Background: Human milk lactadherin protects breastfed infants against symptomatic rotavirus infections.
Results: A 20-aa peptide (namely pDGE-RGD) derived from equine lactahderin inhibits human rotavirus infectivity.
Conclusion: pDGE-RGD interacts specifically with α2β1 integrin, thus hindering the rotavirus-cell attachment process.
Significance: The discovery of the pDGE-RGD peptide may prove useful in the development of inhibitors of receptor recognition by rotavirus or other integrin-using viruses.
Keywords: antiviral agent, integrin, peptides, proteomics, RNA virus, donkey, lactadherin, milk, rotavirus
Abstract
Human rotavirus is the leading cause of severe gastroenteritis in infants and children under the age of 5 years in both developed and developing countries. Human lactadherin, a milk fat globule membrane glycoprotein, inhibits human rotavirus infection in vitro, whereas bovine lactadherin is not active. Moreover, it protects breastfed infants against symptomatic rotavirus infections. To explore the potential antiviral activity of lactadherin sourced by equines, we undertook a proteomic analysis of milk fat globule membrane proteins from donkey milk and elucidated its amino acid sequence. Alignment of the human, bovine, and donkey lactadherin sequences revealed the presence of an Asp-Gly-Glu (DGE) α2β1 integrin-binding motif in the N-terminal domain of donkey sequence only. Because integrin α2β1 plays a critical role during early steps of rotavirus host cell adhesion, we tested a minilibrary of donkey lactadherin-derived peptides containing DGE sequence for anti-rotavirus activity. A 20-amino acid peptide containing both DGE and RGD motifs (named pDGE-RGD) showed the greatest activity, and its mechanism of antiviral action was characterized; pDGE-RGD binds to integrin α2β1 by means of the DGE motif and inhibits rotavirus attachment to the cell surface. These findings suggest the potential anti-rotavirus activity of equine lactadherin and support the feasibility of developing an anti-rotavirus peptide that acts by hindering virus-receptor binding.
Introduction
Human rotavirus is a double-stranded RNA virus belonging to the Reoviridae family. It is transmitted by the fecal-oral route, causing severe gastroenteritis in infants and children under the age of 5 years in both developed and developing countries, although mortality occurs mainly in developing countries. As a result of rotavirus infection, 114 million episodes of diarrhea, 25 million clinic visits, 2.4 million hospital admissions, and more than 500,000 deaths in children up to age 5 occur worldwide annually (1). Groups at increased risk for rotavirus infection include children who are hospitalized or in community care centers and undernourished and/or immunodeficient children (2). Moreover, because exclusive breastfeeding was found to be associated with a lower incidence of rotavirus gastroenteritis (3–6), non-exclusively breastfed children are considered an additional group more vulnerable to rotavirus infections. The mature virion is a triple-layered particle of about 100 nm in diameter; the most external layer is composed of two viral proteins (VPs),3 VP7 (∼34 kDa) and VP4 (87 kDa) (7, 8), with VP4 being the major determinant of tropism and receptor binding (9–12). Trimeric spikes of VP4 are anchored into the intermediate VP6 layer, whereas the trimeric calcium-binding protein VP7 covers the virion surface, locking VP4 spikes into place. The proteolytic cleavage of VP4 by trypsin is essential for optimum rotavirus infectivity and produces two subunits, VP5* (60 kDa) and VP8* (28 kDa), which remain associated with the virion (13–15). Initial cell attachment by rotaviruses is mediated by VP8* binding to host cell glycans (16). Infection of permissive cells by many rotaviruses, including human (e.g. Wa and K8), monkey (RRV and SA11), and bovine (NCDV) strains, also depends on virus binding to particular integrins, a family of cell surface proteins that recognize extracellular matrix proteins (e.g. collagen), cell surface ligands (e.g. vascular cell adhesion molecule-1) (17), growth factors (e.g. fibroblast growth factor-1) (18), and viral proteins (e.g. rotavirus). VP5* recognition of the collagen-binding α2β1 integrin is a key event in rotavirus binding and entry into cells, which is followed by the interaction of VP7 with integrins αxβ2, α4β1, and αvβ3 (9, 19–24). The VP5* subunits of almost all group A rotaviruses contain the Asp-Gly-Glu (DGE) sequence at aa 308–310, a motif that has been implicated in α2β1 recognition by type I collagen (17). Mutation of the putative α2β1 ligand sequence DGE abrogates binding of truncated VP5* to the integrin α2 subunit I domain (α2I) and VP5* competition with RRV cell binding and infectivity (9, 25). In addition, DGE-containing peptides, such as Asp-Gly-Glu-Ala (DGEA), specifically inhibit rotavirus-cell binding and infection mediated by α2β1 (9, 20, 21, 25). Binding by infectious monkey (SA11 and RRV) and human (Wa) rotaviruses to recombinant α2β1 expressed on K562 cells was specifically inhibited by DG-containing peptides and a function-blocking antibody to the α2I domain (9, 21, 23). Therefore, the interaction of rotavirus with α2β1 integrin can be considered a target for the development of antiviral agents aimed at preventing or reducing rotavirus infection.
Bioactive components in milk are an important research focus (26). In vitro, lactadherin, a milk fat globule membrane (MFGM) glycoprotein, inhibits rotavirus infection when sourced from humans, whereas bovine lactadherin is not active against human rotavirus (27). Moreover, it has been reported that human milk lactadherin protects breastfed infants against symptomatic rotavirus infections (6). Lactadherins isolated from different animal species are characterized by two N-terminal epidermal growth factor (EGF)-like domains, the second of which contains an Arg-Gly-Asp (RGD) integrin-binding motif that engages αvβ3/5 integrins and two C-terminal discoidin/F5/8C domain repeats (28). Bovine lactadherin consists of 427 amino acid residues, including an N-terminal signal sequence of 18 amino acids that is cleaved during intracellular processing. In contrast, human lactadherin is a protein of 387 amino acids, including an N-terminal signal sequence of 23 residues. When separated by two-dimensional electrophoresis, each of these proteins shows an apparent molecular mass above the theoretical value, with the difference due to glycosylation (29–31). MFGM-associated proteins from water buffalo, sheep, and horse also have been analyzed (32–34). Comparative species-to-species structural studies among MFGM proteins showed high variation, especially for lactadherin (35). It is proposed that these differences could explain the divergent effects of bovine and human lactadherin on human rotavirus infection. In order to test if the lack of rotavirus inhibition by bovine lactadherin is atypical or represents a shared property among lactadherins of related domesticated animals, we undertook a proteomic analysis of MFGM proteins from equine milk and elucidated the amino acid sequence of donkey lactadherin. Alignment of the human, bovine, and donkey lactadherin sequences revealed the presence of a DGE integrin-binding sequence motif in the donkey sequence only, suggesting a potential anti-rotavirus activity. To test this hypothesis, a minilibrary of donkey lactadherin-derived peptides containing DGE sequence was tested for anti-rotavirus activity. A 20-amino acid peptide containing both DGE and RGD sequence showed the greatest activity, and its mechanism of antiviral action was characterized. These findings shed light on the mechanisms of equine lactadherin inhibition of rotavirus infection and support the feasibility of developing an anti-rotavirus peptide that acts through inhibition of virus-receptor binding.
EXPERIMENTAL PROCEDURES
Preparation of Fat Globules from Donkey Milk
Milk was collected from several donkeys at a single farm. Protease inhibitors (Complete; Roche Applied Science) were added, and the milk was immediately stored at −20 °C. Thawed milk was centrifuged at 5000 × g for 30 min at 10 °C, and the pellet was discarded. The cream layer and skimmed milk were centrifuged at 189,000 × g for 70 min at 6 °C. Fat globules were recovered in the supernatant and washed three times with 0.9% (w/v) NaCl.
Sample Protein Preparation and Two-dimensional Electrophoresis
Washed fat globules were incubated at 4 °C overnight in 20 mm Tris-HCl, pH 8.6, containing 1% (w/v) ASB-14, 1% (v/v) Triton X-100, 7 m urea, and 2 m thiourea to extract the proteins associated with fat globule membranes. After centrifugation at 18,400 × g for 10 min at 10 °C, the floating cream layer was discarded. Proteins were precipitated from the supernatant with methanol and chloroform, as described previously (36). Pellets containing proteins were solubilized in 20 mm Tris-HCl, pH 8.6, containing 7 m urea, 2 m thiourea, 1% (w/v) ASB-14, 1% (v/v) DTT, and 0.5% (v/v) IPG buffer. Total protein was quantified using the 2-D Quant Kit (GE Healthcare). Extracted proteins (200 μg) were loaded onto 13-cm pH 3–10 NLIPG strips (GE Healthcare). Isoelectric focusing was carried out on an IPGphor unit (GE Healthcare) at 20 °C and 8000 V for a total of 70,000 V-h. Strips were incubated at room temperature in 50 mm Tris-HCl, pH 8.6, containing 6 m urea, 30% (v/v) glycerol, 2% (w/v) SDS, enriched with 2% (w/v) DTT for 20 min and afterward with 4.5% (w/v) iodoacetamide for 20 min. SDS-polyacrylamide gel electrophoresis was carried out on homogeneous running gels with 11.7% total acrylamide concentration and a 2.6% grade of cross-linking (Ettan DALT II system, GE Healthcare) at 400 V and 50 mA per gel for ∼3 h. Gels were stained using the Processor Plus (GE Healthcare) with Blue Coomassie Colloidal stain (37) and scanned with a GS-800 densitometer (Bio-Rad).
Enzymatic Digestion of Proteins
In-gel multiple enzymatic digestion on selected two-dimensional electrophoresis spots was performed according to the published method (38). Briefly, excised spots were destained for 2 h in 40% (v/v) ethanol containing 50 mm NH4HCO3, dehydrated three times (10 min each) in acetonitrile, and dried in a vacuum concentrator for 30 min. Rehydration was performed at room temperature for 5 min with 5 μl of 75 ng/μl trypsin in 25 mm NH4HCO3 (Promega Corp., Madison, WI), 5 μl of 75 ng/μl Asp-N solution (Sigma), or 5 μl of 75 ng/μl chymotrypsin in 25 mm NH4HCO3 (Sigma). Digestion continued at 37 °C. Asp-N peptides were collected after 2 h of digestion and submitted to mass spectrometry (MS). Trypsin and chymotrypsin digestions were performed overnight after the addition of 15 μl of 25 mm NH4HCO3, 10% (v/v) acetonitrile. These supernatants were collected and submitted for MS analysis on the following day.
Mass Spectrometric Analysis
For MALDI-TOF mass spectrometry, 0.5 μl of each peptide mixture was applied to a target disk and allowed to air-dry. Subsequently, 0.5 μl of matrix solution (1% (w/v) α-cyano-4-hydroxycinnamic acid in 30% (v/v) acetonitrile and 0.1% (v/v) trifluoroacetic acid were applied to the dried sample and dried under vacuum. Spectra were collected on an Ultraflex II MALDI-TOF-TOF (Bruker Daltonik GmbH, Germany). The reported mass spectra are the result of signals of ∼800 laser shots. The spectra were externally calibrated using the standard calibration mixture from Bruker Daltonik. Manual/visual estimation of the mass spectra was performed using Flex Analysis software (Bruker Daltonik). For LC-MS/MS experiments, an Agilent 1100 series liquid chromatograph and LC/MSD XCT series ion trap mass spectrometer equipped with nano-electrospray ionization source were used. Peptide mixtures were separated on an RP C18 column (Zorbax 300SB-C18; 3.5 μm 150 × 0.075 mm) (CPS) equilibrated with 0.1% (v/v) formic acid in water. Peptides were eluted over 55 min with a linear gradient of 5–70% (v/v) of 0.5% water, 0.1% formic acid in acetonitrile. The flow rate was 300 nl/min. The mass spectrometer was operated in positive ion mode. Data were processed using DataAnalysis software (Agilent Technologies, Santa Clara, CA).
Spectra Interpretation
For peptide mass fingerprinting (PMF) identification, the MS-Fit software package in ProteinProspector version 5.10.9 was used for interpretation of spectra (39). Data were searched against the Horse database UniProtKB.2012.03.21.
The parameters used for the search were as follows: S-carbamidomethyl derivate on cysteine as fixed modification, oxidation on methionine as variable modification, and two missed cleavage sites for trypsin digestion. The peptide mass tolerance was 30 ppm.
The LC-MS/MS and MALDI-TOF/TOF spectra were analyzed using the MASCOT MS/MS Ions search engine against the UniProtKB.2012.03.21 database. The parameters used for the search were as follows: S-carbamidomethyl derivate on cysteine as fixed modification, oxidation on methionine as variable modification, two missed cleavage sites for each enzymatic digestion, and default selection of charge states ions (+2, +3). The peptide mass tolerance was set to 0.6 Da, and the fragment mass tolerance was set to 0.8 Da. Protein hits were validated if the protein scores were above the MASCOT default significance threshold (p ≤ 0.05) and a minimum of two unique peptides matched.
N-terminal Sequencing
Spots were passively eluted from gels as described elsewhere (40), and the membrane was analyzed by direct microsequencing on a Procise 492 protein sequencer (Applied Biosystems). All chemicals were purchased from Applied Biosystems.
Cell Lines and Viruses
African green monkey kidney epithelial cells (MA104) and human epithelial colorectal adenocarcinoma cells (Caco-2) were propagated, respectively, in Eagle's minimum essential medium (MEM) and Dulbecco's modified Eagle's medium (DMEM; Sigma). Both media were supplemented with 1% (v/v) Zell Shield (Minerva Biolabs, Berlin, Germany) and heat-inactivated, 10% (v/v) fetal bovine serum (Sigma).
Chinese hamster ovary (CHO-K1) cells were cultured in high glucose DMEM (PAA, Pasching, Austria) supplemented as above. The derivation of CHO K1 cell transfectants expressing human α2 integrin subunit (CHO α2) or the PBJ-1 empty vector (CHO K1 PBJ-1) has been described previously (41–45). CHO cell transfectants were grown as for the parental cell line, except that G418 sulfate (Sigma) at 0.1–0.8 μg/ml was included in the growth medium.
Human rotavirus strain Wa was obtained from the ATCC; the origin of porcine rotavirus P9 G3 strain CRW-8 has been described previously (46). Wa and CRW-8 infectivity was activated with 5 μg/ml porcine pancreatic trypsin type IX (Sigma) for 30 min at 37 °C and propagated in MA104 cells in MEM containing 0.5 μg/ml trypsin as described previously (47).
Antibodies
Mouse monoclonal antibodies (mAbs) directed to human rotavirus VP6 (0036), α2β1 integrin (AK7), and αvβ3 integrin (LM609) were purchased from Covalab (Villeurbanne, France), Abcam (Cambridge, UK), and Millipore (Molsheim, France), respectively.
Peptides
Synthetic peptides, pRGD-Hu (EVRGDVFP), pRGD-Bo (SHRGDVFI) (derived from human and bovine lactadherin, respectively), pRGD-Eq (SHRGDVFT), pDGE (QNDGECHV) (derived from equine lactadherin), pRGE (SHRGEVFT), and pAGE (QNAGECHV) were purchased from Eurogentec (Liege, Belgium). Synthetic peptides pDGE-RGDmut1 (NNDGECHVIDDSHRGDVFTQ), pDGE-RGDmut2 (QNDGECHVIDDSDRGDVFTQ), pDGE-RGDmut3 (QNDGECHVIDDSHRGDVFSQ), pAGE-RGD (QNAGECHVIDDSHRGDVFTQ), and pDGE-RGE (QNDGECHVIDDSHRGEVFTQ) were purchased from Proteogenix (Valparc, France). pDGE-RGD19 (NDGECHVIDDSHRGDVFTQ) and pDGEA (DGEA) were purchased from the PolyPeptide Group (Strasbourg, France). pAGE-RGE (QNAGECHVIDDSHRGEVFQT) was purchased from the CASLO Laboratory (Technical University of Denmark, Lyngby, Denmark). pDGE-RGD peptide (QNDGECHVIDDSHRGDVFTQ), derived from equine lactadherin, was purchased from Eurogentec, PolyPeptide Group, Proteogenix, and CASLO Laboratory. Biotinylated peptides Biot-pDGE (Biotin-QNDGECHV), Biot-pAGE (Biotin-QNAGECHV), and Biot-pDGE-RGD (Biotin-QNDGECHVIDDSHRGDVFTQ) were purchased from Proteogenix. MALDI-TOF mass spectrometry analysis confirmed that pDGE-RGD, the peptide of major interest for this study, was soluble and monomeric (data not shown).
Rotavirus Infectivity Assays
Peptide blockade of virus infectivity was determined by a focus reduction assay using confluent MA104 cell monolayers in 96-well trays, as described previously (21). Unless otherwise stated, cells were treated for 1 h at 37 °C with peptides at 0.01–500 μm in serum-free medium prior to the virus addition. Rotavirus infection was performed at a multiplicity of infection (MOI) of 0.02, unless otherwise indicated, for 1 h at 37 °C, in the presence of the peptide unless otherwise stated. Infected cells were washed with serum-free medium and incubated in this medium at 37 °C in a humidified incubator in 5% (v/v) CO2, 95% (v/v) air. In some experiments (as indicated), peptide pDGE-RGD was included with these washed, infected cells for the duration of their incubation. After 16 h of incubation, infected cells were fixed with cold acetone/methanol (50:50), and viral titers were determined by indirect immunostaining, by using the UltraTech HRP Streptavidin-Biotin detection system (Beckman Coulter) according to the manufacturer's instructions. Peptide blockade of viral infectivity is expressed as a mean percentage ± S.D. Where possible, antiviral effective concentration (IC50) values were calculated by regression analysis using the dose-response curves generated from the experimental data, using PRISM 4 (GraphPad Software, San Diego, CA). IC50 values were compared using the sum-of-squares F test.
Assay of Virus Yield
To test the ability of pDGE-RGD to inhibit multiple cycles of viral replication, confluent MA104 cells in 24-well trays were infected with trypsin-activated Wa rotavirus (MOI = 0.02) for 1 h at 37 °C and washed as above. Cells were incubated in medium supplemented with 0.5 μg/ml porcine trypsin and 30, 100, or 300 μm pDGE-RGD, with fresh pDGE-RGD added to peptide-treated wells at 16 h after infection and at later fixed intervals and an equal volume of medium added to untreated controls at these times. Infected cells and cell supernatants were harvested at 48 h postinfection, and virus titers were determined by indirect immunostaining of MA104 cell monolayers inoculated with serial dilutions of the samples. The assay was performed in triplicate.
Rotavirus-Cell Binding Assay
Confluent MA104 cell monolayers in 24-well trays were washed, incubated with peptides or antibodies for 1 h at 37 °C, and cooled on ice for 20 min in the continuing presence of peptides or antibodies. Trypsin-activated virus, which had been cooled to 4 °C, was allowed to attach to cells on ice for 1 h (MOI = 3) in the presence of peptides or antibodies. Cells were washed with cold MEM; cold MEM was added. Cells were subjected to two rounds of freeze-thawing and incubated at 37 °C for 30 min with 10 μg/ml porcine trypsin to release bound virus, and the lysate was clarified by low speed centrifugation for 10 min. Cell-bound virus titers were determined by indirect immunostaining as above.
Rotavirus-Cell Entry Assay
Confluent MA104 cell monolayers in 96-well trays were washed twice with MEM and cooled on ice for 20 min. Trypsin-activated virus, which had been cooled to 4 °C, was adsorbed to cells on ice for 1 h at 4 °C (MOI = 0.02). Peptide pDGE-RGD was added to washed cells at 0.01–500 μm for 1 h at 37 °C. Rotavirus entry was stopped by the removal of surface-bound virions with two quick washes with 3 mm EGTA in PBS, which removes the virion outer capsid (48, 49). Cells were incubated in MEM for 16 h at 37 °C, fixed, and stained, and titers were determined as described above for the infectivity assay.
Virucidal Assay
Trypsin-activated Wa rotavirus was reacted with 100 μm pDGE-RGD peptide or diluent for 1 h at 37 °C and then titrated. Viral yields were determined by indirect immunostaining, by counting the number of infectious foci at 1:1,024, 1:2,048, and 1:4,096 dilutions.
Peptide-Cell Binding Assay
Confluent CHO K1 and CHO α2 cells in 96-well trays were washed twice with DMEM containing 1% (w/v) BSA and incubated for 1 h at 37 °C with 0.3, 3, or 30 μm Biotin-pDGE, Biotin-pAGE, or Biotin-pDGE-RGD peptides in the same DMEM diluent. Washed cells were incubated with streptavidin-HRP (Beckman Coulter) for 10 min at room temperature, washed twice with PBS, and reacted with ABTS reagent (ThermoScientific) for 30 min at room temperature. The reaction was stopped with PBS containing 5% (w/v) SDS, and the absorbance was determined at λ = 405 nm.
Cell Viability Assay
The cytotoxicity of peptide pDGE-RGD was assessed in MA104 and CaCo2 cells seeded in 96-well plates at a density of 1.2 × 104 cells/well. After 24 h of culture, cells were treated with peptide at 0.0044–880 μm. Cell viability was determined 48 h later with the CellTiter 96 proliferation assay kit (Promega) according to the manufacturer's instructions. Absorbances were measured using a model 680 microplate reader (Bio-Rad) at 490 nm. Viability in treated cells was expressed as a percentage relative to cells incubated with culture medium alone. The 50% cytotoxic concentration (CC50) was determined using logarithmic viability curves. Where possible, a selectivity index (SI) was calculated by dividing the CC50 by the IC50 value.
Statistical Analysis
One-way analysis of variance, in some cases followed by the Bonferroni test, was used to assess the statistical significance of differences in virus titers and absorbance data. Significance was set at the 95% level.
RESULTS
Proteomic Analysis and Protein Sequencing Indicate the Presence of Two Putative Integrin Ligand Motifs in Equine Lactadherin
The two-dimensional electrophoresis map of donkey milk fat globule membrane proteins was determined (Fig. 1). The two-dimensional electrophoresis map showed a pattern very similar to that previously published for horse MFGM proteins, where lactadherin was the major protein and was identified in several spots at different molecular masses (34). The spots 1–4, corresponding to the molecular masses of horse lactadherin, were excised, and the proteins were identified by MALDI-TOF MS and PMF as the horse milk fat globule-EGF factor 8 splice variant, also known as horse lactadherin (Table 1). To understand the nature of the difference between the ∼45- and ∼50-kDa proteins, their N-terminal amino acid sequences were determined (Table 2). The sequences of proteins 1 and 2 matched the N-terminal amino acid sequence of horse lactadherin (UniProt F7B0S3) with an identity of ≥95%. Proteins 3 and 4 corresponded to the same horse lactadherin sequence, commencing from the amino acid residue at position 44. Because the 45-kDa form was most similar to the human counterpart, its amino acid sequence was examined further. The results of multiple enzymatic digestions of this 45-kDa protein with trypsin, Asp-N, and chymotrypsin, followed by MALDI-TOF/TOF and nano-LC-electrospray ionization MS/MS analyses (Table 3), together with the N-terminal amino acid sequence, gave an (almost) complete coverage of the sequence of this form of donkey lactadherin (Fig. 2). The sequence was submitted to the UniProt database, and the accession number C0HJR4 was assigned.
FIGURE 1.
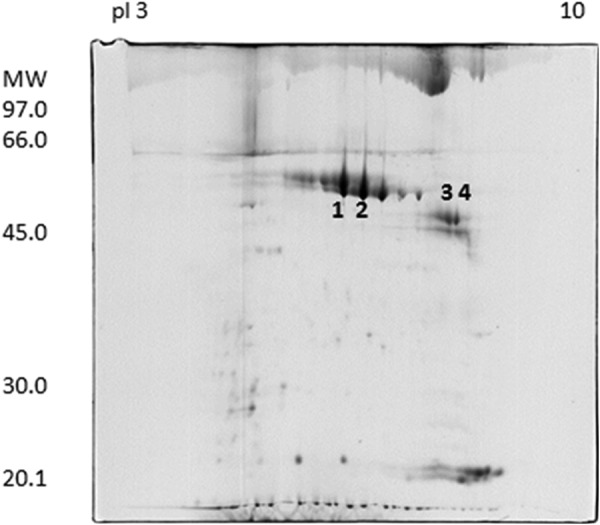
Two-dimensional electrophoresis map of donkey milk fat globule membrane proteins. Identified proteins are marked with numbers. The spots numbered 1–4 were excised from the gel and analyzed by MALDI-TOF MS and PMF. All were identified as horse milk fat globule-EGF factor 8 splice variant, also known as horse lactadherin.
TABLE 1.
Identification of donkey milk fat globule membrane proteins by PMF
| Spot no. (from Fig. 1) | Protein name | UniProt KB/TrEmbl entry | MOWSE scorea | Experimental/nominal molecular mass | PMF number of signals m/z matched/number of signals m/z collected (% sequence coverage) |
|---|---|---|---|---|---|
| Da | |||||
| 1 | Milk fat globule-EGF factor 8 splice variant | F5CEP2 | 5.25e + 8 | 50,000/43,308 | 24/78 (66.2%) |
| 2 | Milk fat globule-EGF factor 8 splice variant | F5CEP2 | 2.26e + 9 | 50,000/43,308 | 23/59 (61.1%) |
| 3 | Milk fat globule-EGF factor 8 splice variant | F5CEP2 | 4.74e + 6 | 45,000/43,308 | 17/60 (54.1%) |
| 4 | Milk fat globule-EGF factor 8 splice variant | F5CEP2 | 1.44e + 7 | 45,000/43,308 | 18/54 (54.1%) |
a MOWSE score is obtained by the MS-Fit search and concerns the highest scored protein.
TABLE 2.
N-terminal amino acid sequences and identification of donkey milk fat globule membrane proteins
| Spot number (Fig. 1) | N-terminal amino acid sequence by Edman degradation | Comparison with horse lactadherin amino acid sequence (F7B0S3) | Identity |
|---|---|---|---|
| % | |||
| 1 | ASGDFCDSSQCLNGGTCLLGQ | 1AIAGDFCDSSQCLNGGTCLLGQ22 | 95% |
| 2 | ASGDFCDSSQCLNGGTCLLGQDDL | 1AIAGDFCDSSQCLNGGTCLLGQDDL25 | 96% |
| 3 | ASGPCFPNPCQNDGECHVIDDSHR | 44GPCFPNPCQNDGECHVIDDSHR65 | 96% |
| 4 | ASGPCFPNPCQNDGECHVID | 44GPCFPNPCQNDGECHVID62 | 90% |
TABLE 3.
Identification of the 45-kDa donkey lactadherin short form by enzymatic digestions and mass spectrometry techniques
| Enzyme | PMF by MALDI-TOF |
LC-MS/MS |
||||
|---|---|---|---|---|---|---|
| Position | m/z, single charge | Sequencea | Position | m/z, charge state | Sequencea | |
| Trypsin | 127–134 | 838.4305 | GGTAEYLK | 89–102 | 770.3600 (+2) | TGIVNAWTASNYDK |
| 138–144 | 866.4367 | VAYSVDGR | 289–300 | 706.2300 (+2) | DFGHIQYVAAYK | |
| 116–126 | 1074.5902 | VTGVVTQGASR | 25–37 | 801.7400 (+2) | GDVFTQYICSCPR | |
| 349–357 | 1105.6265 | ILPVAWHNR | 278–288 | 558.2200 (+2) | QVTGVVTQGAR | |
| 358–367 | 1173.6660 | ITLRVELLGCam | 160–171 | 624.6400 (+2) | VFVGNVDNSGLK | |
| 336–345 | 1225.6034 | NMFETPFLAR | 116–126 | 537.7100 (+2) | VTGVVTQGASR | |
| 336–345 | 1241.5983 | NMoxFETPFLAR | 199–224 | 959.2800 (+3) | FELLGCEVNAGCAEPLGLEDNSIPDR | |
| 289–300 | 1411.7005 | DFGHIQYVAAYK | 349–357 | 553.1500 (+2) | ILPVAWHNR | |
| 146–157 | 1411.7328 | KFQFIRDAGDSK | 173–185 | 854.8500 (+2) | VNMFDVPLEVQYVR | |
| 323–335 | 1483.7625 | IFPGNLDNNSHKK | 255–277 | 856.5900 (+3) | FNAWTAQSNSASEWLQVDLGSQK | |
| 186–198 | 1519.7620 | LVPVACamHHGCamTLR | 186–198 | 760.2300 (+2) | LVPVACHHGCTLR | |
| 25–37 | 1602.7039 | GDVFTQYICamSCamPR | 336–345 | 613.2900 (+2) | NMFETPFLAR | |
| 172–185 | 1708.8727 | VNMFDVPLEVQYVR | 323–334 | 678.1600 (+2) | IFPGNLDNNSHK | |
| 103–115 | 1717.8989 | NPWIQVNLMRKMR | 79–85 | 435.6300 (+2) | WVPELAR | |
| 172–185 | 1724.8676 | VNMFDVPLEVQYVR | ||||
| 234–248 | 1878.8962 | TWGLNAFSWYPFYAR | ||||
| 255–277 | 2567.2161 | FNAWTAQSNSASEWLQVDLGSQK | ||||
| 199–224 | 2875.3237 | FELLGCamEVNAGCamAEPLGLEDNSIPDR | ||||
| Chymotrypsin | 347–354 | 953.5931 | VRILPVAW | |||
| 233–240 | 964.4999 | RTWGLNAF | ||||
| 32–39 | 1012.4339 | ICamSCamPRGY | ||||
| 87–95 | 1053.5588 | HRTGIVNAW | ||||
| 71–79 | 1157.5561 | FGFMGLQRW | ||||
| 246–255 | 1225.6688 | YARLDKQGKF | ||||
| 76–86 | 1267.7270 | QRWVPELARL | ||||
| 29–39 | 1404.6035 | TQYICamSCamPRGY | ||||
| 246–258 | 1596.8281 | YARLDKQGKFNAW | ||||
| 296–310 | 1618.7608 | VAAYKVSHSNDGANW | ||||
| 184–197 | 1618.8305 | VRLVPVACamHHGCamTL | ||||
| 148–161 | 1625.8282 | QFIRDAGDSKDKVF | ||||
| 329–342 | 1708.7748 | DNNSHKKNMFETPF | ||||
| 215–232 | 1966.9352 | GLEDNSIPDRQITASSTY | ||||
| 162–183 | 2453.2017 | VGNVDNSGLKVNMFDVPLEVQY | ||||
| 40–71 | 3393.4742 | TGTHCamETTCamAMPLGMETGAIADAQISASSVYF | ||||
| 314–342 | 3393.6393 | RDQRAADSKIFPGNLDNNSHKKNMFETPF | ||||
| 101–132 | 3578.8319 | DKNPWIQVNLMRKMRVTGVVTQGASRGGTAEY | ||||
| Asp-N | 155–165 | 1207.6317 | DSKDKVFVGNV | 272–288 | 872.3800 (+2) | DLGSQKQVTGVVTQGAR |
| 166–175 | 1140.5354 | DNSGLKVNMF | 289–305 | 968.8900 (+2) | DFGHIQYVAAYKVSHSN | |
| 180–198 | 814.3400 (+3) | EVQYVRLVPVACHHGCTLRF | ||||
| 131–142 | 724.3000 (+2) | EYLKTFKVAYSV | ||||
| 166–175 | 570.7000 (+2) | DNSGLKVNMoxF | ||||
| 26–44 | 774.8400 (+3) | DVFTQYICSCPRGYTGTHC | ||||
| 158–165 | 439.2100 (+2) | DKVFVGNV | ||||
| 166–175 | 562.6800 (+2) | DNSGLKVNMF | ||||
| 320–328 | 495.7300 (+2) | DSKIFPGNL | ||||
| 45–60 | 842.7800 (+2) | ETTCAMoxPLGMoxETGAIA | ||||
| 143–151 | 583.7300 (+2) | DGRKFQFIR | ||||
| 13–19 | 415.1600 (+2) | DGECHVI | ||||
| 250–266 | 920.3900 (+2) | DKQGKFNAWTAQSNSAS | ||||
| 1–12 | 674.6700 (+2) | ASGPCFPNPCQN | ||||
| 45–60 | 834.8300 (+2) | ETTCAMoxPLGMETGAIA | ||||
a Cam, carbamidomethylated cysteine; Mox, oxidized methionine.
FIGURE 2.

Amino acid sequence of the 45-kDa form of donkey lactadherin obtained by N-terminal sequencing, PMF by MALDI-TOF, and LC-MS/MS. The sequence of the 45-kDa form of donkey lactadherin obtained (UniProt entry C0HJR4) is aligned with two isoforms of Equus caballus lactadherin (UniProt entries F7B0S3 and F5CEP2). Amino acids of the 45-kDa form of donkey lactadherin that differ from the other lactadherins are given in boldface type and underlined.
The sequence alignment of this equine (EQ) lactadherin with the bovine (BO) and human (HU) lactadherins revealed some very interesting features in the N-terminal domains (Fig. 3). An RGD motif is conserved in the three proteins, being present at amino acid residues 23–25 in equine and human lactadherin and at residues 67–69 in bovine lactadherin. RGD is an integrin-binding motif that engages αvβ3, αvβ5, α3β1, and α5β1 integrins to facilitate cell adhesion. Surprisingly, the equine lactadherin was shown to contain an additional integrin binding motif, the DGE sequence that can specifically bind the α2β1 integrin, which is present at residues 12–14.
FIGURE 3.

Sequence alignment of the equine (EQ Lact45), bovine (BO Lactad), and human (HU Lactad) lactadherins. The integrin-binding motifs RGD and DGE are given in boldface type and boxed.
Functional Roles of the Equine Lactadherin RGD and DGE Sequences in Inhibition of Rotavirus Infection
In order to assess the importance of the RGD sequence (shared by human, bovine, and equine lactadherin) and the DGE motif (present in equine lactadherin only) for inhibition of rotaviral infection, the effect on Wa infection of cellular treatment with the 8-amino acid peptides pRGD-Hu, pRGD-Bo, pRGD-Eq, and pDGE was determined. These human, bovine, and equine RGD peptides inhibited Wa infection of MA104 cells in a dose-dependent fashion to maxima at 500 μm of 57, 50, and 47%, respectively (Fig. 4A). In contrast, MA104 cell treatment with an 8-amino acid RGE peptide harboring a single amino acid substitution (Asp → Glu) known to inhibit the ability of the RGD triplet to bind integrin (pRGE) had no effect on virus infectivity even at high concentrations (Fig. 4A). Notably, the DGE-containing peptide pDGE derived from equine lactadherin inhibited Wa infectivity to a maximum of 84% (Fig. 4B), making it possible to assess its IC50 at 17 μm (Table 4). As expected, an AGE peptide (pAGE), despite its ability to inhibit rotaviral infectivity to a maximum of 80%, showed a significantly higher IC50 (257 μm) than pDGE (p < 0.0001; Table 4). As a positive control, we tested the Type I collagen-derived peptide Asp-Gly-Glu-Ala (pDGEA); this α2β1 integrin ligand showed an antiviral activity comparable with the one previously reported (21), as depicted in Fig. 4B.
FIGURE 4.
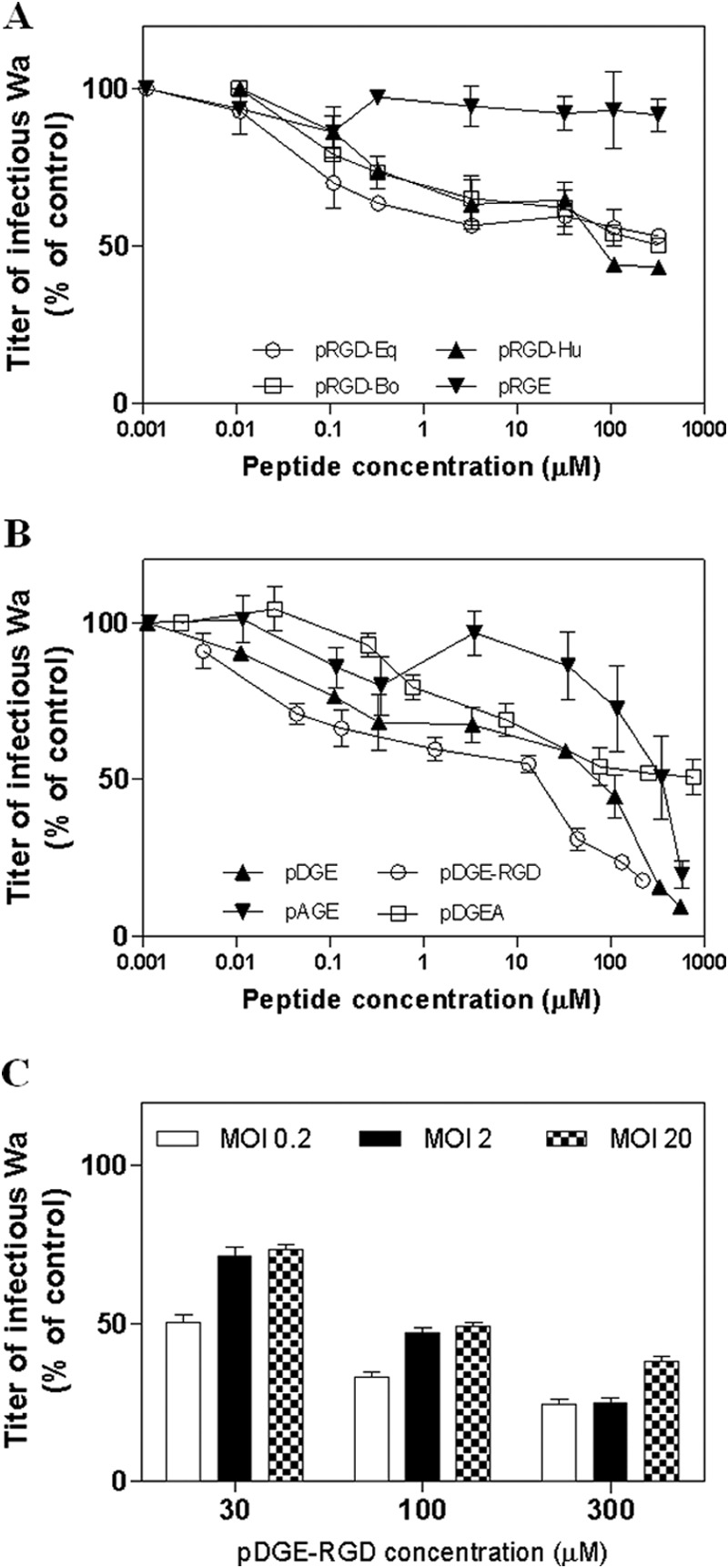
A and B, blockade of Wa infection in MA104 cells by peptides pRGD-Eq, pRGD-Hu, pRGD-Bo, and pRGE (A) and pDGE-RGD, pDGE, pAGE, and pDGEA (B). C, blockade of Wa infection in MA104 cells by pDGE-RGD at different MOIs. The infectivity titer of virus in the treated sample is expressed as a percentage of the titer obtained in the absence of peptide (untreated control). Error bars, S.D. of three independent experiments.
TABLE 4.
Effect of lactadherin-derived peptides on cell viability and Wa rotavirus infectivity in MA104 cells
| Peptides | Features | IC50 | 95% confidence Interval | CC50 | SI |
|---|---|---|---|---|---|
| μm | μm | ||||
| pDGE | peptide DGE, 8 aa, equine | 17.3 | 6.9–43.5 | >880 | >50.9 |
| pAGE | aa substitution in position 3 | 256.6 | 177.7–370.3 | >880 | >3.4 |
| pDGE-RGD | Wild type peptide | 4.4 | 2.7–7.1 | >880 | >200 |
| pAGE-RGD | aa substitution in position 3 | >220 | NAa | >880 | >4 |
| pDGE-RGE | aa substitution in position 16 | >220 | NA | >880 | >4 |
| pAGE-RGE | aa substitutions in position 3 and 16 | >220 | NA | >880 | >4 |
| pDGE-RGDmut1 | aa substitution in position 1 | 13.5 | 5.1–35.7 | >880 | >65.2 |
| pDGE-RGDmut2 | aa substitution in position 13 | 13.1 | 7.5–22.9 | >880 | >67.2 |
| pDGE-RGDmut3 | aa substitution in position 19 | 6.7 | 2.2–20.4 | >880 | >131.3 |
a n.a., not assessable.
A 20-Amino Acid Region of Equine Lactadherin Has Elevated Specific Anti-rotaviral Activity in Epithelial Cell Cultures That Depends on Its RGD and DGE Sequences
Interestingly, the 20-amino acid peptide pDGE-RGD, corresponding to amino acids 10–29 of equine lactadherin and harboring both the RGD and DGE motifs, showed the highest anti-rotaviral efficacy (IC50 = 4.4 μm; Table 4). pDGE-RGD was therefore selected as the lead compound for further studies. Of note, the pDGE-RGD peptide maintained a significant (p < 0.05) inhibitory activity against high MOIs of Wa (Fig. 4C). The stage of the rotavirus replication cycle inhibited by peptide pDGE-RGD was examined. Peptide addition 1 h before or immediately after infection did not inhibit Wa infectivity. The greatest infectivity inhibition was achieved when the peptide was added during infection (Fig. 5), as performed in the experiments described above and in the following studies. To assess the role played by each motif in inhibition of rotavirus infectivity, the antiviral activity of several mutant peptides derived from pRGD-DGE that harbored mutations in DGE (Asp → Ala, pAGE-RGD), RGD (Asp → Glu, pDGE-RGE), or both triplets (pAGE-RGE) was tested. These peptides failed to show any antiviral activity even at high concentrations, demonstrating the importance of these motifs for anti-rotavirus activity (Table 4). In contrast, peptides with single amino acid substitutions outside these functional groups, namely pDGE-RGDmut1, pDGE-RGDmut2, and pDGE-RGDmut3, retained the ability to inhibit Wa infection (Table 4), at levels comparable with those of pDGE-RGD. No significant difference was seen between the IC50 of pDGE-RGD and these peptides with amino acid substitutions outside the DGE and RGD functional groups (p > 0.05). The antiviral efficacy of pDGE-RGD and its AGE and RGE mutants was also evaluated in the Caco-2 intestinal cell line (Table 5). Whereas pDGE-RGD showed antiviral activity in Caco-2 cells (IC50 = 8.8 μm) comparable with that in MA104 cells, pDGE-RGE, pAGE-RGD, and pAGE-RGE exhibited a significant loss of activity (p < 0.0001). Interestingly, these Caco-2 cell studies identified significant differences in antiviral activity between these three peptides (0.002 < p < 0.0194), with pDGE-RGE showing a lower IC50 (46 μm) than pAGE-RGD (IC50 = 101 μm) and pAGE-RGE (IC50 = 152 μm). This suggests that the DGE sequence plays a major role in the process leading to rotavirus inhibition. In order to rule out any nonspecific effect due to loss of cell viability, MTS cell viability assays were performed after incubation of Wa-infected and control MA104 and Caco-2 cell cultures for 72 h with a range of peptide concentrations. The tested peptides did not impair cell viability at any concentration tested on MA104 cells, showing a 50% cytotoxic concentration (CC50) of >880 μm (Tables 4 and 5). In Caco-2 cells, the pDGE-RGD CC50 was 945 μm. CC50 values were used to calculate the SI of pDGE-RGD, which was >100 for both cell lines (Tables 4 and 5).
FIGURE 5.
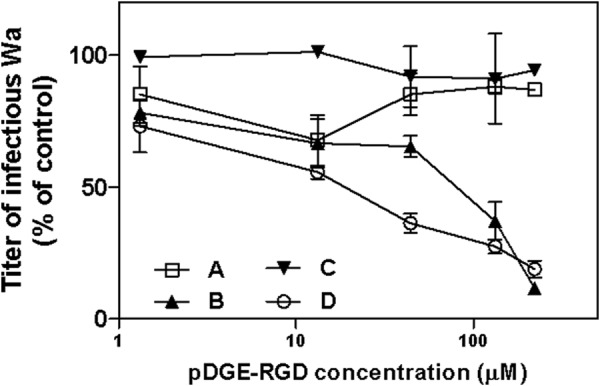
A–C, time-of-addition experiments were performed by treating cells with pDGE-RGD for 1 h before infection (A) or for 1 h during infection (B) or by adding the peptide immediately after infection (C). Control experiments were performed by treating cells both before and during viral adsorption (D). The infectivity titers of virus in the treated samples are expressed as a percentage of the titer obtained in the absence of peptide (untreated control). Error bars, S.D.
TABLE 5.
Effect of lactadherin-derived peptides on cell viability and Wa rotavirus infectivity in Caco-2 cells
| Peptides | Features | IC50 | 95% confidence interval | CC50 | SI |
|---|---|---|---|---|---|
| μm | μm | ||||
| pDGE-RGD | Wild type peptide | 8.8 | 6.1–12.5 | 945.1 | 102.7 |
| pDGE-RGE | aa substitution in position 16 | 46.0 | 24.9–84.9 | >880 | 19.1 |
| pAGE-RGD | aa substitution in position 3 | 101.7 | 78.6–131.6 | >880 | 8.7 |
| pAGE-RGE | aa substitutions in position 3 and 16 | 152.5 | 123.3–188.6 | >880 | 5.8 |
Infectious Rotavirus Yield Is Reduced by the 20-Amino Acid Lactadherin Peptide
The antiviral activity of pDGE-RGD was evaluated in a virus yield assay, a more stringent test that allows multiple cycles of viral replication to occur before measuring the titer of infectious virus produced. Wa rotavirus yield was reduced in a dose-dependent manner in cells treated with 100 or 300 μm pDGE-RGD (p < 0.05), showing 50 and 84% inhibition, respectively (Fig. 6).
FIGURE 6.
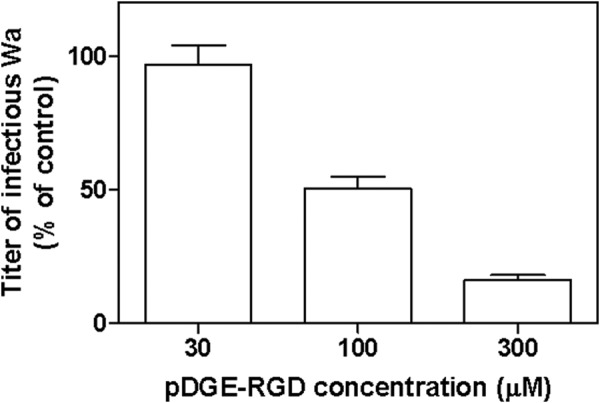
Effect of peptide pDGE-RGD on multiple cycles of Wa replication. Cells were treated after virus infection at the given peptide concentrations. The titer of rotavirus in the treated samples is expressed as a percentage of the titer in the untreated control. Error bars, S.D.
The 20-Amino Acid Lactadherin Peptide Binds α2β1 Integrin in a DGE-dependent Manner
The results described above suggested that pDGE-RGD may inhibit rotavirus infectivity by binding the α2β1 integrin, thereby blocking viral binding to this cellular receptor. The ability of equine lactadherin peptides to specifically bind α2β1 was tested by treating CHO cells expressing surface α2β1 with biotinylated pDGE-RGD, pDGE, and pAGE peptides. Peptides pDGE-RGD and pDGE bound to the surface of α2β1-expressing CHO cells in a dose-dependent manner, whereas pAGE did not show this ability (Fig. 7A). Peptide pDGE-RGD bound to a greater extent than pDGE, because pDGE-RGD binding was significant greater than that of pDGE at 3 and 30 μm (0.01 < p < 0.02). In contrast, pDGE binding to the CHO parental cell line (CHO-K1) (Fig. 7B) was reduced compared with α2β1-expressing CHO cells at all concentrations tested (p < 0.0006). No difference was detected between pDGE and pAGE binding to CHO-K1 cells (p > 0.05). Interestingly, substantial binding of pDGE-RGD to CHO-K1 cells was evident, indicating binding to a cell surface component(s) other than α2β1. Consistently, at 30 μm, pDGE-RGD binding to the surface of α2β1-expressing CHO cells was significantly greater than to the parental cell line (p < 0.05).
FIGURE 7.
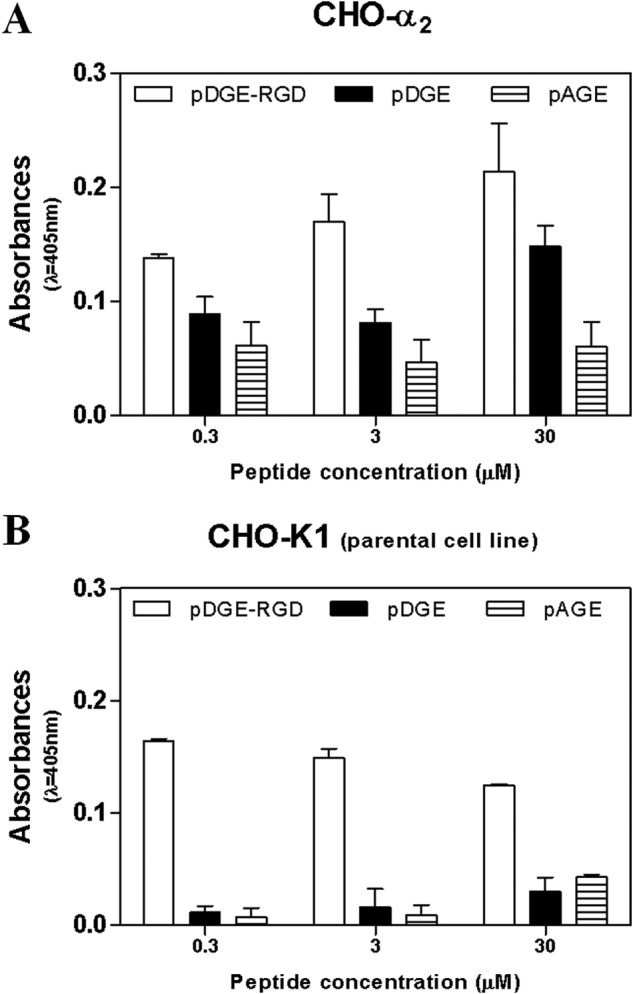
Evaluation of the ability of biotinylated pDGE-RGD, pDGE, and pAGE to interact with the α2β1 integrin. Experiments were performed with CHO cells expressing α2β1 (CHO-α2; A) and the parental CHO-K1 cells lacking this integrin (B). Levels of peptide-cell binding are expressed in absorbance units (λ = 405 nm). Error bars, S.D. of three independent experiments.
The 20-Amino Acid Lactadherin Peptide Preferentially Inhibits Rotavirus-Cell Attachment Rather than Entry in a DGE-dependent Manner
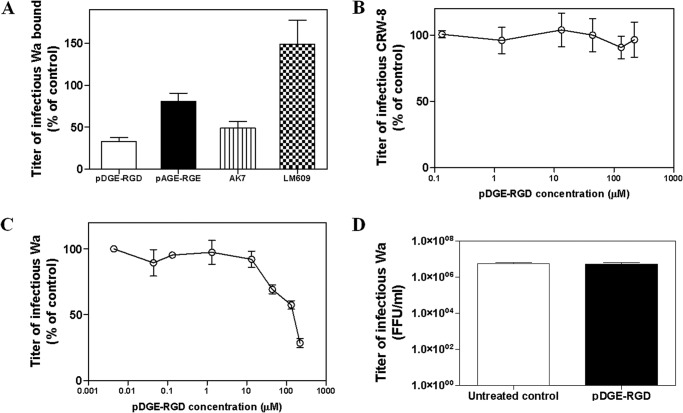
In order to evaluate the ability of pDGE-RGD to inhibit rotavirus-cell attachment, we measured the titer of infectious Wa rotavirus particles attached to pDGE-RGD-treated and untreated MA104 cells. pDGE-RGD significantly reduced Wa binding by a maximum of 67% (p < 0.0001), whereas pAGE-RGE did not show any inhibitory activity (p > 0.05; Fig. 8A). As found previously (9), the AK7 mAb to integrin α2β1 reduced virus-cell binding to a maximum of 51% (p < 0.05), whereas mAb LM609 to integrin αvβ3 had no significant effect (p > 0.05). In a control experiment, we tested the antiviral efficacy of pDGE-RGD against the integrin-independent rotavirus strain CRW-8. As expected, pDGE-RGD did not inhibit CRW-8 infectivity at any concentration tested (Fig. 8B). The effect of pDGE-RGD on viral entry also was investigated. This peptide significantly inhibited Wa rotavirus-cell entry only when added to cells at very high concentrations (i.e. 300 or 500 μm) (p < 0.05; Fig. 8C). In order to rule out any virucidal effect of pDGE-RGD on rotavirus, Wa was exposed for 1 h to a pDGE-RGD concentration (100 μm) able to inhibit 80% of Wa infectivity. The infectious titer of the pDGE-RGD-treated viral suspension was indistinguishable from that of the untreated control (p > 0.05; Fig. 8D).
FIGURE 8.
Evaluation of the mechanism of action of pDGE-RGD. A, effect of pDGE-RGD on Wa binding to the MA104 cell surface; control experiments were performed by treating cells with pAGE-RGE, anti-α2 antibody AK7 or anti-αvβ3 antibody LM609. On the y axis, the infectious titer of Wa bound to cells is expressed as a percentage of the titer bound to control MA104 cells in the absence of peptides or antibodies. B, effect of pDGE-RGD on CRW-8 infection in MA104 cells. The infectivity titer of virus in the treated sample is expressed as a percentage of the titer obtained in the absence of peptide (untreated control). C, effect of pDGE-RGD on Wa rotavirus entry into MA104 cells; the rotavirus titer measured in the treated samples is expressed as a percentage of the titer obtained in the untreated control. Evaluation of the virucidal effect of pDGE-RGD on infectious rotavirus particles is shown in D. On the y axis, the Wa infectious titers are expressed as focus-forming units/ml (FFU/ml). in the three graphs, error bars represent the S.D. values of three independent experiments.
DISCUSSION
Many human and animal rotaviruses contain the Asp-Gly-Glu (DGE) sequence in VP8*, which is necessary for their usage of the α2β1 integrin to aid infection. In this study, we have identified the putative integrin ligand motifs RGD and DGE in the N-terminal domain of donkey lactadherin. Whereas the RGD motif is common to human, bovine, and donkey lactadherins, the DGE motif is found only in the donkey sequence. Direct evaluation of the antiviral activity of donkey lactadherin would be of interest but has proven to be difficult. Difficulties arise when experiments to purify donkey lactadherin with the same methods used for the human and bovine proteins are attempted, because human and bovine milk contain a much higher amount of fat (∼3.5%) than donkey milk (∼0.3%) from which only a very small amount of lactadherin could be recovered. For this reason, we turned to the recombinant protein. We achieved bacterial expression of full-length donkey lactadherin, but the recombinant His-tagged protein was insoluble and could not be refolded.4 This is consistent with the insolubility of milk-derived lactadherins, which are associated with the milk fat globule membrane (50). In any case, our main focus is on the development of rotavirus inhibitors based on derivative peptides rather than analyzing the full-length protein; one main reason is that it is very unlikely that lactadherin molecules pass through the digestive tract intact, at least not in a relevant amount (e.g. we have easily digested donkey lactadherin with trypsin to solve their primary structure). Moreover, the ability of the DGE integrin binding domain to inhibit rotavirus replication can be addressed much more clearly working with small peptides rather than intact proteins, where the tertiary and eventually quaternary structure of the molecule could mask the biological activity of the inner part of some protein segment. To this end, we have demonstrated that peptides derived from donkey lactadherin, which contain the intact RGD and DGE sequences, specifically and effectively inhibit rotavirus infection at low micromolar levels. We selected a 20-amino acid peptide derived from donkey lactadherin harboring the RGD and a DGE motif (pDGE-RGD), depending on its favorable SI and its ability to limit the production of viral progeny resulting from multiple cycles of viral replication. The differences between MA104 and Caco-2 cells in the effectiveness of pDGE-RGD in preventing rotavirus cell infection parallel the data documented in a previous work (21), showing that a DGEA peptide was a less effective inhibitor of virus binding in Caco-2 cells than in MA104 cells. The higher surface expression of α2β1 on Caco-2 cells than on MA104 cells (51) could explain these findings, because higher levels of peptide would be needed to block all available integrin sites that could bind virus.
During our investigation of the functional roles of the equine lactadherin RGD and DGE sequences in inhibition of rotavirus infection, we found that pAGE-RGD, pDGE-RGE, and pAGE-RGE mutant peptides lack antiviral activity on MA104 cells. Interestingly, we were able to discriminate a different contribution from the two motifs using CaCo2 cells, with the DGE triplet being more critical than RGD in limiting Wa infectivity (IC50 of pAGE-RGD > IC50 of pDGE-RGE). These data, together with the fact that peptides harboring single amino acid substitutions outside these functional motifs retain their antiviral activity, suggest that both triplets are important for the inhibition of rotavirus infectivity. These specific substitutions (i.e. Asp → Ala for DGE and Asp → Glu for RGD) were chosen because an aspartate or glutamate residue is a critical feature of all integrin recognition sites (52). The amino acid substitution in the DGE motif (i.e. Asp → Ala) is known to alter the integrin α2β1 binding ability of VP5* (21, 24, 32), with Asp-308 in VP5* being the major requirement for α2I binding by rotavirus. Overall, these results suggest that the antiviral activity of pDGE-RGD is dependent on its ability to interact with α2β1 integrin. This notion is supported further by the finding that infection by integrin-independent virus CRW-8 was unaffected by pDGE-RGD.
The demonstration of the antiviral efficacy of pDGE (an 8-aa peptide harboring only the DGE triplet) extends data reported previously (21), which showed that individual peptide polymers containing the collagen-binding motif DGEA inhibit rotavirus infectivity by more than 60% in Caco-2 and MA104 cells and provided additional evidence of the importance of α2β1 in the rotavirus-cell attachment and entry process. We also tested the antiviral efficacy of DGEA peptide in our experimental systems, confirming the efficacy previously reported against the Wa strain (21) and showing that pDGE has a greater inhibitory capacity. Notably, an 8-aa mutant peptide (pAGE) showed a significant anti-rotaviral activity only at high concentrations. Taken together, these results confirm that the antiviral activity of pDGE-RGD peptide is mostly dependent on the integrity of the DGE triplet. Nevertheless, pDGE shows a less effective antiviral activity than pDGE-RGD, suggesting that a second region of this peptide provides an additional contribution in inhibiting rotavirus infectivity. To this end, we showed that 8-aa RGD peptides (namely pRGD-Hu, pRGD-Bo, and pRGD-Eq derived from human, bovine, or equine lactadherin, respectively) are endowed with a slight antiviral efficacy. Published studies provide evidence that rotavirus infectivity is not inhibited by RGD peptide, and function-blocking antibodies to RGD-binding integrins do not inhibit rotavirus infection (20, 23, 53). No evidence for Wa binding to integrins via RGD is available. We speculate that the slight antiviral efficacy of RGD peptides may be due to an indirect effect on rotavirus cell binding, perhaps by anchoring pDGE-RGD peptide to the cell surface and sterically blocking Wa virus access to receptors.
The results of the virucidal assay rule out the possibility of a direct effect of peptide treatment on rotavirus particles, whereas the time of addition assays suggest that pDGE-RGD might function by hindering the attachment of the virus to the host cell. The results of rotavirus cell-binding experiments confirm this conclusion; the titer of infective particles attached on the surface of pDGE-RGD-treated cells is significantly reduced compared with the untreated control. Moreover, pAGE-RGE peptide does not reduce the number of infectious particles attached to the cell surface, showing that this ability is strictly dependent on the integrity of the DGE or RGD sequences. Consistent with these considerations, we demonstrated that both pDGE and pDGE-RGD bind α2β1 integrin in a DGE-dependent manner, because pAGE does not interact with α2β1-expressing CHO-K1 cells. The pDGE and pDGE-RGD peptides interact in a dose-dependent manner with α2β1 expressed on the surface of CHO-K1 cells. Importantly, pDGE-RGD bound the cellular surface to a greater extent than pDGE, which parallels the higher antiviral activity of the longer peptide. Moreover, we show that pDGE-RGD (unlike pDGE) is able to interact with the surface of the CHO-K1 parental cell line that does not express α2β1 on the cellular surface. This result suggests that pDGE-RGD interacts with a second surface determinant, a clue that could explain the greater antiviral activity of pDGE-RGD compared with pDGE. Of the known RGD-binding integrins, CHO-K1 cells express α5β1 and αvβ5 but not α3β1 or αvβ3 (54, 55). It is likely that pDGE-RGD binds α5β1 or αvβ5 on CHO-K1 cells and α2β1-expressing CHO-K1 cells as well as binding α2β1 on α2β1-expressing CHO-K1 cells. The close proximity of the RGD and DGE motifs in pDGE-RGD is expected to preclude binding to both α2β1 and an RGD-recognizing integrin simultaneously by a single peptide molecule. However, peptide anchorage to an RGD integrin like α5β1, which is abundant on CHO-K1 cells, could prevent virus access to α2β1 and possibly other receptors. Interestingly, rotavirus-cell entry experiments showed that pDGE-RGD is able to inhibit rotavirus infectivity when added after virus-cell attachment, even if only slightly and at high concentrations. One possibility is that high concentrations of pDGE-RGD could interfere with virus-cell attachment, even after it took place, by mediating detachment of part of the viral population from the cell surface. It is also possible that integrin-bound pDGE-RGD is endocytosed and inhibits virion conversion into the transcriptionally active, double-layered particle, which for Wa has been proposed to occur during exit from late endosomes (56). Interestingly, Wa VP8* binds the GM1 ganglioside, which can associate with integrins such as α5β1 and α2β1 in membrane microdomains (57, 58) and has been proposed to classify cargo for uptake and trafficking in late endosomes (59). Binding of pDGE-RGD to β1 integrins internalized in endosomes also might interfere with Wa access to the cytoplasm, which is essential for replication (60).
Overall, the information obtained in the present study indicates that a specific 20-aa peptide from the N-terminal domain of donkey lactadherin is able to block rotavirus cell adhesion and infectivity in a DGE-dependent manner. Previously reported data (21) show that polymerization increases the effectiveness of peptide polymers containing the collagen-binding motif DGEA as rotavirus inhibitors by up to 10-fold, suggesting that it might be possible to develop more effective inhibitors of rotavirus-integrin interactions. In our experimental system, we also tested the in vitro antiviral activity of a dendrimeric peptide harboring multiple DGE motifs; this molecule displays an IC50 (3.6 μm) that is not significantly different from the pDGE-RGD IC50 (p > 0.05; data not shown). Currently, no antivirals are commercially available for rotavirus. The discovery of the pDGE-RGD peptide may prove useful in the development of inhibitors of receptor recognition by rotavirus or other viruses that use α2β1 integrin during cell attachment and entry like human echovirus 1 (61, 62).
This work was supported by a grant from the Piedmont Regional Call for Innovation Poles 2007–2013 (POR-FESR Asse I-I.1.3 Innovation and SMEs): “Development of a food supplement based on equine milk for the prevention of Rotavirus infections” received through Rotalactis s.r.l. A. C. and D. L. are founders of the start-up company Rotalactis that partially funded the research.
A. Civra, M. G. Giuffrida, M. Donalisio, L. Napolitano, Y. Takada, B. S. Coulson, A. Conti, and D. Lembo, unpublished observations.
- VP
- viral protein
- aa
- amino acid(s)
- MFGM
- milk fat globule membrane
- MEM
- minimum essential medium
- MOI
- multiplicity of infection
- PMF
- peptide mass fingerprinting
- SI
- selectivity index.
REFERENCES
- 1. Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., Wu Y., Sow S. O., Sur D., Breiman R. F., Faruque A. S. G., Zaidi A. K. M., Saha D., Alonso P. L., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ochieng J. B., Omore R., Oundo J. O., Hossain A., Das S. K., Ahmed S., Qureshi S., Quadri F., Adegbola R. A., Antonio M., Hossain M. J., Akinsola A., Mandomando I., Nhampossa T., Acácio S., Biswas K., O'Reilly C. E., Mintz E. D., Berkeley L. Y., Muhsen K., Sommerfelt H., Robins-Browne R. M., Levine M. M. (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222 [DOI] [PubMed] [Google Scholar]
- 2. Gleizes O., Desselberger U., Tatochenko V., Rodrigo C., Salman N., Mezner Z., Giaquinto C., Grimprel E. (2006) Nosocomial rotavirus infection in European countries: a review of the epidemiology, severity and economic burden of hospital-acquired rotavirus disease. Pediatr. Infect. Dis. J. 25, S12–S21 [DOI] [PubMed] [Google Scholar]
- 3. Prameela K. K., Vijaya L. R. (2012) The importance of breastfeeding in rotaviral diarrhoeas. Malays. J. Nutr. 18, 103–111 [PubMed] [Google Scholar]
- 4. Morrow A. L., Rangel J. M. (2004) Human milk protection against infectious diarrhea: implications for prevention and clinical care. Semin. Pediatr. Infect. Dis. 15, 221–228 [DOI] [PubMed] [Google Scholar]
- 5. Kurugöl Z., Geylani S., Karaca Y., Umay F., Erensoy S., Vardar F., Bak M., Yaprak I., Ozkinay F., Ozkinay C. (2003) Rotavirus gastroenteritis among children under five years of age in Izmir, Turkey. Turk. J. Pediatr. 45, 290–294 [PubMed] [Google Scholar]
- 6. Newburg D. S., Peterson J. A., Ruiz-Palacios G. M., Matson D. O., Morrow A. L., Shults J., Guerrero M. L., Chaturvedi P., Newburg S. O., Scallan C. D., Taylor M. R., Ceriani R. L., Pickering L. K. (1998) Role of human-milk lactadherin in protection against symptomatic rotavirus infection. Lancet 351, 1160–1164 [DOI] [PubMed] [Google Scholar]
- 7. Shaw A. L., Rothnagel R., Zeng C. Q., Lawton J. A., Ramig R. F., Estes M. K., Prasad B. V. (1996) Rotavirus structure: interactions between the structural proteins. Arch. Virol. Suppl. 12, 21–27 [DOI] [PubMed] [Google Scholar]
- 8. Yeager M., Berriman J. A., Baker T. S., Bellamy A. R. (1994) Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graham K. L., Halasz P., Tan Y., Hewish M. J., Takada Y., Mackow E. R., Robinson M. K., Coulson B. S. (2003) Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and recognize αXβ2 and αVβ3 by using VP7 during cell entry. J. Virol. 77, 9969–9978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ludert J. E., Feng N., Yu J. H., Broome R. L., Hoshino Y., Greenberg H. B. (1996) Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 70, 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bass D. M., Mackow E. R., Greenberg H. B. (1991) Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology 183, 602–610 [DOI] [PubMed] [Google Scholar]
- 12. Kirkwood C. D., Bishop R. F., Coulson B. S. (1998) Attachment and growth of human rotaviruses RV-3 and S12/85 in Caco-2 cells depend on VP4. J. Virol. 72, 9348–9352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Espejo R. T., López S., Arias C. (1981) Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37, 156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Estes M. K., Graham D. Y., Mason B. B. (1981) Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39, 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilbert J. M., Greenberg H. B. (1998) Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J. Virol. 72, 5323–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fleming F. E., Böhm R., Dang V. T., Holloway G., Haselhorst T., Madge P. D., Deveryshetty J., Yu X., Blanchard H., von Itzstein M., Coulson B. S. (2014) Evaluation of specificity and effects of monoclonal antibodies submitted to the Eighth Human Leucocyte Differentiation Antigen Workshop on rotavirus-cell attachment and entry. J. Virol. 88, 4558–457124501414 [Google Scholar]
- 17. Takada Y., Ye X., Simon S. (2007) The integrins. Genome Biol. 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mori S., Takada Y. (2013) Crosstalk between fibroblast growth factor (FGF) receptor and integrin through direct integrin binding to FGF and resulting integrin-FGF-FGFR ternary complex formation. Med. Sci. 10.3390/medsci1010020 [DOI] [Google Scholar]
- 19. Ciarlet M., Crawford S. E., Cheng E., Blutt S. E., Rice D. A., Bergelson J. M., Estes M. K. (2002) VLA-2 (α2β1) integrin promotes rotavirus entry into cells but is not necessary for rotavirus attachment. J. Virol. 76, 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coulson B. S., Londrigan S. L., Lee D. J. (1997) Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. U.S.A. 94, 5389–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Graham K. L., Zeng W., Takada Y., Jackson D. C., Coulson B. S. (2004) Effects on rotavirus cell binding and infection of monomeric and polymeric peptides containing α2β1 and αxβ2 integrin ligand sequences. J. Virol. 78, 11786–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guerrero C. A., Méndez E., Zárate S., Isa P., López S., Arias C. F. (2000) Integrin α(v)β(3) mediates rotavirus cell entry. Proc. Natl. Acad. Sci. U.S.A. 97, 14644–14649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hewish M. J., Takada Y., Coulson B. S. (2000) Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74, 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zárate S., Espinosa R., Romero P., Méndez E., Arias C. F., López S. (2000) The VP5 domain of VP4 can mediate attachment of rotaviruses to cells. J. Virol. 74, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Graham K. L., Takada Y., Coulson B. S. (2006) Rotavirus spike protein VP5* binds alpha2beta1 integrin on the cell surface and competes with virus for cell binding and infectivity. J. Gen. Virol. 87, 1275–1283 [DOI] [PubMed] [Google Scholar]
- 26. López Alvarez M. J. (2007) Proteins in human milk. Breastfeed Rev. 15, 5–16 [PubMed] [Google Scholar]
- 27. Kvistgaard A. S., Pallesen L. T., Arias C. F., López S., Petersen T. E., Heegaard C. W., Rasmussen J. T. (2004) Inhibitory effects of human and bovine milk constituents on rotavirus infections. J. Dairy Sci. 87, 4088–4096 [DOI] [PubMed] [Google Scholar]
- 28. Raymond A., Ensslin M. A., Shur B. D. (2009) SED1/MFG-E8: a bi-motif protein that orchestrates diverse cellular interactions. J. Cell Biochem. 106, 957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spertino S., Cipriani V., De Angelis C., Giuffrida M. G., Marsano F., Cavaletto M. (2012) Proteome profile and biological activity of caprine, bovine and human milk fat globules. Mol. Biosyst. 8, 967–974 [DOI] [PubMed] [Google Scholar]
- 30. Fortunato D., Giuffrida M. G., Cavaletto M., Garoffo L. P., Dellavalle G., Napolitano L., Giunta C., Fabris C., Bertino E., Coscia A., Conti A. (2003) Structural proteome of human colostral fat globule membrane proteins. Proteomics 3, 897–905 [DOI] [PubMed] [Google Scholar]
- 31. Cavaletto M., Giuffrida M. G., Conti A. (2004) The proteomic approach to analysis of human milk fat globule membrane. Clin. Chim. Acta 347, 41–48 [DOI] [PubMed] [Google Scholar]
- 32. D'Ambrosio C., Arena S., Salzano A. M., Renzone G., Ledda L., Scaloni A. (2008) A proteomic characterization of water buffalo milk fractions describing PTM of major species and the identification of minor components involved in nutrient delivery and defense against pathogens. Proteomics 17, 3657–3666 [DOI] [PubMed] [Google Scholar]
- 33. Pisanu S., Ghisaura S., Pagnozzi D., Biosa G., Tanca A., Roggio T., Uzzau S., Addis M. F. (2011) The sheep milk fat globule membrane proteome. J. Proteomics 74, 350–358 [DOI] [PubMed] [Google Scholar]
- 34. Barello C., Garoffo L. P., Montorfano G., Zava S., Berra B., Conti A., Giuffrida M. G. (2008) Analysis of major proteins and fat fractions associated with mare's milk fat globules. Mol. Nutr. Food Res. 52, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 35. Cebo C., Rebours E., Henry C., Makhzami S., Cosette P., Martin P. (2012) Identification of major milk fat globule membrane proteins from pony mare milk highlights the molecular diversity of lactadherin across species. J. Dairy Sci. 95, 1085–1098 [DOI] [PubMed] [Google Scholar]
- 36. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 37. Lambri M., Dordoni R., Giribaldi M., Riva Violetta M., Giuffrida M. G. (2012) Heat-unstable protein removal by different bentonite labels in white wines. LWT Food Sci. Technol. 10.1016/j.lwt.2011.11.022 [DOI] [Google Scholar]
- 38. Hellmann U., Wernstedt C., Góñez J., Heldin C. H. (1995) Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224, 451–455 [DOI] [PubMed] [Google Scholar]
- 39. Pappin D. J., Hojrup P., Bleasby A. J. (1993) Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3, 327–332; Erratum (1993) Curr. Biol. 3, 487 [DOI] [PubMed] [Google Scholar]
- 40. Reuter A., Fortunato D., Garoffo L. P., Napolitano L., Scheurer S., Giuffrida M. G., Vieths S., Conti A. (2005) Novel isoforms of Pru av 1 with diverging immunoglobulin E binding properties identified by a synergistic combination of molecular biology and proteomics. Proteomics 5, 282–289 [DOI] [PubMed] [Google Scholar]
- 41. Kamata T., Takada Y. (1994) Direct binding of collagen to the I domain of integrin α2β1 (VLA-2, CD49b/CD29) in a divalent cation-independent manner. J. Biol. Chem. 269, 26006–26010 [PubMed] [Google Scholar]
- 42. Kamata T., Wright R., Takada Y. (1995) Critical threonine and aspartic acid residues within the I domains of β2 integrins for interactions with intercellular adhesion molecule 1 (ICAM-1) and C3bi. J. Biol. Chem. 270, 12531–12535 [DOI] [PubMed] [Google Scholar]
- 43. King S. L., Kamata T., Cunningham J. A., Emsley J., Liddington R. C., Takada Y., Bergelson J. M. (1997) Echovirus 1 interaction with the human very late antigen-2 (integrin α2β1) I domain. Identification of two independent virus contact sites distinct from the metal ion-dependent adhesion site. J. Biol. Chem. 272, 28518–28522 [DOI] [PubMed] [Google Scholar]
- 44. Takada Y., Hemler M. E. (1989) The primary structure of the VLA-2/collagen receptor α 2 subunit (platelet GPIa): homology to other integrins and the presence of a possible collagen-binding domain. J. Cell Biol. 109, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takada Y., Ylänne J., Mandelman D., Puzon W., Ginsberg M. H. (1992) A point mutation of integrin β1 subunit blocks binding of α5β1 to fibronectin and invasin but not recruitment to adhesion plaques. J. Cell Biol. 119, 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nagesha H. S., Holmes I. H. (1988) New porcine rotavirus serotype antigenically related to human rotavirus serotype 3. J. Clin. Microbiol. 26, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coulson B. S., Fowler K. J., Bishop R. F., Cotton R. G. (1985) Neutralizing monoclonal antibodies to human rotavirus and indications of antigenic drift among strains from neonates. J. Virol. 54, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cohen J., Laporte J., Charpilienne A., Scherrer R. (1979) Activation of rotavirus RNA polymerase by calcium chelation. Arch. Virol. 60, 177–186 [DOI] [PubMed] [Google Scholar]
- 49. Estes M. K., Graham D. Y., Smith E. M., Gerba C. P. (1979) Rotavirus stability and inactivation. J. Gen. Virol. 43, 403–409 [DOI] [PubMed] [Google Scholar]
- 50. Taylor M. R., Couto J. R., Scallan C. D., Ceriani R. L., Peterson J. A. (1997) Lactadherin (formerly BA46), a membrane-associated glycoprotein expressed in human milk and breast carcinomas, promotes Arg-Gly-Asp (RGD)-dependent cell adhesion. DNA Cell Biol. 16, 861–869 [DOI] [PubMed] [Google Scholar]
- 51. Londrigan S. L., Hewish M. J., Thomson M. J., Sanders G. M., Mustafa H., Coulson B. S. (2000) Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 81, 2203–2213 [DOI] [PubMed] [Google Scholar]
- 52. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 53. Coulson B. S. (1997) in Leucocyte Typing VI (Kishimoto T., Kikutani H., von dem Borne A. E. G. K., Goyert S. M., Mason D. Y., Miyasaka M., Moretta L., Okumura K., Shaw S., Springer T. A., Sugamura K., Zola H., eds) pp. 391–393, Garland Publishing Inc., New York [Google Scholar]
- 54. Ahrens I. G., Moran N., Aylward K., Meade G., Moser M., Assefa D., Fitzgerald D. J., Bode C., Peter K. (2006) Evidence for a differential functional regulation of the two β(3)-integrins α(V)β(3) and α(IIb)β(3). Exp. Cell Res. 312, 925–937 [DOI] [PubMed] [Google Scholar]
- 55. Azab W., Osterrieder N. (2012) Glycoproteins D of equine herpesvirus type 1 (EHV-1) and EHV-4 determine cellular tropism independently of integrins. J. Virol. 86, 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Díaz-Salinas M. A., Silva-Ayala D., López S., Arias C. F. (2014) Rotaviruses reach late endosomes and require the cation-dependent mannose-6-phosphate receptor and the activity of cathepsin proteases to enter the cell. J. Virol. 88, 4389–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffmann C., Berking A., Agerer F., Buntru A., Neske F., Chhatwal G. S., Ohlsen K., Hauck C. R. (2010) Caveolin limits membrane microdomain mobility and integrin-mediated uptake of fibronectin-binding pathogens. J. Cell Sci. 123, 4280–4291 [DOI] [PubMed] [Google Scholar]
- 58. Ning Y., Buranda T., Hudson L. G. (2007) Activated epidermal growth factor receptor induces integrin α2 internalization via caveolae/raft-dependent endocytic pathway. J. Biol. Chem. 282, 6380–6387 [DOI] [PubMed] [Google Scholar]
- 59. Ravindran M. S., Tanner L. B., Wenk M. R. (2013) Sialic acid linkage in glycosphingolipids is a molecular correlate for trafficking and delivery of extracellular cargo. Traffic 14, 1182–1191 [DOI] [PubMed] [Google Scholar]
- 60. Desselberger U. (2014) Rotaviruses. Virus Res. 190, 75–96 [DOI] [PubMed] [Google Scholar]
- 61. Bergelson J. M., St John N. F., Kawaguchi S., Pasqualini R., Berdichevsky F., Hemler M. E., Finberg R. W. (1994) The I domain is essential for echovirus 1 interaction with VLA-2. Cell Adhes. Commun. 2, 455–464 [DOI] [PubMed] [Google Scholar]
- 62. Xing L., Huhtala M., Pietiäinen V., Käpylä J., Vuorinen K., Marjomäki V., Heino J., Johnson M. S., Hyypiä T., Cheng R. H. (2004) Structural and functional analysis of integrin α2I domain interaction with echovirus 1. J. Biol. Chem. 279, 11632–11638 [DOI] [PubMed] [Google Scholar]