Background: Orc1 is the largest subunit of the origin recognition complex that promotes genome duplication.
Results: We studied the dynamics of Orc1 during the cell division cycle.
Conclusion: Orc1 binds to mitotic chromosomes, and during G1 phase in the daughter cells it then forms spatial-temporal patterns in the nucleus.
Significance: The large subunit of ORC orchestrates the earliest stages of chromosome inheritance.
Keywords: Cell Cycle, DNA-binding Protein, DNA Replication, Mitosis, Protein-DNA Interaction, FOXA1, Origin Recognition Complex
Abstract
Replication of eukaryotic chromosomes occurs once every cell division cycle in normal cells and is a tightly controlled process that ensures complete genome duplication. The origin recognition complex (ORC) plays a key role during the initiation of DNA replication. In human cells, the level of Orc1, the largest subunit of ORC, is regulated during the cell division cycle, and thus ORC is a dynamic complex. Upon S phase entry, Orc1 is ubiquitinated and targeted for destruction, with subsequent dissociation of ORC from chromosomes. Time lapse and live cell images of human cells expressing fluorescently tagged Orc1 show that Orc1 re-localizes to condensing chromatin during early mitosis and then displays different nuclear localization patterns at different times during G1 phase, remaining associated with late replicating regions of the genome in late G1 phase. The initial binding of Orc1 to mitotic chromosomes requires C-terminal amino acid sequences that are similar to mitotic chromosome-binding sequences in the transcriptional pioneer protein FOXA1. Depletion of Orc1 causes concomitant loss of the mini-chromosome maintenance (Mcm2–7) helicase proteins on chromatin. The data suggest that Orc1 acts as a nucleating center for ORC assembly and then pre-replication complex assembly by binding to mitotic chromosomes, followed by gradual removal from chromatin during the G1 phase.
Introduction
In eukaryotic cells, the genome is duplicated once during the S phase of each cell division cycle and is a highly regulated process (1). Initiation of DNA replication requires ordered assembly of a pre-replication complex (pre-RC)3 at all origins of DNA replication, a process that requires the origin recognition complex (ORC), Cdc6, and Cdt1 bound to the hexameric Mcm2–7 complex (2–4). The process in human cells is thought to begin with binding of the six-subunit ORC to chromatin, a prerequisite for pre-RC assembly; however, human cell ORC is not a stable complex during the cell division cycle. In Saccharomyces cerevisiae and Schizosaccharomyces pombe, all six subunits of ORC have been reported to remain associated with the chromatin throughout the cell division cycle, although phosphorylation plays a key role in regulating ORC activity (5–12). In Xenopus, ORC is not associated with chromatin during S phase and is absent from the mitotic chromatin (13–17). The expression of Orc1 in human cells and in Drosophila is regulated by E2F (18, 19). Therefore, the assembly of pre-RCs at all origins depends on the E2F/Rb pathway with ORC activity being regulated by Orc1 expression (12, 19), but this is particularly important in cells entering the cell division cycle following a period of quiescence. In Drosophila, Orc1 is degraded at the end of M phase by the anaphase-promoting complex activated by Fzr/Cdh1 (20). In contrast, in human cells Orc1 is ubiquitinated and then degraded during the G1 to S phase transition and is reloaded at the M-G1 transition when new pre-RCs are formed (21–26). Thus, Orc1 re-binding to chromatin is an obligatory step for pre-RC assembly during G1. The Mcm2–7 hetero-hexamer is destined to become part of the active helicase following initiation of DNA replication at each origin. Recruitment of the Mcm2–7 helicase complex to ORC-Cdc6 complexes completes the formation of the pre-RC, a step mediated by the Mcm2–7-binding partner Cdt1.
The initiation of DNA replication occurs at origin sites whose density along the chromosome is critical for complete genome duplication. In all eukaryotic cells, origins of replication are located in replication domains, i.e. regions of chromosomes that replicate at defined times during S phase and are spatially organized within the nucleus (27–29). The spatiotemporal replication pattern is inherited from mother to daughter nuclei in a cell type-specific manner (30–32). It has been suggested from studies in budding yeast (33) and in mammalian cells (34, 35) that the establishment of the temporal program of DNA replication during S phase occurs during early G1 (36). Following assembly of pre-RCs either during exit from mitosis or during early G1, establishment of the pattern of origin distribution along chromosomes (called the origin decision point) and a separate replication timing decision point occur concurrent with the organization of chromosomes into distinct nuclear domains (28, 30, 34–39).
Maps of chromatin interactions determined by chromosome conformation capture technologies reveal the most definitive correlation with DNA replication timing profiles, indicating that clusters of replicons form a domain in a chromosome that is replicated at a characteristic time during S phase, and the domain is spatially compartmentalized into the visible replication foci in cells (40–43). This has been elegantly demonstrated at the single molecule level in S. pombe where early origins are activated at specific sites in the genome, but late firing origins derive from stochastic clusters of origins that form foci of replication sites in the nucleus (44). There are, however, few molecular insights into how spatiotemporal patterning of DNA replication occurs (45), but it is thought not to involve specific DNA sequences at the origins of DNA replication (33). In fission yeast, it has been shown that ORC binding to chromosomes during the M/G1 period of the cell division cycle pre-determines DNA replication origin usage and their efficiency of utilization during S phase, and it is also related to the timing of pre-RC assembly during G1 (46). In Drosophila, high density ORC binding to chromosomal sites is a determinant of early replication of those sites, and ORC is found to be associated with H3.3-enriched open chromatin sites (47).
To address the role of Orc1 protein in human cells during the M/G1 transition and G1, synchronized cells were used, and the dynamics of ORC complex assembly was followed throughout the cell division cycle. Orc1 associated with mitotic chromosomes dependent upon amino acid sequences in Orc1 that are related in sequence to the mitotic chromosome-binding sequences of a transcriptional pioneer protein FOXA1. Following mitosis, Orc1 was present in the G1 nuclei, but it displayed varied spatial distributions during G1 phase before it was removed at the G1/S phase boundary. Other ORC subunits, such as Orc2, were assembled onto chromatin after Orc1 and remained chromatin-bound, but it dissociated from Orc1 during late G1 and early S phase. Orc1 patterning in the G1 nucleus was related to the spatiotemporal pattering of DNA replication in the nucleus during S phase.
EXPERIMENTAL PROCEDURES
Raising Monoclonal Antibodies against Human Orc1 Protein
For production of specific antibodies capable of immunoprecipitation of native Orc1 protein, GST-Orc1ΔN400, containing Orc1 amino acid sequences 401–861 fused to the C terminus of glutathione S-transferase was expressed and purified from Escherichia coli BL21 (DE3) cells as described previously (24). The Orc1ΔN400 protein was separated from the GST tag by treatment with PreScission Protease (GE Healthcare) and used as an antigen for monoclonal antibody production using protocols described previously (48). The hybridomas were screened by an enzyme-linked immunosorbent assay, and positive clones were screened further for the ability to immunoprecipitate soluble GST- or MBP-tagged Orc1. Positive clones were screened further to test their ability to immunoprecipitate endogenous native Orc1 protein from HeLa whole cell extracts. The clone used in this study was Orc1 78-1-172. MBP-tagged Orc1 was purified as described previously (49).
Epitope-tagged Orc1 Construct and Mutant Orc1 Construction
Human Orc1 cDNA was cloned into mammalian expression vectors pEYFP-C1, pEYFP-N1, and pEGFP-C1 and expressed from a CMV promoter (Clontech.). Electroporation was performed on trypsinized cells resuspended in 250 μl of growth medium and transferred to cuvettes containing 2 μg of YFP-Orc1 protein plasmid plus 20 μg of salmon sperm DNA. Cells were seeded onto acid-washed coverslips and processed for immunofluorescence localization or live cell imaging. A U2OS stable cell line containing the pEYFP-Orc1 was generated by transfection and clonal selection and was maintained in DMEM (high glucose) with 10% fetal bovine serum (FBS) and 0.5 mg/ml G418 (Invitrogen). Tetracycline-inducible U2OS GFP-Orc1 cells were maintained and induced as described previously (49). Orc1 mutants were generated using the site-directed mutagenesis kit (Stratagene) as per the supplier's specifications. Orc1 fragments to study the FOXA1-related sequences were cloned into the pAcGFP1-Nuc vector (Clontech). U2OS cells were transfected with 1 μg of plasmid using X-tremeGENE HP (Roche Applied Science) according to the manufacturer's instructions, and cells were visualized 24 h post-transfection. HeLa cells were transfected with EGFP-Orc1, and live cells imaging was done.
Live Cell Microscopy
Human cells stably expressing YFP-Orc1 or cells transiently transfected with 2 μg of EYFP-Orc1 and/or enhanced CFP-PCNA, GFP-Orc1 fragments, or a tetracycline-inducible GFP-Orc1 U2OS cell line were used for live cell imaging. The cells were transferred to an FCS2 live cell chamber (Bioptechs Inc., Butler, PA) mounted onto the stage of a Delta Vision optical sectioning deconvolution instrument (Applied Precision) on an Olympus or Nikon microscope and kept at 37 °C in L-15 medium (minus phenol red) containing 30% FBS or DMEM with 10% FBS. Time-lapse images acquired with ×63 or ×100 objective lens were captured with a Coolsnap CCD camera or the PerkinElmer Life Sciences spinning disk acquisition setup.
Cell Culture, RNA Interference, and Extract Preparation
HeLa and U2OS human cells were grown in DMEM containing high glucose (Gibco) supplemented with penicillin/streptomycin and 10% FBS. MCF7 and IMR-90 diploid cells were grown in DMEM supplemented with 10% FBS. HeLa cells were grown in suspension in Joklik Modified Eagle's Essential Minimal Medium supplemented with 5% bovine serum. Successive salt fractionation was done as described previously (50). HeLa suspension cells were synchronized by double thymidine block and release as described previously (24). Nuclear extracts were prepared by resuspending the cells in Buffer A (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.4% Nonidet P-40, 5 mm MgOAc, 10% glycerol, 1 mm DTT, 20 μm MG132, 1 mm ATP); the suspension was homogenized using a Dounce B pestle. The pelleted nuclei were washed, resuspended in Buffer A + 0.5 mm CaCl2, and treated with DNase I (Invitrogen) and Benzonase ultra (Sigma) for 1 h. The salt concentration was then brought up to 400 mm NaCl and incubated for 30 min; the salt was then adjusted to 200 mm. Immunoprecipitations were carried out using antibody cross-linked protein G Dynabeads (Invitrogen). RNA interference was carried out as described previously (51). The small interfering RNAs (siRNA) targeted to Orc1 were Orc1-1 (coding region) CUGCACUACCAAACCUAUA (26) and Orc1-2 (3′UTR) GGAAUUGUUGUAACAAAGU). Control siRNA was targeted to luciferase (CUUACGCUGAGUACUUCGA (51)); all siRNAs were synthesized by Dharmacon Inc (Lafayette, CO). siRNA was delivered into the cells at a final concentration of 100 nm using Oligofectamine (Invitrogen). Cells were transfected twice at a gap of 24–30 h and analyzed for immunoblotting and immunofluorescence.
A second extract preparation procedure was used for immunoprecipitation of Orc1 from a nuclear extract (Fig. 1C). GFP-Orc1 U2OS TREx stable cell lines were grown in DMEM containing high glucose (Gibco) supplemented with penicillin/streptomycin and 10% FBS. The cells were induced with 1 μg/ml tetracycline and harvested 24 h after the induction. The cells were washed in PBS and suspended in Buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 20 μm MG132 and protease and phosphatase inhibitor tablets from Roche Applied Science) with 0.4% of Nonidet P-40. The cells were incubated for 10 min with end-to-end shaking at 4 °C. The cytoplasmic content was removed after spinning at 1300 × g for 5 min. The cells were washed again with Buffer A and centrifuged. The nuclei pellet was suspended in buffer containing 20 mm Tris-HCl, pH 7.5, 400 mm NaCl, 0.4% Nonidet P-40, 5 mm MgCl2, 0.1 mm EDTA, 10% glycerol, 1 mm DTT, 1 mm CaCl2, 20 μm MG132/ and protease as well as phosphatase inhibitor tablets from Roche Applied Science. Benzonase was added to the buffer, and the suspension incubated for 30 min on ice with intermittent mixing. The concentration of NaCl and Nonidet P-40 was brought down to 200 mm and 0.2%, respectively, with dilution buffer after half an hour incubation on ice. The extract was centrifuged in an Eppendorf microcentrifuge at 14,000 rpm (20,800 × g) for 15 min, and supernatant containing mostly the chromatin-bound proteins was used for Orc1 immunoprecipitation. The control mouse serum as well as the Orc1 mouse ascites were cross-linked to protein G Dynabeads (Invitrogen), and the antibody cross-linked beads were suspended in PBS in 1:1 ratio. The extract was incubated with increasing amounts of beads for 5 h at 4 °C with end-to end shaking. The beads were washed three times with washing buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.15% Nonidet P-40, 5 mm MgCl2, 0.1 mm EDTA, 10% glycerol, 1 mm DTT, and protease as well as phosphatase inhibitor tablets from Roche Applied Science. Finally, the washed beads were suspended in Laemmli sample buffer, and 8% SDS-polyacrylamide gels were run for silver staining as well as for immunoblot.
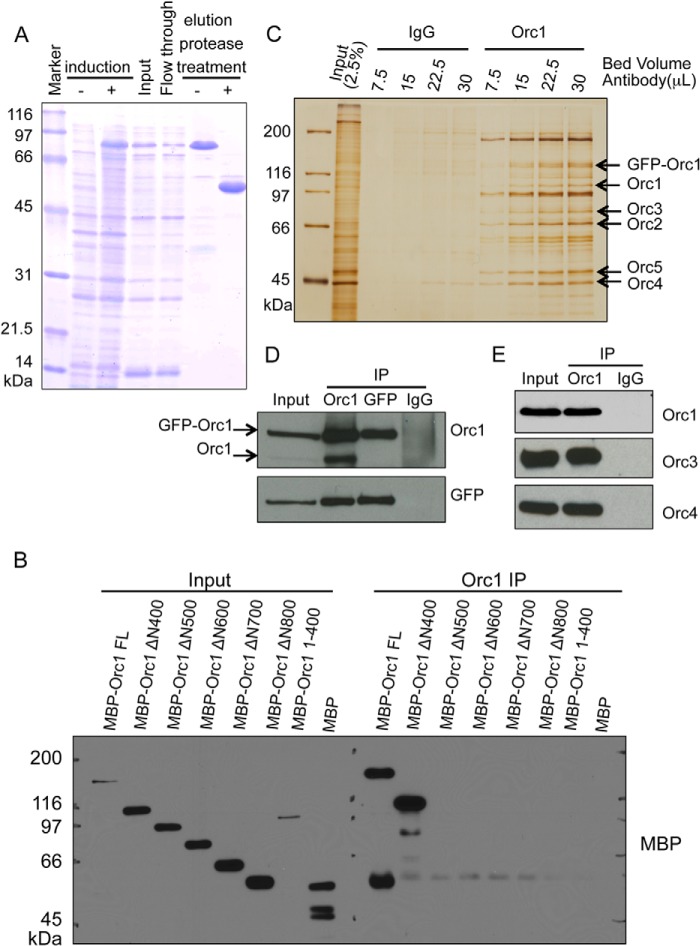
FIGURE 1.
Characterization of a monoclonal antibody targeting human Orc1. A, Coomassie Brilliant Blue-stained gel showing step for purification of the Orc1-ΔN400 fragment from E. coli cells. B, epitope mapping for the Orc1 antigen. The fragments of MBP-fused Orc1 were subjected to immunoprecipitation (IP) using the Orc1 78-1-172 monoclonal antibody, and then Western blots were performed with anti-MBP antibody. Only the full-length (FL) MBP-Orc1 and MBP-Orc1-ΔN400 versions but not other N-terminal truncated mutants were immunoprecipitated. C, silver-stained gel showing that increasing amounts of beads cross-linked to Orc1 78-1-172 monoclonal antibody, but not IgG, can immunoprecipitate endogenous Orc1 protein as well as the exogenously expressed GFP-Orc1 fusion protein from HeLa cells expressing GFP-Orc1. D, immunoblot detecting Orc1 and GFP in immunoprecipitates similar to those shown in C. E, Orc1 78-1-172 monoclonal antibody can co-immunoprecipitate other subunits of ORC, including Orc3 and Orc4 as detected by immunoblot.
Antibodies
For immunoprecipitations, the following mouse monoclonal antibodies were used: Orc1 78-1-172 (this study), Orc2 920-2-44 (24), and Orc3 PKS1-16 (52). For immunofluorescence and Western blots, rabbit polyclonal anti-Orc1 (Bethyl Labs), rabbit polyclonal anti-Orc2 CS205-5 (pAb205 (52)), rabbit polyclonal anti-Orc3 CS1890 (24), and goat polyclonal anti-Orc4 (Abcam) were used.
Immunofluorescence
For visualizing the proliferating cell nuclear antigen (PCNA (53)) and Mcm2–7 on chromatin, cells were pre-extracted in 0.5% Triton X-100 in Cytoskeletal buffer (10 mm PIPES, pH 7, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2) for 5 min on ice to remove soluble proteins, then fixed in 1.7% paraformaldehyde, and chilled, and methanol was extracted. Immunofluorescence was carried out using standard procedures (48). Cells were examined using a Zeiss Axioplan 2i fluorescence microscope (Carl Zeiss Inc., Thornwood, NY) equipped with Chroma filters (Chroma Technology, Brattleboro, VT). OpenLab software (Improvision, Boston, MA) was used to collect digital images from a Hamamatsu ORCA cooled CCD camera. Antibodies used were as follows: PCNA PC10 mAb, Mcm3 738 pAb, and Orc2 205 pAb (all from the Cold Spring Harbor Laboratory Antibody Shared Resource). Anti-GFP mAb was from Roche Applied Science, and anti-BrdU mAb was from Molecular Probes.
dUTP Incorporation
12 μl of PBS and 3 μl of FuGENE 6 reagent were added into a tube and then incubated for 5 min at 4 °C. To this, 1.5 μl of 100 μm dUTP (final 10 μm Chroma tide and Alexa Fluor dUTP 594, Molecular Probes and Invitrogen Detection Technologies) was added and incubated for an additional 20 min at 4 °C. The entire mixture was layered over the coverslip with cells that were stably expressing YFP-Orc1 and incubated for 15 min at 4 °C, following which cells were washed with fresh DMEM and left for 30 min in the incubator for entire consumption of dUTP. Media were changed again, and cells were left in the incubator for an additional 12–14 h following which cells were used for live cell imaging.
RESULTS
Generation and Characterization of Monoclonal Antibodies against Human Orc1 Protein
We studied the dynamics of the ORC complex in vivo, and for this purpose new monoclonal antibodies were raised that immunoprecipitated the native Orc1 protein from human cells. Purified Orc1-ΔN400 fragment was used as an antigen to immunize the mice (Fig. 1A). One of the clones (Orc1 78-1-172) was selected for further characterization. The antibody was able to immunoprecipitate the MBP-tagged full-length Orc1 and an N-terminal truncated version Orc1-ΔN400 but not other N-terminal truncated proteins. Thus, the antibody recognized the 400–500- amino acid region in MBP-tagged Orc1 protein (Fig. 1B). The antibody precipitated both endogenous Orc1 and GFP-tagged Orc1 from a U2OS cell line that expressed an inducible GFP-Orc1 (Fig. 1C). It could also co-immunoprecipitate other endogenous subunits of ORC, including Orc3 and Orc4 (Fig. 1, D and E), suggesting its utility for studying ORC dynamics throughout the cell division cycle. The antibody only detected the native form of the protein as confirmed by the ability to immunoprecipitate Orc1 (Fig. 1) and the inability to detect Orc1 in Western blots (result not shown).
Dynamics of ORC throughout the Cell Division Cycle
To better understand the dynamics of ORC on chromatin, we tested various chromatin fractions. Nuclei were first briefly treated with micrococcal nuclease, and then the nuclei were subjected to successive extractions with increasing salt concentrations (50). Of all the ORC subunits tested, Orc1 bound to chromatin or a nuclear structure in higher salt concentrations than all of the other ORC subunits (results not shown). This observation was consistent with a previous study using fluorescence recovery after photobleaching analysis with YFP-tagged proteins in human cells that showed YFP-Orc1 was not recovered after photobleaching, whereas YFP-Orc2, YFP-Orc3, and YFP-HP1α were readily recovered, indicating Orc1 was more tightly bound to some structures compared with other ORC subunits (54).
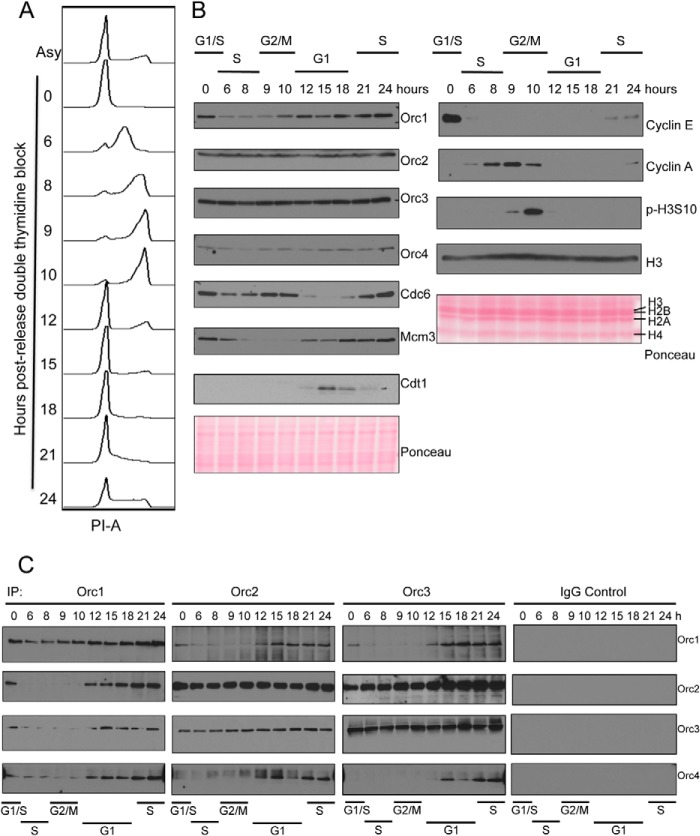
Unlike yeast cells, human cells have a dynamic ORC cycle throughout the cell division cycle. Formation of ORC begins with the binding of Orc1 to chromatin, which is followed by recruitment of other ORC subunits during early G1 phase. As the cells enter S phase, the Orc1 subunit is ubiquitinated and targeted for degradation, and ORC becomes dispensable for DNA replication (22–24, 26). We studied the dynamics of ORC assembly and chromatin binding along with other pre-RC components in double thymidine blocked and released HeLa suspension cells. Because ORC is bound to chromatin tightly, especially Orc1, we have adapted a nuclear fractionation method that effectively solubilizes ORC in nuclei by digesting both RNA and DNA to near completion. HeLa cells were synchronized at the G1/S border using a double thymidine block and released, and the cell cycle profile was analyzed by flow cytometry of DNA content (Fig. 2A). The cells started entering S phase as they were released from the block as seen by an immediate decrease in cyclin E levels (Fig. 2B). Cyclin A levels accumulated as the cells progressed through S phase and entered G2 phase. After 9 h, cells started to become mitotic as evidenced by the appearance of phospho-H3S10 on mitotic chromosomes. Levels of Cdc6 in nuclei fluctuate during the cell cycle, and it has been shown that Cdc6 is targeted for degradation by APC-CDH1-mediated proteolysis during early G1 (55), also Cdc6 phosphorylation by cyclin A/CDK2 leads to its re-localization to cytoplasm during S phase (56). Our results show that Cdc6 levels were low during S and early G1 phase in nuclei (Fig. 2B) consistent with previous observations. Our results also demonstrated that the levels of Orc1 dramatically decreased as the cells entered S phase, although the levels of other ORC subunits, including Orc2, Orc3, and Orc4, remained fairly constant. Orc1 levels started to increase as the cells entered the mitotic phase. Mcm2–7 exists as chromatin-bound and unbound forms in human cells, and the chromatin-bound form is known to be dissociated as cells progress through S phase (57), whereas the chromatin bound Mcm2–7 levels increase during G1 (58). Our results were consistent because Mcm3 that was tightly associated with nuclei decreased during S phase, and the levels increased as the cells progressed through G1 (Fig. 2B).
FIGURE 2.
Cell cycle dynamics of ORC assembly. A, flow cytometry profile of DNA content of double thymidine blocked and released HeLa suspension cells. Cell cycle profile of asynchronous and synchronized cells at the indicated hours after release are shown; x axis shows propidium iodide-area levels, and y axis shows cell count at different time points after release. B, expression levels of pre-RC proteins and cell cycle markers in synchronized HeLa nuclear extracts were studied by Western blot. Ponceau S-stained blots are shown as loading controls. C, immunoprecipitation of Orc1, Orc2, Orc3, or control IgG from double thymidine block and release synchronized HeLa S3 nuclear extracts. Co-immunoprecipitation of Orc1, Orc2, Orc3, and Orc4 were investigated by Western blots as indicated.
We then performed immunoprecipitations using synchronized HeLa cell nuclear extracts with monoclonal antibodies against Orc1, Orc2 and Orc3 or a control IgG (Fig. 2C). The immunoprecipitated proteins were detected with specific antibodies. The results show that the amount of Orc1 immunoprecipitated with Orc1 antibody decreased in S phase cells, concomitant with a decrease in levels of Orc1 during that time. Similarly, when either Orc2 or Orc3 antibodies were used for immunoprecipitation, the amount of Orc1 that was co-immunoprecipitated was lower in S phase cells; the interaction was detectable by the beginning of G1, which corresponds to the 12-h time point. Orc1 was not bound to Orc2 or Orc3 during mitosis. Orc2 and Orc3 interacted with each other throughout the cell cycle, consistent with previous reports (24, 52, 59). However, Orc4 was found to be associated with Orc2 and Orc3 only when Orc1 was also present in the complex, and the interaction was lost when Orc1 was degraded during S phase. These results suggest that recruitment of Orc4 to the full ORC was stabilized and/or promoted in the presence of Orc1, consistent with earlier reports on assembly of ORC both in vitro and in vivo (24, 26).
Mapping of an Orc1 Domain Required for Association with Mitotic Chromosomes
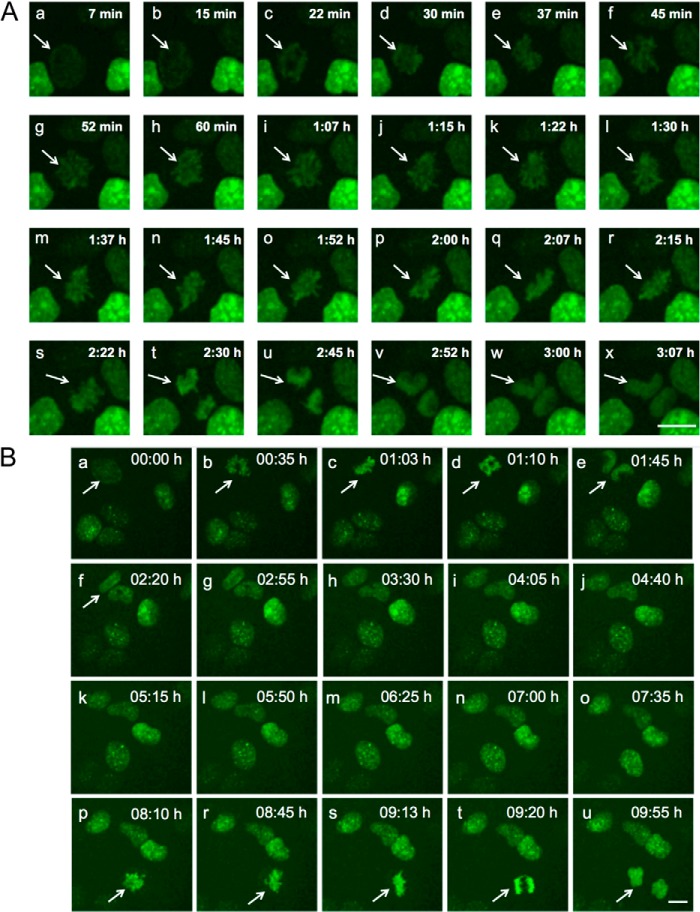
We investigated the dynamics of Orc1 chromatin localization using a tetracycline-inducible GFP-Orc1 present in a U2OS-derived cell line with live cell imaging. Orc1 levels build up as cells prepared to enter into mitosis, and the Orc1 protein was loaded onto chromatin during prophase, simultaneous with chromatin condensation (Fig. 3, A, panels a–s, and B, panels a–u, and supplemental movie 1). Orc1 was observed on chromatin throughout mitosis, and after the cells underwent telophase, the Orc1 signal became diffuse as the nuclear membrane formed in the daughter cells, and chromatin decondensation occurred (Fig. 3, A, panels t–x, and B, panel u). Mitotic association of Orc1 was also seen in YFP-Orc1 U2OS stable cells and GFP-Orc1 HeLa cells (results not shown). Unlike our observations, a previous study observed that Orc1 only localized to telophase chromosomes and was diffusely located in prophase, metaphase, and anaphase cells and that the BAH domain of Orc1 facilitated chromosome association in telophase (60). However, we note that immunofluorescence labeling of formalin-fixed cells was used in that study to examine Orc1 chromosome association, although in this study GFP-Orc1-expressing live cells were used. The expression levels of GFP-Orc1 were comparable with that of endogenous Orc1 (49). Thus, it is possible in the earlier study that the method of fixing cells before antibody staining and/or the methods for cell permeabilization might have caused dissociation of Orc1 from chromosomes in early mitosis or the antibody failed to recognize tagged Orc1 during early mitosis due to possible structural hindrance. An earlier study in Chinese hamster ovary (CHO) cells also showed mitotic association of GFP-CgOrc1 in live cells, although paraformaldehyde fixation prevented the detection of Orc1 binding to mitotic chromosomes (39).
FIGURE 3.
Orc1 associates with mitotic chromosomes. Tetracycline-inducible GFP-Orc1 U2OS cell line was followed by time-lapse live cell imaging through mitosis. Highlighted by an arrow is a cell followed through mitosis (panels a–x in A and panels a–u in B) at indicated time points (min means minutes, and h means hours). Orc1 is loaded onto chromatin during prophase and stays on chromatin throughout mitosis, after telophase Orc1 becomes diffusely distributed in daughter nuclei (panels v–x). Scale bar denotes 5 μm. A and B represent images from two separate videos.
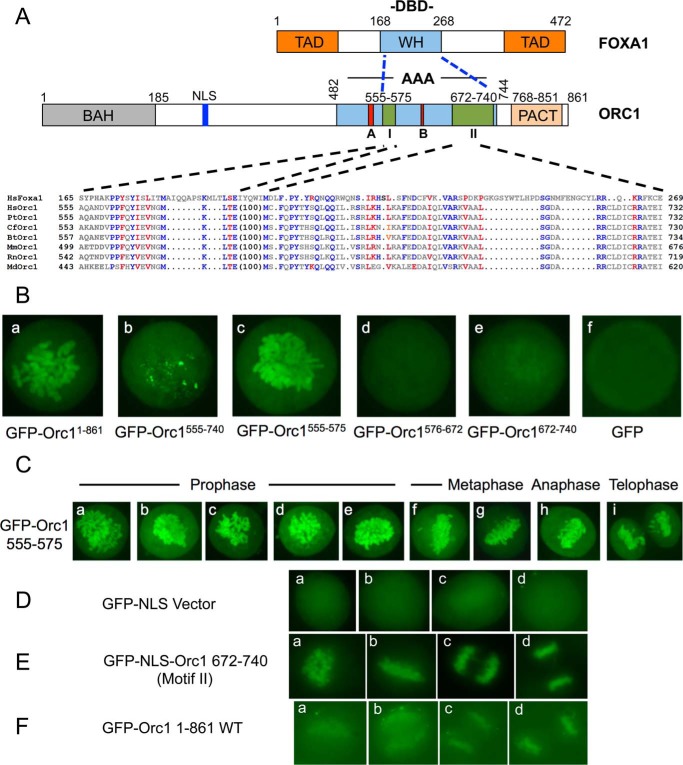
The region of Orc1 involved in mitotic chromosome association was investigated. We noticed that human Orc1 shares partial sequence similarity with the human FOXA1. FOXA1 is a pioneer transcription factor that binds to mitotic chromosomes and enables other factors to bind (61, 62). The DBD of FOXA1 protein binds specific DNA and is required for its localization to mitotic chromosomes only when a nuclear localization signal is present (63, 64). Interestingly, we have found a sequence in Orc1 that is similar to the chromatin-binding domain of the FOXA1 protein. The sequence is conserved among vertebrate Orc1 proteins and is split into two motifs by an intervening stretch in Orc1 of 100 amino acids (Fig. 4A). The motifs are buried inside the AAA+ domain of Orc1, which is involved in its ATPase activity, DNA binding, and DNA replication initiation functions in yeast, Drosophila, and human Orc1 proteins (65–68). Studies in archaea have demonstrated that Orc1 has DNA binding activity that is mediated by the C-terminal winged helix domain linked to its AAA+ domain (66). Thus it is possible that this region in Orc1 is responsible for chromatin or DNA binding. We cloned Orc1 motif I (Orc1(555–575)), Motif II (Orc1(672–740)), the intervening sequence (Orc1(575–672)), and a fragment spanning the whole region (Orc1(555–740)) into separate GFP tag vectors and checked mitotic chromosome association in live U2OS cells; however, none of the fragments were associated with mitotic chromosomes (results not shown). Orc1 has a nuclear localization signal (NLS) near its N terminus (Fig. 4A); thus, in the expressed fragments NLS was not present. To circumvent the issue, we used an NLS-containing vector for expression of these C-terminal Orc1 fragments, just as was done for FOXA1. Using live cell imaging, the results showed that in the presence of NLS, motif I was sufficient for mitotic chromosome association (Fig. 4B, panel c) similar to full-length Orc1 (Fig. 4B, panel a), although motif II caused weak but nonetheless obvious association with mitotic chromosomes (Fig. 4B, panel e). However, when cells were fixed in methanol and stained by indirect immunofluorescence with anti-GFP antibodies, both full-length Orc1 and motif II, but not GFP alone, were readily observed to bind mitotic chromatin (Fig. 4, D–F). By live cell imaging, the Orc1(575–672) fragment consisting of the intervening sequence did not associate with mitotic chromosomes, and it did not have any sequence similarity with the FOXA1 DBD (Fig. 4B, panel d). Interestingly, the Orc1(555–740) fragment spanning motif I, the intervening sequence, and motif II did not show localization to the entire genome; instead, it appeared spotty on chromosomes (Fig. 4B, panel b). The inability of this fragment to localize to mitotic chromosomes might be due to abnormal secondary structure of the fragment, but in full-length Orc1, the intervening sequence is sequestered by other Orc1 sequences. Moreover, in the presence of NLS, motifs I and II individually were sufficient to associate with mitotic chromosomes in prophase, metaphase, anaphase, and telophase (Fig. 4, C, panels a–i, and E), recapitulating the mitotic chromosome association of full-length Orc1.
FIGURE 4.
Orc1 domains associate with mitotic chromosomes. A, domain structures of human FOXA1 and Orc1 proteins. Sequence alignment of FOXA1 DBD and Orc1 is shown. TAD, transactivation domain; WH, winged helix domain; A, Walker A motif; B, Walker B motif; I, motif I; II, motif II. Hs, Homo sapiens (human); Pan troglodytes (chimpanzee); Cf, Canis familiaris (dog); Bt, Bos taurus (bovine); Mm, Mus musculus (mouse); Rt, Rattus norvegicus (rat); Md, Monodelphis domestica (opossum) (100). In Orc1, sequences mean Orc1 protein sequences have 100 amino acid intervening residues. It suggests that Orc1 contains two motifs separated by 100 amino acid residues. Blue residues represent identity between FOXA1 and Orc1, and red residues represent similarity. B, localization of GFP-Orc1 or different GFP-NLS-Orc1 fragments in mitotic U2OS cells as indicated. U2OS cells were transiently transfected, and live cell images were captured. Each of the constructs in panels b–f have an NLS present. C, localization of GFP-NLS-Orc1(555–575) (motif I) during mitosis using live cell imaging of transfected U2OS cells. D, localization of GFP-NLS in methanol-fixed U2OS cells. Following transfections are with a plasmid expressing the protein: E same as D, except GFP-NLS-Orc1(672–740) (motif II). F same as D except GFP-Orc1(1–861).
Orc1 Shows Extensive Mitotic Chromosome Association Prior to Orc2 and Knockdown of Orc1 Causes Loss of Mcm2–7 from Chromatin
It is a possibility that binding of Orc1 to the chromatin during mitosis creates a nucleating center for assembly of ORC and then full pre-RC assembly and defines the locations within chromosomes that could serve as origins of DNA replication. During mitotic exit, Orc2 was distributed diffusely in the cell, although Orc1 was associated with chromatin in telophase (Fig. 5A, panel a). Orc1 appeared in the daughter nuclei prior to Orc2, suggesting that Orc1 chromatin association is one of the first events in pre-RC assembly. After mitosis, both proteins appeared in the nucleus during G1 (Fig. 5A, panel b). A previous study demonstrated that during mitosis, Orc2 and Orc3 chromosome interactions were restricted to centromeres and did not show extensive mitotic chromosome association (52). It is known that depletion of Orc1 results in reduced chromatin association of Orc2 (26). Mcm2–7 is also known to associate with chromatin during very late M and during the G1 phase (58). These results indicate that Orc1 is the first ORC subunit to bind to chromatin extensively during mitosis and that it facilitates recruitment of other ORC subunits to chromatin.
FIGURE 5.
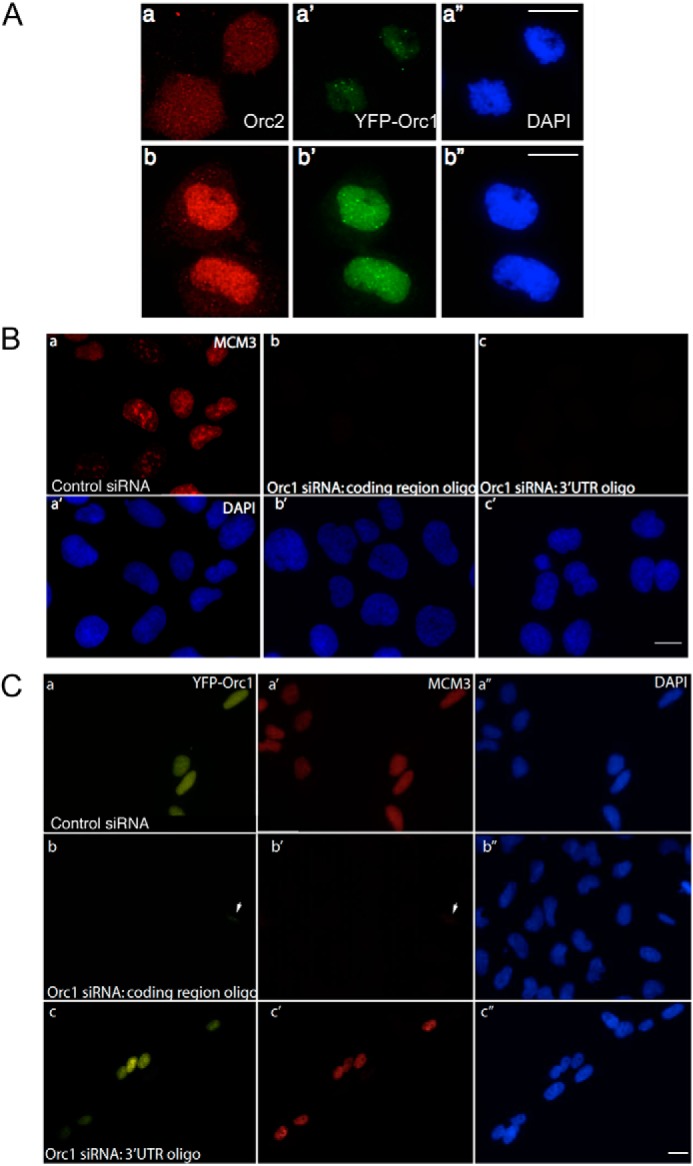
Orc1 shows extensive mitotic chromosome association before Orc2, and depletion of Orc1 causes loss of Mcm3 from chromatin. A, YFP-Orc1 localization relative to Orc2 localization in U2OS cells. In late mitosis YFP-Orc1 appears on chromatin in the daughter nuclei prior to Orc2, which remains in the cytoplasm (panels a–a″), and then Orc2 localizes to nuclei after telophase (panels b–b″). Orc2 was detected using the pAb205 antibody. Scale bar equals 5 μm. B, prominent MCM3 staining (red) on chromatin in control (luciferase) siRNA-treated cells (panel a) as compared with loss of MCM3 staining in Orc1 siRNA-treated U2OS cells using siRNA targeting the coding region (panel b) or the 3′UTR of endogenous Orc1 (panel c). DNA was stained with DAPI. Scale bar represents 5 μm. C, labeling of MCM3 on chromatin in control (luciferase siRNA)-treated U2OS cells stably expressing YFP-Orc1 (panels a and a′). YFP-Orc1 cell line treated with Orc1 siRNA targeting the coding region of endogenous and exogenous Orc1 shows loss of MCM3 from chromatin (panels b and b′). The arrowhead denotes a cell showing partial knockdown of Orc1 (as seen by YFP-Orc1) showing low levels of MCM3 loading. YFP-Orc1 cell line treated with Orc1 siRNA targeting the 3′UTR of endogenous Orc1 (panels c and c′). Note the rescue of MCM3 loading onto chromatin showing that siRNA-resistant exogenous YFP-Orc1 can functionally complement endogenous Orc1.
To address the function of Orc1 in pre-RC recruitment, an siRNA approach was used to knock down Orc1. siRNA oligonucleotides targeted against either the coding region of Orc1 or the 3′UTR of Orc1 were used in addition to a control nontargeting siRNA targeted against a non-human cell protein luciferase. More than 85% knockdown of Orc1 was achieved in HeLa and U2OS cells using oligonucleotides targeting either the coding region or the 3′UTR of endogenous Orc1 (data not shown). In U2OS cells, depletion of Orc1 either with siRNA against the coding region of Orc1 or the 3′UTR of endogenous Orc1 resulted in loss of Mcm3 from chromatin (Fig. 5B), suggesting loading of Mcm3 protein on chromatin requires Orc1, consistent with earlier findings (26, 69).
To address whether an siRNA-resistant YFP-Orc1 could functionally complement the loss of endogenous Orc1, U2OS cells stably expressing a YFP-Orc1 that was resistant to the siRNA that targeted the 3′UTR of Orc1 were treated with siRNAs. In these cells only depletion of endogenous Orc1 but not YFP-Orc1 was observed (data not shown). Mcm3 loading was normal when the siRNA targeting the 3′UTR was used, suggesting YFP-Orc1 could functionally complement endogenous Orc1 and rescue the MCM3 (minichromosome maintenance complex 3) loading defect (Fig. 5C, panels c–c″). In cells treated with control siRNA, Orc1, and Mcm3, chromatin loading was normal (Fig. 5C, panels a–a″). However, in cells treated with siRNA against coding region of Orc1, depletion of both endogenous Orc1 and YFP-Orc1 resulted in the loss of Mcm3 protein from the chromatin (Fig. 5C, panels b–b″). These results demonstrate that YFP-Orc1 behaves like endogenous Orc1 in terms of its cell cycle degradation (supplemental movie 2) and its ability to load Mcm2–7 onto the chromatin.
Orc1 Shows Dynamic Nuclear Localization in Human Cells
In human cells, biochemical studies have demonstrated that the binding of Orc1 protein to chromatin is lost during the G1/S transition, and Orc1 is degraded as cells enter S phase (21, 22, 26). We confirmed these observations using an in vivo imaging approach to study the dynamics of human Orc1 in living cells. Fluorescent-tagged human Orc1 (EYFP-Orc1 C1, Orc1-EYFP N1, or GFP-Orc1) was used either for transient transfection experiments in a variety of cells or to generate stable cell lines in human U2OS cells. Although we confirmed that Orc1 was degraded at the G1-S phase transition, the results also revealed an unexpected pattern of nuclear localization of GFP-Orc1 in cervical cancer HeLa cells (Fig. 6A) or YFP-Orc1 in mammary epithelial MCF7 cells (Fig. 6B) and osteosarcoma U2OS cells (results not shown). In a subset of Orc1-expressing cells, Orc1 showed punctate nuclear staining throughout the nucleus in early G1 phase (Fig. 6, A, panel a and B, panels a–c), whereas other cells that were later in G1 phase showed that Orc1 was restricted to large and discrete foci, some of which surrounded nucleoli (Fig. 6, A, panels b–i, and B, panels d–i). Both YFP-Orc1 and Orc1-YFP showed a similar distribution and were readily incorporated in the ORC complex (data not shown, but see Fig 1, C and D). The distribution of GFP-Orc1 in live HeLa cells fell into different categories, including punctate throughout the nucleus (Fig. 6A, panels a–c), perinuclear, perinucleolar, and/or intranucleolar regions (Fig. 6A, panels d–g), or a smaller number of large irregularly sized foci at the nuclear periphery or in interior nuclear regions (Fig. 6B, panels h and i). Interestingly, these patterns formed by Orc1 during G1 phase show resemblance to DNA replication patterns seen in S phase, which can be categorized into three main patterns in their order of appearance as follows: small discrete foci scattered throughout the nucleus; the nuclear and nucleolar periphery and nucleolar interior; and a smaller number of heterogeneously sized foci (31, 34, 70–73). Time-lapse live imaging studies using fluorescently tagged YFP-Orc1 in human MCF7 (Fig. 6B and supplemental movies 2 and 3) revealed its cell cycle regulated distribution. Orc1 first showed a punctate distribution throughout the nucleus in early G1 phase (Fig. 6B, panels a–c), but it then was found at larger foci around the nucleoli (Fig. 6B, panels d–j) and the nuclear periphery in late G1 phase, reminiscent of the pattern of DNA replication in S phase (supplemental movies 2 and 3). By 10–14 h after the YFP-Orc1 pattern was predominantly punctate throughout the nucleus, Orc1 almost disappeared from the nucleus (Fig. 6B, panel l). Thus, in human cell lines studied, Orc1 showed dynamic spatiotemporal patterns, after which it was degraded.
FIGURE 6.
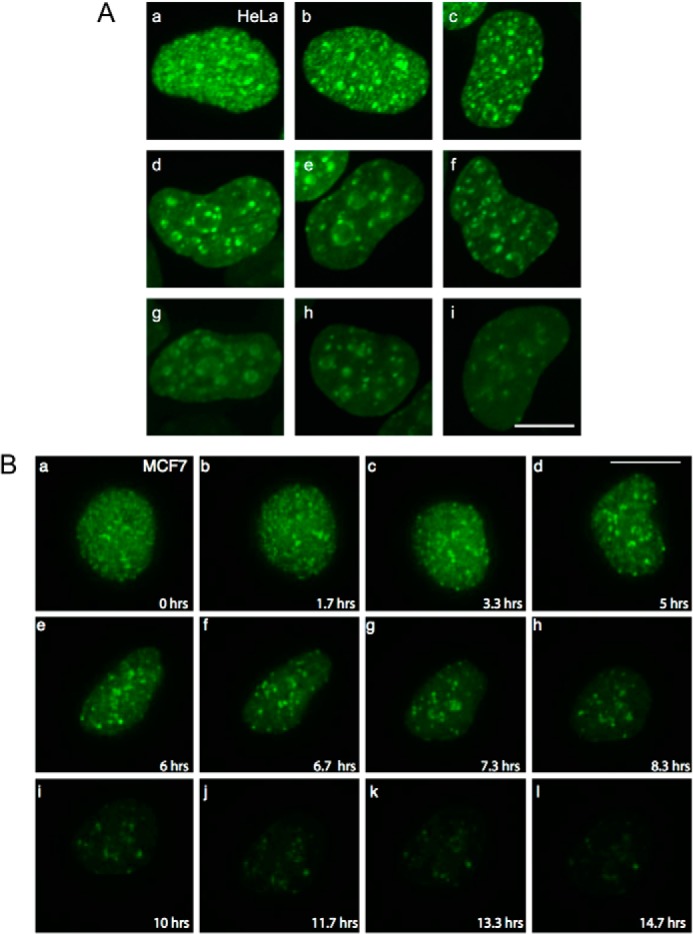
Orc1 shows differential patterning in human cells. A, GFP-Orc1 localization in HeLa cells. Cells were transiently transfected with GFP-Orc1, and localization pattern was investigated in live HeLa cells. Image stacks were collected and deconvolved. Scale bar denotes 7 μm. B, visualization of YFP-Orc1 in living cells (MCF7 cells). Image stacks of cells expressing YFP-Orc1 were collected and deconvolved (from supplemental movie 3). Scale bar is equal to 10 μm. Note the change in the Orc1 pattern, initially appearing as uniformly distributed punctate spots (panels a–c) and later relocalizing to specific large foci. Images were captured every 20 min for ∼15 h.
Spatiotemporal Patterning of Orc1 Protein Occurs during G1
Even though Mcm3 and PCNA proteins are present in all cells throughout the cell division cycle, when asynchronously growing MCF7 cells were extracted with detergent and fixed with methanol, only chromatin bound Mcm3 or PCNA was detected by immunofluorescence (Fig. 7). Chromatin-bound Mcm3 was detected in cells in G1 phase or S phase but not in G2 cells, whereas chromatin-bound PCNA was detected only in the S phase. Thus, combining these two immunofluorescence patterns enables cells to be uniquely assigned to G1, S phase, or G2 phase. We expressed YFP-Orc1 in MCF7 cells and performed triple labeling for DNA replication proteins or dual color live cell imaging.
FIGURE 7.
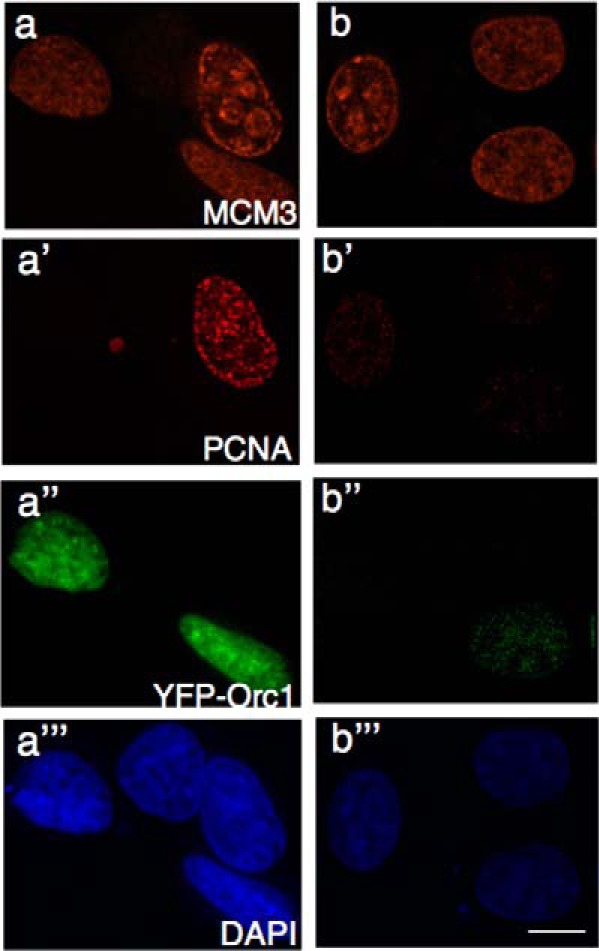
Spatial patterning of Orc1 occurs during G1 phase. Asynchronously growing MCF7 cells expressing YFP-Orc1 were fixed and immunostained for MCM3 (orange, a and b) or PCNA (red, a′, and b′) or visualized for YFP-Orc1 (green, a″ and b″). Nuclei were detected by DAPI straining (blue, a‴, and b‴). Orc1 is present only in G1 cells (MCM3-positive and PCNA-negative). Scale bar denotes 5 μm.
Immunolabeling of Mcm3 and PCNA in YFP-Orc1 expressing MCF7 cells showed that Orc1 was present only in cells that were PCNA-negative and had a homogeneous Mcm3 distribution, indicating G1 cells (Fig. 7, a–a‴). In cells where very faint PCNA staining was observed, indicating very early S phase, Orc1 staining was also very weak (Fig. 7, b–b‴). Live cell imaging in cells expressing YFP-Orc1 and CFP-PCNA showed that the disappearance of Orc1 was concomitant with the appearance of PCNA on chromatin (supplemental movie 4). These results suggest that temporal patterns formed by Orc1 are restricted to G1 nuclei only. However, it is still possible that a small fraction of Orc1 remains bound to chromatin beyond the S phase. These observations support the earlier findings that Orc1 levels are cell cycle-regulated and degraded at the G1/S transition (22, 23).
Orc1 Localization in G1 Phase Overlaps with Temporal Patterning of DNA Replication in S Phase
The finding that the spatiotemporal dynamics of Orc1 during G1 phase were similar to some of the global DNA replication patterns, particularly the late replication patterns seen in S phase, raised the question whether Orc1 temporal dynamics might reflect temporal replication patterns. Specifically, the Orc1 patterns late in G1 phase resemble the late replication patterns of DNA replication foci observed in S phase. Previous studies show that, in mammalian cells, the spatiotemporal patterns of DNA replication are inherited from mother nuclei to daughter nuclei (30–32). Therefore, we investigated whether the Orc1 replication-like patterns in G1 phase in daughter nuclei coincided with the S phase patterns of DNA replication from the previous cell cycle. To investigate this possibility, U2OS cells stably expressing YFP-Orc1 were pulse-labeled with Alexa dUTP 594 (∼15-min pulse by transfection of the nucleoside triphosphate) in the S phase of generation 1, followed by three washes to remove nucleotides; then the cells were allowed to go through mitosis and enter G1 of the next generation and were monitored by live cell imaging (Fig. 8A). Imaging for both Alexa dUTP 594 (red) and YFP-Orc1 (green) patterns revealed overlap of the patterns in 6% of the G1 phase cells (Fig. 8B, panels a and b). In other words, DNA replication patterns from the previous generation showed overlap with Orc1 patterns in the G1 phase of the next generation. Similar results were obtained when cells were labeled with BrdU in S phase and allowed to go through mitosis, and the YFP-Orc1 pattern was investigated in fixed cells (Fig. 8C).
FIGURE 8.
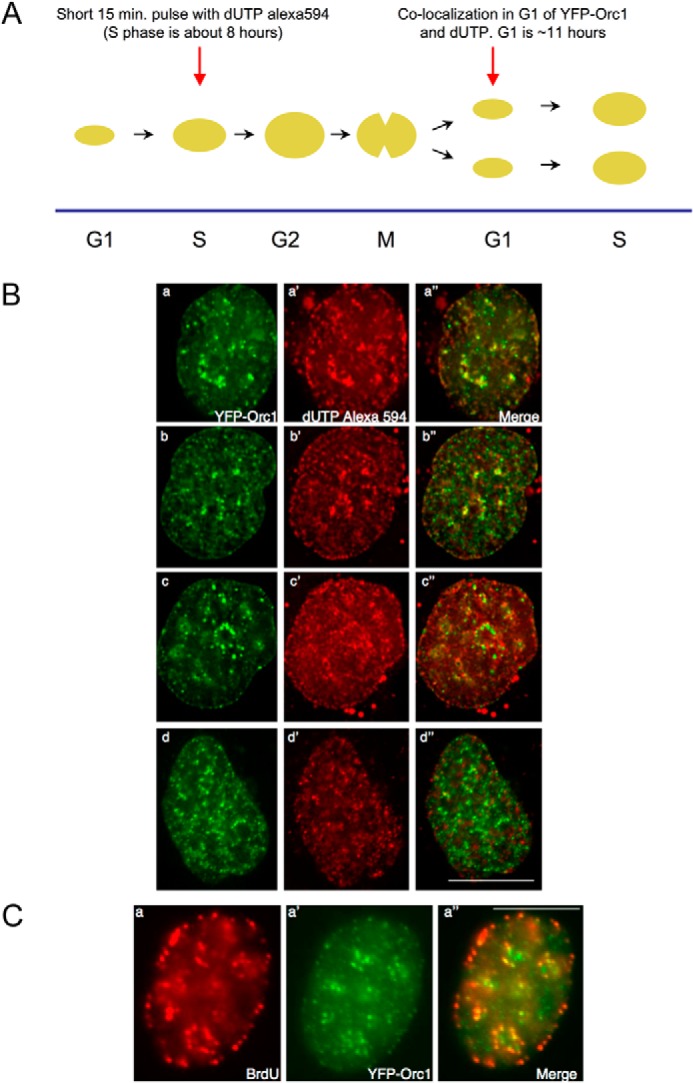
Orc1 localization in G1 phase overlaps with an inherited spatiotemporal pattern of DNA replication. A, scheme for the experiment. Human U2OS cells expressing YFP-Orc1 (green) were transfected for 15 min with dUTP Alexa 594 (red) (see “Experimental Procedures”). Following washing out the label and incubating for ∼12–14 h (ensuring the cells progress from S phase in generation 1 to G1 of generation 2), live cells were imaged every 5 min using a Delta Vision optical sectioning deconvolution instrument (Applied Precision) on a Olympus microscope with a ×63 1.4NA objective. B, top two panels represent significant overlap of YFP-Orc1 with the dUTP Alexa 594, both in nuclear and nucleolar periphery (merge is panels a and b). YFP-Orc1 and dUTP Alexa 594 do not show any overlap but are immediately adjacent to each other (panels c–c″), and (panels d–d″) show the expected most common situation of no overlap between YFP-Orc1 and dUTP Alexa 594. Scale bar denotes 5 μm. Overlap was seen in 6% of all YFP-Orc1-positive cells. C, cells expressing YFP-Orc1 were incubated for 10 min with BrdU, following which BrdU was extensively washed out. Following ∼12–14 h incubation at 37 °C (ensuring the cells enter from S phase in generation 1 to G1 of generation 2), cells were fixed in 2% formaldehyde. BrdU was detected using anti-BrdU mouse mAb (red) in YFP-Orc1 (green)-positive cells. Orc1 was observed both in the nuclear and nucleolar periphery. In a population of cells Orc1 showed significant overlap with BrdU-positive regions. Scale bar denotes 5 μm.
To determine the significance of this overlap, the YFP-Orc1 spots and the Alexa dUTP 594 spots were scanned using a vision optical sectioning deconvolution instrument (Applied Precision) on an Olympus microscope with a 63 × 1.4NA objective. Overlap, based on a Pearson coefficient of correlation, was binned into two classes (high overlap of >0.70 and <0.70). Only 6% of the cells had multiple spots with a high coefficient of correlation, and these spots tended to be larger foci, reminiscent of late replicating foci.
The overlap of YFP-Orc1 and the replication pattern were most evident in late S phase patterns. In some cells, the YFP-Orc1 pattern did not overlap with the Alexa dUTP pattern; instead, the patterns were adjacent to each other (Fig. 8B, panel c). In most of the cells, no co-localization between YFP-Orc1 and DNA replication pattern was observed (Fig. 8B, panel d); however, this result was expected because matching a snapshot of a 15-min S phase pattern that lasts about 6 h to a snapshot of a YFP-Orc1 pattern in G1 that changes over the course of a much longer G1 has very low probability.
DISCUSSION
The initiation of DNA replication involves the ordered and highly regulated assembly on chromatin of a pre-RC that requires ORC, Cdc6, and Cdt1 to load the hexameric Mcm2–7 complex at all potential replication origins (2, 12, 74–76). In human cells, Orc2–5 forms the core ORC, and Orc1 is known to be transiently associated with this complex (12, 22, 24, 77, 78). Orc1 associated with chromatin beginning in early mitosis and is the first among pre-RC components to bind to chromatin. Orc1 levels gradually increased as the cells progressed through mitosis, and during G1 phase the highest levels of Orc1 on chromatin were present. Levels of other ORC subunits, including Orc2, Orc3, and Orc4, were fairly constant throughout the cell division cycle; and interactions between Orc1 and Orc2, Orc1 and Orc3, Orc1 and Orc4, Orc2 and Orc4, and Orc3 with Orc4 were detectable by the beginning of G1. Association of Orc4 with the complex occurred only when Orc1 was also present suggesting that the recruitment of Orc4 to ORC was facilitated or stabilized by Orc1, consistent with earlier reports (24, 26). Interestingly, even though Orc1 was present at earlier time points during mitosis, the interaction between Orc1 with Orc2 and Orc3 or Orc4 occurred only in G1 phase, suggesting a controlled assembly of ORC.
Live cell imaging studies showed that Orc1 associated with mitotic chromosomes extensively throughout mitosis. The sequence similarity between Orc1 and the DBD of the pioneer transcription factor FOXA1, coupled with the observation that motifs 1 and II of this conserved sequence were individually sufficient for mitotic chromosome binding when NLS was present, suggests a mechanism for Orc1 binding to mitotic chromosomes. FOXA1 is a well characterized pioneer factor, which is a transcription factor that can bind to silent chromatin and facilitate other factors to bind (62). FOXA proteins have the ability to bind compacted chromatin, mediated through a high affinity DNA-binding site and interaction with histones H3 and H4 (61). FOXA1 is a member of the FOXA subfamily of proteins, and it has been recently shown that FOXA1 shows slow recovery kinetics in interphase nuclei by fluorescence recovery after photobleaching compared with mitotic cells and has more extensive chromatin binding during mitosis compared with interphase cells (63). Interestingly, some of these characteristics are similar to Orc1, which also shows extensive mitotic chromosome binding, slow recovery kinetics in interphase nuclei (54), and interaction with histones through its BAH domain (79). Another important feature of FOXA pioneer factors is the resemblance of their DBD to the linker histone DBD. There is structural similarity between the winged helix motif DBDs of FOXA and the linker histone 5 (80). FOXA and linker histone bind along the minor grooves of DNA through their winged helix motifs (80, 81). Interestingly, the bacterial initiator protein DnaA and archaeal Cdc6/Orc1 both have a winged helix motif linked to their AAA+ domain (66, 82, 83). Orc1 is a member of the AAA+ family of ATPases and contains Walker A and Walker B motifs and a BAH domain mediating protein-protein interactions (60). Binding of ATP to Orc1 is essential for the ATPase, DNA binding, and replication initiation activities of the yeast, Drosophila, and human ORC (65–68). The stretch of amino acids in Orc1 (motif I-NLS) that has sequence similarity with the FOXA1 DBD is sufficient for mitotic chromosome association. Interestingly, this motif is buried in the AAA+ domain of Orc1 and contains the box IV motif of the AAA+ domain (84, 85). Box IV is a weakly conserved motif compared with Walker A and Walker B motifs, which are important for ATP binding in AAA+ proteins. Thus, motif I plays a dual role in Orc1, for mitotic chromosome association, and as part of the AAA+ module. Motif II, however, is present in a region of the AAA+ domain of Orc1 that protrudes out of the Orc1 structure present in a recent crystal structure of ORC from Drosophila (86). The structure of this Drosophila ORC, particularly Orc1, is in a conformation that would not allow ATPase activity and thus is most likely not the conformation of ORC that recruits Cdc6 and pre-RC proteins. We suggest that one reason Drosophila ORC may exist in this conformation is to allow recruitment to chromatin, and only later does Orc1 move to be adjacent to Orc4 for pre-RC assembly. Of course, Orc1 in human cells is different from Orc1 in Drosophila because is not associated with the other ORC subunits during early mitosis. Based on these new observations, we suggest that human cell Orc1 binds to mitotic chromosomes and recruits other proteins, including other ORC subunits in the next G1 phase in preparation for initiation of DNA replication. Unlike true pioneer transcription factors, however, we do not think that Orc1 binds specific DNA sequences.
Orc1 shows extensive mitotic chromosome association before Orc2, although it is known that Orc2 only shows restricted centromeric association during mitosis (52). In Drosophila, binding of Orc2 to mitotic chromosomes post-anaphase is dependent on cessation of mitotic CDK activity (87). These results are consistent with Orc1 being the first determinant for chromatin binding and origin recognition. This finding is consistent with the evolutionary perspective that in archaea there is only Orc1- and Cdc6-like proteins for origin recognition, and the other ORC subunits do not exist (66, 88), although we do not suggest that human Orc1 binding to mitotic chromosomes is sequence-specific. We also demonstrated that Orc1 recruits other components of ORC and the pre-RC to chromatin only during G1 phase. A previous study in CHO cells also observed mitotic chromosome association of CgOrc1 as well as CgOrc4 (39); however, it must be noted that Orc1 shows different cell cycle regulation in these cells compared with human cells. In CHO cells, CgOrc1 is not degraded and stays associated with chromosomes throughout the cell cycle (39); binding of CgOrc1 is weaker during mitosis and stronger in G1 (89), and inhibition of CDK activity is necessary for CgOrc1 binding to mitotic chromosomes (90). We suggest that the difference between the CHO results and our results with human cells reflects real species-specific differences and not cell line to cell line variation because we have observed similar Orc1 dynamics in multiple human cells lines, including diploid cells.
Once human cells enter into the G1 phase, the nuclear patterns displayed by Orc1 show similarity to spatiotemporal patterns of DNA replication during S phase, particularly the patterns late in G1 phase that resemble late replication patterns. DNA replication shows a global level of spatiotemporal patterning throughout S phase that can be categorized into early, mid, and late S phase patterns based on the distribution of DNA replication foci (31, 34, 70, 72, 73). Replication foci are discrete sub-nuclear sites where multiprotein complexes involved in DNA replication assemble at replication forks (91). Replication patterns can be visualized by fluorescent-tagged nucleotide analogs or proteins localized to the replication fork such as PCNA (91–93). Patterning of Mcm2–7 proteins occurs from G1 to the S phase of the cell cycle (94).
The spatiotemporal regulation of DNA replication patterns suggests that clusters of replication origins within any given domain of the chromosome are activated during a specific time point in S phase. In mammalian cells replication domains are formed by segments of chromatin with different replication timing profiles (29). Whole genome studies of replication timing in mammalian cells have shown that the timing of DNA replication correlates with the organization and proximity of these regions, as revealed by chromosome conformation capture technologies, suggesting that early and late replication takes place in spatially separate nuclear compartments (40–43). We suggest that the dynamic patterns of Orc1, particularly the patterns notable during late G1 phase, reflect the heritable patterns of DNA replication found in late S phase. The spatial overlap between BrdU patterns in S phase and Orc1 patterns in G1 phase also suggests that replication timing of various chromosomal domains is predetermined in G1 phase.
Several factors are involved in governing spatial-temporal regulation of DNA replication, including chromatin structure and modifications, rate-limiting DNA replication factors, and chromosomal position and organization (45, 95). Selection of new replication sites occurs in each cycle at a specific time point in G1, called the origin decision point (96). At a later timing decision point during G1, the global temporal order of DNA replication program is predetermined (34, 35). G2 nuclei lack these determinants suggesting that they need to be established every cell cycle for proper spatiotemporal regulation of DNA replication (97). We suggest that Orc1 binding to mitotic chromosomes sets these events in place.
The patterning of Orc1 during G1 phase could be due to two different mechanisms. Orc1 may bind to all chromatin during mitosis and be removed and degraded from sites on chromatin as cells progress through G1 phase, first from early replicating domains of chromatin in early G1 phase and later from late-replicating regions of chromatin in late G1 phase. Another possibility is that the dynamic patterns formed by Orc1 in G1 nuclei are dependent on accessibility of chromatin, its three-dimensional chromosomal organization, and epigenetic marks. In early G1, Orc1 may be localized to regions with higher accessibility and forms pre-RC at those sites, and it then dissociates from those sites to assemble pre-RCs at later replicating domains. Accessibility of a particular region on the chromosome can be governed by several factors, including primary DNA sequence, epigenetic modifications on DNA and histones, and the presence of other DNA-binding elements, including transcription factors or chromatin remodeling factors, and insulating elements. Orc1 also shows differential histone binding activity through its BAH domain (79), has affinity to histone H4K20me2, and co-localizes with Orc2 that also co-localizes with histone H3K9me3 (54). In budding yeast, the BAH domain is involved in stable association of ORC with origins and with origin activity (98). In human cells, Orc1 and other ORC subunits have also been shown to be associated with H3K9me3, H3K27me3, and H4K20me3 marks (99–101). Therefore, it is possible that post-translational modifications on histones might be one of the contributing marks that cause the dynamic Orc1 patterning during G1, due to differential affinity of Orc1 to different histone modifications. It is possible that the earlier Orc1 is bound to a particular site in the genome the earlier a pre-RC can form at that site. This is consistent with the finding that in fission yeast timing of ORC binding determines the timing of pre-RC formation and timing of replication (46).
Overall, the results indicate that Orc1 acts as a nucleating center for pre-RC formation by binding to compact chromatin during mitosis and later during G1 phase recruiting components of ORC and pre-RC. ORC associates with chromatin strongly, and particularly, Orc1 shows tight association. Formation of ORC occurs during G1 phase; even though Orc4 is present throughout cell cycle, it only associates with chromatin only when Orc1 is present, suggesting that Orc1 might stabilize or facilitate recruitment of other ORC subunits. It still needs to be determined how the dynamic Orc1 patterns anticipate the spatiotemporal dynamics of DNA replication. The spatial organization of the genome, nuclear architecture, and epigenetic marks might all play a role in dynamic Orc1 binding and spatiotemporal regulation of DNA replication. It is possible that either Orc1 is contributing or reading some inherited pattern present in the chromatin.
Supplementary Material
Acknowledgments
We are grateful to Patricia Wendel for technical assistance and to Carmelita Bautista and Margaret Falkowski for help with the antibody screen and maintenance of cell lines. We thank Stephen Hearn for assistance in microscopic imaging.
This work was supported, in whole or in part, by National Institutes of Health Program Project Grant CA13106 from the NCI and Support Grant CA45508 (to Cold Spring Harbor Laboratory Cancer Center).

This article contains supplemental Movies 1–4.
- pre-RC
- pre-replication complex
- ORC
- origin recognition complex
- PCNA
- proliferating cell nuclear antigen
- NLS
- nuclear localization signal
- CDK
- cyclin-dependent kinase
- DBD
- DNA binding domain
- AAA+
- ATPase associated with diverse cellular activity
- BAH
- bromo-associated homology
- MBP
- maltose-binding protein.
REFERENCES
- 1. O'Donnell M., Langston L., Stillman B. (2013) Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5, a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell S. P., Dutta A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 3. Bell S. D., Botchan M. R. (2013) The minichromosome maintenance replicative helicase. Cold Spring Harb. Perspect. Biol. 5, a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bell S. P., Kaguni J. M. (2013) Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5, a010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78, 303–316 [DOI] [PubMed] [Google Scholar]
- 6. Kong D., DePamphilis M. L. (2001) Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol. 21, 8095–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang C., Stillman B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11, 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lygerou Z., Nurse P. (1999) The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 112, 3703–3712 [DOI] [PubMed] [Google Scholar]
- 9. Nguyen V. Q., Co C., Li J. J. (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 10. Grallert B., Nurse P. (1996) The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 10, 2644–2654 [DOI] [PubMed] [Google Scholar]
- 11. Ogawa Y., Takahashi T., Masukata H. (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 19, 7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DePamphilis M. L. (2003) The 'ORC cycle': a novel pathway for regulating eukaryotic DNA replication. Gene 310, 1–15 [DOI] [PubMed] [Google Scholar]
- 13. Coleman T. R., Carpenter P. B., Dunphy W. G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87, 53–63 [DOI] [PubMed] [Google Scholar]
- 14. Hua X. H., Newport J. (1998) Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 140, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romanowski P., Madine M. A., Laskey R. A. (1996) XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc. Natl. Acad. Sci. U.S.A. 93, 10189–10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rowles A., Tada S., Blow J. J. (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 112, 2011–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun W. H., Coleman T. R., DePamphilis M. L. (2002) Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J. 21, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Asano M., Wharton R. P. (1999) E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 18, 2435–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohtani K., DeGregori J., Leone G., Herendeen D. R., Kelly T. J., Nevins J. R. (1996) Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 16, 6977–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Araki M., Yu H., Asano M. (2005) A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev. 19, 2458–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreitz S., Ritzi M., Baack M., Knippers R. (2001) The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276, 6337–6342 [DOI] [PubMed] [Google Scholar]
- 22. Méndez J., Zou-Yang X. H., Kim S. Y., Hidaka M., Tansey W. P., Stillman B. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9, 481–491 [DOI] [PubMed] [Google Scholar]
- 23. Tatsumi Y., Ohta S., Kimura H., Tsurimoto T., Obuse C. (2003) The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J. Biol. Chem. 278, 41528–41534 [DOI] [PubMed] [Google Scholar]
- 24. Siddiqui K., Stillman B. (2007) ATP dependent assembly of the human origin recognition complex. J. Biol. Chem. 282, 32370-32383 [DOI] [PubMed] [Google Scholar]
- 25. Abdurashidova G., Danailov M. B., Ochem A., Triolo G., Djeliova V., Radulescu S., Vindigni A., Riva S., Falaschi A. (2003) Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 22, 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohta S., Tatsumi Y., Fujita M., Tsurimoto T., Obuse C. (2003) The ORC1 cycle in human cells: II. Dynamic changes in the human ORC complex during the cell cycle. J. Biol. Chem. 278, 41535–41540 [DOI] [PubMed] [Google Scholar]
- 27. Jackson D., Wang X., Rudner D. Z. (2012) Spatio-temporal organization of replication in bacteria and eukaryotes (nucleoids and nuclei). Cold Spring Harb. Perspect. Biol. 4, a010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rhind N., Gilbert D. M. (2013) DNA replication timing. Cold Spring Harb. Perspect. Biol. 3, a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C. W., Lyou Y., Townes T. M., Schübeler D., Gilbert D. M. (2008) Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li F., Chen J., Solessio E., Gilbert D. M. (2003) Spatial distribution and specification of mammalian replication origins during G1 phase. J. Cell Biol. 161, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma H., Samarabandu J., Devdhar R. S., Acharya R., Cheng P. C., Meng C., Berezney R. (1998) Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 143, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sadoni N., Cardoso M. C., Stelzer E. H., Leonhardt H., Zink D. (2004) Stable chromosomal units determine the spatial and temporal organization of DNA replication. J. Cell Sci. 117, 5353–5365 [DOI] [PubMed] [Google Scholar]
- 33. Raghuraman M. K., Brewer B. J., Fangman W. L. (1997) Cell cycle-dependent establishment of a late replication program. Science 276, 806–809 [DOI] [PubMed] [Google Scholar]
- 34. Dimitrova D. S., Gilbert D. M. (1999) The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4, 983–993 [DOI] [PubMed] [Google Scholar]
- 35. Li F., Chen J., Izumi M., Butler M. C., Keezer S. M., Gilbert D. M. (2001) The replication timing program of the Chinese hamster β-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J. Cell Biol. 154, 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gilbert D. M. (2001) Making sense of eukaryotic DNA replication origins. Science 294, 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McNairn A. J., Gilbert D. M. (2003) Epigenomic replication: linking epigenetics to DNA replication. BioEssays 25, 647–656 [DOI] [PubMed] [Google Scholar]
- 38. Wu R., Singh P. B., Gilbert D. M. (2006) Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 174, 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okuno Y., McNairn A. J., den Elzen N., Pines J., Gilbert D. M. (2001) Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20, 4263–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moindrot B., Audit B., Klous P., Baker A., Thermes C., de Laat W., Bouvet P., Mongelard F., Arneodo A. (2012) 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res. 40, 9470–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., Schulz T. C., Robins A. J., Dalton S., Gilbert D. M. (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 20, 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takebayashi S., Ryba T., Gilbert D. M. (2012) Developmental control of replication timing defines a new breed of chromosomal domains with a novel mechanism of chromatin unfolding. Nucleus 3, 500–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yaffe E., Farkash-Amar S., Polten A., Yakhini Z., Tanay A., Simon I. (2010) Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 6, e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaykov A., Nurse P. (2015) The spatial and temporal organization of origin firing during the S phase of fission yeast. Genome Res. 25, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aparicio O. M. (2013) Location, location, location: it's all in the timing for replication origins. Genes Dev. 27, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu P. Y., Nurse P. (2009) Establishing the program of origin firing during S phase in fission yeast. Cell 136, 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacAlpine H. K., Gordân R., Powell S. K., Hartemink A. J., MacAlpine D. M. (2010) Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 49. Hossain M., Stillman B. (2012) Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 26, 1797–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sanders M. M. (1978) Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J. Cell Biol. 79, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- 52. Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23, 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martini E., Roche D. M., Marheineke K., Verreault A., Almouzni G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 143, 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prasanth S. G., Shen Z., Prasanth K. V., Stillman B. (2010) Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc. Natl. Acad. Sci. U.S.A. 107, 15093–15098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petersen B. O., Wagener C., Marinoni F., Kramer E. R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J. M., Helin K. (2000) Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14, 2330–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petersen B. O., Lukas J., Sørensen C. S., Bartek J., Helin K. (1999) Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18, 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fujita M., Yamada C., Tsurumi T., Hanaoka F., Matsuzawa K., Inagaki M. (1998) Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J. Biol. Chem. 273, 17095–17101 [DOI] [PubMed] [Google Scholar]
- 58. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ranjan A., Gossen M. (2006) A structural role for ATP in the formation and stability of the human origin recognition complex. Proc. Natl. Acad. Sci. U.S.A. 103, 4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Noguchi K., Vassilev A., Ghosh S., Yates J. L., DePamphilis M. L. (2006) The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 25, 5372–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., Zaret K. S. (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 [DOI] [PubMed] [Google Scholar]
- 62. Zaret K. S., Carroll J. S. (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Caravaca J. M., Donahue G., Becker J. S., He X., Vinson C., Zaret K. S. (2013) Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 27, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cirillo L. A., Zaret K. S. (2007) Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J. Mol. Biol. 366, 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chesnokov I., Remus D., Botchan M. (2001) Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. U.S.A. 98, 11997–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gaudier M., Schuwirth B. S., Westcott S. L., Wigley D. B. (2007) Structural basis of DNA replication origin recognition by an ORC protein. Science 317, 1213–1216 [DOI] [PubMed] [Google Scholar]
- 67. Giordano-Coltart J., Ying C. Y., Gautier J., Hurwitz J. (2005) Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc. Natl. Acad. Sci. U.S.A. 102, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klemm R. D., Austin R. J., Bell S. P. (1997) Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 88, 493–502 [DOI] [PubMed] [Google Scholar]
- 69. Romanowski P., Madine M. A., Rowles A., Blow J. J., Laskey R. A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6, 1416–1425 [DOI] [PubMed] [Google Scholar]
- 70. Dimitrova D. S., Berezney R. (2002) The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115, 4037–4051 [DOI] [PubMed] [Google Scholar]
- 71. Humbert C., Usson Y. (1992) Eukaryotic DNA replication is a topographically ordered process. Cytometry 13, 603–614 [DOI] [PubMed] [Google Scholar]
- 72. Nakayasu H., Berezney R. (1989) Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 108, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'Keefe R. T., Henderson S. C., Spector D. L. (1992) Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 116, 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kelly T. J., Brown G. W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem. 69, 829–880 [DOI] [PubMed] [Google Scholar]
- 75. Méndez J., Stillman B. (2003) Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. BioEssays 25, 1158–1167 [DOI] [PubMed] [Google Scholar]
- 76. Aparicio O. M., Weinstein D. M., Bell S. P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69 [DOI] [PubMed] [Google Scholar]
- 77. Dhar S. K., Delmolino L., Dutta A. (2001) Architecture of the human origin recognition complex. J. Biol. Chem. 276, 29067–29071 [DOI] [PubMed] [Google Scholar]
- 78. Vashee S., Simancek P., Challberg M. D., Kelly T. J. (2001) Assembly of the human origin recognition complex. J. Biol. Chem. 276, 26666–26673 [DOI] [PubMed] [Google Scholar]
- 79. Kuo A. J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J. K., Patel D. J., Gozani O. (2012) The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Clark K. L., Halay E. D., Lai E., Burley S. K. (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 [DOI] [PubMed] [Google Scholar]
- 81. Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219–223 [DOI] [PubMed] [Google Scholar]
- 82. Erzberger J. P., Pirruccello M. M., Berger J. M. (2002) The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liu J., Smith C. L., DeRyckere D., DeAngelis K., Martin G. S., Berger J. M. (2000) Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell 6, 637–648 [DOI] [PubMed] [Google Scholar]
- 84. Cullmann G., Fien K., Kobayashi R., Stillman B. (1995) Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 4661–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 86. Bleichert F., Botchan M. R., Berger J. M. (2015) Crystal structure of the eukaryotic origin recognition complex. Nature 519, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Baldinger T., Gossen M. (2009) Binding of Drosophila ORC proteins to anaphase chromosomes requires cessation of mitotic cyclin-dependent kinase activity. Mol. Cell. Biol. 29, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dueber E. L., Corn J. E., Bell S. D., Berger J. M. (2007) Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317, 1210–1213 [DOI] [PubMed] [Google Scholar]
- 89. Natale D. A., Li C. J., Sun W. H., DePamphilis M. L. (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 19, 2728–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li C. J., Vassilev A., DePamphilis M. L. (2004) Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex's largest subunit (Orc1) from binding to chromatin during mitosis. Mol. Cell. Biol. 24, 5875–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leonhardt H., Rahn H. P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M. C. (2000) Dynamics of DNA replication factories in living cells. J. Cell Biol. 149, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bravo R., Macdonald-Bravo H. (1987) Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J. Cell Biol. 105, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Celis J. E., Celis A. (1985) Cell cycle-dependent variations in the distribution of the nuclear protein cyclin proliferating cell nuclear antigen in cultured cells: subdivision of S phase. Proc. Natl. Acad. Sci. U.S.A. 82, 3262–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Prasanth S. G., Méndez J., Prasanth K. V., Stillman B. (2004) Dynamics of pre-replication complex proteins during the cell division cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Aladjem M. I. (2007) Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 8, 588–600 [DOI] [PubMed] [Google Scholar]
- 96. Wu J. R., Gilbert D. M. (1997) The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1 phase event that precedes restriction point control. Mol. Cell. Biol. 17, 4312–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lu J., Li F., Murphy C. S., Davidson M. W., Gilbert D. M. (2010) G2 phase chromatin lacks determinants of replication timing. J. Cell Biol. 189, 967–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Müller P., Park S., Shor E., Huebert D. J., Warren C. L., Ansari A. Z., Weinreich M., Eaton M. L., MacAlpine D. M., Fox C. A. (2010) The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 24, 1418–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bartke T., Vermeulen M., Xhemalce B., Robson S. C., Mann M., Kouzarides T. (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 101. Shen Z., Sathyan K. M., Geng Y., Zheng R., Chakraborty A., Freeman B., Wang F., Prasanth K. V., Prasanth S. G. (2010) A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell 40, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.