 67, 2316–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu S., Peigneur S., Gao B., Luo L., Jin D., Zhao Y., Tytgat J. (2011) Molecular diversity and functional evolution of scorpion potassium channel toxins. Mol. Cell. Proteomics 10, M110.002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao B., Peigneur S., Tytgat J., Zhu S. (2010) A potent potassium channel blocker from Mesobuthus eupeus scorpion venom. Biochimie 92, 1847–1853 [DOI] [PubMed] [Google Scholar]
- 48. Gao B., Peigneur S., Dalziel J., Tytgat J., Zhu S. (2011) Molecular divergence of two orthologous scorpion toxins affecting potassium channels. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 159, 313–321 [DOI] [PubMed] [Google Scholar]
- 49. Zhu S., Gao B., Aumelas A., del Carmen Rodríguez M., Lanz-Mendoza H., Peigneur S., Diego-Garcia E., Martin-Eauclaire M.-F., Tytgat J., Possani L. D. (2010) MeuTXKβ1, a scorpion venom-derived two-domain potassium channel toxin-like peptide with cytolytic activity. Biochim. Biophys. Acta 1804, 872–883 [DOI] [PubMed] [Google Scholar]
- 50. Filippov A. K., Kozlov S. A., Pluzhnikov K. A., Grishin E. V., Brown D. A. (1996) M-type K+ current inhibition by a toxin fron the scorpion Buthus eupeus. FEBS Lett. 384, 277–280 [DOI] [PubMed] [Google Scholar]
- 51. Romi-Lebrun R., Lebrun B., Martin-Eauclaire M. F., Ishiguro M., Escoubas P., Wu F. Q., Hisada M., Pongs O., Nakajima T. (1997) Purification, characterization, and synthesis of three novel toxins from the Chinese scorpion Buthus martensi, which act on K+ channels. Biochemistry 36, 13473–13482 [DOI] [PubMed] [Google Scholar]
- 52. Mille B. G., Peigneur S., Diego-García E., Predel R., Tytgat J. (2014) Partial transcriptomic profiling of toxins from the venom gland of the scorpion Parabuthus stridulus. Toxicon 83, 75–83 [DOI] [PubMed] [Google Scholar]
- 53. Luna-Ramírez K., Quintero-Hernández V., Vargas-Jaimes L., Batista C. V., Winkel K. D., Possani L. D. (2013) Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon 63, 44–54 [DOI] [PubMed] [Google Scholar]
- 54. Abdel-Rahman M. A., Quintero-Hernandez V., Possani L. D. (2013) Venom proteomic and venomous glands transcriptomic analysis of the Egyptian scorpion Scorpio maurus palmatus (Arachnida: Scorpionidae). Toxicon 74, 193–207 [DOI] [PubMed] [Google Scholar]
- 55. Almeida D. D., Scortecci K. C., Kobashi L. S., Agnez-Lima L. F., Medeiros S. R., Silva-Junior A. A., Junqueira-de-Azevedo Ide L., Fernandes-Pedrosa Mde F. (2012) Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genomics 13, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. He Y., Zhao R., Di Z., Li Z., Xu X., Hong W., Wu Y., Zhao H., Li W., Cao Z. (2013) Molecular diversity of Chaerilidae venom peptides reveals the dynamic evolution of scorpion venom components from Buthidae to non-Buthidae. J. Proteomics 89, 1–14 [DOI] [PubMed] [Google Scholar]
- 57. Mozhaeva G. N., Naumov A. P., Soldatov N. M., Grishin E. V. (1979) Effect of toxins from the scorpion Buthus eupeus on the sodium channels of the membranes of nodes of Ranvier. Biofizika 24, 235–241 [PubMed] [Google Scholar]
- 58. Eskandari G., Jolodar A., Seyfiabad Shapouri M. R., Bahmainmehr A., Navidpour S. (2014) Production of recombinant α neurotoxin of scorpion venom Mesobuthus eupeus and analysis of its immunogenicity. Iran. Red Crescent Med. J. 16, e9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kharrat R., Mansuelle P., Sampieri F., Crest M., Oughideni R., Van Rietschoten J., Martin-Eauclaire M. F., Rochat H., El Ayeb M. (1997) Maurotoxin, a four disulfide bridge toxin from Scorpio maurus venom: purification, structure and action on potassium channels. FEBS Lett. 406, 284–290 [DOI] [PubMed] [Google Scholar]
- 60. Beeton C., Wulff H., Singh S., Botsko S., Crossley G., Gutman G. A., Cahalan M. D., Pennington M., Chandy K. G. (2003) A novel fluorescent toxin to detect and investigate Kv1.3 channel up-regulation in chronically activated T lymphocytes. J. Biol. Chem. 278, 9928–9937 [DOI] [PubMed] [Google Scholar]
- 61. Freudenthaler G., Axmann M., Schindler H., Pragl B., Knaus H.-G., Schütz G. J. (2002) Ultrasensitive pharmacological characterisation of the voltage-gated potassium channel K(V)1.3 studied by single-molecule fluorescence microscopy. Histochem. Cell Biol. 117, 197–202 [DOI] [PubMed] [Google Scholar]
- 62. Terstappen G. C., Roncarati R., Dunlop J., Peri R. (2010) Screening technologies for ion channel drug discovery. Future Med. Chem. 2, 715–730 [DOI] [PubMed] [Google Scholar]
- 63. Crest M., Jacquet G., Gola M., Zerrouk H., Benslimane A., Rochat H., Mansuelle P., Martin-Eauclaire M. F. (1992) Kaliotoxin, a novel peptidyl inhibitor of neuronal BK-type Ca2+-activated K+ channels characterized from Androctonus mauretanicus mauretanicus venom. J. Biol. Chem. 267, 1640–1647 [PubMed] [Google Scholar]
- 64. Abbas N., Belghazi M., Abdel-Mottaleb Y., Tytgat J., Bougis P. E., Martin-Eauclaire M.-F. (2008) A new Kaliotoxin selective toward Kv1.3 and Kv1.2 but not Kv1.1 channels expressed in oocytes. Biochem. Biophys. Res. Commun. 376, 525–530 [DOI] [PubMed] [Google Scholar]
- 65. Shijin Y., Hong Y., Yibao M., Zongyun C., Han S., Yingliang W., Zhijian C., Wenxin L. (2008) Characterization of a new Kv1.3 channel-specific blocker, J123, from the scorpion Buthus martensii Karsch. Peptides 29, 1514–1520 [DOI] [PubMed] [Google Scholar]
- 66. Mouhat S., Jouirou B., Mosbah A., De Waard M., Sabatier J.-M. (2004) Diversity of folds in animal toxins acting on ion channels. Biochem. J. 378, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rodríguez de la Vega R. C., Merino E., Becerril B., Possani L. D. (2003) Novel interactions between K+ channels and scorpion toxins. Trends Pharmacol. Sci. 24, 222–227 [DOI] [PubMed] [Google Scholar]
- 68. Darbon H., Blanc E., Sabatier J. M. (1999) in Perspectives in Drug Discovery and Design: Animal Toxins and Potassium Channels (Darbon H., Sabatier J. M., eds) Vol. 15/16, pii: e00047 Kluwer/Escom, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 69. Dai L., Wu J. J., Gu Y. H., Lan Z. D., Ling M. H., Chi C. W. (2000) Genomic organization of three novel toxins from the scorpion Buthus martensi Karsch that are active on potassium channels. Biochem. J. 346, 805–809 [PMC free article] [PubMed] [Google Scholar]
- 70. Diego-García E., Peigneur S., Debaveye S., Gheldof E., Tytgat J., Caliskan F. (2013) Novel potassium channel blocker venom peptides from Mesobuthus gibbosus (Scorpiones: Buthidae). Toxicon 61, 72–82 [DOI] [PubMed] [Google Scholar]
- 71. Holaday S. K., Jr., Martin B. M., Fletcher P. L., Jr., Krishna N. R. (2000) NMR solution structure of butantoxin. Arch. Biochem. Biophys. 379, 18–27 [DOI] [PubMed] [Google Scholar]
- 72. Mouhat S., Visan V., Ananthakrishnan S., Wulff H., Andreotti N., Grissmer S., Darbon H., De Waard M., Sabatier J.-M. (2005) K+ channel types targeted by synthetic OSK1, a toxin from Orthochirus scrobiculosus scorpion venom. Biochem. J. 385, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grissmer S., Nguyen A. N., Aiyar J., Hanson D. C., Mather R. J., Gutman G. A., Karmilowicz M. J., Auperin D. D., Chandy K. G. (1994) Pharmacological characterization of five cloned voltage-gated K+ channels, types Kv1.1, 1.2, 1.3, 1.5, and 3.1, stably expressed in mammalian cell lines. Mol. Pharmacol. 45, 1227–1234 [PubMed] [Google Scholar]