Background: Many transmembrane anti-sigma factors, including P. aeruginosa FoxR, undergo periplasmic self-cleavage by an unknown mechanism.
Results: Presence of an OH or SH group at +1 of the cleavage site is essential for FoxR self-cleavage.
Conclusion: FoxR self-cleavage is mediated by an N-O acyl rearrangement.
Significance: The complex proteolytic network that modulates alternative sigma factor activity in Gram-negative bacteria also includes an enzyme-independent step.
Keywords: Cell Signaling, Gene Regulation, Iron, Post-translational Modification (PTM), Proteolysis, Pseudomonas aeruginosa (P. aeruginosa), Siderophore, Signal Transduction
Abstract
The Fox system of Pseudomonas aeruginosa is a cell-surface signaling (CSS) pathway employed by the bacterium to sense and respond to the presence of the heterologous siderophore ferrioxamine in the environment. This regulatory pathway controls the transcription of the foxA ferrioxamine receptor gene through the extracytoplasmic function sigma factor σFoxI. In the absence of ferrioxamine, the activity of σFoxI is inhibited by the transmembrane anti-sigma factor FoxR. Upon binding of ferrioxamine by the FoxA receptor, FoxR is processed by a complex proteolytic cascade leading to the release and activation of σFoxI. Interestingly, we have recently shown that FoxR undergoes self-cleavage between the periplasmic Gly-191 and Thr-192 residues independent of the perception of ferrioxamine. This autoproteolytic event, which is widespread among CSS anti-sigma factors, produces two distinct domains that interact and function together to transduce the presence of the signal. In this work, we provide evidence that the self-cleavage of FoxR is not an enzyme-dependent process but is induced by an N-O acyl rearrangement. Mutation analysis showed that the nucleophilic side chain of the Thr-192 residue at +1 of the cleavage site is required for an attack on the preceding Gly-191, after which the resulting ester bond is likely hydrolyzed. Because the cleavage site is well preserved and the hydrolysis of periplasmic CSS anti-sigma factors is widely observed, we hypothesize that cleavage via an N-O acyl rearrangement is a conserved feature of these proteins.
Introduction
The Fox system of the Gram-negative bacterium Pseudomonas aeruginosa is a signal transduction system used by the bacterium to respond to and regulate the uptake of the siderophore ferrioxamine (1). Siderophores are high affinity iron-chelating compounds that are produced and secreted by bacteria to solubilize the minute amounts of bioavailable iron present in the environment (2, 3). P. aeruginosa produces the two siderophores pyoverdine and pyochelin but is also very efficient in using siderophores produced by other bacterial or fungal species (referred to as xeno- or heterologous siderophores), such as ferrioxamine (1). In Gram-negative bacteria, ferri-siderophore complexes are transported into the bacterial cells by specific TonB-dependent receptors in the outer membrane (4). These proteins form a large 22-stranded β-barrel, which is occluded by a plug domain when the substrate is not present (4). Production of siderophore receptors is an energetically costly process and generally only occurs when the cognate siderophore is present in the environment (1, 5, 6). This process is usually controlled by a trans-envelope regulatory signal transduction pathway known as cell-surface signaling (CSS)2 (7–9). This regulatory cascade involves three proteins: the siderophore receptor itself, an anti-sigma factor located at the cytoplasmic membrane, and an extracytoplasmic function (ECF) sigma factor (σECF) in the cytosol. Sigma factors are small subunits that associate with the RNA polymerase core enzyme, allowing promoter recognition and initiation of gene transcription. Apart from a primary sigma factor that controls expression of genes required for general functions, bacteria contain a variable number of alternative sigma factors of which the σECF constitute the largest group (10, 11). σECF are usually co-expressed with anti-sigma factors that bind to and sequester the sigma factor to keep it in an inactive state (10, 11). In Gram-negative bacteria, these anti-sigma factors are typically cytoplasmic membrane proteins that contain a short cytosolic N-terminal domain of 85–90 amino acids that binds the σECF linked to a larger periplasmic C-terminal region by a single transmembrane segment (8) (see Fig. 1). The N-terminal domains of most anti-sigma factors exhibit structural homology despite a low sequence similarity (12). A common structural motif, termed the ASD (for anti-sigma domain), is responsible for the interaction with the σECF, thereby shielding the DNA and RNA polymerase core enzyme-binding determinants (12–15).
FIGURE 1.
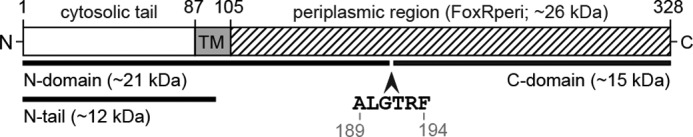
Schematic representation of the P. aeruginosa FoxR protein. The P. aeruginosa FoxR protein has been drawn to scale, and the cytosolic, transmembrane, and periplasmic (FoxRperi) regions of the protein are detailed. The site where the self-cleavage of FoxR occurs (between Gly-191 and Thr-192; GT) and surrounding residues have been indicated. The numbers indicate amino acid positions in the FoxR protein. The N- and C-domains resulting from self-cleavage are illustrated. The exact cleavage sites of the Prc and RseP proteases are unknown.
Activation of σECF normally only occurs in response to a specific inducing signal, such as the presence of the heterologous siderophore ferrioxamine in the P. aeruginosa environment. The presence of this siderophore in the extracellular milieu is sensed by the outer membrane receptor FoxA, which transduces the signal to the FoxR anti-sigma factor and thereby induces the activity of the ECF sigma factor σFoxI in the cytosol (1). Upon activation, σFoxI initiates transcription of the foxA gene, thereby increasing the amount of the ferrioxamine receptor in the outer membrane and the capacity of the bacterium to transport ferrioxamine (1). Receptors involved in both siderophore transport and signaling contain an additional N-terminal periplasmic domain of 70–80 amino acids long referred to as the signaling domain (16–18). This domain, which is composed of two α-helices sandwiched by two antiparallel β-sheets (19, 20), determines the specificity of the signal transduction pathway but has no effect on the transport function of the CSS receptor (19–21). In the current model of CSS, the signaling domain of FoxA interacts with the periplasmic domain of FoxR upon binding of ferrioxamine. As a result, FoxR is subjected to a complex proteolytic cascade, leading to the release and activation of σFoxI (22). Both the C-terminal processing protease Prc and the transmembrane protease RseP play a role in this process (23, 24), although not all proteases involved in FoxR proteolysis have been identified yet (22). Remarkably, the FoxR protein has both anti-sigma and pro-sigma activity. In addition to inhibiting σFoxI in absence of ferrioxamine, FoxR is also required for σFoxI activity in response to this siderophore (25). The pro-sigma function of FoxR is conveyed by its σFoxI-binding cytoplasmic tail (FoxR1–93) (25). We have recently shown that upon perception of ferrioxamine, processing of FoxR results in the production of a ∼12-kDa N-terminal fragment that is associated with activity of σFoxI. The RseP protease is responsible for the generation of this N-tail in vivo by cleaving in or near the FoxR transmembrane segment (22).
Interestingly, the FoxR anti-sigma factor is also subjected to post-translational processing prior to induction of the Fox CSS system by ferrioxamine (22, 24). We have recently shown that upon production, FoxR undergoes self-cleavage between the Gly-191 and Thr-192 residues located in the periplasmic region of the protein (see Fig. 1) (22). This autoproteolytic event, termed initial cleavage, occurs independent of the presence or absence of ferrioxamine. The self-cleavage produces a stable FoxR N-domain of ∼21 kDa, which comprises the cytosolic N-tail, the transmembrane segment, and 85 residues of the periplasmic region, and a stable periplasmic C-domain of ∼15 kDa (see Fig. 1) (22). These two domains of FoxR are functional and interact in the periplasm (22). The N-domain contains the pro-sigma activity of FoxR, whereas the C-domain contains the anti-sigma properties of the protein (22). In our previous work, we have also demonstrated that the initial cleavage is widespread in anti-sigma factors involved in CSS, although the process itself is not essential for protein functionality (22). To gain more insight into this autoproteolytic event of CSS anti-sigma factors, we aimed at determining the mechanism responsible for the self-cleavage of P. aeruginosa FoxR (PaFoxR). Our results strongly indicate that PaFoxR autocleavage is an enzyme-independent mechanism that occurs through an intramolecular N-O acyl rearrangement.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains used in this study are listed in Table 1. Escherichia coli and P. aeruginosa were cultured at 37 °C on a rotary shaker operated at 200 rpm in liquid LB medium (26). For induction experiments, P. aeruginosa was grown in liquid CAS medium (1) supplemented with 400 μm of the iron chelator 2,2′-bipyridyl and without or with 1 μm iron-free ferrioxamine B (Sigma-Aldrich). For Western blot analyses, 1 mm isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium to induce full expression from the pMMB67EH Ptac promoter. When required, antibiotics were used at the following final concentrations: 100 μg ml−1 ampicillin, 25 μg ml−1 piperacillin, and 12.5 μg ml−1 tetracycline for E. coli and 20 μg ml−1 tetracycline for P. aeruginosa.
TABLE 1.
Bacterial strains and plasmids used in this study
ApR, KmR, NalR, and TcR indicate resistance to ampicillin, kanamycin, nalidixic acid, and tetracycline, respectively.
| Strain | Relevant characteristics | Reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 Δ (lacZYA-argF)U169 φ80 lacZDM15 hsdR17 (rK− mK+) recA1 endA1 gyrA96 thi1 relA1; NalR | Ref. 53 |
| TOP10F′ | F′[lacIq, Tn10 (TetR)] mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG; TcR | Invitrogen |
| P. aeruginosa | ||
| PAO1 pvdF ΔfoxR | PAO1 pvdF::KmR with a deletion of amino acids 12–295 of the FoxR (PA2467) protein, KmR | Ref. 25 |
| Plasmid | ||
| pMMB67EH | IncQ broad host range plasmid, lacIq; ApR | Ref. 27 |
| pMMB/HA-FoxR | pMMB67EH carrying the P. aeruginosa foxR gene (PA2467), which has been N-terminally HA-tagged; ApR | Ref. 22 |
| pMMB/HA-FoxR-S166A | pMMB/HA-FoxR in which serine 166 has been mutated to an alanine; ApR | This study |
| pMMB/HA-FoxR-D169A | pMMB/HA-FoxR in which aspartic acid 169 has been mutated to an alanine; ApR | This study |
| pMMB/HA-FoxR-S172A | pMMB/HA-FoxR in which serine 172 has been mutated to an alanine; ApR | This study |
| pMMB/HA-FoxR-H175A | pMMB/HA-FoxR in which histidine 175 has been mutated to an alanine; ApR | This study |
| pMMB/HA-FoxR-A189G/L190G | pMMB/HA-FoxR in which alanine 189 and leucine 190 have both been mutated to glycine; ApR | This study |
| pMMB/HA-FoxR-T192S | pMMB/HA-FoxR in which threonine-192 has been mutated to a serine; ApR | This study |
| pMMB/HA-FoxR-T192C | pMMB/HA-FoxR in which threonine 192 has been mutated to a cysteine; ApR | This study |
| pMMB/HA-FoxR-T192V | pMMB/HA-FoxR in which threonine 192 has been mutated to a valine; ApR | This study |
| pMMB/HA-FoxR-S208A | pMMB/HA-FoxR in which serine 208 has been mutated to an alanine; ApR | This study |
| pMMB/HA-FoxR-S211A | pMMB/HA-FoxR in which serine 211 has been mutated to an alanine; ApR | This study |
| pMPR8b | pMP220 carrying the P. aeruginosa foxA::lacZ transcriptional fusion; TcR | Ref. 1 |
Plasmid Construction
Plasmids used in this study are described in Table 1. In short, the P. aeruginosa foxR (PA2467) wild-type gene and its mutant derivatives were cloned in the EcoRI-XbaI sites of the pMMB67EH plasmid (27) following PCR amplifications using Phusion® hot start high fidelity DNA polymerase (Finnzymes). Point mutations in the foxR gene were introduced by nested PCR with appropriate primers (primer sequences are available upon request). All constructs were validated by DNA sequencing and transferred to P. aeruginosa by electroporation (28).
Enzyme Assay
β-Galactosidase activities in soluble P. aeruginosa cell extracts were measured using the o-nitrophenyl-β-d-galactopyranoside substrate (Sigma-Aldrich) as described previously (1). Activity is represented by Miller units (MU). Assays were performed at least three times in duplicate, and the data given are the averages. The error bars in each graph show S.D.
SDS-PAGE and Immunoblot
SDS-PAGE and immunoblot were performed as described previously (22). In brief, bacterial cultures were grown to late log phase in iron-restricted medium with 1 mm IPTG in the absence or presence of 1 μm ferrioxamine B. Samples were normalized according to the A660 of the culture. Following separation by SDS-PAGE containing 15% (w/v) acrylamide, proteins were transferred to nitrocellulose membranes (Millipore). Ponceau S staining was performed as a loading control prior to immunodetection using a monoclonal antibody against the influenza hemagglutinin epitope (HA.11; Covance) or the MA4-4 monoclonal antibody against the P. aeruginosa OprF protein (29).
Expression of FoxR Using the PURExpress® System
In vitro production of the periplasmic region of FoxR with an N-terminal (FoxRperi-NHA) or a C-terminal HA tag (FoxRperi-CHA) was performed as described previously (22). When indicated, the following protease inhibitors were added to the reaction mixture: 1× cOmplete protease inhibitor mixture (Roche), 1 mm Pefabloc SC (Biomol GmbH), 1 μm Pepstatin A (Sigma), 5 mm EDTA (Sigma), or 5 mm EGTA (Sigma). Reactions were incubated for 2 h at 37 °C and analyzed on SDS-PAGE containing 15% (w/v) acrylamide followed by anti-HA immunoblot as described above.
Computer-assisted Analyses
Pseudomonas sequences were accessed at the Pseudomonas Genome Database (30). Multiple sequence alignments were performed with ClustalW (31), whereas pairwise sequence alignments were performed using the EMBOSS algorithm. Secondary structure prediction was performed with the Phyre2 program (32). The quality of the model was assessed by comparing it with its solved structure homologue BDI_1681 using DALI (33) and TM-align (34). These sites compare structural models, rather than sequences, and return the root mean square deviation of the aligned structures. Two-tailed t-tests were used to calculate p values with GraphPad Prism version 5.01 for Windows.
RESULTS
Structural Modeling of the Periplasmic Region of P. aeruginosa FoxR
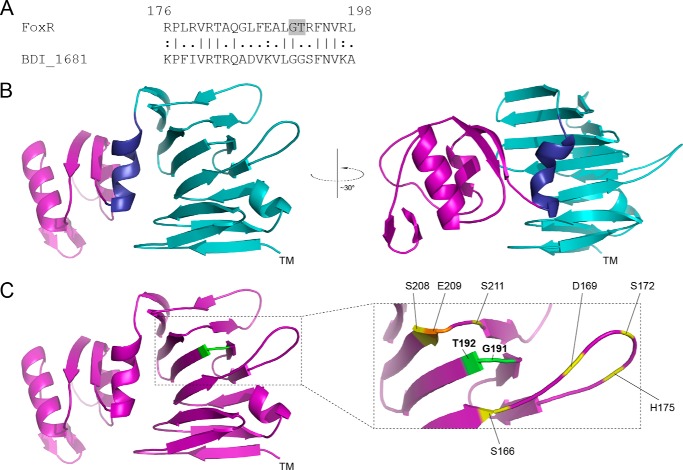
The P. aeruginosa FoxR CSS anti-sigma factor undergoes autoproteolytic cleavage by an unknown mechanism (22). Protein sequence comparisons of P. aeruginosa FoxR (PaFoxR) with other Pseudomonas CSS anti-sigma factors did not reveal any notable differences between proteins that undergo autoproteolysis and those that do not, in addition to the presence of the Gly-Thr residues in the periplasmic region (22). The structure of PaFoxR has not been resolved by x-ray crystallography yet. Therefore, to examine the mechanism behind FoxR autoproteolysis in more detail, we first generated a structural model of this protein. Because processing occurs between Gly-Thr residues located in the periplasmic region of PaFoxR, and the presence of the cytosolic N-tail and transmembrane segment of the protein is not required for self-cleavage to occur (22), only this part of the protein (FoxRperi, residues 105–328) (Fig. 1) was modeled using the Protein Homology/analogy Recognition Engine (Phyre) server (32). This yielded a model (Fig. 2) that is based upon the crystal structure of the putative anti-sigma factor BDI_1681 from Parabacteroides distasonis ATCC8503 (Protein Data Bank accession number 4M0H), in which ∼95% of the submitted sequence could be assigned. The gene coding for this putative anti-sigma factor is located adjacent to a predicted σECF gene (BDI_1680) and a CSS receptor gene (BDI_1682), which strongly suggests that the resulting protein is, like FoxR, part of a CSS system. Indeed, although FoxR and BDI_1681 only show ∼20% identity and ∼43% similarity in the modeled region in both sequence-based and secondary structure-based alignments, the confidence that the two proteins are homologues was predicted to be 100%. Of note, these comparisons revealed that residues 176–198 of FoxR, which contain the Gly-Thr cleavage site, were highly similar to the equivalent region in BDI_1681 (Fig. 2A). To further assess the quality of the Phyre2 model, we submitted it to the DALI (33) and TM-align (34) servers, where algorithms calculated a root mean square deviation of 1.5 and 1.9, respectively. Furthermore, TM-align returned a TM score of 0.89, which is considered to represent models of high confidence. Overall, these analyses indicated that we could address structure function relations in the FoxRperi protein with some confidence based on the structural model obtained. The structural model of P. aeruginosa FoxRperi predicts that this region forms two distinct domains separated by a single α-helix (dark blue; residues 246–255) (Fig. 2B). The N terminus of FoxRperi (light blue; residues 114–245) is calculated to mainly consist of stacked β-strands, whereas the C terminus (purple; residues 256–328) is predicted to contain two α-helices flanked by β-strands on each side (Fig. 2B). Interestingly, this part of the model shows strong structural homology to the periplasmic domains of outer membrane CSS receptors (19, 35, 36). The GT site where the autocleavage of PaFoxR occurs (green; residues 191–192) is predicted to be located in a small, and, based upon the model, tight loop between β-strands 8 and 9 of the N terminus of FoxRperi (Fig. 2C).
FIGURE 2.
Structural modeling of PaFoxR. A, a pair-wise alignment of the P. aeruginosa FoxR protein (residues 176–198) is shown with its homologue BDI_1681 of P. distasonis. The GT cleavage site in FoxR is shaded. B, a cartoon representation of the structural model of the periplasmic region of FoxR (FoxRperi) created using the Phyre program (32) is shown. The location of the transmembrane segment is indicated by TM. The C terminus of FoxRperi (residues 256–328) has been colored purple, and the N terminus of FoxRperi (residues 114–245) is in light blue. The helix separating these distinct regions (residues 246–255) is shown in dark blue. C, the residues flanking the self-cleavage site (Gly-191 and Thr-192) have been colored green. Residues that could be part of a putative proteolytic active site (Ser, etc.) are indicated in yellow, and Glu-209 is shown in orange (see text).
Mutational Analysis of Potential Active Site Residues of PaFoxR
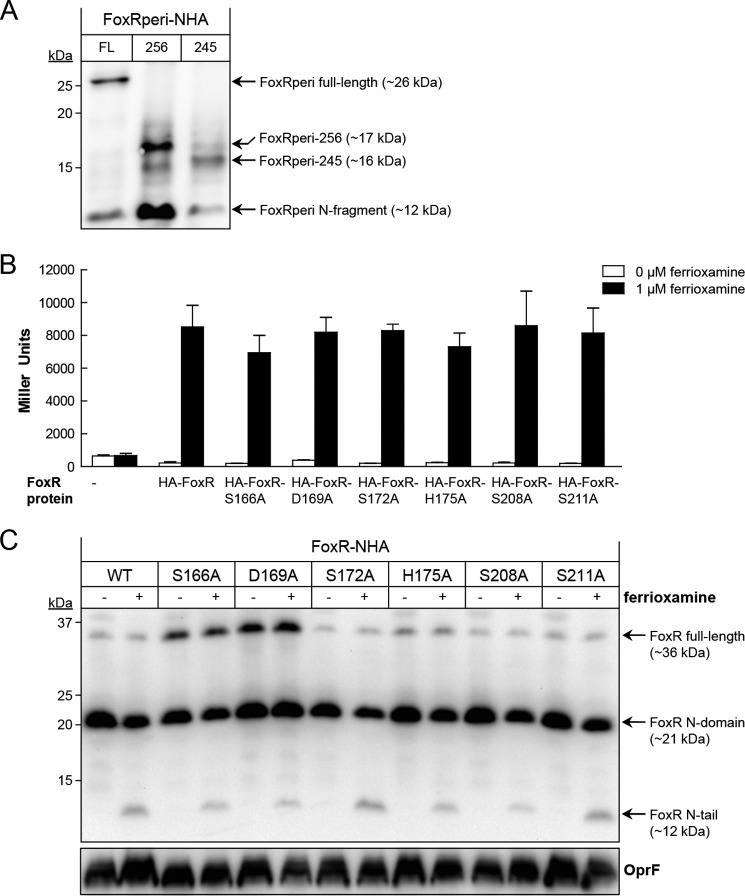
To identify potential active site residues responsible for the self-cleavage of FoxRperi, we first examined whether the C terminus of this protein contained any determinants important for the autoproteolytic reaction. An N-terminally HA-tagged FoxRperi (FoxRperi-NHA) protein was expressed using the PURExpress® system, and cleavage was monitored by Western blot. The PURExpress® system is a protease-free in vitro transcription/translation platform that we have previously successfully used to show that PaFoxR undergoes self-cleavage (22). Using this system, we produced a full-length FoxRperi-NHA as a ∼26-kDa product, and we observed that a large portion of the protein is processed into a ∼12-kDa N-fragment (Fig. 3A). Interestingly, deletion of the entire C terminus of FoxRperi (the purple domain in Fig. 2B; FoxRperi-NHA-256 protein) does not block the production of the N-fragment (Fig. 3A). Likewise, deletion of both the C terminus and the α-helix connecting the N and C terminus of FoxRperi (the dark blue α-helix in Fig. 2B; FoxRperi-NHA-245 protein) also allowed FoxR self-cleavage to occur (Fig. 3A). These results indicate that the determinants for autocleavage reside in the N terminus of FoxRperi (light blue domain in Fig. 2B). These results underline the two-domain composition of FoxRperi, because deletion of the C terminus of this protein does not affect the folding and cleavage of the N terminus. Interestingly, based on the structural model of FoxRperi, we detected a number of residues in the N terminus that could be part of a proteolytic active site (i.e. serine, aspartic acid, and histidine residues), which were located in close proximity to the GT site (Fig. 2C, yellow). To analyze the role of these potential active site residues, we constructed six FoxR variants in which these amino acids were substituted for alanine. The FoxR-S166A, -D169A, -S172A, -H175A, -S208A, and -S211A mutations were introduced in the pMMB/HA-FoxR plasmid, a pMMB67EH derivative in which an N-terminally HA-tagged FoxR is expressed from the IPTG-inducible Ptac promoter (22). We used a P. aeruginosa ΔfoxR mutant that is not able to respond to ferrioxamine (25) to evaluate the functionality of these FoxR variants by measuring the enzymatic activity of a σFoxI-dependent foxA::lacZ transcriptional fusion (1). All six PaFoxR variants were fully active, i.e. they were able to inhibit σFoxI in absence of ferrioxamine and activate σFoxI in response to the siderophore to a similar extent as the wild-type protein (Fig. 3A). This indicates that changing any of these residues did not affect the function of FoxR, regardless whether self-cleavage of the protein was affected or not. This is in line with our previous observation that blocking the autoproteolytic event in PaFoxR by mutating the GT motif does not result in major loss of activity of the protein (22). To assess whether the mutations disturbed the self-cleavage, we analyzed the FoxR mutant variants by Western blot. Autocleavage of most FoxR variants was not affected, but it was partially compromised in the FoxR-S166A and -D169A mutants because a higher amount of the full-length protein was detected (Fig. 3B). However, because the total amount of these proteins seemed to be increased and the majority of FoxR-S166A and -D169A was still present in the processed form (Fig. 3B), it is not likely that these residues are part of a putative proteolytic active site of FoxR. In agreement with the β-galactosidase results, Western blot analyses also showed that the FoxR N-tail, the presence of which is associated with σFoxI activity in response to ferrioxamine (22), was produced for all FoxR protein variants in the presence of the siderophore (Fig. 3B).
FIGURE 3.
Mutational analysis of putative active site residues of PaFoxR. A, the FoxRperi protein with an N-terminal HA tag (FoxRperi-NHA-FL; amino acids 107–328) was synthesized using the PURExpress® in vitro transcription/translation system. In addition, a FoxRperi-NHA truncate lacking the complete C terminus (FoxRperi-NHA-256; amino acids 107–256) and a truncate also lacking the α-helix (FoxRperi-NHA-245; amino acids 107–245) were produced. Reactions were analyzed by anti-HA immunoblot. B, β-galactosidase activity of the P. aeruginosa PAO1 pvdF ΔfoxR mutant bearing the pMPR8b plasmid (foxA::lacZ transcriptional fusion) and the pMMB67EH (empty), pMMB/HA-FoxR (WT), pMMB/HA-FoxR-S166A, pMMB/HA-FoxR-D169A, pMMB/HA-FoxR-S172A, pMMB/HA-FoxR-H175A, pMMB/HA-FoxR-S208A, or pMMB/HA-FoxR-S211A plasmid expressing the corresponding N-terminally HA-tagged FoxR protein. Bacteria were grown in iron-restricted CAS medium without or with 1 μm ferrioxamine. C, Western blot analysis of the P. aeruginosa pvdF ΔfoxR mutant bearing the pMMB/HA-FoxR (WT) plasmid or one of its derivative plasmids expressing one of the FoxR-S166A, -D169A, -S172A, H175A, or -S211A mutant variants. Bacteria were grown in iron-restricted medium with 1 mm IPTG in the absence (−) or presence (+) of 1 μm ferrioxamine. Proteins were detected using a monoclonal anti-HA tag antibody. As a loading control, a monoclonal antibody against the OprF protein was used. The positions of the molecular size marker (in kDa) and the FoxR protein fragments are indicated.
Addition of Protease Inhibitors Does Not Inhibit PaFoxR Autocleavage
Because we were unable to identify active site residues around the GT cleavage site of PaFoxR by site-directed mutagenesis, we decided to use a broader approach to establish which class of protease activity PaFoxR exhibits. To do that, we tried to block FoxR processing using specific protease inhibitors. Autocleavage of FoxR was examined by expressing the C-terminally HA-tagged FoxRperi (FoxRperi-CHA) protein using the PURExpress® system. The addition of a mix of serine and cysteine protease inhibitors (cOmplete protease inhibitor mixture), a serine protease inhibitor (Pefabloc), or an aspartic acid protease inhibitor (Pepstatin A) to the reaction did not affect the synthesis or the self-cleavage of FoxRperi (Fig. 4). Metalloprotease activity could not be determined well because the presence of EDTA in the PURExpress® system completely blocked production of FoxRperi (Fig. 4), most likely because of the sequestering of Mg2+ ions required by components in the kit. The addition of EGTA did also affect protein production to some extent but did not alter the proportion of FoxRperi that was being processed (Fig. 4). Together, these results suggest that PaFoxR does not function as a serine, cysteine, aspartic acid protease and probably also not as a metalloprotease.
FIGURE 4.
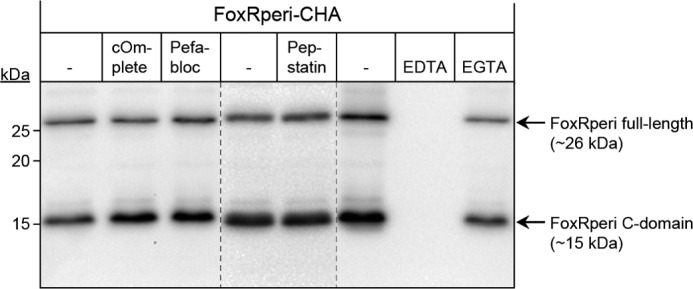
Effect of protease inhibitors on PaFoxR self-cleavage. FoxRperi with a C-terminal HA tag (FoxRperi-CHA) was produced using the PURExpress® in vitro transcription/translation system in the presence of protease inhibitors. Subsequently, the reaction products were analyzed by Western blot using a monoclonal anti-HA antibody. The positions of the molecular size marker (in kDa) and the FoxR protein fragments are indicated.
A Hydroxyl (OH) or Sulfhydryl (SH) Group at the +1 of the Cleavage Site Is Required for PaFoxR Autocleavage
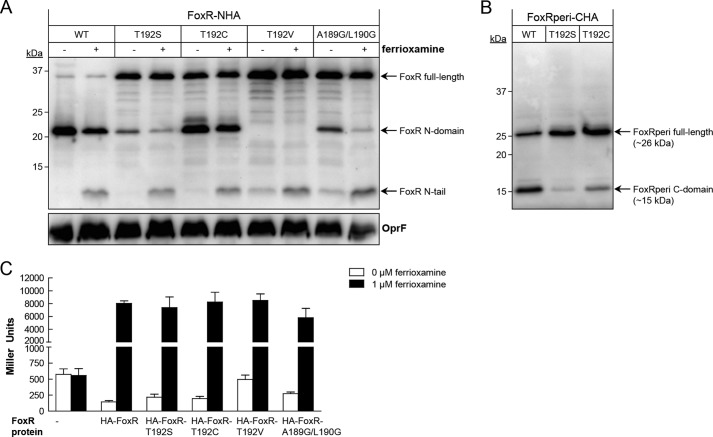
The results obtained thus far indicated that the autoproteolytic activity of PaFoxR cannot be blocked by mutating putative active site residues or by using protease inhibitors. This suggests that the self-cleavage of FoxR may occur through an enzyme-independent mechanism. Interestingly, it has been reported that the cell wall protein CwpV of Clostridium difficile undergoes enzyme-independent autoproteolysis between a Gly and a Thr residue via an intramolecular N-O acyl rearrangement and subsequent ester hydrolysis (37). This spontaneous cleavage is responsible for the autoprocessing of a variety of proteins, including the well studied glycosylasparaginases (38–40), and requires a threonine, serine, or cysteine residue located at the +1 of the cleavage site. The nucleophilic hydroxyl (OH; Thr and Ser) or sulfhydryl (SH; Cys) group on the side chains of these residues performs a nucleophilic attack on the α-carbonyl carbon of the preceding amino acid. After collapse of the resulting tetrahedral intermediate, an ester intermediate is formed, which is then hydrolyzed, thus producing two separate peptide chains. The proteolysis resulting from this sequence can be inhibited by mutating the +1 Thr, Ser, or Cys residue to another non-nucleophilic amino acid (39). We have previously shown that substituting the Thr-192 (+1 of the cleavage site) of PaFoxR for Ala or Gln produces a protein that is functional but not able to undergo autocleavage (22). To examine whether an N-O acyl rearrangement could play a role in the self-cleavage of FoxR, the Thr-192 was changed to either a Ser or Cys residue, which both still contain nucleophilic OH and SH groups, respectively, or to Val, which is very similar to Thr but contains a methyl group instead of the OH group. Western blot analyses of these N-terminally HA-tagged proteins showed that mutation of the Thr-192 to Val (FoxR-T192V) completely blocked autocleavage because only the full-length form of the protein could be detected (Fig. 5A). In contrast, mutation of Thr-192 to either Ser (FoxR-T192S) or Cys (FoxR-T192C) allowed autocleavage of FoxR and production of the N-domain. This occurred, however, to a lesser extent than in the WT protein, especially for the T192S mutant protein in which the self-cleavage happened but was less efficient (Fig. 5A). Importantly, processing of the FoxRperi-T192S and -T192C protein variants still occurred upon expression of these variants in the PURExpress® system (Fig. 5B), indicating that these mutant proteins do also not require additional proteases for their cleavage. This suggests that the OH and SH groups, respectively, of the Ser and Cys residues can mimic the function of Thr-192 in the proteolytic reaction and indicates that the OH group of Thr-192 is important for promoting the autocleavage of FoxR.
FIGURE 5.
Potential role of an N-O acyl rearrangement in PaFoxR self-cleavage. A, anti-HA tag immunoblot of P. aeruginosa pvdF ΔfoxR carrying the pMMB/HA-FoxR (WT), pMMB/HA-FoxR-T192S, pMMB/HA-FoxR-T192C, pMMB/HA-FoxR-T192V, or pMMB/HA-FoxR-A189G/L190G plasmid. Bacteria were grown under iron-restricted conditions in the presence of 1 mm IPTG without (−) and with (+) 1 μm ferrioxamine. B, Western blot of FoxRperi-CHA incorporating either the T192S or T192V mutation during expression in the PURExpress® system. Proteins were detected using a monoclonal anti-HA tag antibody. The positions of the molecular size marker (in kDa) and the FoxR protein fragments in A and B are indicated. As a loading control in A, a monoclonal antibody against the OprF protein was used. C, β-galactosidase activity of the P. aeruginosa PAO1 pvdF ΔfoxR mutant bearing the pMPR8b plasmid (foxA::lacZ transcriptional fusion) and the pMMB67EH (empty), pMMB/HA-FoxR (WT), pMMB/HA-FoxR-T192S, pMMB/HA-FoxR-T192C, pMMB/HA-FoxR-T192V, or pMMB/HA-FoxR-A189G/L190G plasmid. Bacteria were grown in iron-restricted medium in the absence or presence of 1 μm ferrioxamine.
Not all Gly-Thr bonds are susceptible to autocleavage. Proteins that undergo self-cleavage via an N-O acyl rearrangement have an exceptionally strained backbone conformation near the scissile peptide bond to position all the functional groups correctly (38, 41–43). In our structural model, the Gly-Thr autocleavage site of PaFoxR is predicted to be located in a small, tight loop between two β-strands (Fig. 2C), similar to the position of the cleavage site in the C. difficile CwpV protein (37). To examine whether the conformational strain in the loop of PaFoxR is important for the cleavage reaction, we attempted to reduce the rigidity around the scissile peptide bond by replacing the two amino acids preceding the cleavage site (Ala-189 and Leu-190) with Gly, which is often found in flexible protein loops. Autocleavage of the FoxR-A189G/L190G mutant still occurred but was less efficient than that of the wild-type protein, similar to the result obtained with the FoxR-T192S mutant protein (Fig. 5A). This suggests that these neighboring residues are important for the self-cleavage process, likely by maintaining a correct and strained backbone conformation of FoxR. Altogether, these results are consistent with spontaneous autocleavage caused by a sequence of an N-O acyl rearrangement followed by ester hydrolysis.
Next we assayed whether these amino acid substitutions disturbed the functionality of PaFoxR. As shown in Fig. 5C, none of the mutations affected the ability of PaFoxR to activate σFoxI in presence of ferrioxamine. This is consistent with our previous work, in which we have shown that both cleaved and noncleaved PaFoxR proteins are able to respond to the siderophore (22). In agreement with this result, the PaFoxR N-tail was detected for each protein variant in the presence of ferrioxamine (Fig. 5A). This protein band was also detected for the FoxR-T192V and the FoxR-A189G/L190G proteins in absence of the siderophore (Fig. 5A), which correlates with a slight increase of σFoxI activity in this condition when compared with the wild-type protein (FoxR-T192V: 494 MU versus 145 MU (p < 0.0001), and FoxR-A189G/L190G: 247 MU versus 145 MU (p < 0.0001)) (Fig. 5C). This is in accordance with our previous results showing that blocking the self-cleavage of PaFoxR results in a protein that has lost part of its anti-sigma activity, which produces a small but significant increase of σFoxI activity in the absence of ferrioxamine (22).
DISCUSSION
Proteases are divided in four main classes, i.e. metalloproteases, aspartic acid, serine, and cysteine proteases. Whereas members of the latter two families contain a nucleophilic residue (Ser or Cys, respectively) in the active site of the enzyme that attacks the scissile peptide bond, the first two use a metal ion or an Asp residue, respectively, to induce the nucleophilic attack of a water molecule. Interestingly, some proteins that cannot be classified as proteases based on homology can achieve nonenzymatic autocleavage independent of co-factors via chemical sequences initiated by N-O or N-S acyl shifts (40). Central to this self-catalyzed acyl shift is a nucleophilic residue at +1 of the cleavage site (i.e. Thr, Ser, or Cys) that uses either its hydroxyl (OH; Thr or Ser) or sulfhydryl (SH; Cys) group to attack the α-carbonyl carbon of the preceding residue. The resulting fleeting tetrahedral intermediate (hydroxyoxazolidine) decomposes to an ester, which is subsequently hydrolyzed. The chemical basis for the N-O and related N-S acyl rearrangements has been studied extensively, also because of their roles in protein splicing (44, 45) in which an internal region of a polypeptide chain (i.e. the intein) is excised post-translationally and the flanking fragments (i.e. the N- and C-extein) are religated to create a functional protein (46). Chemical studies have shown, among others, that local conformational strain can accelerate the N-O shift by destabilizing the amide moiety and that nearby residues can participate through acid/base catalysis (47–49). N-O or N-S acyl rearrangements are responsible for the nonenzymatic cleavage of a variety of other proteins (40), including the superfamily of N-terminal nucleophile Ntn-hydrolases, which are produced as inactive precursors and activated following post-translational autoprocessing (38, 50). A well studied example is the Flavobacterium meningosepticum glycosylasparaginase (39, 48, 51). In the current study, we propose for the first time the autocleavage via an N-O acyl shift of a bacterial transmembrane regulatory protein, the P. aeruginosa FoxR CSS anti-sigma factor. This protein is self-processed between the periplasmic Gly-191 and Thr-192 residues, which generates two separated domains (22). Mutation of threonine-192 to alanine, glutamine, or valine inhibits cleavage, whereas mutation to serine or, most notably, cysteine maintained a level of autoproteolytic activity (Fig. 5A and Ref. 22). This indicates that an appropriately nucleophilic character of the side group of these residues is essential to induce the cleavage reaction, which is in line with the accepted mechanism for the N-O acyl shift. Resolved crystal structures of Ntn-hydrolases have revealed that the cleavage region shows a distorted in trans conformation, in which this domain is highly strained and turned to precisely align all the chemical groups involved in the autocleavage reaction (38, 41–43, 52). In this work, we have used a structural model of FoxR to determine the location of the GT cleavage site (Fig. 2C), which is predicted to be located in a small, presumably tight loop similar to the cleavage site in the CwpV protein (37). Mutation of the Ala-189 and Leu-190 residues preceding the GT cleavage site reduced the efficiency of self-cleavage, indicating that they may be required to hold the cleavage region in a distorted conformation. Based on the body of chemical literature on N-O acyl rearrangements and on the similar features shared by FoxR with the proteins subjected to the nonenzymatic self-cleavage described before, we propose that the mechanism targeting the PaFoxR self-cleavage involves an N-O acyl rearrangement. We therefore suggest a model (Fig. 6) in which the hydroxyl group of Thr-192 of FoxR attacks the carbonyl carbon of Gly-191, thereby generating a tetrahedral intermediate that collapses to an ester. This ester is then hydrolyzed returning two peptide chains: one with a glycine at its C terminus and one with a threonine at its N terminus.
FIGURE 6.
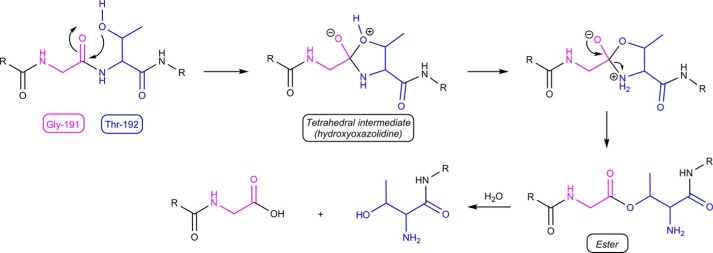
Proposed sequence for self-cleavage of PaFoxR. An initial N-O acyl rearrangement is followed by ester hydrolysis. The two R labels indicate the remainders of the protein chain. The cores of Gly-191 and Thr-192 have been colored for clarity.
To efficiently initiate an N-O acyl rearrangement, the hydroxyl group of the +1 residue may be activated through deprotonation to make it more nucleophilic (48). Loss of the general base responsible for this process should block the activation of the nucleophile and consequently the self-cleavage of the protein. We used the structural model of FoxRperi to identify residues with a basic group (e.g. COO−) near the GT cleavage site that could be responsible for this step of the self-cleavage process. A Glu (Glu-209) with possible access to Thr-192 is located in the loop between β-strand 10 and 11 (Fig. 2C, orange). However, mutation of this residue had no effect on the self-cleavage of FoxR (data not shown). In contrast, mutation of Asp-169 (Asn in BDI_1681) appears to hinder FoxR autocleavage (Fig. 3B). Therefore, it is possible that this residue is responsible for the deprotonation of Thr-192, although based on the position of Asp-169 in our model (Fig. 2C), it seems more likely that its mutation leads to structural changes resulting in suboptimal positioning of the moieties required for the N-O rearrangement. To be able to determine exactly how the proposed N-O acyl shift responsible for the autocleavage of FoxR occurs and which other residues are involved, it is essential that the structure of FoxR is first resolved.
The self-cleavage of CSS anti-sigma factors is a widespread mechanism that is linked to the presence of a conserved GT cleavage site in the periplasmic domain of these proteins (22). However, the biological function of separating CSS anti-sigma factors in two domains is still not completely understood. For P. aeruginosa FoxR, we have previously shown that the resulting N- and C-domains produced upon autocleavage interact and function together to transduce the CSS signal generated by ferrioxamine (22). This results in the production of an N-terminal FoxR fragment (the N-tail), which is thought to stimulate σFoxI activity (22). However, whereas the autocleavage of Ntn-hydrolases through an N-O or N-S acyl rearrangement generally reflects a post-translational activation process, the self-cleavage of FoxR is not required to produce an active protein. The complete blockage of this process still produces a functional protein and generation of the pro-sigma N-tail in response to the signal (Fig. 5 and Ref. 22). The only noticeable difference we have detected is a slight but significant increase of σFoxI activity with these FoxR mutant proteins (Fig. 5 and Ref. 22). This suggests that these mutant proteins have partially lost their anti-sigma activity and that the self-cleavage is therefore necessary to tightly regulate the activity of the sigma factor. This might be related to the intrinsic stability of either the anti-sigma factor itself or the σECF, and the self-cleavage might be necessary to protect the anti-sigma factor from degradation. Alternatively, the self-cleavage of the anti-sigma factor could provide a faster CSS response to the presence of the inducing signal. Further analyses are necessary to address the advantage that this widespread process confers to the CSS signal transduction cascade. In summary, in this study we propose that the self-cleavage event between the periplasmic Gly-191 and Thr-192 residues of the P. aeruginosa CSS anti-sigma factor FoxR is mediated by an N-O acyl rearrangement, reflecting a nonenzymatic process independent of co-factors.
Acknowledgments
We thank M. Glas and T. Krell for helpful discussions.
This work was supported by Netherlands Organization for Scientific Research through ECHO Grant 2951201 (to K. C. B.), Marie Curie Career Integration Grant (CIG) 3038130 from the European Union, and Ramon&Cajal Grant RYC2011-08874 and Plan Nacional Grant SAF2012-31919 from the Spanish Ministry of Economy (to M. A. L.).
- CSS
- cell-surface signaling
- ECF
- extracytoplasmic function
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MU
- Miller units
- Phyre
- Protein Homology/Analogy Recognition Engine.
REFERENCES
- 1. Llamas M. A., Sparrius M., Kloet R., Jiménez C. R., Vandenbroucke-Grauls C., Bitter W. (2006) The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188, 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ratledge C., Dover L. G. (2000) Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941 [DOI] [PubMed] [Google Scholar]
- 3. Wandersman C., Delepelaire P. (2004) Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58, 611–647 [DOI] [PubMed] [Google Scholar]
- 4. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poole K., McKay G. A. (2003) Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8, d661–d686 [DOI] [PubMed] [Google Scholar]
- 6. Visca P., Leoni L., Wilson M. J., Lamont I. L. (2002) Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45, 1177–1190 [DOI] [PubMed] [Google Scholar]
- 7. Koebnik R. (2005) TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13, 343–347 [DOI] [PubMed] [Google Scholar]
- 8. Llamas M. A., Imperi F., Visca P., Lamont I. L. (2014) Cell-surface signaling in Pseudomonas: stress responses, iron transport and pathogenicity. FEMS Microbiol. Rev. 38, 569–597 [DOI] [PubMed] [Google Scholar]
- 9. Braun V., Mahren S., Sauter A. (2006) Gene regulation by transmembrane signaling. Biometals 19, 103–113 [DOI] [PubMed] [Google Scholar]
- 10. Bastiaansen K. C., Bitter W., Llamas M. A. (2012) ECF sigma factors: from stress management to iron uptake. In Bacterial regulatory networks (Filloux A., ed) pp. 59–86, Caister Academic Press, Norfolk, UK [Google Scholar]
- 11. Staroń A., Sofia H. J., Dietrich S., Ulrich L. E., Liesegang H., Mascher T. (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 74, 557–581 [DOI] [PubMed] [Google Scholar]
- 12. Campbell E. A., Greenwell R., Anthony J. R., Wang S., Lim L., Das K., Sofia H. J., Donohue T. J., Darst S. A. (2007) A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 27, 793–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campbell E. A., Tupy J. L., Gruber T. M., Wang S., Sharp M. M., Gross C. A., Darst S. A. (2003) Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-σ RseA. Mol. Cell 11, 1067–1078 [DOI] [PubMed] [Google Scholar]
- 14. Shukla J., Gupta R., Thakur K. G., Gokhale R., Gopal B. (2014) Structural basis for the redox sensitivity of the Mycobacterium tuberculosis SigK-RskA σ-anti-σ complex. Acta Crystallogr. D 70, 1026–1036 [DOI] [PubMed] [Google Scholar]
- 15. Maillard A. P., Girard E., Ziani W., Petit-Härtlein I., Kahn R., Covès J. (2014) The crystal structure of the anti-σ factor CnrY in complex with the σ factor CnrH shows a new structural class of anti-σ factors targeting extracytoplasmic function σ factors. J. Mol. Biol. 426, 2313–2327 [DOI] [PubMed] [Google Scholar]
- 16. Koster M., van de Vossenberg J., Leong J., Weisbeek P. J. (1993) Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol. Microbiol. 8, 591–601 [DOI] [PubMed] [Google Scholar]
- 17. Braun V., Mahren S., Ogierman M. (2003) Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6, 173–180 [DOI] [PubMed] [Google Scholar]
- 18. Schalk I. J., Yue W. W., Buchanan S. K. (2004) Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 54, 14–22 [DOI] [PubMed] [Google Scholar]
- 19. Ferguson A. D., Amezcua C. A., Halabi N. M., Chelliah Y., Rosen M. K., Ranganathan R., Deisenhofer J. (2007) Signal transduction pathway of TonB-dependent transporters. Proc. Natl. Acad. Sci. U.S.A. 104, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia-Herrero A., Vogel H. J. (2005) Nuclear magnetic resonance solution structure of the periplasmic signalling domain of the TonB-dependent outer membrane transporter FecA from Escherichia coli. Mol. Microbiol. 58, 1226–1237 [DOI] [PubMed] [Google Scholar]
- 21. Schalk I. J., Lamont I. L., Cobessi D. (2009) Structure-function relationships in the bifunctional ferrisiderophore FpvA receptor from Pseudomonas aeruginosa. Biometals 22, 671–678 [DOI] [PubMed] [Google Scholar]
- 22. Bastiaansen K. C., Otero-Asman J., Luirink J., Bitter W., Llamas M. A. (2015) Processing of cell-surface signalling anti-sigma factors prior to signal recognition is a conserved autoproteolytic mechanism that produces two functional domains. Environ. Microbiol. 10.1111/1462-2920.12776 [DOI] [PubMed] [Google Scholar]
- 23. Bastiaansen K. C., Ibañez A., Ramos J. L., Bitter W., Llamas M. A. (2014) The Prc and RseP proteases control bacterial cell-surface signalling activity. Environ. Microbiol. 16, 2433–2443 [DOI] [PubMed] [Google Scholar]
- 24. Draper R. C., Martin L. W., Beare P. A., Lamont I. L. (2011) Differential proteolysis of sigma regulators controls cell-surface signalling in Pseudomonas aeruginosa. Mol. Microbiol. 82, 1444–1453 [DOI] [PubMed] [Google Scholar]
- 25. Mettrick K. A., Lamont I. L. (2009) Different roles for anti-sigma factors in siderophore signalling pathways of Pseudomonas aeruginosa. Mol. Microbiol. 74, 1257–1271 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27. Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. (1986) Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131 [DOI] [PubMed] [Google Scholar]
- 28. Choi K. H., Kumar A., Schweizer H. P. (2006) A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 [DOI] [PubMed] [Google Scholar]
- 29. Finnen R. L., Martin N. L., Siehnel R. J., Woodruff W. A., Rosok M., Hancock R. E. (1992) Analysis of the Pseudomonas aeruginosa major outer membrane protein OprF by use of truncated OprF derivatives and monoclonal antibodies. J. Bacteriol. 174, 4977–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., Yu N. Y., Hancock R. E., Brinkman F. S. (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D595–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 33. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Skolnick J. (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malki I., Simenel C., Wojtowicz H., de Amorim G. C., Prochnicka-Chalufour A., Hoos S., Raynal B., England P., Chaffotte A., Delepierre M., Delepelaire P., Izadi-Pruneyre N. (2014) Interaction of a partially disordered antisigma factor with its partner, the signaling domain of the TonB-dependent transporter HasR. PLoS One 9, e89502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferguson A. D., Chakraborty R., Smith B. S., Esser L., van der Helm D., Deisenhofer J. (2002) Structural basis of gating by the outer membrane transporter FecA. Science 295, 1715–1719 [DOI] [PubMed] [Google Scholar]
- 37. Dembek M., Reynolds C. B., Fairweather N. F. (2012) Clostridium difficile cell wall protein CwpV undergoes enzyme-independent intramolecular autoproteolysis. J. Biol. Chem. 287, 1538–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brannigan J. A., Dodson G., Duggleby H. J., Moody P. C., Smith J. L., Tomchick D. R., Murzin A. G. (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378, 416–419 [DOI] [PubMed] [Google Scholar]
- 39. Guan C., Cui T., Rao V., Liao W., Benner J., Lin C.-L., Comb D. (1996) Activation of glycosylasparaginase: formation of active N-terminal threonine by intramolecular autoproteolysis. J. Biol. Chem. 271, 1732–1737 [DOI] [PubMed] [Google Scholar]
- 40. Perler F. B., Xu M.-Q., Paulus H. (1997) Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1, 292–299 [DOI] [PubMed] [Google Scholar]
- 41. Xu Q., Buckley D., Guan C., Guo H.-C. (1999) Structural insights into the mechanism of intramolecular proteolysis. Cell 98, 651–661 [DOI] [PubMed] [Google Scholar]
- 42. Klabunde T., Sharma S., Telenti A., Jacobs W. R., Jr., Sacchettini J. C. (1998) Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 5, 31–36 [DOI] [PubMed] [Google Scholar]
- 43. Ditzel L., Huber R., Mann K., Heinemeyer W., Wolf D. H., Groll M. (1998) Conformational constraints for protein self-cleavage in the proteasome. J. Mol. Biol. 279, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 44. Shao Y., Xu M.-Q., Paulus H. (1996) Protein splicing: evidence for an N-O acyl rearrangement as the initial step in the splicing process. Biochemistry 35, 3810–3815 [DOI] [PubMed] [Google Scholar]
- 45. Paulus H. (1998) The chemical basis of protein splicing. Chem. Soc. Rev. 27, 375–386 [Google Scholar]
- 46. Starokadomskyy P. L. (2007) Protein splicing. Mol. Biol. 41, 278–293 [Google Scholar]
- 47. Johansson D. G., Wallin G., Sandberg A., Macao B., Åqvist J., Härd T. (2009) Protein autoproteolysis: conformational strain linked to the rate of peptide cleavage by the pH dependence of the N → O acyl shift reaction. J. Am. Chem. Soc. 131, 9475–9477 [DOI] [PubMed] [Google Scholar]
- 48. Qian X., Guan C., Guo H.-C. (2003) A dual role for an aspartic acid in glycosylasparaginase autoproteolysis. Structure 11, 997–1003 [DOI] [PubMed] [Google Scholar]
- 49. Du Z., Shemella P. T., Liu Y., McCallum S. A., Pereira B., Nayak S. K., Belfort G., Belfort M., Wang C. (2009) Highly conserved histidine plays a dual catalytic role in protein splicing: a pKa shift mechanism. J. Am. Chem. Soc. 131, 11581–11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oinonen C., Rouvinen J. (2000) Structural comparison of Ntn-hydrolases. Protein Sci. 9, 2329–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guan C., Liu Y., Shao Y., Cui T., Liao W., Ewel A., Whitaker R., Paulus H. (1998) Characterization and functional analysis of the cis-autoproteolysis active center of glycosylasparaginase. J. Biol. Chem. 273, 9695–9702 [DOI] [PubMed] [Google Scholar]
- 52. Oinonen C., Tikkanen R., Rouvinen J., Peltonen L. (1995) Three-dimensional structure of human lysosomal aspartylglucosaminidase. Nat. Struct. Biol. 2, 1102–1108 [DOI] [PubMed] [Google Scholar]
- 53. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]