Background: Treponema pallidum contains a paucity of outer membrane proteins.
Results: The C-terminal domains of TprC/D and TprI are β-barrels with porin function; their N-terminal domains provide periplasmic anchors for the β-barrels.
Conclusion: Full-length TprC subfamily proteins possess a dual domain topology.
Significance: TprC/D are bona fide rare outer membrane proteins and a new class of dual function, bipartite, bacterial outer membrane protein.
Keywords: Electron Microscopy (EM), Microbial Pathogenesis, Outer Membrane, Small Angle X-ray Scattering (SAXS), X-ray Scattering, beta-Barrel, Syphilis, Treponema pallidum, Outer Membrane Proteins, Porins
Abstract
We previously identified Treponema pallidum repeat proteins TprC/D, TprF, and TprI as candidate outer membrane proteins (OMPs) and subsequently demonstrated that TprC is not only a rare OMP but also forms trimers and has porin activity. We also reported that TprC contains N- and C-terminal domains (TprCN and TprCC) orthologous to regions in the major outer sheath protein (MOSPN and MOSPC) of Treponema denticola and that TprCC is solely responsible for β-barrel formation, trimerization, and porin function by the full-length protein. Herein, we show that TprI also possesses bipartite architecture, trimeric structure, and porin function and that the MOSPC-like domains of native TprC and TprI are surface-exposed in T. pallidum, whereas their MOSPN-like domains are tethered within the periplasm. TprF, which does not contain a MOSPC-like domain, lacks amphiphilicity and porin activity, adopts an extended inflexible structure, and, in T. pallidum, is tightly bound to the protoplasmic cylinder. By thermal denaturation, the MOSPN and MOSPC-like domains of TprC and TprI are highly thermostable, endowing the full-length proteins with impressive conformational stability. When expressed in Escherichia coli with PelB signal sequences, TprC and TprI localize to the outer membrane, adopting bipartite topologies, whereas TprF is periplasmic. We propose that the MOSPN-like domains enhance the structural integrity of the cell envelope by anchoring the β-barrels within the periplasm. In addition to being bona fide T. pallidum rare outer membrane proteins, TprC/D and TprI represent a new class of dual function, bipartite bacterial OMP.
Introduction
After years of steady decline in the United States during the 1990s, the new millennium has witnessed a dramatic resurgence in the incidence of syphilis, a multistage, sexually transmitted infection caused by the spirochete Treponema pallidum subsp. pallidum, particularly among men who have sex with men (1). On a global scale, syphilis poses a major threat to public health with an estimated 12 million new cases annually (2). Of great concern, mother-to-child transmission of syphilis occurs in approximately 2 million pregnancies per year, more than any other infectious disease, including human immunodeficiency virus infection (3). These trends have taken root although T. pallidum remains exquisitely sensitive to penicillin G after 7 decades of use (4, 5). The inability of epidemiological approaches to curtail the spread of syphilis underscores the need for stratagems based on better knowledge of the molecular biology of its etiologic agent (6).
T. pallidum is a highly motile, extracellular bacterium renowned for its invasiveness, immunoevasiveness, and persistence, along with its recalcitrance to in vitro propagation and genetic manipulation (7–10). The proteins that assemble into the syphilis spirochete's outer membrane (OM)2 determine the bacterium's ability to obtain nutrients, negotiate its way through tissue and endothelial barriers, fend off host defenses, and accomplish the many other facets of its complex and enigmatic infectivity program (7, 8, 11). Unfortunately, the dearth of information concerning its repertoire of outer membrane proteins (OMPs) has long been a major stumbling block to basic syphilis research and vaccine development (12, 13). It is well established that the physical properties, composition, and molecular architecture of the T. pallidum OM differ considerably from those of Gram-negative bacteria (11). The T. pallidum OM is extremely fragile (14, 15), lacks lipopolysaccharide (16), and has an unusual phospholipid content (17) and a markedly lower (∼1,000-fold) density of membrane-spanning proteins than its Gram-negative counterparts (17–19). The paucity of pathogen-associated molecular patterns and membrane-spanning proteins in the T. pallidum OM is believed to be the ultrastructural basis for the syphilis spirochete's remarkable capacity to evade both innate and adaptive responses in its obligate human host, attributes that have earned it the designation “stealth pathogen” (20, 21). However, efforts to move beyond these general features and broad concepts to a molecular understanding of how this unorthodox OM meets the physiological and virulence-related demands of stealth pathogenicity have been fraught with difficulty (11, 12). Among the many factors hindering progress is the lack of sequence relatedness between prototypical OMPs of Gram-negatives and T. pallidum rare OMPs (16), an indication of the phylogenetic gulf separating spirochetes from proteobacteria (22).
Previously, we used a novel bioinformatics-based approach to identify rare OMPs based upon the premise that they form β-barrels, the structural hallmark of OM-spanning proteins in all diderms as well as the endosymbiotic organelles of eukaryotes (i.e. chloroplasts and mitochondria) derived from them (23–25). The consensus computational framework that we developed (26) yielded ranked clusters of putative β-barrel forming proteins, many of which are members of the paralogous T. pallidum repeat family (Tpr) (7, 11). Among the highest ranked Tpr candidates was the subfamily containing TprC (TP0117) and TprD (TP0131) (which are identical), TprF (TP0316), and TprI (TP0620) (27, 28). In a subsequent report (29), we demonstrated that TprC/D (hereafter referred to as TprC) does, indeed, possess the properties expected of a bona fide rare OMP (i.e. β-barrel structure, amphiphilicity, low abundance, and surface exposure) and, additionally, can form channels in large unilamellar liposomes (LUVs). We also noted that TprC expressed in T. pallidum is stably tethered within the periplasm. Unexpectedly, using the Conserved Domain Database server, we discovered that TprC contains N- and C-terminal domains (TprCN and TprCC, respectively) corresponding to regions in the major outer sheath protein (MOSP) of the oral commensal Treponema denticola, the parental Tpr ortholog (30). Using a battery of biophysical methodologies, we determined that TprCC, but not TprCN, can fold to form an amphiphilic β-barrel with porin activity equivalent to that of the full-length polypeptide. Based on these findings, we proposed that TprCC inserts into the OM bilayer, whereas TprCN anchors the β-barrel within the periplasm.
In the present study, we confirmed the bipartite topology of native TprC and demonstrated that TprI possesses the same dual domain architecture, also forms trimers, displays comparable porin activity, and, like TprC, is both surface-exposed and tethered within the periplasm in T. pallidum. TprF, the fourth member of the subfamily, is truncated and lacks a MOSPC-like domain. Although rich in β-strand secondary structure, recombinant TprF is water-soluble, unable to integrate into membranes, and devoid of porin activity, and, based on small angle x-ray scattering (SAXS) (31), forms an extended, inflexible structure. Moreover, unlike TprC and TprI, native TprF in T. pallidum is entirely periplasmic and tightly bound to the protoplasmic cylinder. By thermal denaturation, both the MOSPN-and MOSPC-like domains of TprC and TprI are highly thermostable, endowing their full-length proteins with a high degree of conformational stability. Interestingly, in contrast to E. coli OmpF, a classical porin in which the monomers form tightly integrated trimers (32–34), the structural stability of full-length TprC and TprI appears to be due predominantly to the conformational integrity of their monomeric β-barrels. It is particularly noteworthy that we have been able to express recombinant forms of TprC and TprI with PelB signal sequences that localize to the Escherichia coli OM and adopt bipartite topologies identical to their native counterparts; as in T. pallidum, TprF expressed in E. coli with a PelB signal sequence resides entirely within the periplasm. We propose that, by anchoring the OM-inserted β-barrels within the periplasm, the MOSPN-like domains of TprC and TprI not only stabilize the OM but also enhance the structural integrity of the entire cell envelope. In addition to being bona fide T. pallidum rare outer membrane proteins, TprC/D and I represent a new class of bipartite bacterial outer membrane protein with porin and cell envelope stabilizing functions.
EXPERIMENTAL PROCEDURES
Propagation and Harvesting of T. pallidum
Animal protocols described in this work strictly follow the recommendations of the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the University of Connecticut Health Center Animal Care Committee under the auspices of Animal Welfare Assurance A347-01. The Nichols strain of T. pallidum subspecies pallidum was propagated by intratesticular inoculation of adult male New Zealand White rabbits with 1 × 108 treponemes/testis and harvested ∼10 days later as described previously (35).
Identification of Polypeptides Specific for TprI and TprI/F
Previously, we used multiple-sequence alignment of TprC, TprF, and TprI, performed using MacVector (version 11.1.0), to identify a stretch of 70 amino acids unique to TprC (TprCSp) (29). Using this same alignment, we identified a 52-amino acid stretch specific for TprI (TprISp) and an 86-amino acid stretch in TprI and TprF (TprI/FSp) not found in TprC.
Cloning of DNAs Encoding TprIFl, TprIN, TprIC, TprF, TprISp, TprI/FSp, E. coli OmpA, and E. coli Skp
Cloning of DNAs encoding full-length TprCFl, TprCN, and TprCC was described previously (29). The codon-optimized full-length tprIFl gene (minus the signal sequence) was synthesized by Genscript (Piscataway, NJ) and cloned into the BamHI and HindIII restriction sites of pET23b (Novagen, San Diego, CA). DNAs encoding TprIN and TprIC were PCR-amplified from tprIFl and cloned into the BamHI and HindIII restriction sites of pET23b (Novagen, San Diego, CA). DNAs encoding TprISp and TprI/FSp were PCR-amplified from T. pallidum DNA and cloned into the NheI and HindIII restriction sites of pET23b. DNAs encoding OmpA and Skp were PCR-amplified from E. coli K12 genomic DNA and cloned into the NheI and HindIII restriction sites of pET23b. All constructs were confirmed by nucleotide sequencing.
Expression, Purification, and Folding of Recombinant Proteins
Expression, purification, and folding of recombinant TprCFl, TprCN, TprCC (29), the polypeptide transport-associated (POTRA) domain of TP0326 (26), E. coli OmpG (36), and E. coli OmpF (37) were described previously. TprI, TprIN, TprIC, TprF, and E. coli OmpA, cloned as described above into pET23b, were expressed in E. coli C41 (DE3) (Agilent Technologies, Inc., Santa Clara, CA). For batch purification, 1 liter of LB was inoculated with 50 ml of overnight culture grown at 37 °C; isopropyl 1-thio-β-d-galactopyranoside (final concentration, 0.1 mm) was added when the culture reached an optical density (600 nm) between 0.2 and 0.3. Cells were grown for an additional 3 h and then harvested by centrifugation at 6,000 × g for 15 min at 4 °C. The pellets were resuspended with 20 ml of 50 mm Tris (pH 7.5), 100 μg of lysozyme, and 100 μl of protease inhibitor mixture (Sigma-Aldrich) and stored at −20 °C. After thawing, the bacterial suspension was lysed by sonication with three 30-s pulses interspersed with 30 s of rest on ice. The pellet was recovered by centrifugation at 20,000 × g for 30 min at 4 °C and then incubated in solubilization buffer (100 mm NaH2PO4 (pH 8.0), 10 mm Tris, 8 m urea) for 30 min at 4 °C; the remaining insoluble material was removed by centrifugation at 20,000 × g for 30 min at 4 °C. The supernatant was added to Ni-NTA-agarose matrix (Qiagen) that had been equilibrated in solubilization buffer, and then the mixture was incubated with shaking at room temperature for 30 min. The matrix was treated with wash buffer (100 mm NaH2PO4 (pH 6.3), 10 mm Tris, 8 m urea) and subsequently elution buffer (100 mm NaH2PO4 (pH 4.5), 10 mm Tris, 8 m urea). SDS-PAGE and immunoblot analysis using the polyhistidine tag mouse monoclonal antibody (Sigma-Aldrich) were employed to identify the protein and assess purity during purification. The purified TprI, TprIC, and OmpA proteins were incubated in 1% n-dodecyl β-d-maltoside (DDM), 100 mm NaCl, 50 mm Tris, pH 7.5 (DDM buffer), 0.5% DDM, 100 mm NaCl, 50 mm Tris, pH 7.5, and 0.5% n-decyl-β-d-maltopyranoside, 50 mm NaCl, 10 mm Tris, pH 8.5, respectively, for 24 h at 4 °C to ensure complete folding of the proteins. The samples were centrifuged at 20,000 × g for 30 min at 4 °C to remove misfolded aggregates. Purified TprF and TprIN were diluted 10-fold and further dialyzed for renaturation in downgraded concentrations (6, 4, and 2 mm) of urea; finally, TprF was dialyzed against 50 mm Tris (pH 7.5) and 50 mm NaCl buffer.
TprISp, TprI/FSp, and Skp were expressed in E. coli C41 (DE3). For purification, harvested cell pellets were resuspended with 20 ml of 50 mm Tris (pH 7.5), 300 mm NaCl, 10 mm imidazole, 10% glycerol, 100 μg of lysozyme, and 100 μl of protease inhibitor mixture and then stored at −20 °C. After thawing, the bacterial suspensions were lysed by sonication for three 30-s pulses interspersed with 30 s of rest on ice. The supernatants were then cleared of cellular debris by centrifugation at 18,000 × g for 20 min at 4 °C and applied to a Superflow Ni-NTA (Qiagen, Valencia, CA) immobilized metal affinity chromatography column, which had been equilibrated with buffer A (50 mm Tris (pH 7.5), 300 mm NaCl, 10% glycerol). The proteins were eluted with buffer A supplemented with 300 mm imidazole. Fractions containing the protein were concentrated using an Amicon-Ultra concentrator (Millipore, Billerica, MA) with a nominal molecular mass cut-off of 10 kDa and dialyzed into phosphate-buffered saline (PBS).
Protein concentrations were determined by measuring A280 in 20 mm sodium phosphate (pH 6.5) and 6 m guanidine hydrochloride (38). The ProtParam tool provided by the ExPASy proteomics server (39) was used to calculate the molar extinction coefficients (m−1 cm−1) of proteins.
Immunologic Reagents
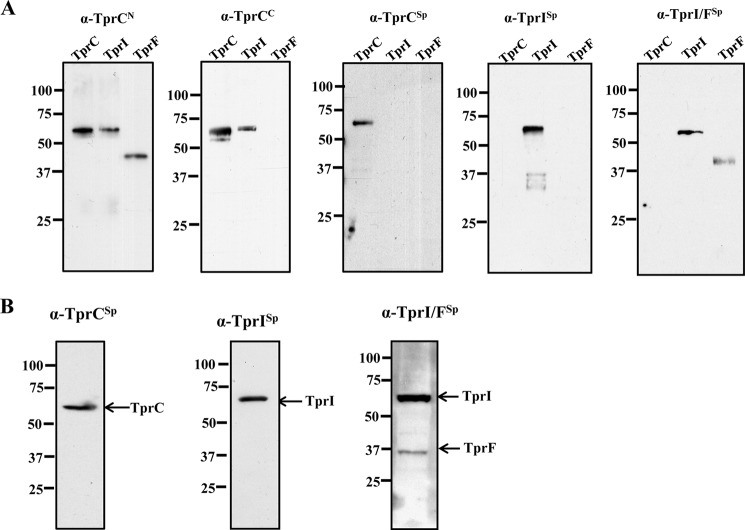
Rat polyclonal antisera directed against FlaA and TprCSp were described previously (40). Rat antisera directed against TprCN, TprCC, TprISp, TprI/FSp, E. coli OmpA, and E. coli Skp were generated in female Sprague-Dawley rats as described previously (29). A mouse monoclonal antibody against the B subunit of E. coli ATP synthase was purchased from Abcam (Cambridge, MA). Polyclonal antibodies to POTRA, TprCN, and TprCC were generated in rabbits by Rockland, Inc. according to their established protocol. Immune rabbit serum was generated as described previously (40). The specificities of the antisera generated with TprCSp, TprISp, and TprI/FSp were confirmed by immunoblot analysis using recombinant proteins and T. pallidum lysates (Fig. 1).
FIGURE 1.
Antigenic reactivities of TprC subfamily-specific antisera. A, immunoblot analysis of 100 ng of recombinant TprCFl, TprIFl, and TprF using rat antisera directed against TprCN, TprCC, TprCSp, TprISp, and TprI/FSp. B, immunoblot analysis of T. pallidum lysates (2 × 108 organisms/lane) using rat antisera directed against TprCSp, TprISp, and TprI/FSp. Molecular mass standards are on the left of each immunoblot.
Circular Dichroism Spectroscopy and Thermal Denaturation of Recombinant Proteins
Far-UV circular dichroism (CD) analyses were performed using a Jasco J-715 spectropolarimeter equipped with a Peltier temperature controller system (Jasco, Easton, MD). Spectra were acquired at room temperature in a 1-mm path length cuvette, with a 1-nm bandwidth, 8-s response time, and scan rate of 20 nm/min. Spectra of each sample, representing the average of nine scans, were baseline-corrected by subtracting the spectral attributes of the buffer. The DICHROWEB server was utilized to assess the secondary structure contents of the proteins from their spectra (41). Thermal denaturation of proteins was performed by following the change in molar ellipticity at 218 nm as a function of temperature (42). The protein samples were heated at a constant rate of 1 °C/min in a 1-mm cell and scanned over a temperature range of 25–90 °C. The thermal curves were fit to a two-state equilibrium unfolding model to determine denaturation temperatures (Tm values).
Protein Thermal Denaturation Assays Using SYPRO® Orange
SYPRO® Orange becomes highly fluorescent when bound to protein hydrophobic sites, which become accessible upon thermal unfolding (43). Thermal denaturation assays using SYPRO® Orange were performed as described previously (43, 44). Briefly, 45 μl of 1 μm TprCN, TprIN, or TprF in Tris buffer (50 mm Tris, 100 mm NaCl at pH 7.5) or 45 μl of buffer alone were added to 5 μl of 200× SYPRO® Orange (Invitrogen) diluted from a 5,000-fold stock solution in DMSO. Each sample was added in triplicate into a 96-well plate. Protein melting experiments were carried out using an iCycler iQ real-time PCR detection system (Bio-Rad) with 465 and 580 nm as the excitation and emission wavelengths, respectively. Melting curve fluorescent signals were acquired between 25 and 90 °C using a ramping rate of 0.2 °C s−1. Experiments were repeated three independent times, and melting temperatures (Tm) were determined in Prism by using a modification of the Boltzmann model.
Heat Modifiability
Recombinant proteins solubilized in SDS sample buffer were subjected to SDS-PAGE with or without boiling (10 min), followed by staining with GelCode® Blue (Thermo Fisher, Rockford, IL). To examine the heat modifiability of native TprI, freshly harvested T. pallidum was incubated overnight at 4 °C with native lysis buffer (50 mm Tris (pH 7.0), 0.5% DDM, and 5% protease inhibitor mixture). Insoluble material was removed from the sample lysed with native lysis buffer by centrifugation at 20,000 × g for 20 min at 4 °C; lysates solubilized in SDS sample buffer were split in half, and one aliquot was boiled for 10 min. Subsequently, the lysates were resolved by SDS-PAGE, and the gel, including the stack, was transferred to nitrocellulose membranes (0.45-μm pore size; GE Healthcare) for immunoblot analysis (29).
Triton X-114 Phase Partitioning
10 μg of recombinant proteins were added to 2% Triton X-114 in PBS supplemented with 0.5% protease inhibitor mixture and incubated overnight at 4 °C. The samples were phase-separated, and the detergent-enriched and aqueous phases were extracted five times with PBS and 2% Triton X-114, respectively (45). All samples were precipitated with 10 volumes of acetone overnight at −80 °C for subsequent SDS-PAGE analysis and staining with GelCode® Blue. Triton X-114 phase partitioning of native TprC, TprI, and TprF was performed with or without preincubation in DDM as described previously (29). The aqueous, detergent, and insoluble fractions were resuspended with 50 μl of 1× Laemmli sample buffer (Bio-Rad) for subsequent SDS-PAGE and immunoblot analysis.
Blue Native PAGE
Blue native PAGE of recombinant and native proteins was performed as described previously (29).
Size Exclusion Chromatography
To assess the relative size of the TprCC construct, we carried out analytical size exclusion chromatography using a Superdex 200 10/300 GL column. The column was equilibrated at room temperature in 50 mm Tris, pH 8.0, 200 mm NaCl, 0.1% lauryldimethylamine oxide (w/v). 25 μl of TprCC at 4.6 mg/ml was loaded onto the column and subsequently run at a flow rate of 0.5 ml/min. The relative size of the TprCC construct was estimated by comparing the retention volume with a standard curve calculated using blue dextran.
Preparation of Liposomes and Performance of Liposome Floatation Assays
Liposomes were prepared and used in floatation assays as described previously (29).
Pore Formation Assays
Pore formation assays were performed using LUVs loaded with the fluorophore Tb(DPA)33− as described previously (29, 30, 37, 46).
Reconstitution of TprCC in Nanodiscs
Denatured TprCC was purified using Ni-NTA resin and refolded in 1% DDM as described above. N-terminal His-tagged MSP1E3D1 (E3D1) nanodisc outer coat protein (47) was expressed in E. coli (DE3) cells with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The protein was purified further and dialyzed using a protocol described elsewhere (48). Nanodiscs were formed using two buffers at pH 7.5: 1) 20 mm Tris-HCl, 0.5 mm EDTA and 2) 20 mm Tris-HCl, 0.5 mm EDTA, 60 mm sodium cholate. The powdered form of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Inc.) was dissolved in buffer 2 and used as the lipid source for the preparation of discs. E3D1 was mixed with TprCC in a ratio of 1:1.5 with a 100-fold molar excess of 1,2-dimyristoyl-sn-glycero-3-phosphocholine. The reaction mixture containing proteins and lipid was kept on a shaker at room temperature for 2 h, following which wet Bio-Beads SM2 (Sigma) were added (2 g/ml of reaction mixture) and left on a shaker for 18 h at room temperature. The solution was separated from the beads using a 0.45-μm syringe and loaded onto a Superdex S-200 column equilibrated with 20 mm Tris-HCl, 150 NaCl, 0.5 mm EDTA (pH 7.5). Fractions containing nanodisc peaks were pooled and concentrated to be further analyzed through SDS-PAGE and transmission electron microscopy (TEM).
Electron Microscopy
Single droplets containing folded TprCC (1 mg/ml) in 0.1% DDM, 100 mm NaCl, 50 mm Tris, pH 7.5, were applied to Parlodion and carbon-coated 400-mesh copper grids (Ted Pella, Tustin, CA). Specimens were negatively stained with 1% uranyl acetate (Sigma) and examined at 60 kV of accelerating voltage on a Hitachi H-765 electron microscope. Approximately 400 single particle projections of TprCC monomers and trimers were selected for single particle analysis using the X-Windows-based microscopy image processing package (Xmipp) (49). Selected single particle projections (72 × 72-pixel frame) were aligned by multireference alignment and reference-free alignment procedures (50, 51). The best projections from each set were averaged after several cycles of multireference alignments, statistical analysis, and classification. For TEM of TprCC embedded in nanodiscs, samples were diluted severalfold in water to obtain a well dispersed population, applied to Parlodion and carbon-coated 400-mesh copper grids, and negatively stained with 0.75% uranyl formate. Images were taken on a Tecnai G2 Spirit BioTWIN microscope (FEI) at an accelerating voltage of 80 kV with a defocus of ∼−1.6. Class averaging was performed by averaging 59–72 discs/image using Xmipp.
SAXS Analysis
SAXS and wide angle x-ray scattering data were collected simultaneously at beamline X9 at the National Synchrotron Light Source (Upton, NY) at 10 °C by two overlapping detectors: a Mar 165 CCD SAXS detector 3.4 m from the sample and a custom built Photonic Science CCD wide angle x-ray scattering detector. The two-dimensional scattering images collected on the CCD detectors were circularly averaged using software developed at the beamline to yield one-dimensional scattering profiles as a function of momentum transfer q (q = 4πsin(θ)/λ, where 2π is the scattering angle and λ is the wavelength). The x-ray wavelength was 0.855 Å, and the angular range collected was 0.008 ≤ q ≥ 1.0060. The sample cell contained a glass capillary sealed across the evacuated chamber. Protein samples and matching buffer solutions were flowed through the capillary during exposure to reduce radiation damage. For data collection, 30 μl of the protein sample at concentrations between 1.0 and 4 mg/ml or matching buffer (50 mm Tris (pH 7.5), 100 mm NaCl) were exposed for 90 s and subdivided into three 30-s exposures of 30 μl. After each measurement, the capillary was washed thoroughly and purged with compressed nitrogen. The raw scattering data were scaled, and the buffer was subtracted using the pyXS program developed at Brookhaven National Laboratory (52). Each individual scattering curve was visually inspected for radiation damage and aggregation prior to averaging by Guinier plot analysis (53). Averaged scattering curves from the SAXS and wide angle x-ray scattering detectors were scaled and merged in PRIMUS (54) to yield a low noise composite curve. The radii of gyration (Rg) were initially calculated using AUTORG (55). P(r) functions were calculated using the program GNOM (56) with a maximum particle diameter of 140 Å for TprF. 20 independent runs in DAMMIN (57) were performed, and then the most probable model was computed with DAMAVER (58).
Indirect Immunofluorescence Analysis of T. pallidum in Gel Microdroplets
Freshly harvested T. pallidum cells were encapsulated in low melting point agarose (Sigma) microdroplets as described previously (14, 26, 35, 40). Encapsulated organisms were probed in a two-step process. In the first step, rat antiserum directed against TprCN, TprCC, or FlaA (each diluted 1:100) was added to the bead suspensions (0.2–0.3 ml) in the presence or absence of 0.1% (v/v) Triton X-100, followed by incubation for 2 h. In the second step, samples were incubated with gentle mixing in a 34 °C water bath for 2 h. The beads were then washed three times by low speed centrifugation (100 × g) and resuspended in CMRL medium followed by incubation for 1 h at 34 °C with 1 μg/ml goat anti-rat Alexa Fluor 488 conjugates (Invitrogen). The beads were then washed three times with CMRL medium and observed with a epifluorescent Olympus BX-41 microscope using a 100× (1.4 numerical aperture) oil immersion objective equipped with a Retiga Exi charge-coupled device (CCD) camera (Q Imaging, Tucson, AZ) and fluorescein isothiocyanate (FITC) and rhodamine ω filter sets. For each sample, three slides were prepared, and ∼100 organisms were scored for labeling.
Opsonophagocytosis Assays
Opsonophagocytosis assays were performed as described previously (29, 59).
Fractionation of E. coli Expressing TprC, TprI, and TprF with a PelB Signal Sequence
The DNAs encoding the tprC, tprI, and tprF genes without the native signal sequences were cloned into the BamHI and HindIII sites of a modified pET20b vector (EMD Millipore) (60) downstream and in frame with an N-terminal PelB signal sequence. The recombinant proteins were expressed in E. coli C41 (DE3) at 20 °C without induction in LB medium supplemented with 50 μg ml−1 ampicillin. Fractionation of E. coli cells was based on protocols published previously (61). Briefly, 1 liter of cells was pelleted and resuspended in 10 ml of 200 mm Tris-HCl (pH 8), 1 m sucrose, 1 mm EDTA; lysozyme was added to a final concentration of 1 mg/ml. The suspension was mixed and incubated for 10 min at room temperature, following which 40 ml of ultrapure deionized water (dH2O) were added before the suspension was placed on ice; the cells were then centrifuged at 200,000 × g for 45 min at 4 °C. The supernatant, which contained the periplasmic fraction, was removed. The pellet, consisting of spheroplasts, was resuspended in 5 ml of ice-cold 20 mm Tris-HCl (pH 7.5), 5 mm EDTA, 0.2 mm DTT supplemented with 50 μl DNase (1 mg/ml). The spheroplasts were ruptured in a French press with three passes at 108 pascals. Unbroken cells were removed by centrifugation at 5,000 × g for 10 min at 4 °C. The supernatant, containing cytosolic and crude membrane fractions, was centrifuged at 300,000 × g for 2 h at 4 °C. The supernatant (cytosolic fraction) was removed, and the pellet (crude membranes) was resuspended in 10 ml of 50 mm Tris-HCl (pH 7.5), 2% (w/v) Triton X-100, 10 mm MgCl2 and centrifuged at 90,000 × g for 30 min at 4 °C. The supernatant (inner membranes) was removed. The pellet (outer membranes) was washed in 2 ml of 50 mm Tris-HCl (pH 7.5), 2% (w/v) Triton X-100, 10 mm MgCl2; centrifuged at 90,000 × g for 30 min at 4 °C; washed three times in 1 ml of dH2O; and frozen at −20 °C prior to SDS-PAGE and immunoblot analysis.
Immunofluorescence Assay of E. coli Expressing Exported TprC, TprI, and TprF
E. coli C41 (DE3) strains expressing exported recombinant TprC, TprI, and TprF were grown to mid-log phase. Aliquots of 1 ml of cells were pelleted, washed, and resuspended in PBS and then fixed in 4% paraformaldehyde for 30 min at room temperature, followed by several washes with PBS. For permeabilization, cells were resuspended in Buffer P (PBS with 0.1% Triton X-100, 10 mm EDTA) with 100 μg/ml lysozyme for 30 min at room temperature. Intact or permeabilized cell samples were resuspended in PBS or Buffer P, respectively, containing 2% BSA prior to incubation with 1:1,000 dilutions of rat anti-TprCN, -TprCC, or -Skp antiserum for 2 h at 4 °C, followed by incubation with a 1:300 dilution of goat anti-rat Alexa Fluor 488 (Invitrogen) for an 1 h. After washing, cells were mounted in Vectashield® antifade reagent (Invitrogen) containing DAPI, and 10-μl drops were applied to glass slides. Fluorescent images were acquired on an epifluorescence Olympus BX-41 microscope using a ×40 (1.4 numerical aperture) oil immersion objective equipped with a Retiga Exi CCD camera (Q Imaging, Tucson, AZ), and the data were analyzed using Cell M (Olympus) and ImageJ.
Statistical Analysis
Statistical analyses were performed in GraphPad Prism using unpaired Student's t tests with two-tailed p values; p values <0.05 were considered significant.
RESULTS
Full-length TprC (TprCFl) and TprI (TprIFl), but Not TprF, Form Amphiphilic β-Barrels with Pore-forming Activity
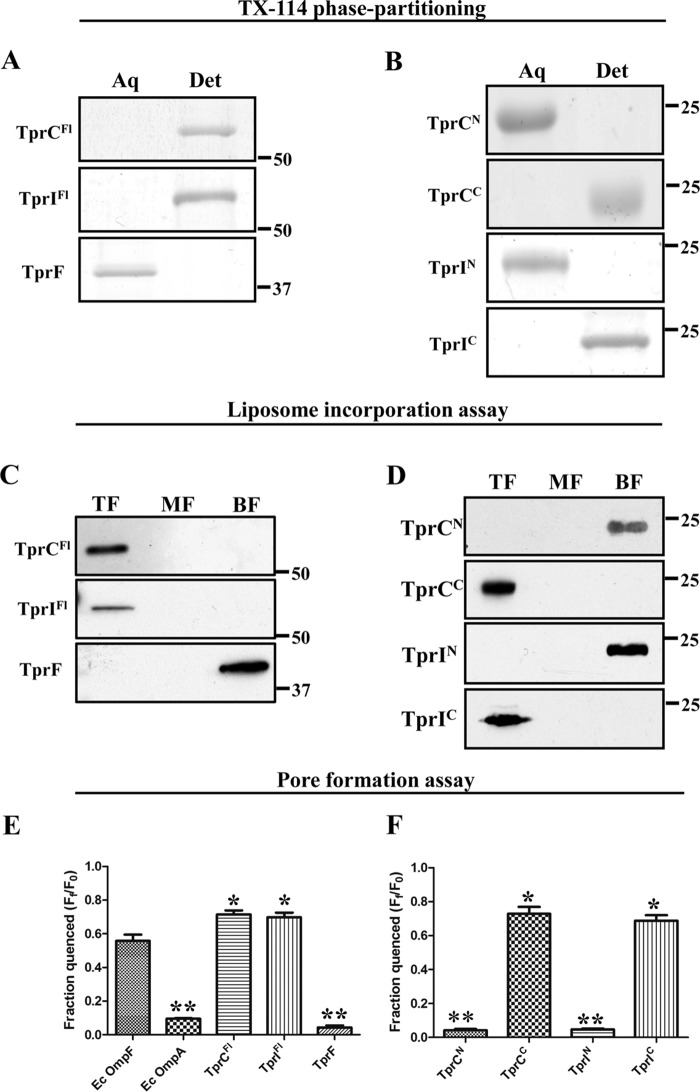
As depicted in Fig. 2, the NCBI Conserved Domain Database server predicts that TprI, like TprC, contains N- and C-terminal domains (TprIN (residues 83–281) and TprIC (409–609), respectively) related to the MOSPN and MOSPC domains of the major outer sheath protein of T. denticola; the truncated TprF, on the other hand, contains only a MOSPN-like domain (TprFN, residues 83–281). This analysis, coupled with our previous studies showing that TprCFl and TprCC, but not TprCN, can form trimeric, amphiphilic β-barrels with channel-forming activity (29), lead to the prediction that TprIFl can fold to form an amphiphilic β-barrel, but TprF cannot. As a first test of this prediction, we assessed the heat modifiability of the recombinant TprC subfamily proteins. β-Barrel-forming proteins characteristically remain folded in SDS at room temperature and, consequently, run with lower apparent molecular masses by SDS-PAGE than when denatured by boiling in SDS, a property termed heat modifiability (62). As controls, we included E. coli OmpG, a single domain, 14-stranded β-barrel (63), and E. coli OmpA, which consists of an N-terminal, 8-stranded β-barrel and a C-terminal peptidoglycan-binding domain (64–66). Fig. 3A shows that OmpG, OmpA, TprCFl, and TprIFl displayed heat modifiability, whereas boiling had no effect on the SDS-PAGE mobility of TprF. Furthermore, consistent with previous results for native TprC (29), native TprI, detected in T. pallidum lysates using a TprI-specific antiserum (see “Experimental Procedures” and Fig. 1B), also displayed heat modifiability (Fig. 3C). We next assessed the amphiphilic properties of the three recombinant Tpr proteins using Triton X-114 phase partitioning and liposome incorporation. Of note, for these experiments, we used LUVs whose phospholipid content simulates the T. pallidum OM (17). As shown in Fig. 4A, TprCFl and TprIFl both partitioned into the Triton X-114 detergent-enriched phase, whereas TprF was recovered in the aqueous phase. Following separation on discontinuous sucrose gradients, TprIFl, like TprCFl, was recovered from the liposome-containing top fractions, whereas TprF sedimented in the bottom fractions containing unincorporated protein (Fig. 4C). Additionally, as shown in Fig. 4E, TprIFl, but not TprF, exhibited porin activity in the Tb(DPA)33− efflux assay comparable with that of TprCFl; channel formation by both TprCFl and TprIFl was significantly greater than that by E. coli OmpF, an archetypal porin, (67). In contrast, OmpA, which exists predominantly in a closed confirmation (67), exhibited poor channel-forming activity (Fig. 4E). Using these same biophysical methodologies, we also confirmed that the bipartite architecture and porin activity of TprI mirror those of TprC. The results in Figs. 3B and 4 (B, D, and F) collectively demonstrate that TprIC alone is capable of forming an amphiphilic β-barrel with porin-like function.
FIGURE 2.
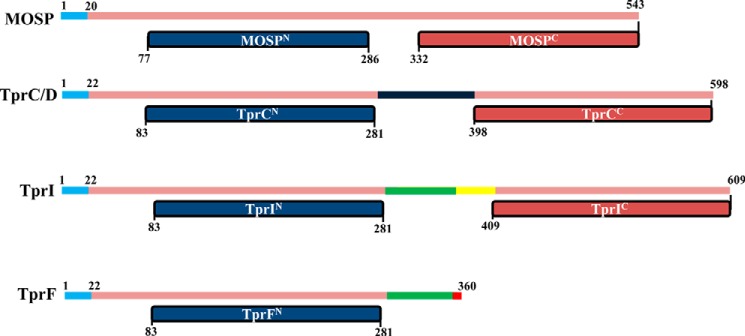
Domain architectures of T. denticola major outer surface protein and TprC subfamily members. The signal sequences of all three proteins are shown in blue. The portions of TprC and TprI colored in black and yellow, respectively, denote the TprC- and TprI-specific regions of each protein (TprCSp and TprISp). The green regions in TprI and TprF denote regions present in TprI and TprF but not TprC (TprI/FSp). The red portion in TprF denotes an 11-amino acid stretch unique to that protein.
FIGURE 3.
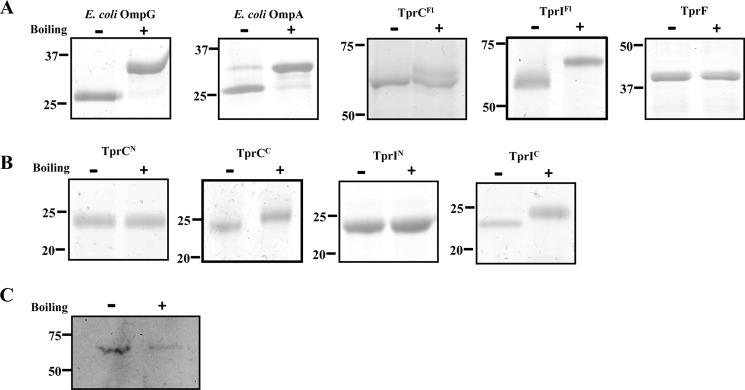
Recombinant TprIFl and TprCFl, but not TprF, are heat-modifiable. Heat modifiability of folded recombinant proteins (A and B) and native TprI in T. pallidum (C). Following SDS-PAGE with (+) or without (−) boiling in sample buffer, proteins were stained with GelCode® Blue (A and B) or transferred to nitrocellulose (C) for immunoblotting with TprI-specific antiserum. Molecular mass standards (kDa) are on the left of each panel.
FIGURE 4.
TrpC and TprI, but not TprF, are amphiphilic, pore-forming proteins. A and B, 10 μg of each recombinant protein were phase-partitioned in Triton X-114, separated by SDS-PAGE, and stained with GelCode® Blue. Lanes show aqueous (Aq) and detergent-enriched (Det) phases. C and D, liposomes were reconstituted with 10 μg of each folded recombinant protein, followed by sucrose density gradient ultracentrifugation. Fractions were subjected to SDS-PAGE followed by immunoblotting with antisera directed against TprCFl (TprC and TprI), TprCN (TprCN, TprIN, and TprF), and TprCC (TprCC and TprIC). Lanes show that top fractions (TF) contain liposome-incorporated material, whereas the middle and bottom fractions (MF and BF, respectively) contain unincorporated material. E and F, quenching of LUVs encapsulating Tb(DPA)33− after incubation with 100 nm recombinant proteins in 50 mm HEPES (pH 7.5), 100 mm NaCl supplemented with 5 mm EDTA. Each bar represents the mean ± S.E. (error bars) for three independent experiments. Statistical significance compared with E. coli OmpF was assigned according to the following scheme: *, p < 0.05; **, p < 0.0001. In A–D, molecular mass markers are shown at the right side of each gel or immunoblot.
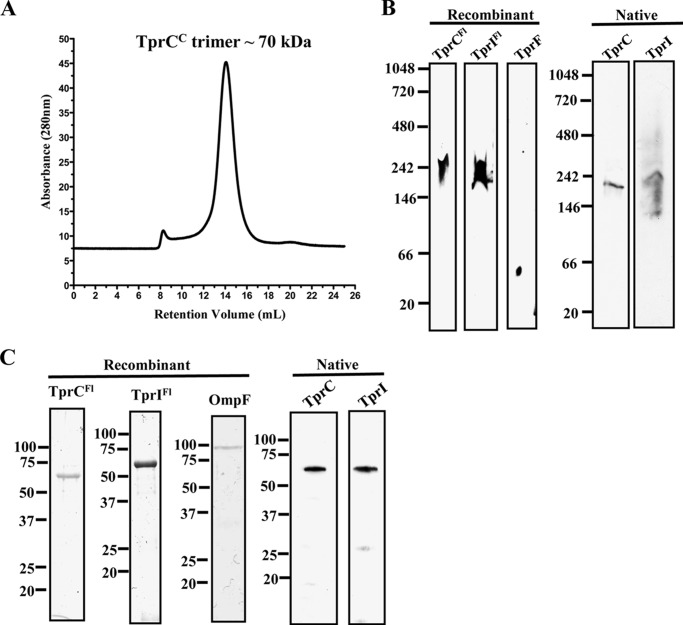
TprCFl and TprIFl Trimers Are Unstable in SDS
Classical porins, such as E. coli OmpF, remain trimeric at room temperature in the presence of SDS (68). Previously, we established that TprCFl and native TprC form trimers when solubilized in the non-denaturing detergent DDM and that the MOSPC-like domain is responsible for trimerization (29). Using size exclusion chromatography, we confirmed that TprCC forms trimers in lauryldimethylamine oxide, another non-denaturing detergent (Fig. 5A). Interestingly, when we examined the heat modifiability of TprCFl, TprCC, and TprIC in this study, we noted that they migrated as monomers in SDS at room temperature (Fig. 3), suggesting that TprC and TprI trimers are less stable than those formed by OmpF. To test this supposition, we directly compared the stability of TprC subfamily and OmpF trimers. Under non-denaturing conditions (i.e. blue native PAGE), TprCFl and TprIFl formed trimers, whereas TprF did not (Fig. 5B). Using antisera specific for TprC (29) and TprI (as described above), native TprC and TprI also formed trimers (Fig. 5B). In contrast, in SDS at room temperature, TprCFl and TprIFl were monomeric, whereas refolded OmpF retained its trimeric state (Fig. 5C). Similarly, native TprC and TprI also migrated as monomers in SDS at room temperature (Fig. 5C). The lack of a TprF-specific antiserum precluded our ability to assess the oligomeric state of native TprF.
FIGURE 5.
TprCFl and TprIFl form SDS-dissociable trimers. A, size exclusion chromatography analysis of TprCC in 50 mm Tris, pH 8, 200 mm NaCl, and 0.1% lauryldimethylamine oxide. The relative size of the TprCC was estimated by comparing the retention volume with a standard curve calculated using blue dextran. B, blue native PAGE. TprCFl, TprIFl, TprF, and T. pallidum lysates solubilized in 0.5% DDM with 50 mm Tris (pH 7.0) were separated by blue native PAGE followed by immunoblot analysis with antisera directed against TprCFl (TprCFl and TprIFl), TprCN (TprF), TprCSp (native TprC), and TprISp (native TprI). C, SDS-PAGE without boiling. TprCFl, TprIFl, OmpF, and T. pallidum lysates were subjected to SDS-PAGE without boiling. Recombinant proteins were stained with GelCode®, whereas T. pallidum lysates were immunoblotted with TprCSp and TprISp antisera. Molecular mass standards (kDa) are shown on the left.
Divergent Secondary Structures of the MOSPN- and MOSPC-like Domains
We next used far-UV CD spectroscopy to assess the α-helix and β-sheet contents of TprCFl, TprIFl, TprF, TprCN, TprCC, TprIN, and TprIC and compare them with the single domain OmpG and the bipartite OmpA. Table 1 contains a summary of the secondary structure data for all of the proteins. In accord with published structures (63–66), OmpG contains only miniscule (4%) α-helix, whereas OmpA, although predominantly β-sheet, contains substantial (22%) α-helix. Similar to OmpA, both TprCFl and TprIFl contain mixtures of β-sheet and α-helix, with the former predominant. As with OmpG, the MOSPC-like domains consist almost entirely of β-sheet; the MOSPN-like domains contribute virtually all of the α-helical content to the full-length proteins. As expected, TprCN, TprIN, and TprF have nearly identical proportions of β-sheet and α-helix.
TABLE 1.
Secondary structures of TprC subfamily constructs, E. coli OmpG, and E. coli OmpA determined by CD spectroscopy
| Construct | β-Sheet | α-Helix | Random coil |
|---|---|---|---|
| % | % | % | |
| E. coli OmpG | 54.5 | 4.5 | 36.9 |
| E. coli OmpA | 56.2 | 22.6 | 21.2 |
| TprCFl | 52.7 | 18.6 | 29.7 |
| TprCN | 36.2 | 25.4 | 38.4 |
| TprCC | 58.6 | 5.1 | 36.3 |
| TprIFl | 53.6 | 16.5 | 29.9 |
| TprIN | 36.5 | 25.6 | 37.9 |
| TprIC | 59.2 | 4.2 | 36.6 |
| TprF | 35.7 | 22.2 | 42.0 |
The TprC Subfamily Members Possess a High Degree of Thermal Stability
The above heat modifiability data suggested that the TprC and TprI β-barrels possess conformational stability comparable with that of their Gram-negative counterparts. To confirm and extend this notion, we next used far-UV CD thermal denaturation, a well established method for assessing protein secondary structure stability (42, 69), to examine the stability of the TprC subfamily members and their MOSPN- and MOSPC-like domains during thermal stress; OmpA and OmpF were included as comparators. The changes in CD ellipticity at 218 nm for each protein, plotted as fraction of unfolded protein versus temperature (data not shown), were utilized to derive the unfolding midpoints (Tm values) (Table 2). Mesophilic proteins with Tm values above 45 °C are generally considered to be thermostable (70, 71); all of the proteins studied herein met this criterion. Of note, the Tm for OmpF is in close agreement with published values determined by both CD and differential scanning calorimetry (72). Each curve shows a sharp unfolding transition, indicating a two-state thermal unfolding, all of which were irreversible upon cooling from 90 to 25 °C. The Tm values of TprCFl and TprIFl were comparable with that of OmpF and well above that of OmpA. Although the thermal stability of the MOSPC-like domains was expected, the stability of the MOSPN-like domains and TprF was not. To confirm these latter results, we measured the Tm values of TprF, TprCN, and TprIN using a fluorescence-based thermal denaturation assay (43, 44). For all three proteins, the Tm values obtained after non-linear fitting were nearly identical to the CD thermal denaturation values (Table 2). Also of interest were the findings that the Tm values of TprCFl and TprIFl were greater than those of either domain, suggesting that both domains contribute to the overall marked stability of the full-length proteins.
TABLE 2.
Thermal denaturation temperatures (Tm values) of TprC subfamily proteins, E. coli OmpA, and E. coli OmpF calculated from far-UV CD transition and SYPRO Orange® dye
Transition data refer to the molar ellipticity at 218 nm.
| Protein | Tma by CD | Tm by SYPRO® Orange |
|---|---|---|
| E. coli OmpF | 79.3 ± 0.7 | |
| E. coli OmpA | 52 ± 1.5 | |
| TprCFl | 71.5 ± 0.2 | |
| TprCN | 50.3 ± 0.4 | 50.0 ± 0.5 |
| TprCC | 62.3 ± 0.5 | |
| TprIFl | 67.2 ± 1.2 | |
| TprIN | 50. 6 ± 0.4 | 50.0 ± 0.5 |
| TprIC | 62.1 ± 0.5 | |
| TprF | 51.4 ± 0.8 | 50.2 ± 0.8 |
a Tm values were obtained by non-linear regression analysis to the CD thermal curves.
TprCC Can Circularize, whereas TprF Forms an Extended, Inflexible Structure
To complement the biophysical evidence that MOSPC-like domains form β-barrels and can trimerize, we used TEM to visualize folded, recombinant TprCC. In the first set of experiments, we examined TprCC folded in DDM. Inspection of the grids revealed evenly dispersed particles in a variety of orientations (Fig. 6A), which, on closer inspection, were a mixture of monomers, trimers, and aggregates. Two-dimensional reconstructions of ∼800 monomers and trimers were generated using Xmipp version 3.0 (49). The image-averaged monomer (Fig. 6A) is a circular structure with an external diameter of ∼5 nm and a central pore of ∼20 Å. The monomers in the image-averaged trimer, containing three negatively stained pits, tilt downward in a rosette-like arrangement with a 3-fold axis of symmetry (Fig. 6A). Additionally, we examined TprCC in self-assembled phospholipid bilayer nanodiscs, which provide a true lipidic environment for protein folding (47). TEM of nanodiscs containing TprCC showed a mixture of monomers and trimers in an approximate ratio of 1:3 (Fig. 6B). As with detergent-folded TprCC, a central pore is readily visible in the monomers and in the monomeric subunits of trimers (Fig. 6B).
FIGURE 6.
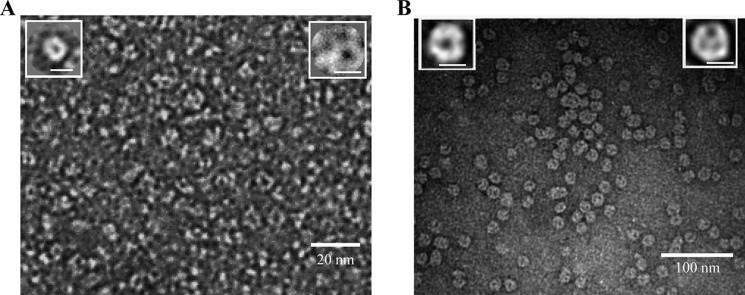
Single particle image analysis confirms that TprCC can circularize. A, electron micrograph of negatively stained TprCC showing average projection maps of monomeric and trimeric forms as insets in the left and right corners, respectively. Scale bar for insets, 5 nm. Samples applied to grids contained folded TprCC (1 mg/ml) in 0.1% DDM, 100 mm NaCl, 50 mm Tris, pH 7.5. B, electron micrograph of negatively stained TprCC reconstituted in E3D1 nanodiscs showing average projections of monomer and trimer as insets in the left and right corners, respectively. Scale bar for insets, 10 nm. For both A and B, single particle image analysis was performed using Xmipp (47).
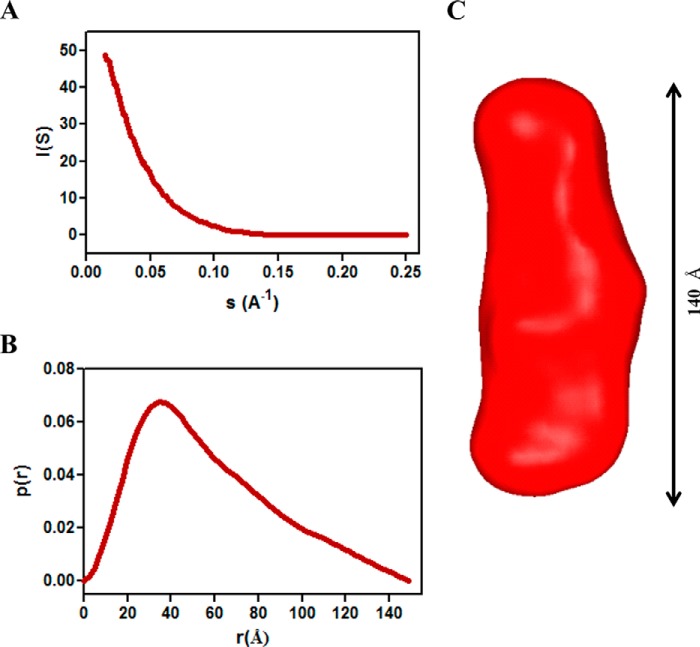
Small angle x-ray scattering is a powerful tool for determining the overall shape and structural flexibility of a protein (73, 74). One relevant example is the demonstration by SAXS of the conformational flexibility between domains 2 and 3 of the periplasmic POTRA arm of E. coli BamA (75). X-ray scattering (31, 76) was employed to define the envelope structure of TprF in solution and, by extension, the MOSPN-like domains of TprC and TprI. Scattering data were scaled and averaged in PRIMUS (54) to yield a low noise composite curve (Fig. 7A). The distance distribution function, P(r), representing the scattering curve in real space, enables delineation of the overall conformation of the scatterer (i.e. TprF); the largest dimension, Dmax, is estimated from the point where the function approaches zero. P(r) was derived using an indirect Fourier transformation of the scattering curve implemented in the program GNOM (56). The P(r) of TprF (Fig. 7B) shows a unimodal peak at 34.7 Å and Dmax at 140 Å, indicating that TprF adopts an extended, inflexible conformation. Fig. 7C depicts a model for the solution structure generated using DAMMIN and DAMAVER (57, 58).
FIGURE 7.
SAXS analysis reveals TprF as an extended, inflexible structure. A, experimental scattering curve for TprF, scaled and averaged in PRIMUS (54). B, P(r) function of TprF calculated from the experimental scattering curve using GNOM (56). C, averaged ab initio reconstruction of the TprF molecular envelope calculated by DAMAVER (58).
TprC and TprI Are Bipartite and Surface-exposed in T. pallidum, whereas TprF Is Periplasmic
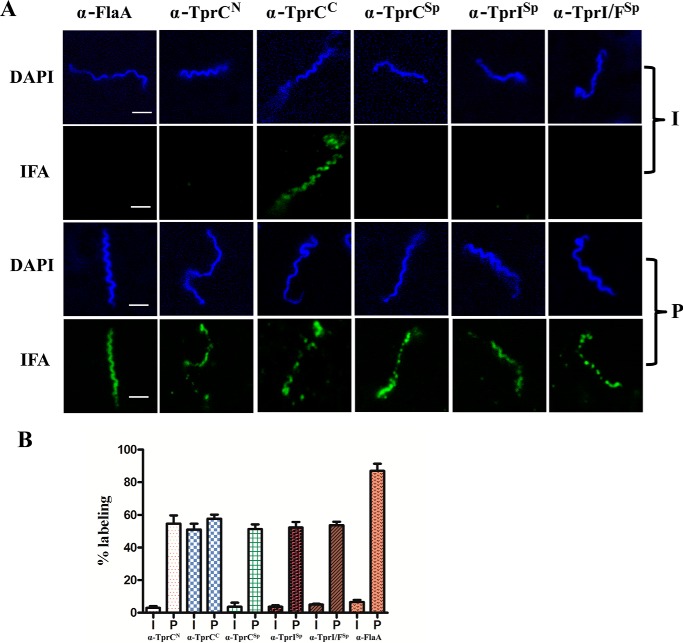
To establish the cellular locations of the MOSPN and MOSPC-like domains of native TprC and TprI, we performed indirect immunofluorescence analysis of treponemes encapsulated in gel microdroplets (14, 26, 35, 40). This sensitive method is ideally suited for this purpose because it not only preserves the integrity of fragile T. pallidum OMs throughout the immunolabeling procedure but also allows for their controlled removal by detergent solubilization (in this case 0.10% Triton X-100) in order to detect periplasmic antigens. Shown in Fig. 8, A and B, respectively, are representative images and corresponding bar graphs for the percentages of labeled organisms in three independent experiments. More than 50% (55 ± 5%) of the treponemes were labeled by antibodies against TprCC in the absence of detergent, far greater than the miniscule percentage (4 ± 2%) labeled with antibodies against TprCN under the same conditions. The result for anti-TprCN is consistent with the small percentage of OM-damaged organisms typically observed in this assay (26, 40) and herein (6 ± 3%) based upon labeling with antibodies against FlaA. Importantly, the percentage of treponemes labeled by the anti-TprCN antiserum increased dramatically in the presence of detergent (56 ± 8%), as was observed for FlaA (87 ± 10%). Indirect immunofluorescence analysis (IFA) experiments also were performed to localize the interdomain regions used to generate the TprCSp-, TprISp-, and TprI/FSp-specific antisera. With all three antisera, labeling above background was observed only with detergent-treated organisms (Fig. 8, A and B). No labeling of intact or permeabilized T. pallidum was observed with normal rat serum (data not shown). Knowing the reactivities of the various antisera and that all three TrpC subfamily proteins are co-expressed as determined by immunoblotting (Fig. 1B), we conclude from the collective IFA results that only the MOSPC-like domains of TprC and TprI are inserted in the T. pallidum OM, whereas the MOSPN-like domains and TprF are periplasmic.
FIGURE 8.
Bipartite topology of native TprC by IFA. A, motile T. pallidum were encapsulated in gel microdroplets and probed with 1:100 dilutions of rat antisera against TprCN, TprCC, TprCSp, TprISp, TprI/FSp, or FlaA without (intact, I) or with (permeabilized, P) removal of OMs by preincubation with 0.10% Triton X-100. Antibody binding was detected with 1 μg/ml goat anti-rat Alexa Fluor 488 (green) conjugate. Scale bars, 3 μm. B, bar graphs showing the percentage of labeling of intact (I) and permeabilized (P) T. pallidum with the various antisera; each bar represents the mean ± S.D. (error bars) for three independent experiments.
MOSPC-like but Not MOSPN-like Domains Are Opsonic Targets
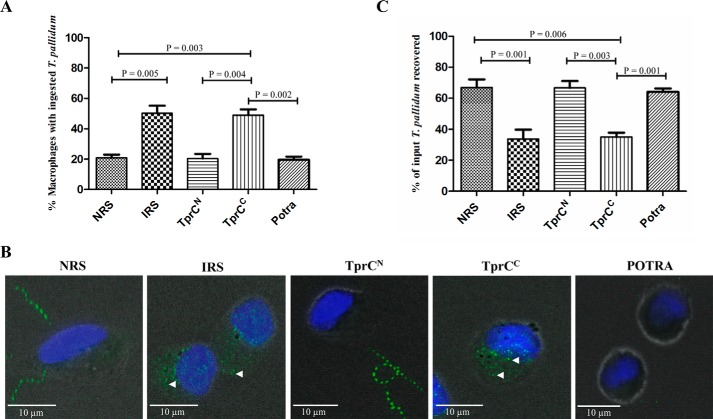
Internalization of treponemes by macrophages is believed to be a primary mechanism for bacterial clearance during syphilitic infection (59, 77). To complement the above IFA results demonstrating that only MOSPC-like domains are surface-exposed in T. pallidum, we assessed opsonophagocytosis of treponemes following incubation with rabbit antisera to TprCN and TprCC. As shown in Fig. 9A (see Fig. 9B for representative micrographs), immune rabbit serum and TprCC antiserum significantly increased uptake of organisms compared with the background levels seen with normal rabbit serum, anti-TprCN, and an antiserum to the periplasmic POTRA domain of Tp0326 (36), the BamA ortholog of T. pallidum. As a qualitative measure of the degree of surface antibody binding by the treponemal populations, we also enumerated spirochetes in supernatants collected following the 4-h incubation periods (Fig. 9C). Consistent with the internalization results, supernatants containing spirochetes incubated with immune rabbit serum and the TprCC antiserum contained significantly lower percentages of organisms (using input values as 100%) than supernatants containing spirochetes incubated with normal rabbit serum, anti-POTRA, or the TprCN antiserum.
FIGURE 9.
Only the MOSPC-like domains are opsonic targets. A, percentages of rabbit peritoneal macrophages containing internalized treponemes. Results shown are means ± S.E. (error bars) for three independent experiments. B, representative micrographs. Each image is a composite of bright field images, nuclei stained with DAPI, and FITC-labeled T. pallidum. Arrowheads, degraded treponemes inside macrophages. C, percentages of treponemes recovered following a 4-h incubation of rabbit peritoneal macrophages and treponemes with the indicated sera. Results shown are means ± S.E. for three independent experiments. p values of <0.05 (Student's t test) were considered significant.
The β-Barrels of Native TprC and TprI Are Tethered within the Periplasm
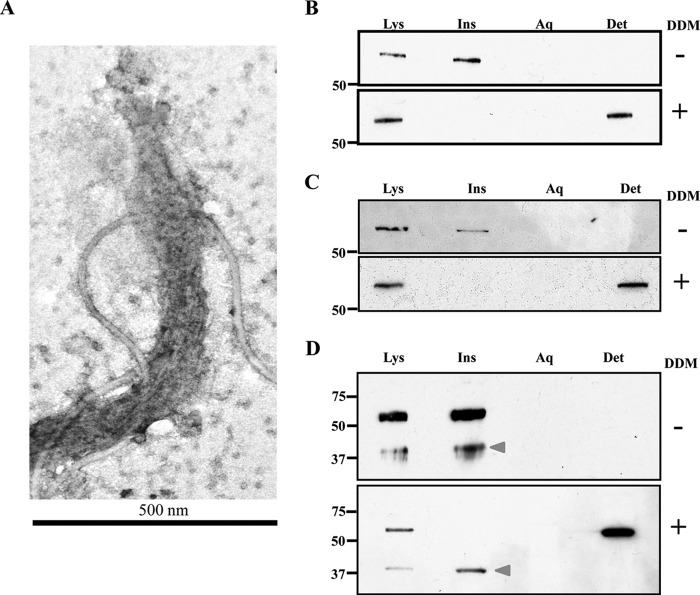
In our previous study (29), we discovered that when T. pallidum was subjected to our standard Triton X-114 phase-partitioning protocol, TprC remained with the detergent-insoluble fraction, which, by TEM, contains the peptidoglycan sacculus (Fig. 10A). However, when phase partitioning was performed on lysates initially solubilized with DDM, TprC was recovered in the detergent-enriched phase. Because the β-barrel is OM-embedded and should be readily solubilized with Triton X-114 based on the phase partitioning results with recombinant TprCFl and TprCC, we interpreted these findings to mean that in T. pallidum, the N-terminal domain tethers the β-barrel within the periplasm. Herein, we repeated these findings for TprC (Fig. 10B) and obtained identical results for TprI (Fig. 10C). Thus, in T. pallidum the β-barrel of TprI also appears to be tethered by the native protein's N-terminal domain. TprF, on the other hand, resisted solubilization with DDM and remained with the Triton X-114-insoluble material (Fig. 10D), indicating that it is more tightly bound within the periplasm than either TprC or TprI.
FIGURE 10.
Native TprC and TprI are amphiphilic but tethered to the peptidoglycan sacculus, whereas TprF is entirely periplasmic and tightly bound. A, extensively washed Triton X-114-insoluble material visualized in negatively stained whole mounts by transmission electron microscopy. Triton X-114 phase partitioning of T. pallidum lysates (1 × 109 organisms) without (−) or with (+) presolubilization with 2% DDM. Whole cells (WC), Triton X-114-insoluble material (Ins), and aqueous and Triton X-114-enriched phases (Aq and Det, respectively) were separated by SDS-PAGE followed by immunoblotting using antisera directed against TprCSp (B), TprISp (C), or TprI/FSp (D). Arrowheads in D, TprF. Molecular mass markers (kDa) are shown at the left of each immunoblot.
TprC and TprI Expressed in E. coli with PelB Signal Sequences Are OM-associated, Surface-exposed, and Bipartite, whereas Exported TprF Is Periplasmic
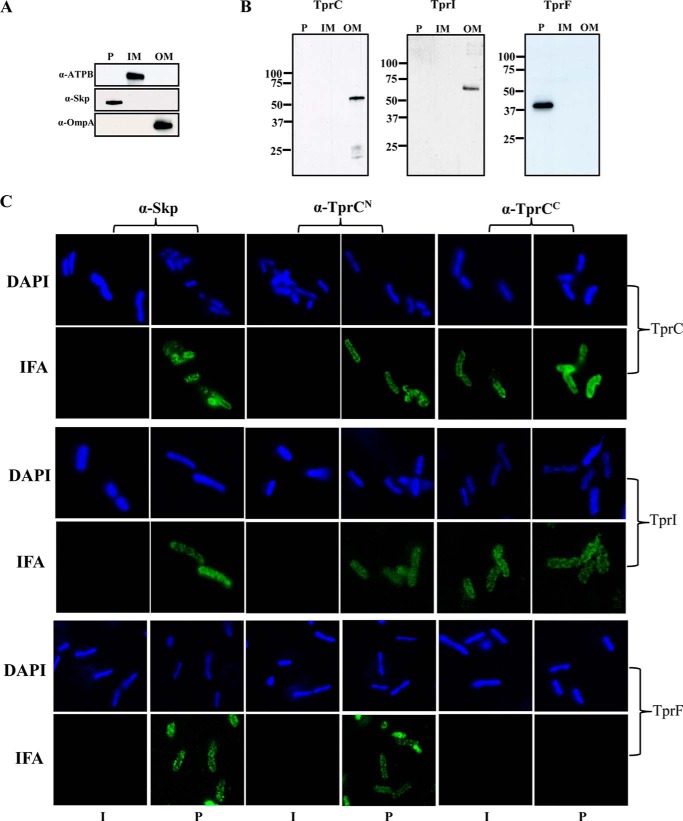
Topologic analysis of individual TprC subfamily proteins in T. pallidum is problematic due to their high degree of sequence similarity compounded by the syphilis spirochete's recalcitrance to cultivation and genetic manipulation. One strategy for circumventing these difficulties is to express these proteins in a genetically malleable surrogate with export signals that direct them to the appropriate cellular compartments. Along these lines, we placed the coding sequences for the processed forms of TprC, TprI, and TprF downstream of a PelB signal sequence and expressed them in E. coli. As shown in Fig. 11B, following fractionation, TprC and TprI were recovered with the OMs, whereas TprF was recovered with the periplasmic proteins. IFA of intact and permeabilized isopropyl 1-thio-β-d-galactopyranoside-induced cells was performed using TprCN and TprCC antisera and antiserum to the periplasmic chaperone Skp (78, 79); representative images are presented in Fig. 11C. Intact E. coli expressing TprC or TprI, but not TprF, were labeled only with antibodies to TprCC, whereas antisera to TprCN and Skp labeled only permeabilized cells. Neither TprC domain antiserum reacted with uninduced intact or permeabilized cells (data not shown).
FIGURE 11.
TprC and TprI, but not TprF, expressed in E. coli with PelB signal sequences fractionate with OMs and display bipartite topology. Comparable portions of periplasmic (P), inner membrane (IM), and outer membrane (OM) fractions, generated as described under “Experimental Procedures,” were subjected to SDS-PAGE and immunoblotting with antisera directed against ATP synthase B subunit (ATPB), Skp, and OmpA as markers for the IM, P, and OM fractions, respectively (A), and TprC (for E. coli expressing TprC and TprI) or TprCN (for E. coli expressing TprF) (B). C, IFA of intact (I) or permeabilized (P) E. coli C41 (DE3) expressing TprC, TprI, or TprF with PelB signal sequence were probed with 1:1,000 dilutions of rat antisera against TprCN, TprCC, and Skp (periplasmic control). Antibody binding was detected with 1 μg/ml goat anti-rat Alexa Fluor 488 conjugate.
DISCUSSION
The discovery of rare outer membrane proteins in T. pallidum dates from the use of freeze-fracture electron microscopy to explain the poor antigenicity of live spirochetes in vitro (18, 19) and the bacterium's well recognized propensity for evading humoral responses during human infection (7, 20). In the pregenomics era, efforts to identify the entities visualized as intramembranous particles were impeded by the fragility of the T. pallidum OM, the limited quantity of OMs that could be isolated from this noncultivatable bacterium, and the low cellular abundance of these presumptive OM-spanning proteins (11, 12, 80). The availability of the genomic sequence in 1998 (16) did not immediately resolve this conundrum because the spirochete lacks orthologs for OMPs that at the time had been well characterized in model proteobacteria, especially E. coli. However, it was noted that the T. pallidum genome encodes a large family of paralogous proteins, the Tprs, related to the MOSP of T. denticola (16). Extrapolating from published studies of MOSP, Fraser et al. (16) conjectured that the Tprs function as adhesins and porins and, given the number of paralogs, might also be a system for antigenic variation. Using a bioinformatics-based approach to identify T. pallidum proteins predicted to form β-barrels, we found that, among the Tprs, the TprC subfamily contains the highest ranked candidate OMPs (26). Subsequently, we reported that TprC is an authentic rare OMP and that it possesses a bipartite topology in which TprCC comprises the β-barrel (29). Herein, we confirmed this architectural model for native TprC expressed by T. pallidum and extended it to the closely related TprI, which also contains MOSPN- and MOSPC-like domains. The truncated TprF, in contrast, was found to be a hydrophilic, extended structure in purified recombinant form and tethered within the periplasm in T. pallidum.
Because the first crystal structure of an OMP, the porin of Rhodobacter capsulatus, was solved in 1991 (81), numerous high resolution structures of OMPs have been reported (82). In the vast majority, regardless of function, the entire polypeptide comprises the β-barrel (82, 83). Three classes of bipartite OMPs, in which a single polypeptide comprises the entire OM-spanning protein, have emerged as exceptions to this generalization: autotransporters (84), BamAs (Omp85 superfamily) (85–88), and OmpAs (65, 66, 89). Whereas BamA and OmpA consist of β-barrels with sizeable periplasmic domains, the autotransporters are distinctive in that the hydrophilic passenger domain extrudes through the barrel to the exterior of the cell. BamA and OmpA, however, differ topologically; the 16-transmembrane strand β-barrel of BamA resides within the C-terminal half of the protein, whereas the converse is true for the 8-stranded barrel of OmpA. The periplasmic portions of these two proteins, although both mixtures of α-helix and β-strand, also differ markedly. The flexible, N-terminal periplasmic portion of BamA consists of multiple POTRA domains (90), which function as a “jointed” arm to intercept newly exported OMP precursors bound to periplasmic chaperones and thread them to the other components of the β-barrel assembly machine (Bam) for insertion into the OM (85, 87, 91). The single N-terminal periplasmic domain of OmpA fulfills a very different functional role, enhancing the stability of the OM by tethering the β-barrel to the murein layer residing beneath the bilayer (65, 89). The data presented here and elsewhere (29, 30) for TprC, TprI, and MOSP of T. denticola reveal that these full-length proteins represent a new class of OMP. Like BamA, they consist of N-terminal periplasmic and C-terminal β-barrel domains. However, the N-terminal domain, a single, extended, highly stable, mixed α/β structure, appears to play a structural role akin to the C-terminal region of OmpA, although we do not know the molecules within the periplasm with which it interacts. Interestingly, the TprC and TprI β-barrels possess features, namely formation of trimers and ostensibly open channels, more reminiscent of OmpF than of OmpA, which has poor conductivity due to a largely occluded channel (92, 93). Previously, we described TprC as a functional analog of E. coli OmpF. Based on the data as a whole, a more accurate analogy for TprC and TprI would seem to be as a functional chimera of OmpF and OmpA.
Gram-negatives, prototypical diderms, have highly impermeable, LPS-containing OMs but also express abundant non-selective porins for diffusion-dependent uptake of substrates present at high concentrations in the extracellular environment; lower copy number, selective OMP transporters are employed for importation of scarce nutrients, such as iron (67, 83). How T. pallidum meets its nutritional requirements, given its unorthodox OM and diminutive content of OMPs, is a longstanding mystery (12). The bacterium's extremely slow doubling time, ∼30 h in rabbits (94), is almost certainly part of the answer; such a slowly growing bacterium must consume nutrients at a dramatically lower rate than organisms with doubling times on the order of 20 min. Lacking LPS (16), the T. pallidum OM also is more permeable than that of Gram-negatives and may be capable of taking up some nutrients, such as long-chain fatty acids, by diffusion across the bilayer (95). Nevertheless, it has long been presumed that the syphilis spirochete, an extreme auxotroph (16), could not survive without porin-like proteins (96). Although initial reports of T. pallidum porins proved to be erroneous (97, 98), we have now provided strong evidence that Tprs with C termini capable of forming amphiphilic β-barrels can fulfill this function. Thus, it appears as if stealth pathogenicity involves striking a delicate balance between the number of OMPs needed to sustain viability and an expression profile of OMPs that limits vulnerability to host antibodies. Given the premium on minimizing the number of surface-exposed antigenic targets, one also would anticipate that the channels formed would be large. This conjecture is supported by the assays performed herein, in which TprC and TprI enhanced permeability of liposomes to a degree greater than OmpF, as well as previous black-lipid conductance studies of T. denticola MOSP (99).
Porins of Gram-negative bacteria are renowned for their resistance to protease digestion and denaturation by heat, low pH, or chaotropic agents (68, 83). The remarkable sturdiness of these molecules in their trimeric state undoubtedly reflects the harsh environments, including the human gastrointestinal tract, in which they have evolved to maintain structure and function. Our collective results reveal that some of these physical traits are dispensable for an extraintestinal, extracellular pathogen, such as T. pallidum, whose surface does not need to withstand a comparable chemical and enzymatic assault under normal in vivo growth conditions. For example, native TprC and TprI expressed by T. pallidum are readily digested by proteinase K, suggesting that their extracellular loops are not as compact as those of conventional porins (33, 100, 101). The trimers of Gram-negative porins resist dissociation in high concentrations of SDS at ambient temperature, a property attributed to the low binding of this denaturing detergent by the closely packed hydrophobic interface that joins the monomers (33, 68, 100, 101). For OmpF and OmpC, additional stability of the trimer is provided by polar and ionic interactions between the L2 loops of each monomer and neighboring subunits (33, 68, 100, 101). The finding that both TrpC and TprI trimers dissociate in SDS at room temperature indicates that the β-barrel domains lack one or more of the structural features that stabilize conventional porin trimers. Analysis of OmpF revealed that dissociation of trimer and unfolding of monomer are cooperative events (68). The instability of TprC and TprI trimers in SDS at ambient temperature suggests that, during thermal stress, trimer dissociation precedes monomer unfolding and that the denaturation curves mainly capture the inherent stability of the monomers.
Our consensus computational framework identified the truncated TprF as one of the higher ranked candidate rare OMPs within the T. pallidum genome (26). An intriguing question is how a Tpr without a C-terminal β-barrel domain could have met bioinformatics criteria as a possible OMP. The answer lies in the CD analysis, which shows that TprF possesses sufficient β-sheet structure to fool some of the algorithms used to generate the consensus. However, biophysical analysis of TprCN, TprIN, and TprF showed that they are soluble proteins with no propensity to insert into lipid bilayers. The predicted compartmental location for soluble exported proteins in T. pallidum and E. coli would be the periplasmic space, exactly as was found. In T. pallidum, TprCN and TprIN anchor their respective β-barrels to the cytoplasmic cylinder, whereas TprF is tethered even more firmly within the periplasm. These results point to interactions of all three proteins with periplasmic constituents that prevent them from being freely soluble. TprF has unique sequences at its C terminus, which could be responsible for its tighter binding than TprCN or TprIN to the cell cylinder. The most logical cellular arrangement for TprCN and TprIN is that they extend downward from the OM-inserted β-barrels, binding near the PG layer, which in T. pallidum is midway in the periplasmic space (102, 103). The length of TprF determined by SAXS, ∼15 nm, accommodates this deduction extremely well. The periplasmic space of T. pallidum has been accurately measured under native conditions by two groups using cryoelectron tomography (102, 103). The width of the space bulges to accommodate the flagellar filaments. However, in regions away from the flagella, the space narrows such that the mean distance between the inner leaflet of the OM and the murein layer is ∼14 nm. Also worth noting is that the intramembranous particles visualized by freeze-fracture electron microscopy are non-mobile and generally located in regions of the OM away from the flagella (104). The thermal denaturation experiments demonstrate that the full-length Tprs form highly stable structures due to the contributions of both the MOSPN- and MOSPC-like domains. By vertically linking the OM and the cytoplasmic cylinder, an extremely strong bipartite molecule would enhance the structural integrity of the entire cell envelope in addition to stabilizing the OM.
Organisms with OMs possess a Bam for integrating newly exported OMPs into the OM lipid bilayer (85, 87). Recognition of a C-terminal motif by POTRA1 of BamA facilitates proper folding and integration of the nascent OMP into the OM (105–107). We previously noted that TprC and TprI have canonical C-terminal recognition motifs (36). Despite the phylogenetic distance separating spirochetes and proteobacteria (22) and the dramatically different subunit compositions of their Bam complexes (36), TprC and TprI expressed in E. coli with signal sequences appear to fold properly within the OM and achieve their correct topology. This finding clearly demonstrates that the bipartite conformation is not dependent on expression of these proteins in a particular lipidic milieu and is consistent with other reports showing that Bam complexes can recognize and fold OMPs from distantly related organisms (108). It also establishes E. coli as a viable heterologous platform for fine topologic mapping of T. pallidum rare OMPs and genetically dissecting their structure-function relationships, objectives that syphilologists have been striving for since the beginning of the molecular era (109).
Acknowledgments
The TEM work was performed at the Biosciences Electron Microscopy Facility of the University of Connecticut. We are indebted to Dr. Melissa J. Caimano for many helpful suggestions with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Public Health Service Grant AI-26756 (to J. D. R.) and NIH PSI: Biology Grant U54 GM094611 (to M. G. M.). This work was also supported by research funds from Connecticut Children's Medical Center.
- OM
- outer membrane
- OMP
- outer membrane protein
- Tpr
- T. pallidum repeat family
- LUV
- large unilamellar vesicle
- MOSP
- major outer sheath protein
- SAXS
- small angle x-ray scattering
- Ni-NTA
- nickel-nitrilotriacetic acid
- DDM
- n-dodecyl β-d-maltoside
- TEM
- transmission electron microscopy
- IFA
- immunofluorescence analysis
- POTRA
- polypeptide transport-associated
- Bam
- β-barrel assembly machine.
REFERENCES
- 1. Patton M. E., Su J. R., Nelson R., Weinstock H., Centers for Disease Control and Prevention (2014) Primary and secondary syphilis: United States, 2005–2013. MMWR Morb. Mortal. Wkly. Rep 63, 402–406 [PMC free article] [PubMed] [Google Scholar]
- 2. Hook E. W., 3rd, Peeling R. W. (2004) Syphilis control: a continuing challenge. N. Engl. J. Med. 351, 122–124 [DOI] [PubMed] [Google Scholar]
- 3. Klausner J. D. (2013) The sound of silence: missing the opportunity to save lives at birth. Bull. World Health Organ. 91, 158–158A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Workowski K. A., Berman S. (2010) Sexually transmitted diseases treatment guidelines, 2010. MMWR Recomm. Rep. 59, 1–110 [PubMed] [Google Scholar]
- 5. Galvao T. F., Silva M. T., Serruya S. J., Newman L. M., Klausner J. D., Pereira M. G., Fescina R. (2013) Safety of benzathine penicillin for preventing congenital syphilis: a systematic review. PLoS One 8, e56463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ho E. L., Lukehart S. A. (2011) Syphilis: using modern approaches to understand an old disease. J. Clin. Invest. 121, 4584–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lafond R. E., Lukehart S. A. (2006) Biological basis for syphilis. Clin. Microbiol. Rev. 19, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radolf J. D., Hazlett K. R. O., Lukehart S. A. (2006) in Pathogenic Treponemes: Cellular and Molecular Biology (Radolf J. D., Lukehart S. A., eds) pp. 197–236, Caister Academic Press, Norfolk, UK [Google Scholar]
- 9. Norris S. J., Cox D. L., Weinstock G. M. (2001) Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J. Mol. Microbiol. Biotechnol. 3, 37–62 [PubMed] [Google Scholar]
- 10. Cox D. L. (1994) Culture of Treponema pallidum. Methods Enzymol. 236, 390–405 [DOI] [PubMed] [Google Scholar]
- 11. Cameron C. E. (2006) in Pathogenic Treponema: Molecular and Cellular Biology (Radolf J. D., Lukehart S. A., eds) pp. 237–266, Caister Academic Press, Norwich, UK [Google Scholar]
- 12. Radolf J. D. (1995) Treponema pallidum and the quest for outer membrane proteins. Mol. Microbiol. 16, 1067–1073 [DOI] [PubMed] [Google Scholar]
- 13. Cameron C. E., Lukehart S. A. (2014) Current status of syphilis vaccine development: need, challenges, prospects. Vaccine 32, 1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox D. L., Akins D. R., Porcella S. F., Norgard M. V., Radolf J. D. (1995) Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol. Microbiol. 15, 1151–1164 [DOI] [PubMed] [Google Scholar]
- 15. Cox D. L., Chang P., McDowall A. W., Radolf J. D. (1992) The outer membrane, not a coat of host proteins, limits antigenicity of virulent Treponema pallidum. Infect. Immun. 60, 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fraser C. M., Norris S. J., Weinstock G. M., White O., Sutton G. G., Dodson R., Gwinn M., Hickey E. K., Clayton R., Ketchum K. A., Sodergren E., Hardham J. M., McLeod M. P., Salzberg S., Peterson J., Khalak H., Richardson D., Howell J. K., Chidambaram M., Utterback T., McDonald L., Artiach P., Bowman C., Cotton M. D., Fujii C., Garland S., Hatch B., Horst K., Roberts K., Sandusky M., Weidman J., Smith H. O., Venter J. C. (1998) Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281, 375–388 [DOI] [PubMed] [Google Scholar]
- 17. Radolf J. D., Robinson E. J., Bourell K. W., Akins D. R., Porcella S. F., Weigel L. M., Jones J. D., Norgard M. V. (1995) Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect. Immun. 63, 4244–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Radolf J. D., Norgard M. V., Schulz W. W. (1989) Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. U.S.A. 86, 2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker E. M., Zampighi G. A., Blanco D. R., Miller J. N., Lovett M. A. (1989) Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J. Bacteriol. 171, 5005–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salazar J. C., Hazlett K. R., Radolf J. D. (2002) The immune response to infection with Treponema pallidum, the stealth pathogen. Microbes Infect. 4, 1133–1140 [DOI] [PubMed] [Google Scholar]
- 21. Radolf J. D., Desrosiers D. C. (2009) Treponema pallidum, the stealth pathogen, changes, but how? Mol. Microbiol. 72, 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paster B. J., Dewhirst F. E. (2006) in Pathogenic Treponema: Molecular and Cellular Biology (Radolf J. D., Lukehart S. A., eds) pp. 9–18, Caister Academic Press, Norwich, UK [Google Scholar]
- 23. Tommassen J. (2010) Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156, 2587–2596 [DOI] [PubMed] [Google Scholar]
- 24. Ulrich T., Gross L. E., Sommer M. S., Schleiff E., Rapaport D. (2012) Chloroplast β-barrel proteins are assembled into the mitochondrial outer membrane in a process that depends on the TOM and TOB complexes. J. Biol. Chem. 287, 27467–27479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silhavy T. J., Kahne D., Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cox D. L., Luthra A., Dunham-Ems S., Desrosiers D. C., Salazar J. C., Caimano M. J., Radolf J. D. (2010) Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect. Immun. 78, 5178–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Centurion-Lara A., Castro C., Barrett L., Cameron C., Mostowfi M., Van Voorhis W. C., Lukehart S. A. (1999) Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun E. S., Molini B. J., Barrett L. K., Centurion-Lara A., Lukehart S. A., Van Voorhis W. C. (2004) Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect. 6, 725–737 [DOI] [PubMed] [Google Scholar]
- 29. Anand A., Luthra A., Dunham-Ems S., Caimano M. J., Karanian C., LeDoyt M., Cruz A. R., Salazar J. C., Radolf J. D. (2012) TprC/D (Tp0117/131), a trimeric, pore-forming rare outer membrane protein of Treponema pallidum, has a bipartite domain structure. J. Bacteriol. 194, 2321–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anand A., Luthra A., Edmond M. E., Ledoyt M., Caimano M. J., Radolf J. D. (2013) The major outer sheath protein (Msp) of Treponema denticola has a bipartite domain architecture and exists as periplasmic and outer membrane-spanning conformers. J. Bacteriol. 195, 2060–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pérez J., Nishino Y. (2012) Advances in x-ray scattering: from solution SAXS to achievements with coherent beams. Curr. Opin. Struct. Biol. 22, 670–678 [DOI] [PubMed] [Google Scholar]
- 32. Cowan S. W., Garavito R. M., Jansonius J. N., Jenkins J. A., Karlsson R., König N., Pai E. F., Pauptit R. A., Rizkallah P. J., Rosenbusch J. P. (1995) The structure of OmpF porin in a tetragonal crystal form. Structure 3, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 33. Yamashita E., Zhalnina M. V., Zakharov S. D., Sharma O., Cramer W. A. (2008) Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 27, 2171–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phale P. S., Philippsen A., Widmer C., Phale V. P., Rosenbusch J. P., Schirmer T. (2001) Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry 40, 6319–6325 [DOI] [PubMed] [Google Scholar]
- 35. Hazlett K. R., Cox D. L., Decaffmeyer M., Bennett M. P., Desrosiers D. C., La Vake C. J., La Vake M. E., Bourell K. W., Robinson E. J., Brasseur R., Radolf J. D. (2005) TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 187, 6499–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Desrosiers D. C., Anand A., Luthra A., Dunham-Ems S. M., LeDoyt M., Cummings M. A., Eshghi A., Cameron C. E., Cruz A. R., Salazar J. C., Caimano M. J., Radolf J. D. (2011) TP0326, a Treponema pallidum β-barrel assembly machinery A (BamA) orthologue and rare outer membrane protein. Mol. Microbiol. 80, 1496–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luthra A., Zhu G., Desrosiers D. C., Eggers C. H., Mulay V., Anand A., McArthur F. A., Romano F. B., Caimano M. J., Heuck A. P., Malkowski M. G., Radolf J. D. (2011) The transition from closed to open conformation of Treponema pallidum outer membrane-associated lipoprotein TP0453 involves membrane sensing and integration by two amphipathic helices. J. Biol. Chem. 286, 41656–41668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edelhoch H. (1967) Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 39. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hazlett K. R., Sellati T. J., Nguyen T. T., Cox D. L., Clawson M. L., Caimano M. J., Radolf J. D. (2001) The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193, 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whitmore L., Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greenfield N. J. (2006) Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 1, 2527–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 44. Lavinder J. J., Hari S. B., Sullivan B. J., Magliery T. J. (2009) High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 131, 3794–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brusca J. S., Radolf J. D. (1994) Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228, 182–193 [DOI] [PubMed] [Google Scholar]
- 46. Heuck A. P., Tweten R. K., Johnson A. E. (2003) Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J. Biol. Chem. 278, 31218–31225 [DOI] [PubMed] [Google Scholar]
- 47. Nath A., Atkins W. M., Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 48. Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., Sligar S. G. (2009) Chapter 11: reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de la Rosa-Trevín J. M., Otón J., Marabini R., Zaldívar A., Vargas J., Carazo J. M., Sorzano C. O. (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 [DOI] [PubMed] [Google Scholar]
- 50. van Heel M., Gowen B., Matadeen R., Orlova E. V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M., Patwardhan A. (2000) Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 [DOI] [PubMed] [Google Scholar]
- 51. Pascual-Montano A., Donate L. E., Valle M., Bárcena M., Pascual-Marqui R. D., Carazo J. M. (2001) A novel neural network technique for analysis and classification of EM single-particle images. J. Struct. Biol. 133, 233–245 [DOI] [PubMed] [Google Scholar]
- 52. Allaire M., Yang L. (2011) Biomolecular solution x-ray scattering at the National Synchrotron Light Source. J. Synchrotron Radiat. 18, 41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guinier A. (1939) La diffraction des rayons X aux tres petits angles: application a l'etude de phenomenes ultramicroscopiques. Ann. Phys. (Paris) 12, 161–237 [Google Scholar]
- 54. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) Primus: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 55. Petoukhov M. V., Svergun D. I. (2007) Analysis of x-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 17, 562–571 [DOI] [PubMed] [Google Scholar]
- 56. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 57. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lukehart S. A., Miller J. N. (1978) Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 121, 2014–2024 [PubMed] [Google Scholar]
- 60. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thein M., Sauer G., Paramasivam N., Grin I., Linke D. (2010) Efficient subfractionation of Gram-negative bacteria for proteomics studies. J. Proteome Res. 9, 6135–6147 [DOI] [PubMed] [Google Scholar]
- 62. Beher M. G., Schnaitman C. A., Pugsley A. P. (1980) Major heat-modifiable outer membrane protein in gram-negative bacteria: comparison with the ompA protein of Escherichia coli. J. Bacteriol. 143, 906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yildiz O., Vinothkumar K. R., Goswami P., Kühlbrandt W. (2006) Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 25, 3702–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang Y. (2002) The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292, 396–401 [DOI] [PubMed] [Google Scholar]
- 65. Smith S. G., Mahon V., Lambert M. A., Fagan R. P. (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 66. Marcoux J., Politis A., Rinehart D., Marshall D. P., Wallace M. I., Tamm L. K., Robinson C. V. (2014) Mass spectrometry defines the C-terminal dimerization domain and enables modeling of the structure of full-length OmpA. Structure 22, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Phale P. S., Philippsen A., Kiefhaber T., Koebnik R., Phale V. P., Schirmer T., Rosenbusch J. P. (1998) Stability of trimeric OmpF porin: the contributions of the latching loop L2. Biochemistry 37, 15663–15670 [DOI] [PubMed] [Google Scholar]
- 69. Pace C. N., Scholtz J. M. (1997) Measuring the Conformational Stability of a Protein. in Protein Structure: A Practical Approach (Creighton T. E., ed) pp. 299–321, Oxford University Press, New York [Google Scholar]
- 70. Mitchell J. C. (2011) Thermostable Proteins: Structure, Stability, and Design, CRC Press, Boca Raton, FL [Google Scholar]
- 71. Kumar S., Tsai C. J., Nussinov R. (2001) Thermodynamic differences among homologous thermophilic and mesophilic proteins. Biochemistry 40, 14152–14165 [DOI] [PubMed] [Google Scholar]
- 72. Visudtiphole V., Thomas M. B., Chalton D. A., Lakey J. H. (2005) Refolding of Escherichia coli outer membrane protein F in detergent creates LPS-free trimers and asymmetric dimers. Biochem. J. 392, 375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pelikan M., Hura G. L., Hammel M. (2009) Structure and flexibility within proteins as identified through small angle x-ray scattering. Gen. Physiol. Biophys. 28, 174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Receveur-Brechot V., Durand D. (2012) How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gatzeva-Topalova P. Z., Warner L. R., Pardi A., Sousa M. C. (2010) Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pauw B. R. (2013) Everything SAXS: small-angle scattering pattern collection and correction. J. Physics Condensed Matter 25, 383201. [DOI] [PubMed] [Google Scholar]
- 77. Radolf J. D., Lukehart S. A. (2006) in Pathogenic Treponemes: Cellular and Molecular Biology (Radolf J. D., Lukehart S. A., eds) pp. 285–322, Caister Academic Press, Norfolk, UK [Google Scholar]
- 78. Sklar J. G., Wu T., Kahne D., Silhavy T. J. (2007) Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Walton T. A., Sandoval C. M., Fowler C. A., Pardi A., Sousa M. C. (2009) The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc. Natl. Acad. Sci. U.S.A. 106, 1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cullen P. A., Cameron C. E. (2006) Progress towards an effective syphilis vaccine: the past, present and future. Expert Rev. Vaccines 5, 67–80 [DOI] [PubMed] [Google Scholar]
- 81. Weiss M. S., Abele U., Weckesser J., Welte W., Schiltz E., Schulz G. E. (1991) Molecular architecture and electrostatic properties of a bacterial porin. Science 254, 1627–1630 [DOI] [PubMed] [Google Scholar]
- 82. Fairman J. W., Noinaj N., Buchanan S. K. (2011) The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr. Opin. Struct. Biol. 21, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Koebnik R., Locher K. P., Van Gelder P. (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37, 239–253 [DOI] [PubMed] [Google Scholar]
- 84. Nicolay T., Vanderleyden J., Spaepen S. (2015) Autotransporter-based cell surface display in Gram-negative bacteria. Crit. Rev. Microbiol. 41, 109–123 [DOI] [PubMed] [Google Scholar]
- 85. Ricci D. P., Silhavy T. J. (2012) The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Noinaj N., Kuszak A. J., Gumbart J. C., Lukacik P., Chang H., Easley N. C., Lithgow T., Buchanan S. K. (2013) Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Selkrig J., Leyton D. L., Webb C. T., Lithgow T. (2014) Assembly of β-barrel proteins into bacterial outer membranes. Biochim. Biophys. Acta 1843, 1542–1550 [DOI] [PubMed] [Google Scholar]
- 88. Heinz E., Lithgow T. (2014) A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Front. Microbiol. 5, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Park J. S., Lee W. C., Yeo K. J., Ryu K. S., Kumarasiri M., Hesek D., Lee M., Mobashery S., Song J. H., Kim S. I., Lee J. C., Cheong C., Jeon Y. H., Kim H. Y. (2012) Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the Gram-negative bacterial outer membrane. FASEB J. 26, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., Kahne D. (2007) Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 [DOI] [PubMed] [Google Scholar]
- 91. Noinaj N., Kuszak A. J., Balusek C., Gumbart J. C., Buchanan S. K. (2014) Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sugawara E., Nikaido H. (1992) Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267, 2507–2511 [PubMed] [Google Scholar]
- 93. Sugawara E., Nikaido H. (1994) OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269, 17981–17987 [PubMed] [Google Scholar]
- 94. Magnuson H. J., Eagle H., Fleischman R. (1948) The minimal infectious inoculum of Spirochaeta pallida (Nichols strain) and a consideration of its rate of multiplication in vivo. Am. J. Syph. Gonorrhea Vener. Dis. 32, 1–18 [PubMed] [Google Scholar]
- 95. Cox D. L., Radolf J. D. (2001) Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology 147, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 96. Blanco D. R., Miller J. N., Lovett M. A. (1997) Surface antigens of the syphilis spirochete and their potential as virulence determinants. Emerg. Infect. Dis. 3, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Blanco D. R., Champion C. I., Exner M. M., Erdjument-Bromage H., Hancock R. E., Tempst P., Miller J. N., Lovett M. A. (1995) Porin activity and sequence analysis of a 31-kilodalton Treponema pallidum subsp. pallidum rare outer membrane protein (Tromp1). J. Bacteriol. 177, 3556–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Blanco D. R., Champion C. I., Exner M. M., Shang E. S., Skare J. T., Hancock R. E., Miller J. N., Lovett M. A. (1996) Recombinant Treponema pallidum rare outer membrane protein 1 (Tromp1) expressed in Escherichia coli has porin activity and surface antigenic exposure. J. Bacteriol. 178, 6685–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Egli C., Leung W. K., Müller K. H., Hancock R. E., McBride B. C. (1993) Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect. Immun. 61, 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. (1992) Crystal structures explain functional properties of two E. coli porins. Nature 358, 727–733 [DOI] [PubMed] [Google Scholar]
- 101. Baslé A., Rummel G., Storici P., Rosenbusch J. P., Schirmer T. (2006) Crystal structure of osmoporin OmpC from E. coli at 2.0 Å. J. Mol. Biol. 362, 933–942 [DOI] [PubMed] [Google Scholar]
- 102. Izard J., Renken C., Hsieh C. E., Desrosiers D. C., Dunham-Ems S., La Vake C., Gebhardt L. L., Limberger R. J., Cox D. L., Marko M., Radolf J. D. (2009) Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 191, 7566–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liu J., Howell J. K., Bradley S. D., Zheng Y., Zhou Z. H., Norris S. J. (2010) Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 403, 546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bourell K. W., Schulz W., Norgard M. V., Radolf J. D. (1994) Treponema pallidum rare outer membrane proteins: analysis of mobility by freeze-fracture electron microscopy. J. Bacteriol. 176, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Struyvé M., Moons M., Tommassen J. (1991) Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218, 141–148 [DOI] [PubMed] [Google Scholar]
- 106. Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., Tommassen J. (2006) Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bennion D., Charlson E. S., Coon E., Misra R. (2010) Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77, 1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ulrich T., Oberhettinger P., Schütz M., Holzer K., Ramms A. S., Linke D., Autenrieth I. B., Rapaport D. (2014) Evolutionary conservation in biogenesis of β-barrel proteins allows mitochondria to assemble a functional bacterial trimeric autotransporter protein. J. Biol. Chem. 289, 29457–29470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Walfield A. M., Hanff P. A., Lovett M. A. (1982) Expression of Treponema pallidum antigens in Escherichia coli. Science 216, 522–523 [DOI] [PubMed] [Google Scholar]