Abstract
An appreciation of the importance of interactions between microbes and multicellular organisms is currently driving research in biology and biomedicine. Many human diseases involve interactions between the host and the microbiota, so investigating the mechanisms involved is important for human health. Although microbial ecology measurements capture considerable diversity of the communities between individuals, this diversity is highly problematic for reproducible experimental animal models that seek to establish the mechanistic basis for interactions within the overall host-microbial superorganism. Conflicting experimental results may be explained away through unknown differences in the microbiota composition between vivaria or between the microenvironment of different isolated cages. In this position paper, we propose standardised criteria for stabilised and defined experimental animal microbiotas to generate reproducible models of human disease that are suitable for systematic experimentation and are reproducible across different institutions.
The issues of modelling host-microbial mutualism
If we look up at the night sky and contemplate perhaps 1024 stars in the Universe, or glance down at the 1027 molecules in our body, a mere 1014 microbes in our lower intestine comes into perspective. Yet the microbiota that populates most animal and plant body surfaces (with various densities and compositions) is a story of big numbers, complex interactions and very multidimensional networks. However trendy it is to think about our bodies as host-microbial superorganisms, composed of eukaryotic and prokaryotic components, these are not new ideas. Louis Pasteur and Harvey Cushing both wrote about these concepts:1, 2 tempered by some scepticism on whether it would be possible to separate the two (microbial/host) components of the living plant or animal ‘superorganisms'. Although the germ-free-breeding program pioneered at Notre Dame University in the early years of the 20th century proved that animals could live without their prokaryotic and other microbial constituents,3, 4, 5 a further 100 years of experiments have shown that a germ-free existence is far from normal. Differences between germ-free and colonised animals can be seen essentially in every organ system,6 and germ-free animals lack a large chunk of the cumulative metabolic pathway map, comprising the synthesis of essential nutrients and vitamins, detoxification, and energy harvesting by metabolism of compounds resistant to the digestive processes of the animal host.7
Whereas observational astronomy has driven theoretical models in physics from the age of Keppler and Newton, the biology of host-microbial interactions remains largely empirical. We are still gazing more at ‘who is there' in our microbiotas than ‘what they are doing'. Yet the renaissance of interest in the microbiota has been driven by much better biological tools to explore the networks. ‘Omics are our equivalent of radio telescopes, spectroscopy, and X-ray astronomy, giving us the beginnings of the detailed insights into the genetics, transcriptional, translational, and metabolic events that must be compiled into generalised theoretical networks.
Our problem as biologists is that whereas astronomical phenomena can be conveniently split up—gravitation, how stars shine, black holes, cosmic inflation, and so on—to find universal laws, we have not yet developed an accepted or defined reductionist approach to understanding host-microbial mutualism. With over 1,000 operational taxonomic units in human feces and a variable composition from person to person, we end up with the equivalent of a different galaxy for every individual, where (for the moment at least) it is hard to generalise universal network laws.8, 9, 10, 11 The situation is hardly better with experimental animals: variability between vivaria is so wide that it is hard for two different research groups to look at the same biological galaxy.12, 13, 14 Even within a vivarium, the modern system of housing experimental animals within individually ventilated isolated cages results in stochastic changes in the microbiota so that every cage becomes its own unique galaxy.15
Handling biological variability is also not new. The trick has been to reduce complex adaptive systems to something manageable. In the conceptual trajectory from molecular structures of crystals, the metabolic reactions of purified proteins, functioning of subcellular organelles, behavior of cell lines, or the experimental animal model, the object is always the same—to understand what happens in an intact animal or human, living in a natural environment. We have been able to handle germ-line genetic variability of animal models, and to share defined models across institutions, by inbreeding programs16 and genetic manipulation of embryos. In experimental animal models, an important part of defining the environment is to ensure that the animals are not infected with any known pathogens for that species (specific pathogen-free: SPF, classified according to the American Association of Laboratory Animal Science or the Federation of European Laboratory Animal Science Associations17, 18). In this article, we discuss the ideas and challenges that we face for equivalent clear annotation and simplification of the microbiota in our experimental animals to allow different biologists to study the same defined phenomena and to start assembling the general laws of host-microbial networks.
Differences in microbiota composition have strong effects on host biology
It is clear that the differences in composition of the microbiota make an immense difference to the biology of the host. Some constituents are known to have especially strong effects—here we will discuss a few examples that illustrate this point.
The clostridial clade of segmented filamentous bacteria (SFB; Candidatus arthromitus) is an excellent example of a species that has such a powerful effect on host immunity that it can even be considered essential to drive the normal formation of the immune system.19 It is intimately attached to ileal intestinal epithelial cells through the eponymous filaments and ‘holdfasts'. SFB induces strong host immune responses including IgA,19, 20 Th17,21, 22 and regulatory T-cell induction,23 whereas colonisation levels are limited by expression of epithelial antimicrobial peptides.24 SFB also drives immunopathology in monocolonised mice including the arthritic phenotype of the K/BxN model25 and induction of experimental allergic encephalomyelitis (EAE).26 Whether or not immunopathology results from strong effects on the immune system by particular members of the microbiota probably depends on the host genetic background that shapes immune repertoires and regulatory mechanisms.25, 27
Serendipitous SFB colonisation differences between mouse lines from two different commercial suppliers led to the discovery of its strong effects on T-cell responses.21 Notwithstanding the acumen involved, this should be a wake-up call for our currently accepted standards of animal supply and experimentation with rather undefined starting conditions.
Sequencing of the SFB genome has shown that the potential metabolic repertoire is actually rather limited. Synthetic enzymes required for the amino acids alanine, threonine, glycine, methionine, arginine, and proline are incomplete. Endogenous synthesis of vitamins and cofactors including riboflavin, porphyrins, and thiamine is also not possible.28, 29 Given that SFB can monocolonise a mouse, it must be able to obtain these compounds from the diet or from effete epithelial cells. This is one illustration of the principle of interlinked metabolism between the diet, the host, and the microbiota.
Molecular mechanisms of host immune stimulation have been shown in a second example of strong effects of a specific bacterium—the human commensal Bacteroides fragilis. This organism expresses a zwitterionic polysaccharide (polysaccharide A; PSA) capable of activating B cells by binding its repetitive structure to the B-cell receptor and of activating T cells through (non-peptide) presentation through major histocompatibility complex Class II.30 Colonisation of germ-free animals with PSA-expressing B. fragilis (but not the PSA-deficient mutant) can normalise the otherwise hypoplastic lymphoid structures and T-cell subsets.31 B. fragilis also can maintain itself in an intimate niche (in this case within the colonic crypts) through direct stimulation by PSA of the Toll-like receptor 2 (TLR2) pathway on FoxP3+ regulatory T cells:32 a regulatory response that offsets activation of Th17 CD4+ cells to clear the organism. PSA can also stimulate TLR2 on plasmacytoid dendritic cell to further enhance interleukin (IL)-10 secretion by CD4+ T cells.33
Another effect comes from microbial production of short-chain fatty acids. Bacterial-derived short-chain fatty acids including butyrate have long been known to be a carbon source for colonic epithelial cells.34 More recently butyrate and propionate have been shown to enhance extrathymic differentiation of regulatory T cells.35 The mechanism is through acetylation and activation of the FoxP3 locus: the carboxylic acids achieve this through inhibition of histone deacetylase enzymes.36, 37 Production of short-chain fatty acids by Clostridia species thus probably accounts for their effectiveness in inducing Treg cells.37, 38
In the preceding paragraphs, we have concentrated on examples of how particular members of the microbiota exert strong effects on the ‘normal' makeup of the host immune system to make the case that studying the interactions at a mechanistic level requires definition and annotation of host-microbial superorganism composition. These examples of strong effects are almost certainly the host-microbial equivalent of monogenic traits. Before progressing to how the problem of interactions in more complex microbiotas may be solved experimentally, we need to mention three further aspects of the multidimensionality of host-microbial superorganism modeling.
Metabolism between the microbes and the host is very integrated—to the extent that the term ‘metabolic handshake' has been coined.39 The combined (bacterial/host) pathways for bile salt synthesis and metabolism provide a prime example of such integration.40
The phenotypes of many animal models of disease depend on exactly which microbiota is being carried. SFB as a trigger of arthritis or EAE was cited above, but this is the tip of the iceberg. For example, colitis can be triggered in immunodeficient mice carrying Helicobacter hepaticus 41, 42, 43, 44 or Helicobacter typhlonius. 45 Metabolic syndrome (including fatty liver and impaired glucose tolerance) is seen in mice with defective inflammasome response pathways carrying Prevotella or TM7.13, 46 More generally, models of obesity,47 autoimmunity,48 cardiovascular disease,49 behavior,50 and tumorigenesis51 are all dependent on microbiota composition.
Diet has a profound effect on the composition of the microbiota.11, 52, 53 Dietary composition not only affects disease susceptibility through direct effects on host metabolism, but also indirectly through changes in the composition and metabolism of intestinal microbes.54
This picture of the host-microbial superorganism is highly complex and variable, especially if the dimension of genetic variability in outbred individuals is added to the complexity of the microbiota.8, 55 The question is now, how realistic is it to define, control, and standardise these factors? Table 1 distinguishes the main interdependent parameters of host and microbial effects on mutualism and describes the potential strategies for standardisation.
Table 1. Spectrum of conditions that are compatible with mutualism.
| Life trajectory | Microbiota | Diet | Niches | Barriers | Metabolism |
|---|---|---|---|---|---|
| Host effects on mutualism | |||||
| In utero ↓ Birth ↓ Neonatal (lactation) ↓ Young adult ↓ Mature adult ↓ Old (±disease episodes) | -Vertical transfer of microbiota from mother or family to baby/infant -alteration of microbial diversity through microbial immigrants (horizontal transfer) -alteration of microbial diversity during mucosal disease | Host-digestible: -Composition of carbohydrates, fats, protein, minerals, vitamins digestible by the host | Host Sites of microbial colonisation: -Oral cavity -Oesophagus -Stomach -Small intestine -Colon -Skin -Genitourinary -Lungs Host provides different microbial niches within the gut: -Mucus -Luminal contents -Position along intestine | Physical: -Mucus -Epithelia Immune: -Anti-bacterial peptide secretion -Mucosal innate and adaptive immunity -Mesenteric lymph nodes and the liver firewalls | Host contributions to mutual metabolism: -bile acid secretion -shedding of mucus and other host molecules Host metabolic responses to microbial molecules |
| Microbiota effects on mutualism | |||||
| Variable composition of the microbiota at different stages of life | Variability in microbiota diversity and microbiota metabolism (fixed axenic, gnotobiotic, or diverse colonisation in experimental animals) | Microbiota salvages host indigestible dietary delivery: -Components of ‘resistant starch' including galactans, arabinans, cellulose, and undigestible fibre. -Carry-over minerals and vitamins | Within different microbial niches there is crossfeeding or competition for: -carbohydrate sources -minerals -electron donors/ acceptors | Microbiota provides colonisation resistance to immigrants and pathogens | Variable microbiota metabolism determined by: -microbiota diversity -niche -metabolic capability |
| Potential for standardisation and manipulation with isobiotic animals | |||||
| -Host developmental program effectively fixed -Microbiota manipulable with diet/lactation | -Microbiota manipulable in gnotobiotic animal models -Variable empirical results of probiotics in humans | Easy to manipulate: microbiota is variable with diet | Unclear: stool transplantation possible modality to change niche colonisation | Barriers variable according to alcohol use, drugs (e.g., nonsteroidal anti-inflammatory drugs) | Use of prebiotics |
The different approaches to solving the problem of microbiota diversity confounding results in experimental animal models
In ecological terms, we have a choice of two broad approaches to avoid diversity of the microbiota56 becoming a critical confounding factor in our animal experiments.
The established method is to take a specific habitat (in the case of mouse experimentation, this can be a single cage) and try to ensure that experimental measurements, and controls are carried out in parallel within this single habitat. In ecological parlance, beta diversity between different habitats57 (the microbiota ‘cage-effect' whereby different experimental and control groups are caged separately15) should be eliminated from the experiment. One can cohouse animals so that representatives of different experimental groups are within a single cage (replicated across a series of cages), so the differences between the groups and the cages can be independently measured.13 In the case of strain-combination experiments, an equivalent approach is to set a heterozygous breeding and compare littermates that are homozygous or heterozygous for the trait in question within a cage. Other measures can supplement cohousing, such as transferring bedding between cages or ventilating open top cages with a single atmosphere (as in a flexible-film isolator). Such experiments have the practical limitation that it is not permitted to overcrowd animals within a cage.
A more important limitation of this approach is that the starting alpha microbiota diversity within each individual animal at the outset is usually uncontrolled and undefined. Therefore, even if the experiment shows a result without confounding cage effects, it may not repeat in a different institution whose animals harbor a different starting microbiota, or at different times within the same institution following changes in the microbiological diversity of the vivarium. Another limitation is that it does not seek to control microbiological beta diversity between different animals in the same cage (this concern is amplified when one considers that there is also diversity between the different niches in the same animal that will not entirely synchronise with each other). These limitations restrict our ability to use replicate animals within the same cage or repeat the measurements in different animals to address the molecular mechanisms of host-microbial interactions.
We believe that there are immense experimental advantages from a second bottom-up approach; to manipulate the starting microbiota in a concerted effort to create standardised defined mouse colonies that can be shared across institutions.
Isogenic and isobiotic mice
In murine animal models, we can certainly manipulate the germ-line DNA by inbreeding and genetic-targeting techniques. As a reasonable approximation, we therefore have colonies of isogenic mice that are identical in the germline and where targeted genetic manipulations can be studied at the level of the intact animal. Enormous numbers of isogenic mice are now available (http://www.findmice.org), either as breeding colonies, as frozen embryos or sperm, or as embryonic stem cells (Figure 1).
Figure 1.
Time line of development of isobiotic mice in comparison with isogenic mice. The panel on the left highlights historical events in the development of gnotobiology. The panel on the right illustrates the huge progress made in the development of isogenic mice.
Although isogenic mice were born out of the breeding programs for amateur ‘mouse fanciers', their impact has been far-reaching.58 Our ideas about mammalian genetics,59 regulation of mammalian development,60, 61 transplantation biology,62 immune restriction,63, 64 infectious susceptibility,65 and tumor biology66 are founded on experiments with inbred animals and later development of the ability to manipulate their germline.67, 68 Quite apart from the ability to relate genes to functional biology, inbred mice reduce experimental variability and increase the inherent power of biological studies generally. One hopes that equivalent advances can be made with similar fixation of the microbiota in isobiotic mice.
Yet in comparison with large numbers of isogenic mouse strains, progress in harmonising the microbiota in isobiotic mice has been modest (Figure 1). It has long been realised that germ-free mice have hypoplastic lymphoid structures and are highly susceptible to intestinal and opportunistic infections.69, 70 In an attempt to normalise the host and limit infectious susceptibility, Russell et al. 71 carried out experiments in which germ-free mice were inoculated with combinations of bacteria from pure culture. To limit the effect of a single bacterial species overgrowing in the intestine, they devised a cocktail of eight species that eventually became popular: Escherichia coli var.mutabilis, Streptococcus faecalis, Lactobacillus acidophilus, Lactobacillus salivarius, group N Streptococcus, Bacteroides distasonis, a Clostridium species and a species of extremely oxygen-sensitive (EOS) spiral-shaped (fusiform) bacteria. Many different variations of similar microbiotas came into use.72 In 1987, the National Cancer Institute of the United States initiated standardisation of various microbiotas, resulting in a revision of this ‘Schaedler flora' to include a Flexistipes fusiform bacterium and three further EOS fusiform species.73, 74
The ‘altered Schaedler flora' (ASF) is still in widespread use today, although very few vivaria strictly control the microbial composition of the mouse colonies. Unfortunately, using standard husbandry, it is extremely easy to introduce immigrant microbial species that diversify the ASF,75 or to diversify other restricted microbiotas, especially if technicians and scientists do not strictly observe aseptic techniques in animal handling and care.76 Diversification is then passed vertically within cohoused interbreeding groups, which complicates interpretations about phenotypic differences between different inbred genetic strains.14 A second disadvantage of the ASF is that the organisms are not freely available (being protected under commercial patent) and recreating the ASF from scratch using pure cultures is very demanding, both in terms of growing the fastidious anaerobic constituents and the inoculation sequence required for them to colonise the intestine. Finally, the ASF is such a minimal microbiota that mice have a phenotype closer to germ-free than to colonised animals in some model systems.12, 76
This means that the ideal practical requirements of isobiotic mice would be (i) a stable microbiota of defined composition that is easy to regenerate using pure culture inoculation of microbes into germ-free recipients; (ii) resistance to immigrant microbes during standard vivaria conditions;72 (iii) metabolism that mirrors that of animals colonised with a diverse microbiota;76 and (iv) it should produce a phenotype in disease models that reflects diverse colonisation conditions.48
Man muß die Dinge so einfach wie möglich machen. Aber nicht einfacher. (One should make things as simple as possible. But no simpler. Albert Einstein)
Discussion of defined microbiotas normally revolves around their prokaryotic constituents. Yet these microbes are themselves carriers of bacteriophage and transposons (insertion sequences), whose movement adds to the inherent genetic variability of even the simplest microbiota. It would be over simplistic to imagine that an isobiotic microbiota does not have inherent genetic instability.
The 19 families of phages are distributed widely in the eubacteria and archaea: in practice, the order Caudovirales (tailed phage carrying ds DNA) and its families Siphoviridae, Podoviridae, and Myoviridae dominate the mammalian intestinal virome. Studies of the human fecal virome have shown differences between individuals (even twins), although within a single individual, the virome was quite stable and composed mainly of temperate (lysogenic) phage. This contrasts with phage in the microbial consortia in sludge and acid-mine effluent that are dominated by lytic bacteriophage behaving in a more predatory fashion.77 Sequence studies of CRISPR/Cas systems (a form of microbial immunity that can target immigrant DNA, leaving a genetic footprint in the spacers of the repetitive elements) also argue against the occurrence of many lytic events.77 Nevertheless, this is a newly emerging area of study, and additional experiments are required to elucidate the role of bacteriophages in shaping the gut microbiota and their contribution to disease (reviewed in ref. 78, 79). Phage attack has been shown to lead to transient changes in community structure and the ability of some bacterial species to acquire phage resistance.80 Emerging data on the identity and ecology of bacteriophages will be germane to developing or modulating isobiotic lines.
Phage particles carry microbial host genes that are relevant to adapting bacteria to the stresses of antibiotic treatment, and possibly also other environmental stress. Antibiotic treatment is an acute form of stressing intestinal bacteria, and certainly results in widely increased phage/bacterial association.81 It is still unclear how far other stresses cause phage-driven adaptation responses in the microbiota.
The upshot is that there is likely sufficient stability in the microbiota to make short/medium-term stability of mouse models using defined isobiotic mice possible, especially with a limited microbiota. However, the genetic background of the microbiota will be subject to drift, both through the endogenous bacterial mutation and movement of genetic elements between bacteria, which may be accelerated under conditions where the microbial consortia are stressed. A simplified defined microbiota where the prophage content and positions can be assessed by genomic sequencing will provide powerful tools to allow us to understand the dynamics and mechanisms of microbiota adaptation in response to different manipulations of the overall superorganism.
If plasticity of the microbiota metagenome is problematic over time, it should also be possible to restore an isobiotic system to defined starting conditions. This can be achieved through recolonisation of fresh germ-free mice with defined archived microbes from pure culture.
The rare biosphere
A further consideration for defined microbiotas is how one handles rare organisms that may have biological importance out of proportion to their frequency—the rare biosphere. Fungi are an important example: although commensal fungi are carried at low levels in healthy organisms, they can bloom or cause opportunistic infections during immune suppression.82 There is also evidence that fungi not only interact metabolically with bacteria and archaea, but also that they shape the succession of recolonisation after antibiotic treatment. Although there is no difficulty in eliminating these organisms from a microbiota when colonising germ-free mice with bacteria from pure culture, experimental inclusion of components of the rare biosphere may pose problems of reproducible low-level colonisation in appropriate niches.
Axenic and gnotobiotic technology
One of the reasons for the development of isolated ventilated cages was to limit the transmission of pathogens or pathobionts within an animal colony.72 Unfortunately, the cages can also provide numerous microenvironments for drift and diversification through the immigration within the same animal-holding room.14
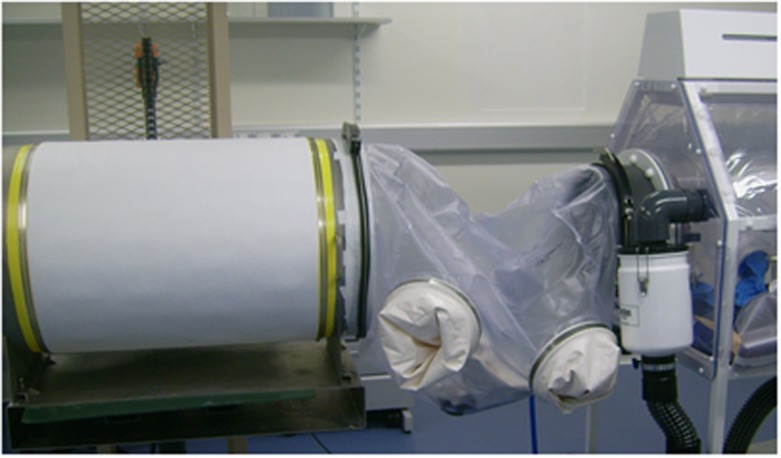
Fortunately, technology for maintaining germ-free or gnotobiotic mice is well established. Flexible film isolators are used that are initially sterilised by an internal mist of 3% peracetic acid, and then ventilated with high-efficiency particulate air-filtered air.6 Sterilised cages, bedding, food, and water can be imported into this environment from large steel drums, which have been passed through an autoclave with an appropriately large sterilisation chamber (Figure 2).
Figure 2.
Sterile connection between a material drum and a flexible film isolator. The large steel drum contains sterile materials to be imported into the isolator, such as food, bedding, or cages. The drum is sealed and autoclaved and then connected to the isolator via a plastic sleeve that is sterilised by spraying with 2% peracetic acid, which then permits transfer of the sterile material into the isolator.
Imports from drums into the isolators are accomplished through plastic sleeves, which are attached to both the port on the isolator and the outside of the drum, then sterilised inside through spraying with 2% peracetic acid. After an hour to allow the peracetic acid chemical sterilisation to be effective, the inner door of the isolator port is opened, the membrane sealing the drum is broken, and the materials are transferred into the isolator using a glove that is set into the plastic sleeve. Germ-free or gnotobiotic animals can be transferred either by a two-stage transfer (first into an empty sterilised drum with a resealable cap, followed by connection to a second isolator) or by directly connecting the isolators with a plastic sleeve.
Our experience has been that animals that are germ-free, or colonised with a limited microbiota, can be maintained over many years using this system, whereas individually ventilated cages are stable over months, but each cage constitutes a microenvironment that is susceptible to microbiota diversification through introduction of new species, probably through minor errors in aseptic handling.
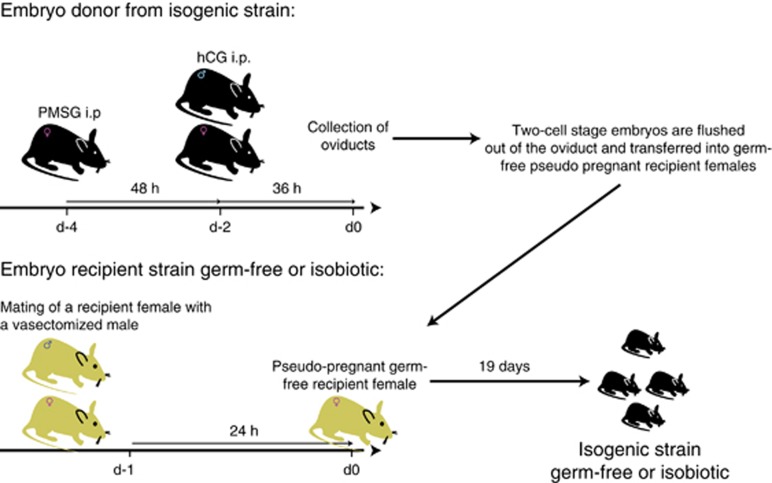
An important adjunct to this technology is the ability to transfer different mouse strains into the desired axenic or gnotobiotic background. This can be done by embryo transfer (Figure 3). Females of the new strain that needs to be imported are superovulated and mated; then the two-cell-stage embryos flushed from their oviducts and meticulously washed in sterile M2 medium. The embryos are then surgically transferred into pseudopregnant germ-free or gnotobiotic females with the required microbiota, by linking their isolator with a sterile microsurgery laminar-flow cabinet. Our experience is that this is a highly reliable and relatively fast way of altering the hygiene status of almost any strain of mice, and in the case of gnotobiotic (isobiotic) transfers, the new strain acquires the microbiota of the foster mothers.
Figure 3.
Experimental protocol for superovulation of embryo donor females followed by embryo transfer into germ-free or isobiotic pseudopregnant recipient females.
One can therefore make a case that the technology of axenic embryo transfer, although specialised, is good for purpose, and that isogenic and isobiotic backgrounds can be manipulated according to experimental need.
The problem of transferring animals that are germ-free or with a defined microbial composition between these flexible-film isolators in different institutions has also been solved using special ‘transport sleeves' (Figure 4). The scientific community is therefore potentially in a position to raise the standards whereby it defines and stabilises 90% of the cellular composition and much of ‘the other genome' in its animal models.
Figure 4.

Specialised transport sleeve. The photo shows an example of a specialised transport sleeve developed to allow shipping of germ-free animals between facilities. The example shown is available from Taconic. Many germ-free facilities have developed their own transport devices that reflect the design of their isolators. Such transport containers allow importation of germ-free or isobiotic mice into isolators at the new facility and ensure hygiene is maintained during transport.
Ethical and scientific advantages of defined stable microbiotas
Ethics require us to reduce refine and replace animal experimentation.83, 84 It is well established that isogenic (genetically inbred) mice have substantially less inherent phenotypic variability across a wide range of experimental protocols than outbred mice.85 Thus, experiments with inbred animals are inherently better powered, requiring fewer per experimental group. This is true not only in immunology, but also in other biological science at the whole-animal level, including behavioral and pharmacological studies. Misguided use of outbred animals to obviate the concern that complex (host genetic) traits are being studied where a single inbred strain may be unrepresentative, decreases experimental power and remains informative only if the animal numbers are considerably escalated. A better solution is usually to use multiple isogenic experimental strains (all of which remain appropriately powered) and a factorial design.85 Translating the factors for complex host genetic traits as differences in microbial composition (or the microbial metagenome), a similar argument can be advanced for isobiotic mice. Here, we can stabilise much of the inherent variability in the microbiota of current vivaria, increasing experimental power, with the potential to simplify the microbial complexity sufficiently to dissect metabolic networks and the mechanisms of effects on the host. By extrapolation from experiments using different isogenic strains to understand complex traits, different isobiotic strains are likely to be required to understand the effects of different microbial consortia.
Of course, it is entirely appropriate to use outbred animal stocks and human populations in high-resolution mapping of complex traits.86 Large numbers are required. For example, 75,000 subjects and controls have been genotyped to detect 163 loci linked to inflammatory bowel disease,87 yet only ∼20% of the variance in disease risk is explained. The complexity of the human microbiome and the distinct consortia, named enterotypes, has been so far accomplished with smaller numbers of human metagenomes.8 Studies of the microbiota composition according to diet or to disease susceptibility have been reported so far with 10s–100s of subjects.9, 11 It remains a challenge that our reference microbial genomes are incomplete.88 Nevertheless, these studies show us the real world complexity of host microbial interactions, and simplified isobiotic microbiotas must be compared with complex colonisations52 to collectively model this complexity, albeit with the key advantage that they leverage sufficient power to dissect the individual mechanistic pathways experimentally.
Proposed criteria for defined microbiotas?
From the previous discussion, although the generation of standard microbiota mice is to some extent empirical, we propose a number of criteria desirable in isobiotic mouse strains (on a wild-type host isogenic background).
Microbial stability over generations on a fixed sterile ‘open-source' diet when maintained under aseptic conditions.
Transferability of the isobiotic microbiota from a pseudopregnant female to pups (of the same or a different isogenic strain) after aseptic two-cell-stage embryo transfer.
The ability consistently to regenerate the particular isobiotic microbiota de novo using germ-free wild-type mice inoculated with the appropriate combination of microbes from cryopreserved pure cultures.
Completeness of overall microbial metabolic pathway representation in the sequenced metagenome.
Distinct microbiota representation (in different isobiotic lines) at species level to avoid direct replication of an individual line: phylogenetic overlap above species level should be expected and tolerated.
No abnormalities in wild-type mice in clinical chemistry, peripheral blood count parameters, gross organ histology, body composition, development or fecundity.
Representative metabolomic and immunological profiles of serum, urine, and organs compared with mice harboring a diverse microbiota.
Normal pathogen colonisation resistance and inflammatory and autoimmune model disease susceptibility.
Relative stability of the isobiotic microbiota under aseptic husbandry within individually ventilated cages.
The experimental approach to making these isobiotic lines can either be (a) through design to obtain representatives of defined (where possible genetically tractable) organisms known to be common in the gastrointestinal (or skin or airways) tract on a phylum/class/order/family basis (e.g., Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria); (b) empirical through cocktail colonisation of germ-free mice and following the resulting microbiota out to stability; or (c) diversification of established isobiotic lines.
In order to ensure reproducibility of experimental data, it is essential that the microbial consortia are stable over time and horizontally and vertically transmissible (see criteria i). Such stability under aseptic conditions will protect from divergence of the microbiota over time. It is clear that it is important to standardise diet for stability of the microbiota.52 As the objective is ultimately to share standardised isobiotic mice with other facilities worldwide, the use of commercially available open-label diets, with 40 kGy sterilisation to eliminate potential microbial immigrants will enable different institutions to reproduce the conditions under which the microbiota is stable. Generating isobiotic lines that are relatively stable will allow researchers to import these lines and house them in individually ventilated cages for specific short-term experiments (see criteria ix), so that the benefits of standardisation are available to those without the resources to establish isolator-based breeding programs.
In order to be able to study different genetically targeted or inbred wild-type mouse strains for comparative work on biological systems under the same isobiotic microbiota conditions, it is important that the microbiotas can be vertically transferred from a pseudopregnant recipient of the required isobiotic background to her embryo-transferred pups (see criteria ii). The technology of ultraclean embryo transfer described above to transfer the isobiotic microbiota has proved successful in our unit.
Isobiotic microbiotas will be analogous to inbred isogenic mouse strains, in that simplification of the traits (in the other genome) will necessarily not be representative of diverse microbiotas. We argued in the previous section that the increase in experimental power and discrimination offsets this disadvantage, especially if multiple isobiotic strains (all of which would remain appropriately powered) and a factorial design are used. However, the outcome does not have to remain entirely empirical, as metagenomic analysis can determine the extent of pathway coverage for each isobiotic microbiota. One can therefore aim to provide complete coverage of the microbial metabolic pathway (see criteria iv) such that each isobiotic consortia at least has the potential to provide a metabolic profile similar to that provided by diverse microbiotas. Moreover, one can assess the consequences of each isobiotic microbiota on (a) the immune phenotype, (b) its phenotype during defined disease models, and (c) the extent to which the intestinal and host metabolome corresponds to mice with a diverse microbiota (see criteria vi, vii, and viii). This also means that different microbiotas need to be maintained in vivaria or deliberately generated from pooled recolonisation of germ-free sentinels, starting from mouse microbial commensal strains from pure culture (see criteria iii).
Sentinels from newly created isobiotic lines need to be sampled across time to provide materials for microbiological, metabolomic, and transcriptomic analyses to determine how increased microbiota diversity impacts on the microbial and host metabolome and host organ function. These readouts need to be benchmarked against phenotypes with diverse microbiotas and microbiotas from commercial breeders.
Standardisation or anarchy?
In this article, we have tried to argue that defined microbiotas should be available in experimental rodents. In practice, strict ‘standardisation' will be impractical because many research groups will wish to manipulate their microbiota for experimental purposes and a single standard cocktail of organisms would not capture the phenotypic variability that we are seeking to model. Indeed, imposing strict standardisation rules would be anarchy and would decrease our ability to discover new correlations. Avoiding anarchy will depend on (a) sharing resources of isobiotic animals and constituent microbes between institutions; (b) avoiding restrictive patents limiting the availability or use of key isobiotic models or their constituent microbes for publicly funded research; and (c) accepting that some questions can be satisfactorily answered with simpler methods such as cohousing. Conversely, the research community needs to agree with standards of biological definition and provide increased reporting of the baseline microbiotas (e.g., through high throughput 16S rRNA or metagenomic gene sequencing) and their consequences in host-microbial superorganism metabolism when using non-standardised microbiotas. This in turn will require making microbiota, microbiome, and metabolome determination more available and more affordable for researchers. Ideally, a diversity of defined isobiotic models will bring a depth of information in reproducible settings that allow us to understand general rules for the networks between prokaryotic and eukaryotic components of the host-microbial superorganism and to relate this to human health at an individual level.
Acknowledgments
A.J.M. received funding from the Swiss National Science Foundation. K.D.M. received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC grant Agreement No. 281785 and the Swiss National Science Foundation.
The authors declare no conflict of interest.
References
- Pasteur L. Observations relatives a la note de M. Duclaux. Compt. Rend. 1885;100:69. [Google Scholar]
- Cushing H, Livingood L.E. Experimental and surgical notes upon the bacteriology of the upper portion of he alimentary canal, with observations on the establishment there of an amicrobic state as a preliminary to operative procedures on the stomach and small intestine. John's Hopkins Hospital Rep. 1900;9:543–549. [Google Scholar]
- Cohendy M. Experiences sur la vie sans microbes. Compt. Rend. 1912;154:533–536. [Google Scholar]
- Cohendy M. Experiences sur la vie sans microbes, élevage aseptique de cobages. Compt. rend. 1914;158:1283–1284. [Google Scholar]
- Glimstedt G. Bakterienfrei Meerschweinchen. Aufzucht, Lebensfähigkeit und Wachstum, nebst Untersuchung über das lymphatische Gewebe. Acta Pathol. Microbiol. Scand. Suppl. 1936;30:1–295. [Google Scholar]
- Smith K., McCoy K.D, Macpherson A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Blottiere H.M., de Vos W.M., Ehrlich S.D, Dore J. Human intestinal metagenomics: state of the art and future. Curr. Opin. Microbiol. 2013;16:232–239. doi: 10.1016/j.mib.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Cotillard A, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Wilson K.H., Brown R.S., Andersen G.L., Tsang J, Sartor B. Comparison of fecal biota from specific pathogen free and feral mice. Anaerobe. 2006;12:249–253. doi: 10.1016/j.anaerobe.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J, et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse H.C.Origins of Inbred Mice Academic Press; (available online at http://www.informatics.jax.org/morsebook/index.shtml ( 1978 [Google Scholar]
- Guillen J. FELASA guidelines and recommendations. J. Am. Assoc. Lab. Anim. Sci. 2012;51:311–321. [PMC free article] [PubMed] [Google Scholar]
- Mahler M, et al. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab. Anim. 2014;48:178–192. doi: 10.1177/0023677213516312. [DOI] [PubMed] [Google Scholar]
- Talham G.L., Jiang H.Q., Bos N.A, Cebra J.J. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen H.L, et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Salzman N.H, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.K., Menezes J.S., Umesaki Y, Mazmanian S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA. 2011;108 (Suppl 1:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering C, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–568. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczesnak A, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp S.J., Harrington E.D., Quake S.R., Relman D.A, Blainey P.C. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S.K, Kasper D.L. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Liu C.H., Tzianabos A.O, Kasper D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Round J.L, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S., Erturk-Hasdemir D., Ochoa-Reparaz J., Reinecker H.C, Kasper D.L. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15:413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger W.E. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Holmes E., Li J.V., Athanasiou T., Ashrafian H, Nicholson J.K. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Swann J.R, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl Acad. Sci. USA. 2011;108 (Suppl 1:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg M.C, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg M.C, et al. Bacteria-triggered CD4+ T Regulatory Cells Suppress Helicobacter hepaticus-induced Colitis. J. Exp. Med. 2002;196:505–515. doi: 10.1084/jem.20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy K.J., Salaun L., Cahill R., Dougan G., Saunders N.J, Powrie F. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 2003;197:111–119. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell N, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J.G, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J.F, Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Sears C.L, Garrett W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A.L, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Backhed F., Fulton L, Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith J.R, Sartor R.B. The role of diet on intestinal microbiota metabolism: downstream impacts on host immune function and health, and therapeutic implications. J. Gastroenterol. 2014;49:785–798. doi: 10.1007/s00535-014-0953-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E.K., Stagaman K., Dethlefsen L., Bohannan B.J, Relman D.A. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertenstein M., Vintersten K, Behringer R. Manipulating the Mouse Embryo. Cold Spring Harbor Laboratory Press: Cold Spring Harbor: New York; 2003. Spenectomy; pp. 280–281. [Google Scholar]
- Russell E.S. One man's influence: a tribute to William Ernest Castle. J. Hered. 1954;45:210–213. [Google Scholar]
- Gluecksohn-Schoenheimer S. The development of two tailless mutants in the house mouse. Genetics. 1938;23:573–584. doi: 10.1093/genetics/23.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluecksohn-Schoenheimer S. The effect of an early lethal (to) in the house mouse. Genetics. 1940;25:391–400. doi: 10.1093/genetics/25.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingham R.E., Brent L, Medawar P.B. Quantitative Studies on Tissue Transplantation Immunity. III. Actively Acquired Tolerance. Phil. Trans. R. Soc. Lond. B. 1956;239:357–414. [Google Scholar]
- Zinkernagel R.M, Doherty P.C. Immunological surveillance against altered self components by sensitised T lymphocytes in lymphocytic choriomeningitis. Nature. 1974;251:547–548. doi: 10.1038/251547a0. [DOI] [PubMed] [Google Scholar]
- Zinkernagel R.M, Doherty P.C. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- Webster L.T. Microbic Virulence and Host Susceptibility in Paratyphoid-Enteritidis Infection of White Mice: Iv. The Effect of Selective Breeding on Host Resistance. J. Exp. Med. 1924;39:879–886. doi: 10.1084/jem.39.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorer P.A., Lyman S, Snell G.D. Studies on the genetic and antigenic basis of tumour transplantation. Linkage between a histocompatibility gene and 'fused' in mice. Proc. Roy. Soc. Lond. B. 1948;135:499–505. [Google Scholar]
- Evans M.J, Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa M. The lymphatic system of germ free guinea pigs. Ann. N Y Acad. Sci. 1959;78:221–236. doi: 10.1111/j.1749-6632.1959.tb53105.x. [DOI] [PubMed] [Google Scholar]
- Dubos R, Schaedler R.W. The effect of the intestinal flora on the growth rate of mice, and on their susceptibility to experimental infections. J. Exp. Med. 1960;111:407–411. doi: 10.1084/jem.111.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler R.W., Dubos R, Costello R. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 1965;122:77–83. doi: 10.1084/jem.122.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trexler P.C, Orcutt R.P. 50 years of laboratory animal science. American Association for Laboratory Animal Science: New York; 1999. Development of Gnotobiotics and Contamination Control in Laboratory Animal Science; pp. 121–128. [Google Scholar]
- Orcutt R.P., Gianni F.J, Judge R.J. Development of an 'Altered Schaedler flora' for NCI gnotobiotic rodents. Microecol. Ther. 1987;17:59. [Google Scholar]
- Dewhirst F.E, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehr M, et al. Charles River altered Schaedler flora (CRASF) remained stable for four years in a mouse colony housed in individually ventilated cages. Lab. Anim. 2009;43:362–370. doi: 10.1258/la.2009.0080075. [DOI] [PubMed] [Google Scholar]
- Norin E, Midtvedt T. Intestinal microflora functions in laboratory mice claimed to harbor a ‘normal' intestinal microflora. Is the SPF concept running out of date. Anaerobe. 2010;16:311–313. doi: 10.1016/j.anaerobe.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S., Shanahan F., Stanton C., Hill C., Coffey A, Ross R.P. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut microbes. 2013;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe M., Leclerc M., Tinsley C.R, Petit M.A. Bacteriophages: an underestimated role in human and animal health. Front. Cell. Infect Microbe. 2014;4:39. doi: 10.3389/fcimb.2014.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Wu M., McNulty N.P., Rohwer F.L, Gordon J.I. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl Acad. Sci. USA. 2013;110:20236–20241. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S.R., Lee H.H., Spina C.S, Collins J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagel G.B, Noverr M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W.M.S, Birch R.L. The Principles of Humane Experimental Technique. Methuen: London; 1959. [Google Scholar]
- Wells D.J. Animal welfare and the 3Rs in European biomedical research. Ann. N Y Acad. Sci. 2011;1245:14–16. doi: 10.1111/j.1749-6632.2011.06335.x. [DOI] [PubMed] [Google Scholar]
- Chia R., Achilli F., Festing M.F, Fisher E.M. The origins and uses of mouse outbred stocks. Nat. Genet. 2005;37:1181–1186. doi: 10.1038/ng1665. [DOI] [PubMed] [Google Scholar]
- Flint J., Valdar W., Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat. methods. 2013;10:1196–1199. doi: 10.1038/nmeth.2693. [DOI] [PubMed] [Google Scholar]