Abstract
Isolated lymphoid follicles (ILFs) develop after birth in the small and large intestines (SI and LI) and represent a dynamic response of the gut immune system to the microbiota. Despite their similarities, ILF development in the SI and LI differs on a number of levels. We show that unlike ILF in the SI, the microbiota inhibits ILF development in the colon as conventionalization of germ-free mice reduced colonic ILFs. From this, we identified a novel mechanism regulating colonic ILF development through the action of interleukin (IL)-25 on IL-23 and its ability to modulate T regulatory cell (Treg) differentiation. Colonic ILF develop in the absence of a number of factors required for the development of their SI counterparts and can be specifically suppressed by factors other than IL-25. However, IL-23 is the only factor identified that specifically promotes colonic ILFs without affecting SI-ILF development. Both IL-23 and ILFs are associated with inflammatory bowel disease, suggesting that disruption to this pathway may have an important role in the breakdown of microbiota-immune homeostasis.
INTRODUCTION
The intestinal microbiota has a profound effect on the host, highlighted by the fact that alterations have been associated with a number of pathologies and less favorable outcomes in disease models.1 Central to this effect is the homeostasis that is reached between the intestinal microbiota and the host immune system. Cross-talk between the host immune system and the microbiota is critical for the development of the gut immune system and the gut-associated lymphoid tissue that in turn regulates the microbiota.2 Disturbance of this balance has been implicated in the development of intestinal pathology and intestinal lymphoid tissues have been associated with human inflammatory bowel disease and increased pathology in mouse models.3, 4, 5, 6 One pronounced effect of the microbiota on the host is the dynamic regulation of isolated lymphoid follicle (ILF) development,7, 8 single B-cell follicles that act as inductive sites for immunoglobulin A (IgA) production.9
The gut-associated lymphoid tissue consists of multifollicular Peyer's patches and similar structures within the large intestine (LI) that develop in utero,10 in addition to a continuum of smaller structures, referred to as solitary isolated lymphoid tissue that develops after birth.7, 8 The smallest of these, termed cryptopatches (CPs), consist primarily of retinoic acid-related orphan receptor (ROR)-γt+ innate lymphoid cells (ILCs).11, 12 CPs are considered to develop into single B-cell follicle-containing ILFs. ILFs exist in a range of sizes, with the larger displaying germinal center characteristics (e.g., follicular dendritic cell networks) and referred to as mature ILFs (mILFs).8, 13, 14 Although lymphotoxin-α/β (LTα1/β2) is required for Peyer's patches and ILF development, the postnatal development of CPs and ILFs requires additional environmental signals. CPs are reduced in mice lacking the aryl hydrocarbon receptor or fed a diet lacking the natural ligands for this receptor,15 but are present in germ-free mice. ILFs are reduced in the small intestines (SI) of germ-free mice and microbial colonization induces their development.8 Antibiotic treatment also reduces ILFs in the SI, supporting a central role for the microbiota in both the induction and maintenance of SI-ILFs.16, 17 An additional microbiota-dependent, but CP-independent, B-cell aggregate has been described in the colon of LTα−/− and RORγt−/− mice, referred to as tertiary lymphoid tissues (tLTs).6, 18
ILF development in the SI is dependent on a number of factors involved in lympho-organogenesis.7, 14, 19, 20 ILFs are also reduced or absent in mice lacking microbial recognition molecules, such as myeloid differentiation primary response gene 88, nucleotide-binding oligomerization domain-containing protein-2 (NOD2) and NOD1.16 Interestingly, although some of these factors also block ILF development in the LI, mice that lack RANKL (receptor activator of NF-κB ligand),19 or CXCL13,21 still develop ILF-like structures in the colon. Colonic ILFs also develop earlier than their SI counterparts and have been observed in germ-free and antibiotic-treated mice, suggesting that the regulation of LI-ILF development differs significantly from SI-ILFs.16, 17, 21 The basis for this differential regulation is undetermined.
Here, we have identified a novel colon-specific regulatory loop of ILF development in the steady state mediated by interleukin (IL)-25 and IL-23. Our observation that conventionalization of germ-free mice specifically reduced LI-ILFs, and that IL-25−/− and IL-23p19−/− mice have alterations in colonic ILFs, suggest that the microbiota regulates ILFs in the LI by modulating expression of these cytokines. Thus, our data suggest that the reciprocal regulation of colonic ILF development by IL-25 and IL-23 has important implications for the understanding of immune-microbiota homeostasis and the development of intestinal pathology.
RESULTS
LI-ILF development and maturation is inhibited by the microbiota
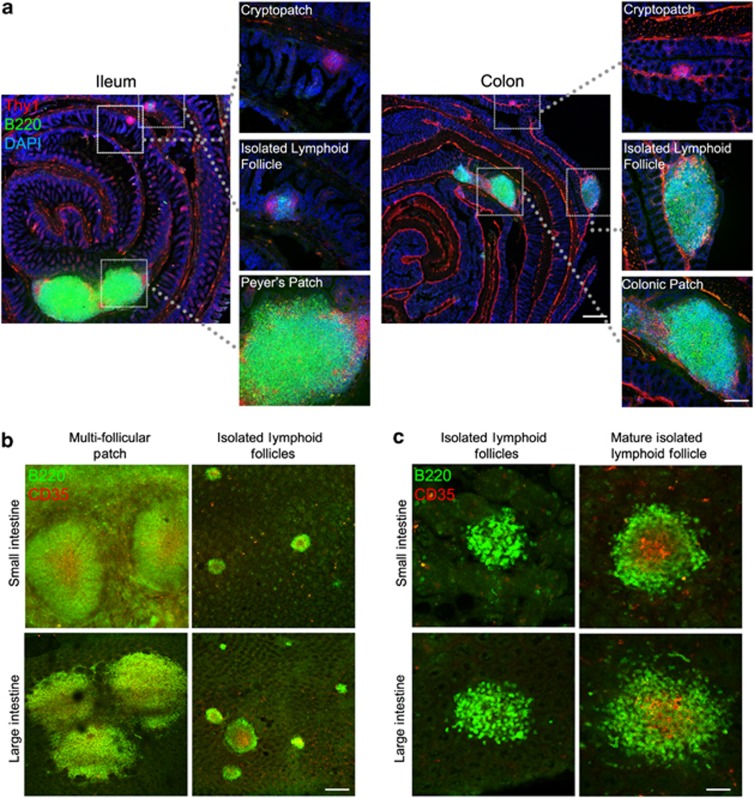
Murine intestines contain a number of organized lymphoid tissues with differential developmental origins. Multifollicular Peyer's patches and their equivalents in the LI develop in utero, whereas solitary B-cell aggregates, referred to as ILFs, develop after birth from CPs. Colonic tLTs may also develop after birth, but these currently cannot be differentiated from ILFs in mice with CPs by immunohistochemistry alone.18 LI-ILFs can be easily differentiated from patches in tissue sections. For example, colonic patches are multifollicular, extend into the submucosa, and contain defined T-cell areas, whereas LI-ILFs are solitary, are located only in the lamina propria (LP), and lack defined T-cell areas21 (Figure 1a). Although these same criteria cannot be applied after whole-mount immunostaining of B cells, patches can be readily differentiated by whole-mount immunostaining (Figure 1b) by their multifollicular nature that may not always be apparent in tissue sections. Despite this, some colonic patches may be single domed and thus difficult to differentiate from large ILFs. However, these are few in number21 and vastly outweighed by the larger numbers of ILFs. Analysis by whole-mount immunostaining remains a superior method to analyze total ILF numbers by reducing sample bias and the effects of ILF enlargement. A small number of ILFs contain follicular dendritic cell networks, visualized by staining with anti-CD35 monoclonal antibody (mAb), and are considered to be mILFs (Figure 1c). CPs consist of ILCs and can be visualized by immunostaining for Thy1 (Figure 1a). The development of CPs is independent of the microbiota, whereas SI-ILF development is predominantly microbiota dependent.8, 14, 16, 17 The effect of the microbiota on LI-ILFs is more variable, as unlike the reduced ILFs observed in the SI of germ-free and antibiotic-treated mice, colonic ILFs have been reported to develop similarly in specific pathogen-free (SPF) and germ-free mice and to be less affected by antibiotic treatment.17, 21
Figure 1.
Gastrointestinal lymphoid tissue of the small and large intestines. (a) Sections of small and large intestine were stained for Thy1 (red), B220 (green), and nuclei (blue) to detect cryptopatches, isolated lymphoid follicles, and multifollicular patches (Peyer's patches in the small intestine and colonic patches in the large intestine). Bar=500 μm in low power panels and 200 μm in higher power panels. (b) Intestinal segments were whole-mount stained for B220 (green) and CD35 (red) to detect multifollicular patches and isolated lymphoid follicles. Bar=200 μm. (c) Mature isolated lymphoid follicles can be differentiated by CD35 staining (red), visualizing follicular dendritic cell networks. Bar=50 μm.
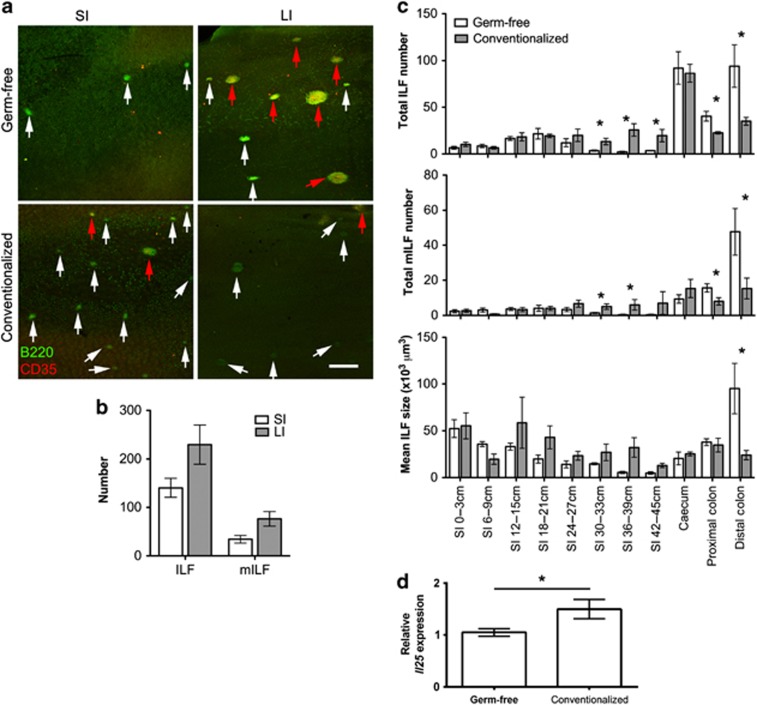
Using whole-mount immunostaining with anti-B220 mAb to detect B-cell aggregates (ILFs) and anti-CD35 mAb to detect follicular dendritic cell networks (mILFs), our own analysis of total ILF and mILF numbers in germ-free mice supported the microbial independence of ILF development and maturation in the LI. In germ-free mice, ILFs were rarely detected in the SI, whereas the LI contained numerous ILFs and mILFs (Figure 2a,b). To show that no further ILF development occurred in the presence of the microbiota, germ-free mice were transferred to conventional housing and their intestines whole-mount immunostained as above. As expected, conventionalization induced a significant increase in ileal ILFs and mILFs (Figure 2c). Interestingly, not only did no further ILF development occur in the cecum and colon of conventionalized mice, a significant reduction in both colonic ILFs and mILFs was observed, associated with a significantly decreased ILF size in the distal colon. IL-25 is produced by the intestinal epithelium in response to the microbiota resulting in reduced ILC responses and IL-23 expression22, 23 and thus may be involved in ILF regulation. Our analysis confirmed that Il25 mRNA was significantly upregulated in the colon of conventionalized mice, correlating with the reduction in ILFs (Figure 2d). These data demonstrate that colonic ILFs, in contrast to their SI counterparts, can be negatively regulated by the microbiota.
Figure 2.
Isolated lymphoid follicle (ILF) development and maturation in the large intestine (LI) of germ-free mice is suppressed by the microbiota. (a) Alternate 3 cm sections of small intestine (SI) and the whole LI were whole-mount immunostained for B220 (green) to detect ILFs and CD35 (red) to detect mature ILFs (mILFs). Representative images of SI and LI are shown. White arrowheads highlight immature ILFs and red arrowheads highlight mILFs. Bar=500 μm. (b) The total number of ILFs and mILFs in the SI and LI of germ-free mice was determined by microscopy (n=3). SI numbers were estimated from representative sections. (c) The number of ILFs and mILFs and the mean ILF size were determined in the intestines of germ-free and conventionalized germ-free mice by microscopy (n=3). (d) The relative level of Il25 mRNA expression in the colon of germ-free and conventionalized mice was determined by quantitative real-time reverse-transcriptase–PCR (qRT–PCR; n=3). Statistical differences were determined by Student's t-test (*P<0.05).
IL-25−/− mice have increased LI-ILFs
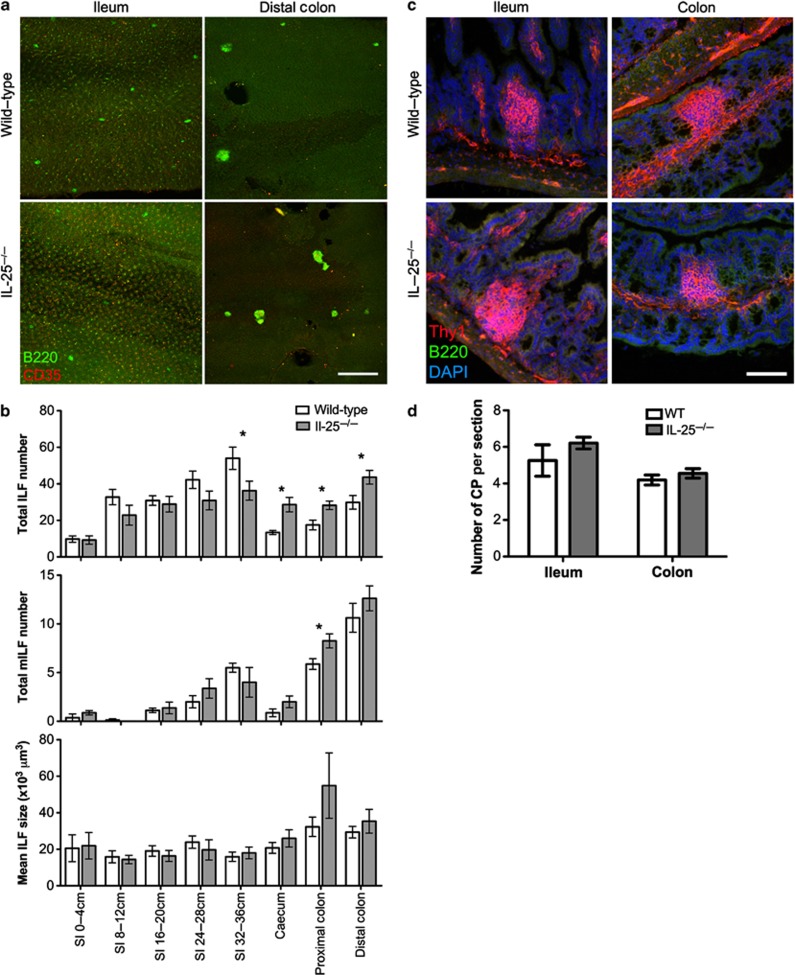
To test whether IL-25 had a role in colonic ILF development, ILFs and mILFs were enumerated in the intestines of wild-type (WT) mice and IL-25−/− mice (Figure 3a). IL-25−/− mice had decreased SI-ILFs, particularly in the ileum (Figure 3b). In contrast, the LI contained significantly higher ILFs. SI-mILF numbers were unaltered, whereas the LI had significantly higher mILFs. There was no significant difference in CP numbers in the ileum or colon of WT and IL-25−/− mice (Figure 3c,d), suggesting IL-25 acts during CP to ILF transition.
Figure 3.
Interleukin (IL)-25−/− mice have increased large intestinal (LI) isolated lymphoid follicles (ILFs). (a) Alternate 4 cm sections of small intestine (SI) and the whole LI of wild-type (WT) and IL-25−/− mice were whole-mount immunostained for B220 (green) to detect ILFs and CD35 (red) to detect mature ILFs (mILFs). Representative images of ileum and distal colon are shown. Bar=500 μm. (b) Total ILF and mILF numbers and mean ILF size were determined by microscopy (n=8). Results are representative of two independent experiments. (c) Sections of ileum and colon were stained for Thy1 (red), B220 (green), and nuclei (blue) to detect cryptopatches (CPs). Bar=50 μm. (d) CP numbers in 20 sections (100 μm apart) of 10 cm of terminal ileum or the whole colon were determined by microscopy and converted to CP/section (n=4). Statistical differences were determined by Student's t-test (*P<0.05).
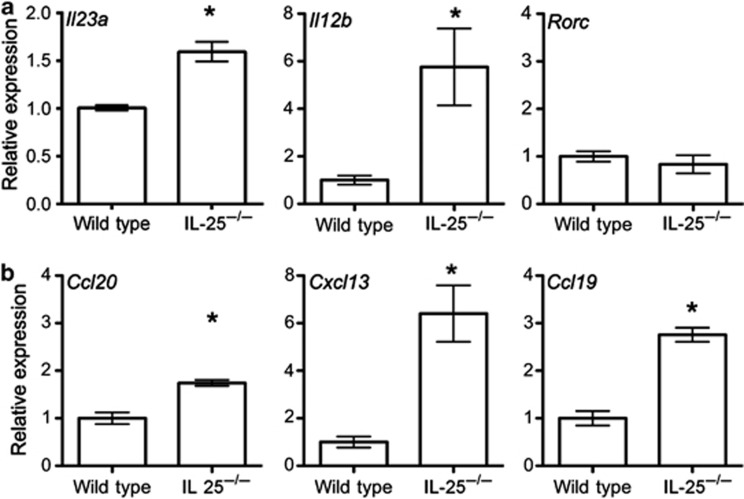
IL-23 is composed of the IL-23p19 and IL-12p40 subunits and, in accordance with previous studies, expression of Il23a and Il12b mRNAs were significantly increased in the colon of IL-25−/− mice23 (Figure 4a). Although IL-25−/− mice have been shown to harbor an increased population of RORγt+ ILCs in the SI,22 no difference in Rorc mRNA expression was observed in the colon. Consistent with the increased numbers of ILFs, significant increases in the expression of chemokines important for lympho-organogenesis and lymphocyte recruitment (Ccl20, Cxcl13, and Ccl19) were observed in the colon of IL-25−/− mice (Figure 4b). These data suggest that the inhibitory effect of the microbiota on ILF development in the colon was because of increased IL-25 production.
Figure 4.
The expression of Il23a, Il12b, Ccl20, Cxcl13, and Ccl19 mRNA is increased in the colon of interleukin (IL)-25−/− mice. (a) The relative levels of Il23a, Il12b, and Rorc mRNA expression in the colon of wild-type and IL-25−/− mice were determined by quantitative real-time reverse-transcriptase–PCR (qRT–PCR; n=3). (b) The relative levels of Ccl20, Cxcl13, and Ccl19 mRNA expression in the colon of wild-type and IL-25−/− mice were determined by qRT–PCR (n=3). Results are representative of two independent experiments. Statistical differences were determined by Student's t-test (*P<0.05).
IL-25−/− mice have increased colonic B cells, IgA production, and Th1 cells
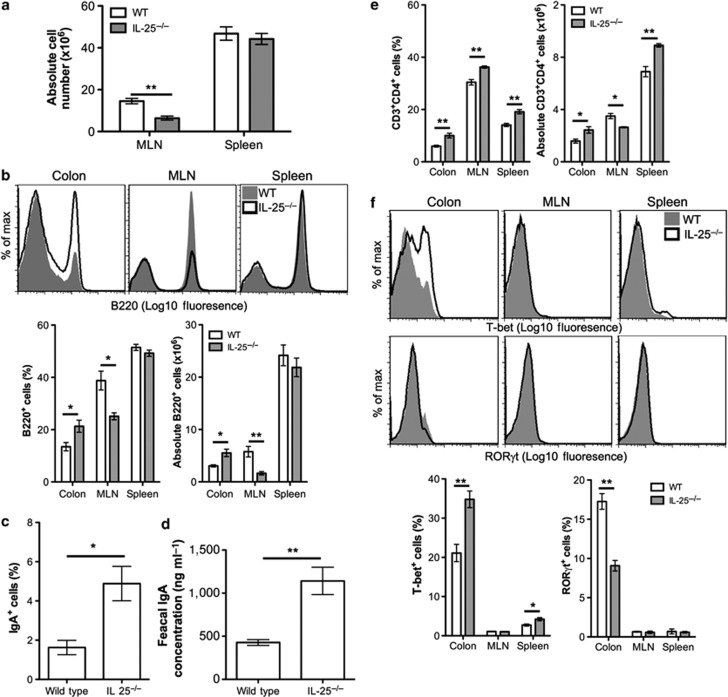
Increased colonic ILFs would likely alter lymphocyte retention and differentiation in the colon. To determine the effect of increased colonic ILFs in IL-25−/− mice on lymphocytes, colonic LP, mesenteric lymph node (MLN), and spleen cells were isolated from WT and IL-25−/− mice and analyzed by flow cytometry. Although splenic cellularity was similar in WT and IL-25−/− mice, a significant reduction in MLN cellularity was observed (Figure 5a). No difference in the total colonic LP cells was observed (data not shown). IL-25−/− mice had a significant increase in the proportion and absolute number of B220+ B cells in the colonic LP (Figure 5b). ILFs are sites of IgA induction9 and, accordingly, IL-25−/− mice also had a significant increase in the proportion of B220+ cells that were IgA+ in the colonic LP (Figure 5c) and a significant increase in fecal IgA (Figure 5d). These data suggest that as a consequence of increased ILFs in IL-25−/− mice, B cells were being retained within the colon, driving higher levels of IgA production.
Figure 5.
Interleukin (IL)-25−/− mice have increased colonic B cells, immunoglobulin A (IgA) responses, and T helper type 1 (Th1) cells. (a) Mesenteric lymph node (MLN) and spleen cells from wild-type (WT) and IL-25−/− mice were isolated and counted (n=4). (b) The relative frequency of B220+ B cells in the colonic lamina propria (LP), MLN, and spleen of WT and IL-25−/− mice was determined by flow cytometry and the absolute cell number calculated (n=4). Representative histograms are shown. (c) The proportion of B220+ cells that express IgA in the colonic LP was determined by flow cytometry (n=3). (d) Homogenates of fecal pellets (10%) from wild-type and IL-25−/− mice were prepared in phosphate-buffered saline (PBS) and IgA levels determined by enzyme-linked immunosorbent assay (ELISA; n=4). Absolute values were calculated from a standard curve of control mouse IgA. (e) The relative frequency of CD3+CD4+ T cells in the colonic LP, MLN, and spleen of WT and IL-25−/− mice was determined by flow cytometry and the absolute cell number calculated (n=4). (f) The relative frequency of T-bet+ and RORγt+ cells in CD3+CD4+ T-cell population from the colonic LP, MLN, and spleen of WT and IL-25−/− mice. Representative histograms are shown. Results are representative of three independent experiments. Statistical differences were determined by Student's t-test (*P<0.05, **P<0.01).
The proportion of CD3+CD4+ T cells within the colonic LP, MLN, and spleen was also significantly increased in IL-25−/− mice (Figure 5e). The absolute CD3+CD4+ T-cell number was significantly increased in the colonic LP and spleen, but not the MLN. Within this population, a significant increase in T-bet+ T helper type 1 (Th1) cells and a significant decrease in RORγt+ Th17 cells was observed in the colonic LP (Figure 5f). T-bet+ Th1 cells were unchanged in the MLN, but a significant increase was observed in the spleen. RORγt+ Th17 cells were not readily detectable in the MLN and spleen. Therefore, increased ILFs in IL-25−/− mice are associated with an expansion of Th1 cells in the colonic LP.
IL-23p19−/− mice have a colon-specific reduction in ILFs
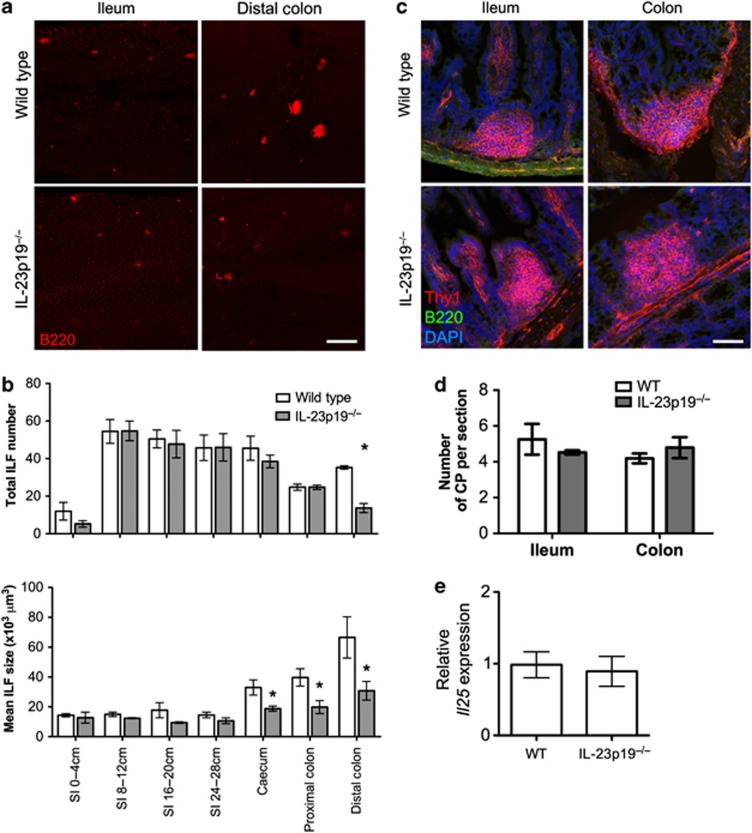
IL-23 is implicated in the pathogenesis of a number of inflammatory diseases, particularly colitis,24 and is increased in the colon of germ-free and IL-25−/− mice.23 Microbiota-induced IL-25 reduces colonic IL-23 expression in germ-free mice following colonization.23 IL-23 induces IL-17 and IL-22 from ILCs,25 both of which may have roles in lympho-organogenesis,26, 27 suggesting the effect of IL-25 on ILFs may be its ability to reduce IL-23 production. To test this, the intestines of IL-23p19−/− and WT mice were whole-mount immunostained and ILF counted (Figure 6a). The SI, cecum, and proximal colon contained equivalent numbers of ILFs (Figure 6b). However, a significant reduction in distal colonic ILFs was observed in IL-23p19−/− mice. ILF size was also reduced throughout the intestines, particularly in the distal colon. Equivalent numbers of CPs were observed in the ileum and colon of WT and IL-23p19−/− mice (Figure 6c,d), suggesting the effect of IL-23 was occurring during CP to ILF transition, correlating with the effect of IL-25. Colonic Il25 mRNA expression was also equivalent (Figure 6e), suggesting the effect on ILFs was not because of increased IL-25 and that the effect of IL-23 on ILF development was downstream of the effect of IL-25.
Figure 6.
Interleukin (IL)-23p19−/− mice have a colon-specific reduction in isolated lymphoid follicles (ILFs). (a) Alternate 4 cm sections of small intestine (SI) and the whole large intestine from wild-type (WT) or IL-23p19−/− mice were whole-mount immunostained for B220 (red) to detect ILFs. Bar=500 μm. (b) ILF numbers and mean ILF size in WT or IL-23p19−/− mice was determined by microscopy (n=4). (c) Sections of ileum and colon were stained for Thy1 (red), B220 (green), and nuclei (blue) to detect cryptopatches (CPs). Bar=50 μm. (d) CP numbers in 20 sections (100 μm apart) of 10 cm of terminal ileum or the whole colon were determined by microscopy and converted to CP/section (n=4). (e) The relative level of Il25 mRNA expression in the colon of WT and IL-23p19−/− mice was determined by quantitative real-time reverse-transcriptase–PCR (qRT–PCR; n=4). Statistical differences were determined by Student's t-test (*P<0.05).
The changes in colonic ILFs in both IL-25−/− and IL-23p19−/− mice may have been the result of altered microbial burdens and/or altered microbiota composition. Total bacteria and the relative proportions of major phyla were analyzed in fecal samples by quantitative real-time reverse-transcriptase–PCR using primers specific for bacterial 16S rRNA. No difference in the total fecal microbial burden was observed between IL-25−/− or IL-23p19−/− mice and respective WT mice (Supplementary Figure S1 online). Furthermore, no significant changes were observed in the composition of the microbiota, with the exception of segmented filamentous bacteria (SFB), that was significantly increased in IL-25−/− mice. This suggests that the differential numbers of ILFs observed in these models was not because of major alterations in the size or composition of the microbiota.
To determine whether the effect of IL-23 was dependent on IL-17 or IL-22, mice treated with LTβR-Ig in utero, which blocks Peyer's patch development, but not CP and ILFs,16 were given neutralizing anti-IL-17 or anti-IL-22 (or isotype-matched controls) between days 14 and 28 after birth. Higher ILF numbers develop in LTβR-Ig-treated mice allowing a greater range for the effect of cytokine neutralization in young mice and excluding confusion with underdeveloped patches. The intestines were whole-mount immunostained and ILFs and mILFs enumerated as before. Blocking IL-17 or IL-22 did not significantly alter the total ILF or mILF number or size in the SI or LI (Supplementary Figure 2a and b). This suggests that IL-17 and IL-22 were not the primary downstream factors of IL-23-mediated regulation of ILFs, implying that IL-23 has additional roles.
Depletion of CD25+ T regulatory cells (Tregs) increases ILF density
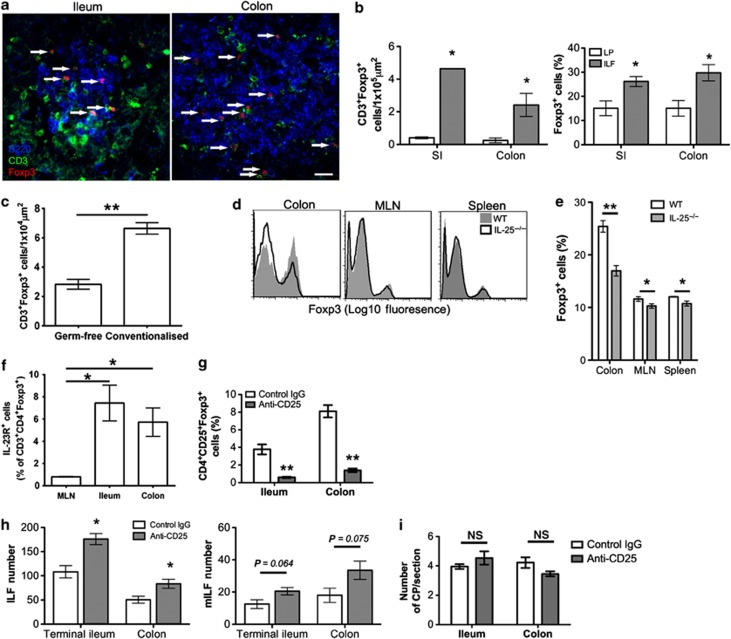
Germ-free mice have reduced colonic Tregs that is reversed by colonization, whereas SI Tregs are unaffected.28 IL-23 may a play a role in this as T cells transferred into RAG1−/−IL-23p19−/− mice develop higher numbers of Tregs, suggesting that IL-23 restrains Treg development.29 Following transfer, T cells migrate to colonic CPs before the development of colitis5 and colonic ILFs contain higher Foxp3 mRNA than the LP.4 Our immunohistochemical analysis showed that CD3+Foxp3+ cells were detectable within ileal and colonic ILFs (Figure 7a) and quantification revealed a significant enrichment in ILFs (Figure 7b). The percentage of Foxp3+ cells was significantly higher in ILFs compared with LP, suggesting enrichment was not because of higher T-cell densities. Analysis of ILFs from germ-free and conventionalized mice showed that the density of CD3+Foxp3+ cells within ILFs significantly increased following colonization (Figure 7c). Furthermore, the proportion of CD3+CD4+ T cells expressing Foxp3 was significantly reduced in the colonic LP of IL-25−/− mice (Figure 7d,e) similar to germ-free mice. These data suggest that although Tregs are enriched in colonic ILFs, the proportion of Tregs negatively correlates with the number of colonic ILFs, implying a role for Tregs in ILF development. Some Tregs have been reported to express IL-23 receptor (IL-23R).30 Analysis of CD3+CD4+Foxp3+ cells from the MLN, SI-LP, and colonic LP showed that a subpopulation of intestinal Tregs express IL-23R that was virtually absent in the MLN (Figure 7f), suggesting that IL-23 could directly modulate Treg differentiation in the intestines.
Figure 7.
Anti-CD25 treatment enhances isolated lymphoid follicle (ILF) numbers in the ileum and colon. (a) Sections of small intestine (SI) and large intestine (LI) were immunostained for B220 (blue), CD3 (green), and Foxp3 (red). Representative images of ILFs from the ileum and colon are shown. Bar=20 μm. (b) The number of CD3+Foxp3+ cells and the proportion of CD3+ cells expressing Foxp3 within ILFs and adjacent areas of lamina propria (LP) were determined by microscopy (n=3). (c) The number of CD3+Foxp3+ within ILFs of germ-free and conventionalized mice (n=3) (d, e) The relative frequency of Foxp3+ cells in the CD3+CD4+ T-cell population of the colonic LP, mesenteric lymph node (MLN), and spleen of wild-type (WT) and interleukin (IL)-25−/− mice was determined by flow cytometry. Representative histograms are shown (n=4). (f) The relative frequency of IL-23R+ cells within the CD4+Foxp3+ T cell population of the MLN, ileum, and colon was determined by flow cytometry (n=3). (g) Mice were treated with anti-CD25 or control Ig. At 7 days after final treatment, mice were killed and the proportion of CD25+Foxp3+ cells within the CD4+ population of the ileal and colonic LP were determined by flow cytometry (n=4). (h) Portions of terminal ileum (8 cm) and the whole colon of anti-CD25 or control Ig-treated mice were whole-mount immunostained for B220 and CD35 to detect ILFs and mature ILFs (mILFs), respectively. ILF and mILF numbers were determined by microscopy (n=4). (i) Sections of ileum and colon were stained for Thy1 (red) and B220 (green) to detect cryptopatches (CPs). CP numbers in 12 sections (100 μm apart) of 10 cm of terminal ileum or the whole colon were determined by microscopy and converted to CP/section (n=4). Statistical differences were determined by Student's t-test (*P<0.05, **P<0.01, NS, not significant).
To determine the role of Tregs in ILF development, CD25+ Tregs were depleted with anti-CD25 mAb. After 14 days, the proportion of CD4+CD25+Foxp3+ cells in the LP were determined by flow cytometry. Anti-CD25 mAb treatment effectively depleted CD4+CD25+Foxp3+ cells in the LP of the SI and colon (Figure 7g). ILFs were determined by whole-mount immunostaining as before in 8 cm sections of terminal ileum and the colon. Anti-CD25 mAb depletion induced a significant increase in ileal and colonic ILFs (Figure 7h). Increased mILFs were also observed, but this was not significant. Anti-CD25 mAb treatment did not significantly alter CP numbers in the SI or colon (Figure 7i). Therefore, depletion of CD25+ Tregs increased numbers of ILFs, suggesting that Tregs regulate CP to ILF transition.
IL-23p19+ cells are enriched in colonic ILFs
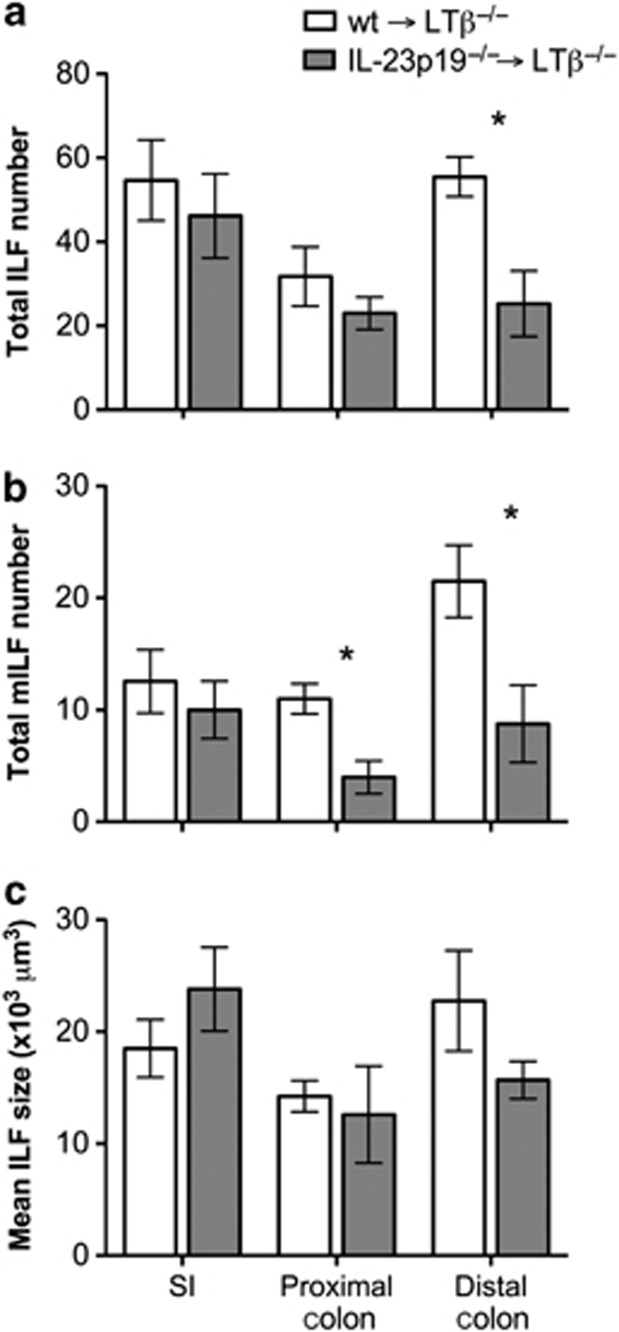
Treg depletion affected both SI- and LI-ILF development (Figure 7h). However, colonization of germ-free mice only inhibited colon ILF (Figure 2c). Colonic, but not SI, Tregs have been shown to increase following colonization,28 suggesting that a colon-specific factor may underlie both of these effects. Microbiota-induced IL-25 is expressed throughout the intestines, whereas IL-23 expression in the colon mirrors the effect on ILFs, in that it is specifically higher in the colon of germ-free mice and reduced upon colonization.23 To determine the cellular source of IL-23 that affects ILF development, irradiated LTβ−/− mice were reconstituted with bone marrow (BM) from either WT or IL-23p19−/− mice (WT→LTβ−/− and IL-23p19−/−→LTβ−/− mice) and ILFs and mILFs were enumerated in the intestines as above. LTβ−/− mice lack gut-associated lymphoid tissues; however, reconstitution with LTβ-sufficient BM induces CP and ILF development.31 No difference in ILF numbers was observed in the SI or the proximal colon of WT→LTβ−/− or IL-23p19−/−→LTβ−/− mice, whereas a significant reduction was observed in the distal colon of IL-23p19−/−→LTβ−/− mice (Figure 8a). This was associated with a significant reduction in colonic mILFs (Figure 8b) and a reduction in distal colonic ILF size (Figure 8c), paralleling the phenotype of IL-23p19−/− mice (Figure 6b). This showed that the effect of IL-23 deficiency was because of production by a cell of hematopoietic origin in the colon.
Figure 8.
LTβ−/− mice reconstituted with interleukin (IL)-23p19−/− bone marrow have a specific reduction in colonic isolated lymphoid follicles (ILFs). LTβ−/− were lethally irradiated and reconstituted with wild-type (wt) or IL-23p19−/− bone marrow. After 10 weeks, alternate 4 cm sections of small intestine (SI) and colon were whole-mount immunostained for B220 and CD35 to detect ILFs and mature ILFs (mILFs), respectively. (a) Total ILF and (b) mILF numbers were determined by microscopy (n=4). SI values are the mean ILF or mILF numbers present in a 4-cm section of SI (5 sections/mouse). (c) Mean ILF size was also determined. Statistical differences were determined by Student's t-test (*P<0.05).
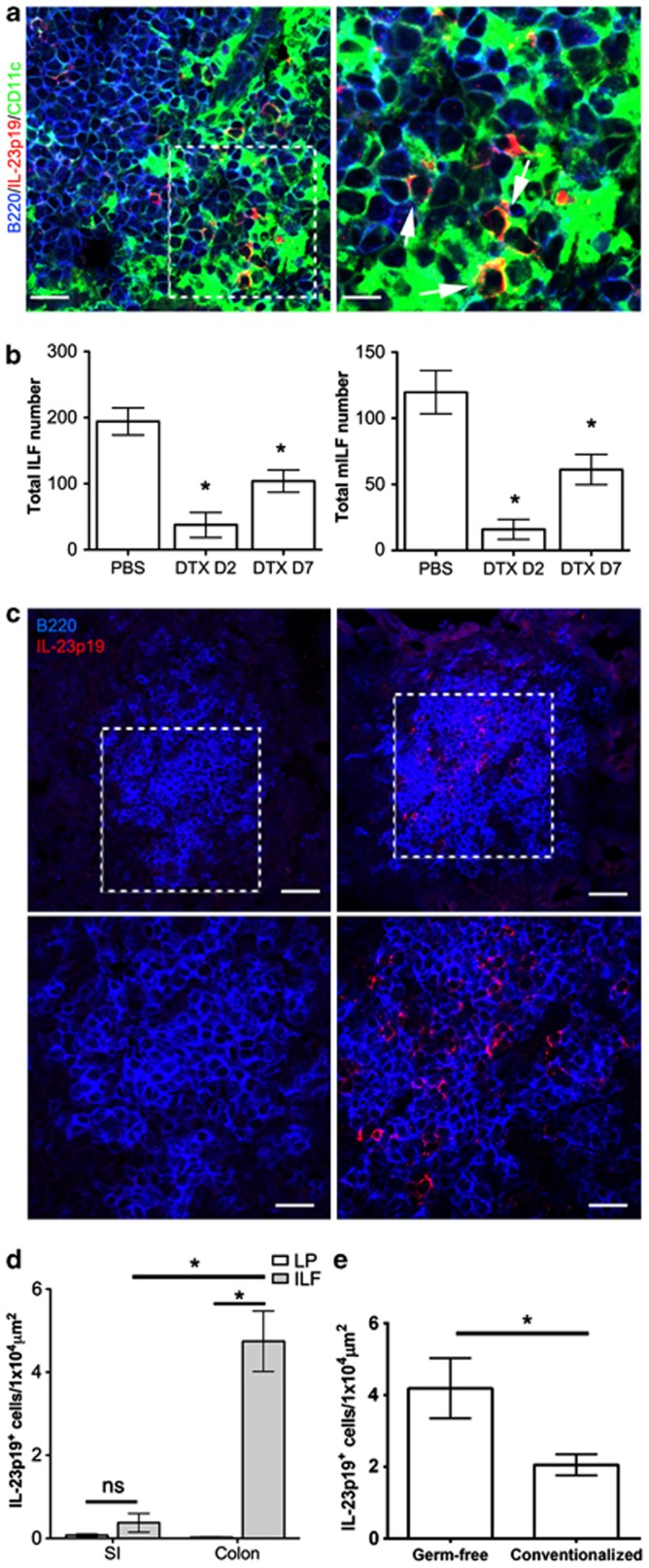
IL-23p19 is thought to be expressed by mononuclear phagocytes in the colon.32, 33 Immunohistochemical analysis showed that IL-23p19+ cells in colonic ILFs coexpressed CD11c, suggesting a mononuclear phagocyte origin for colonic IL-23 (Figure 9a). This was supported by analysis of a large range of microarray data sets (>200) representing 95 distinct murine tissues or cell lineages that confirmed that expression of Il23a, encoding IL-23p19, was restricted to CD11c-epxressing cells (Supplementary Figure S3). CD11c+ cells are critical for SI-ILF development.34 To determine their role in LI-ILF development, irradiated LTβ−/− mice were reconstituted with BM from CD11c-DTR mice. Mice were treated with diphtheria toxin to deplete CD11c-expressing cells and 2 or 7 days later the colons were whole-mount immunostained to detect ILFs and mILFs as before. CD11c+ cell depletion induced a significant reduction in total ILFs and mILFs in the colon (Figure 9b), affirming the critical role for this population in LI-ILF development.
Figure 9.
Interleukin (IL)-23p19+ cells are enriched in colonic isolated lymphoid follicles (ILFs). (a) Sections of colonic ILFs were also immunostained for B220 (blue), IL-23p19 (red), and CD11c (green). Bar=20 μm in the left panel and 5 μm in the right panel. (b) LTβ−/− were irradiated and reconstituted with bone marrow from CD11c-DTR mice. Mice were treated with diphtheria toxin (DTX) and culled 2 or 7 days after treatment. Phosphate-buffered saline (PBS)-treated mice culled 7 days after treatment were included as controls. The colon of each mouse was whole-mount immunostained for B220 to detect ILFs and CD35 to detect mature ILFs (mILFs). ILF and mILF numbers were determined by microscopy (n=3–4). (c) Sections of small intestine (SI) and colon from wild-type mice were immunostained for B220 (blue) and IL-23p19 (red). Representative images of ILF are shown. Bar=40 μm in the upper panel and 20 μm in the lower panel. (d) IL-23p19+ cells within ILFs or adjacent areas of lamina propria (LP) were enumerated and the density of IL-23p19+ cells determined (n=3). (e) IL-23p19+ cells within ILFs of germ-free and conventionalized mice were enumerated and the density of IL-23p19+ cells determined (n=3). Statistical differences were determined by Student's t-test (*P<0.05).
Interestingly, although IL-23p19+ cells were detectable in the LP of the SI, they were rarely within ILFs (Figure 9c). In contrast, IL-23p19+ cells were readily detected in colonic ILFs. Quantification revealed that colonic ILFs contained significantly higher densities of IL-23p19+ cells than SI-ILFs (Figure 9d). Furthermore, enumeration of IL-23p19+ cells within colonic ILFs of germ-free and conventionalized mice revealed a significant decrease in this population upon microbial colonization (Figure 9e), consistent with previous observations23 and supporting a role for modulation of this population in the regulation of colonic ILFs. Therefore, CD11c+IL-23p19+ cells are specifically enriched in colonic ILFs and may underlie the colon-specific regulation of ILF development by IL-25, IL-23, and Tregs.
DISCUSSION
In this study, a novel system regulating ILF development in the colon has been identified. Unlike the SI, where the microbiota promotes ILF development, our studies show that conversely, the microbiota can inhibit colonic ILF development. These contrasting microbiota-driven changes led us to consider the roles of IL-25 and IL-23, both regulated by the microbiota, in colonic ILF development. We found that in the absence of IL-25, colonic ILF development was enhanced. This was associated with increased colonic IL-23 expression23 and, accordingly, IL-23p19−/− mice had a colon-specific decrease in ILFs. The specificity is likely because of the enrichment of IL-23p19+CD11c+ cells within colonic ILFs and their ability to modulate Treg differentiation. Colonic ILFs develop in the absence of a number of factors required for the development of their SI counterparts.18, 19, 21 In addition, colonic ILFs, but not SI-ILFs, can also be suppressed by thymic stromal lymphopoietin (TSLP)35 in addition to IL-25. However, IL-23 is the only factor identified to date that specifically promotes colonic ILFs without affecting ILFs in the SI. Given the important role of IL-23 in inflammatory bowel disease,24 we believe that this may represent a novel function for IL-23 in the pathogenesis of colitis.
The model these data suggest is that in absence of microbiota, IL-25 production is diminished,22, 23 leading to increased IL-23 secretion by CD11c+ cells (Supplementary Figure S4a). Increased IL-23 may suppress colonic Treg development,29 leading to reduced Treg suppression of ILF development and increased ILF numbers. Following colonization, both microbial16 and endogenous stimulation drive ILF development (Supplementary Figure S4b); however, this is now associated with increased microbiota-induced IL-25.23 The increased IL-25 leads to reduced IL-23 secretion by CD11c+ cells, permitting the expansion of colonic Tregs and suppressed colonic ILF development. SI-ILFs are not regulated by IL-25, likely because of the rarity of CD11c+ cells expressing IL-23 within SI-ILFs.
In addition to ILFs, the colon may contain tLTs that develop in response to inflammation independent of CPs.24 It is conceivable that some of the increased ILFs in germ-free and IL-25−/− mice were tLTs. These are indistinguishable from ILFs histologically in that they are single B-cell follicles without defined T-cell zones and can only be defined in mice that lack CPs. To determine the contribution of tLTs, RORγt−/− mice,18 which lack CPs, or mice treated with LTβR-Ig during early life,16 which blocks CP development, could be used. However, the extent to which tLT development in these models is a consequence of CP/ILF absence and/or the loss of other cell populations (e.g., Th17 cells in RORγt−/− mice) is unclear. These complications mean that it is unknown how many tLTs contribute to total colonic B-cell aggregates in CP-containing mice. The pathways controlling inducible bronchus-associated lymphoid tissue development, which can also form independent of RORγt+ cells,26 show some similarities to colonic B-cell aggregates in that they both require LT and CD11c+ cells,26, 36 can be inhibited by Tregs,37 and show some dependence on IL-23.26 This suggests similarities exist in the pathways that regulate RORγt-dependent and -independent structures, confounding the difficulties in delineating the roles of each structure.
Consistent with previous studies, we observed minimal ileal ILFs in germ-free mice.8, 16 Despite this, duodenal and jejunal ILFs were present, although in low numbers, and were unaltered by conventionalization. Our own study of the timing of ILF development after birth showed a similar pattern, with higher duodenal and jejunal ILF development at early ages (up to 4 weeks) followed by ileal ILF development (4–6 weeks) (data not shown). This suggests that at least some SI-ILF development is microbiota independent and that SI-ILF development in germ-free mice broadly resembles that of SPF-housed mice. Colonic ILFs develop earlier than ileal ILFs,17 and the number of ILFs in the colon of germ-free mice has been reported to be similar to that of SPF-housed mice,17, 21, 35 suggesting that colonic ILF development also proceeds normally in germ-free mice. It has been reported that the colonic epithelium undergoes irreversible epigenetic changes in the absence of the microbiota,38 and to what extent such changes contribute to ILF development is undetermined.
The role of the microbiota in the regulation of colonic ILF remains controversial. Colonization with restricted flora has been shown to promote colonic ILF development,16 and a number of studies have shown that colonic B-cell aggregates develop in the absence of factors (e.g., RANK,19 CXCL13,21 RORγt,18 and LTα6) necessary for SI-ILF development, suggesting that some form of colonic B-cell aggregate is critically required for microbiota homeostasis. However, the finding in the current and other studies17, 21 that germ-free and antibiotic-treated mice have equivalent or higher numbers of ILFs compared with SPF mice is difficult to reconcile with these data. ILFs are dynamically regulated and show both temporal and spatial variation throughout the colon, meaning that interpretation of these studies is confounded by the differential mouse strains, sampling of the colon, and methodologies used to identify ILFs, as well as extrinsic factors such as the composition of the microbiota. Our study reconciles some of the discrepancies by showing that dramatic changes in colonic ILFs occur following colonization. Colonization diminished but did not ablate colonic ILFs, supporting the notion that microbiota-independent ILF development is an inappropriate and exaggerated response that is ill-equipped to mediate homeostasis. Therefore, a combination of both pro- and anti-ILF signals from the microbiota are likely required to develop an appropriate ILF response and homeostasis with the microbiota. Given the association between colonic lymphoid aggregates and enhanced pathology,3, 6, 39 understanding how this exaggerated response is regulated by nonmicrobial factors (e.g., IL-23) may aid our understanding of susceptibility to colonic inflammation.
No differences in the overall fecal microbial burden were observed in IL-25−/− or IL-23−/− mice, suggesting that the differences in ILF development were not because of alterations in microbial burden. Altered microbiota composition could drive differential responses (e.g., increased IL-25), but microbiota profiling revealed similar levels of all phyla measured, with the exception of SFB that was significantly increased in IL-25−/− mice. Mono-colonization with SFB, but not Escherichia coli, has recently been shown to drive the formation of tLTs in SI in the absence of CPs; however, both were equally able to drive ILF development in the presence of CPs.40 However, in this study, ileal ILFs were significantly reduced in IL-25−/− mice, suggesting that changes in ILF development in the absence of IL-25 were not because of SFB. Interestingly, SFB specifically colonizes the epithelium overlying lymphoid follicles,41 and the relative proportion of SFB correlated with the number of colonic ILFs, suggesting that changes in SFB abundance may relate to the number of available follicles for colonization. Microbial colonization also induces TSLP production by colonic CD11c+ cells.35 TSLPR−/− and IL-25−/− mice share many similarities in that they have increased colonic ILFs, altered Treg populations, and increased T cells secreting interferon-γ and IL-17.35 This phenotypic similarity suggests a common mechanism underlying the effect of microbial-induced IL-25 and TSLP. If so, this is unlikely to be because of SFB, as TSLPR−/− mice show no difference in the relative abundance of SFB. Thus, the effect of IL-25 or IL-23 deficiency on ILF development is unlikely to be because of alterations in the composition or the size of the microbiota.
Tregs are preferentially located in colonic ILFs of humans and mice.4, 42 Increased colonic ILFs and IL-23 in both germ-free and IL-25−/− mice was accompanied by reductions in Tregs. IL-23 has been shown to limit Treg numbers in the colon,29 but the mechanism remains unclear. A subpopulation of intestinal Tregs expressed IL-23R, suggesting that IL-23 may directly affect these cells. IL-23R expression is under the control of RORγt,43 and intestinal Foxp3+RORγt+ T cells have been described.44 Therefore, IL-23R expression by Foxp3+ cells may be indicative of a differentiating cell rather than a differentiated cell. In support of this, increased Tregs observed in IL-23p19−/−RAG-1−/− mice were derived from naive CD4+ T cells rather than CD4+Foxp3+ T cells.29 Absence of TSLPR also led to a reduction in induced Tregs without affecting thymically derived Tregs.35 These data imply that IL-23 acts during de novo Treg differentiation, either by repressing Foxp3 or promoting Th17 differentiation.
Our data suggest that Tregs play an important role in ILF regulation. How Tregs influence ILF development is unclear. Tregs are present in the follicles of secondary lymphoid organs where they limit the germinal center response by suppressing other follicular T-cell subsets and limiting nonantigen-specific B-cell recruitment.45 In our study, Tregs were present within immature ILFs and depletion of Tregs only had a marginal effect on ILF maturation (data not shown), suggesting their action might be before germinal center formation. Consistent with this hypothesis, Treg depletion did not affect CP numbers, suggesting they inhibit CP to ILF transition. This requires CD11c+ cells and the production of CXCL13 by this population.34 Tregs are potent inhibitors of CD11c+ cell function, and may directly inhibit CXCL13 production. However, CPs with CD11c+ cells that lack B cells are common, suggesting that additional signals regulate CXCL13 production and Treg inhibition of these may also be important. Tregs have also been suggested to play a role in inducible bronchus-associated lymphoid tissue formation in the lung as more develop in mice containing CCR7−/− Tregs.37 Interestingly, CCR7−/− mice also have a higher proportion of larger solitary intestinal lymphoid tissue structures in the SI,8 suggesting Treg migration in response to CCL19 and/or CCL21 rather than CXCL13 may be critical in this process.
We have identified a novel ILF-regulating mechanism in the colon involving microbiota-induced changes in IL-25 and IL-23 expression. The role of the microbiota as a modulator of health and disease, both mucosal and systemic, is growing in importance. The CP/ILF system is a dynamic way in which the immune system responds to the microbiota. The number, maturation status, and cellular composition of ILFs are influenced by many factors,16, 46, 47 and understanding how perturbation of the CP/ILF system alters intestinal homeostasis may be key to understanding how this breaks down. In the SI, the relationship between microbial stimulation and ILF formation is linear, providing an adaptable response for microbiota control. In the colon, ILF regulation is more complex, with the microbiota providing both stimulatory and inhibitory signals for ILF formation, likely because of homeostasis that has to be reached with the higher microbial burdens present. Disruption of this balance of positive and negative factors regulating ILF development may be involved in the development of colonic pathology, as many of the factors are common to both processes, particularly as IL23R polymorphisms are associated with human inflammatory bowel disease.48 Lymphoid aggregates are a feature of human colonic inflammation3 and have been associated with enhanced pathology in mouse models.4, 5, 6, 18 Furthermore, this mechanism may also be involved in nonmucosal chronic inflammatory diseases associated with the development of tLTs where IL-23 and Tregs are implicated in the pathogenesis.
METHODS
Mice. Mice were maintained under SPF conditions and all experiments were carried out under the authority of a UK Home Office Project Licence within the terms and conditions of the UK Home Office “Animals (scientific procedures) Act 1986.” All LTβ−/−, CD11c-DTR, IL-23p19−/−, and IL-25−/−49 mice used in this study were bred and maintained on a C57BL/6 background, with the exception of germ-free CD1 mice. Germ-free mice were maintained under isolated conditions and routinely checked for microbial contamination. Conventionalization of germ-free mice was achieved by transfer to normal housing and the addition of used bedding from conventionally housed mice. All mice used were either littermates or were co-housed for a minimum of 2 weeks with used bedding exchanged between the cages.
In vivo treatments. For BM reconstitutions, mice were lethally irradiated and reconstituted with appropriate BM via tail vein injection the following day. Reconstituted mice were not used until at least 10 weeks afterwards to permit ILF development. For CD11c+ cell depletion, mice were intraperitoneally injected with 100 ng diphtheria toxin (Sigma, Gillingham, UK) or an equivalent volume of phosphate-buffered saline. For CD25+ cell depletion, mice were treated with 200 μg anti-CD25 mAb (PC61) or control rat IgG1 (eBRG1) (both from Ebioscience, Hatfield, UK) weekly for 2 weeks.
Immunohistochemistry. Whole-mount immunostaining was performed using a modification of a previously published method.50 Briefly, ∼4 cm pieces of intestine were washed in phosphate-buffered saline before incubation in Hank's balanced salt solution containing 5 mM EDTA (both from Life Technologies, Paisley, UK) in shaking incubator at 37 °C. The epithelium was washed off and the intestinal pieces fixed in 10% formal saline (Cellpath, Powys, UK), washed in Tris-buffered saline containing 0.1% Triton X-100 (Sigma), and nonspecific binding blocked with 2.5% normal goat serum (Jackson Immunoresearch, Newmarket, UK). Next, 6 μm cryosections were fixed in acetone before blocking with 5% normal goat serum in Tris-buffered saline containing 0.1% Triton X-100. Intestinal pieces and cryosections were then stained with appropriate combinations of anti-mouse rat anti-CD45-Alexa 488 mAb (RA3-6B2; Life Technologies), hamster anti-CD11c-Alexa 647 mAb (N418; Biolegend, London, UK), and/or rat anti-Foxp3-Alexa 647 mAb (FJK-16s; Ebioscience) or purified rat anti-CD35 mAb (8C12), rat anti-CD21/CD35 mAb (7G6), hamster anti-CD3 mAb (145-2C11), rat anti-Thy1.2 (53-2.1) (all from BD Biosciences, Oxford, UK), and/or polyclonal rabbit anti-IL-23p19 (Abcam, Cambridge, UK) detected with goat anti-rat or anti-rabbit IgG Alexa 594 conjugates (both from Life Technologies). Sections were mounted using fluorescent mounting medium (Dako, Ely, UK). Where appropriate, sections were counterstained with 4',6-diamidino-2-phenylindole (Life Technologies).
Microscopy. Whole-mount immunostained intestinal pieces were visualized on a Nikon EC1 confocal microscope (Nikon, Kingston upon Thames, UK). ILFs and mILFs were enumerated visually along the whole length of the intestinal piece. Where appropriate, total SI-ILFs were estimated from alternate SI pieces. At least three representative images from each intestinal piece were captured and the area of each ILF within these determined using ImageJ (http://rsbweb.nih.gov/ij/), from which the mean ILF size per intestinal piece was determined.
Images of cryosections were obtained using a Zeiss LSM5 Pascal confocal microscope (Zeiss, Welwyn Garden City, UK). Analysis of CP in cryosections was achieved by enumeration in 12–20 nonsequential (100 μm apart) sections of 10 cm of terminal ileum and the whole colon from each mouse. For enumeration of IL-23p19+ and Foxp3+ cells, cells were counted in images of all ILFs and adjacent areas of LP (2–14 per section) from three no-sequential sections.
Flow cytometry. Spleen and MLN cells were isolated by standard mechanical disruption. Ileal and colonic LP cells were isolated by enzymatic digestion as previously described.51 Surface staining was achieved by incubation in anti-mouse CD16/CD32 mAb (2.4G2; BD Biosciences) for 15 min followed by incubation with anti-mouse CD4 FITC-conjugate mAb (H129.19), anti-mouse CD3 Pacific Blue-conjugate mAb (500-A2) (both from BD Biosciences), anti-mouse CD45 (B220) PE-Cy7-conjugate mAb (RA3-6B2; Life Technologies), or anti-mouse IL-23R PE conjugate mAb (753317; R&D, Abingdon, UK) for 30 min where appropriate. Intracellular staining was performed using a mouse Foxp3/Transcription factor staining buffer set (Ebioscience) according to the manufacturer's instructions using anti-mouse Foxp3 Alexa 647-conjugate mAb (FJK-16s), anti-human/mouse T-bet-PE-Cy7 conjugate mAb (ebio4b10), anti-mouse RORγt-PE conjugate mAb (AFKJS-9) (all from Ebioscience) or anti-mouse IgA-PE conjugate mAb (11-44-2; Southern Biotech, Birmingham, AL). Samples were acquired using a CyAn ADP flow cytometer (Dako) and analyzed with Flowjo (Treestar, Ashland, OR). At least 50,000 events per sample were collected.
Quantitative real-time reverse-transcriptase–PCR. Total RNA was extracted from the colon using RNA-bee (AMS Biotechnology, Abingdon, UK) and cDNA generated using First-Strand cDNA synthesis kit (GE Healthcare, Little Chalfont, UK), both according to the manufacturer's instructions. Quantitative real-time reverse-transcriptase–PCR was performed with Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies) according to the manufacturer's instructions using a Stratagene MX3005P (Agilent Technologies, Waldbronn, Germany). Expression levels were determined relative to Actb or Gapdh. No template and no reverse transcription controls were included for each primer. Primer sequences used were Actb (5′-TGACAGGATGCAGAAGGAGA-3′ and 5′-GTACTTGCGCTCAGGAGGAGGAG-3′); Gapdh (5′-GGGTGTGAACCACGAGAAAT-3′ and 5′-CCTTCCACAATGCCAAAGTT-3′); Il17a (5′-TTTAACTCCCTTGGCGCAAAA-3′ and 5′-CTTTCCCTCCGCATTGACAC-3′); Il23a (5′-CAACTTCACACCTCCCTAC-3′ and 5′-CCACTGCTGACTAGAACTC-3′); Il12b (5′-GCAACGTTGGAAAGGAAAGA-3′ and 5′-AAAGCCAACCAAGCAGAAGA-3′); Rorc (5′-GACCCACACCTCACAAATTGA-3′ and 5′-AGTAGGCCACATTACACTGCT); Ccl20 (5′-CGACTGTTGCCTCTCGTACA-3′ and 5′-AGCCCTTTTCACCCAGTTCT-3′); Cxcl13 (5′-ACAGACTCCGAGCTAAAGGTTG-3′ and 5′-AATGGGCTTCCAGAATACCG-3′); Ccl19 (5′-GATCGCATCATCCGAAGACT-3′ and 5′-GAGGCCTGGTCCTCTCTTCTTCT-3′); Lta (5′-CCCTCAGAAGCACTTGACC-3′ and 5′-CAGAGAAAACCACCTGGGAG-3′); and Il25 (5′-CAGCAAAGAGCAAGAACC-3′ and 5′-CCTGTCCAACTCATAGC-3′) (all synthesized by Life Technologies).
Assessment of fecal IgA. Fecal IgA levels were determined by enzyme-linked immunosorbent assay as previously described.50 Briefly, 10% homogenates of fecal samples (in phosphate-buffered saline) were centrifuged at 12,000 g for 10 min and the supernatants collected. Supernatants were added to plates previously coated with polyclonal goat anti-mouse IgA (Southern Biotech) and blocked with 5% bovine serum albumin/phosphate-buffered saline. Bound IgA was detected using a goat anti-mouse IgA alkaline phosphatase-conjugate (Southern Biotech) and visualized using p-nitrophenyl phosphate liquid substrate (Sigma). The optical density was determined at 405 nm using Wallac Victor 2 (PerkinElmer, Cambridge, UK) and converted to ng ml−1 using a standard curve of control mouse IgA (BD Biosciences). Values were corrected for nonspecific binding using fecal samples from severe combined immunodeficient mice.
Statistical analyses. Student's t-test or one-way analysis of variance with Tukey's post-hoc test were performed using Prism 4 (Graphpad Software, San Diego, CA). Results are expressed as mean±s.e.m. and P<0.05 was considered significant.
Acknowledgments
We thank S. Jung (Weizzman Institute of Science, Rehovot, Israel) for providing the CD11c-DTR mice, N. Ghilardi (Genentech, South San Francisco, CA) and A. MacDonald (University of Edinburgh) for providing the IL-23p19−/− mice, and Merck Sharp & Dohme (Whitehouse Station, NJ) for providing the IL-25−/− mice. We thank B. Fleming, F. Laing, S. Cumming, S. Auginer, K. Brown (University of Edinburgh), and A. Mowat (University of Glasgow) for technical advice and assistance. This work was supported by project (BB/G003947-1) and Institute Strategic Programme Grant funding from the Biotechnology and Biological Sciences Research Council.
The authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
Supplementary Material
References
- Blumberg R., Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci. Transl. Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura Y., Kamoi R., Iida M. Pathogenesis of aphthoid ulcers in Crohn's disease: correlative findings by magnifying colonoscopy, electron microscopy, and immunohistochemistry. Gut. 1996;38:724–732. doi: 10.1136/gut.38.5.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithauser F., et al. Foxp3-expressing CD103+ regulatory T cells accumulate in dendritic cell aggregates of the colonic mucosa in murine transfer colitis. Am. J. Pathol. 2006;168:1898–1909. doi: 10.2353/ajpath.2006.050228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithauser F., Trobonjaca Z., Moller P., Reimann J. Clustering of colonic lamina propria CD4(+) T cells to subepithelial dendritic cell aggregates precedes the development of colitis in a murine adoptive transfer model. Lab. Invest. 2001;81:1339–1349. doi: 10.1038/labinvest.3780348. [DOI] [PubMed] [Google Scholar]
- Spahn T.W., et al. Induction of colitis in mice deficient of Peyer's patches and mesenteric lymph nodes is associated with increased disease severity and formation of colonic lymphoid patches. Am. J. Pathol. 2002;161:2273–2282. doi: 10.1016/S0002-9440(10)64503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., et al. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J. Immunol. 2002;168:57–64. doi: 10.4049/jimmunol.168.1.57. [DOI] [PubMed] [Google Scholar]
- Pabst O., et al. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J. Immunol. 2006;177:6824–6832. doi: 10.4049/jimmunol.177.10.6824. [DOI] [PubMed] [Google Scholar]
- Knoop K.A., Newberry R.D. Isolated Lymphoid follicles are dynamic reservoirs for the induction of intestinal IgA. Front. Immunol. 2012;3:84. doi: 10.3389/fimmu.2012.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert S.A., Mebius R.E. New insights into the development of lymphoid tissues. Nat. Rev. Immunol. 2010;10:664–674. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- Kanamori Y., et al. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 1996;184:1449–1459. doi: 10.1084/jem.184.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G., Littman D.R. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- Glaysher B.R., Mabbott N.A. Isolated lymphoid follicle maturation induces the development of follicular dendritic cells. Immunology. 2007;120:336–344. doi: 10.1111/j.1365-2567.2006.02508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R.G., Chaplin D.D., McDonald K.G., McDonough J.S., Newberry R.D. Isolated lymphoid follicle formation is inducible and dependent upon lymphotoxin-sufficient B lymphocytes, lymphotoxin beta receptor, and TNF receptor I function. J. Immunol. 2003;170:5475–5482. doi: 10.4049/jimmunol.170.11.5475. [DOI] [PubMed] [Google Scholar]
- Kiss E.A., et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Bouskra D., et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Kweon M.N., et al. Prenatal blockage of lymphotoxin beta receptor and TNF receptor p55 signaling cascade resulted in the acceleration of tissue genesis for isolated lymphoid follicles in the large intestine. J. Immunol. 2005;174:4365–4372. doi: 10.4049/jimmunol.174.7.4365. [DOI] [PubMed] [Google Scholar]
- Lochner M., et al. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J. Exp. Med. 2011;208:125–134. doi: 10.1084/jem.20100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop K.A., Butler B.R., Kumar N., Newberry R.D., Williams I.R. Distinct developmental requirements for isolated lymphoid follicle formation in the small and large intestine: RANKL is essential only in the small intestine. Am. J. Pathol. 2011;179:1861–1871. doi: 10.1016/j.ajpath.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji M., et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Baptista A.P., et al. Colonic patch and colonic SILT development are independent and differentially regulated events. Mucosal Immunol. 2013;6:511–521. doi: 10.1038/mi.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- Zaph C., et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J. Exp. Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern P.P., Izcue A., Maloy K.J., Powrie F. The interleukin-23 axis in intestinal inflammation. Immunol. Rev. 2008;226:147–159. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- Takatori H., et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Moreno J., et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat. Immunol. 2011;12:639–646. doi: 10.1038/ni.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota N., et al. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat. Immunol. 2011;12:941–948. doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- Atarashi K., et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izcue A., et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–570. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M., et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009;15:114–123. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.T., Lugering A., Newell K.A., Williams I.R. Intestinal cryptopatch formation in mice requires lymphotoxin alpha and the lymphotoxin beta receptor. J. Immunol. 2004;173:7183–7189. doi: 10.4049/jimmunol.173.12.7183. [DOI] [PubMed] [Google Scholar]
- Uhlig H.H., et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Rivollier A., He J., Kole A., Valatas V., Kelsall B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K.G., McDonough J.S., Dieckgraefe B.K., Newberry R.D. Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues. Am. J. Pathol. 2010;176:2367–2377. doi: 10.2353/ajpath.2010.090723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi I., et al. Intestinal bacteria induce TSLP to promote mutualistic T-cell responses. Mucosal Immunol. 2013;6:1157–1167. doi: 10.1038/mi.2013.12. [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., et al. Dendritic cells are crucial for maintenance of tertiary lymphoid structures in the lung of influenza virus-infected mice. J. Exp. Med. 2009;206:2339–2349. doi: 10.1084/jem.20090410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks J.R., Davalos-Misslitz A.C., Hintzen G., Ohl L., Forster R. Regulatory T cells interfere with the development of bronchus-associated lymphoid tissue. J. Exp. Med. 2007;204:723–734. doi: 10.1084/jem.20061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T., et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee E.N., et al. Ectopic lymphoid tissue alters the chemokine gradient, increases lymphocyte retention and exacerbates murine ileitis. Gut. 2013;62:53–62. doi: 10.1136/gutjnl-2011-301272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E., et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Obata T., et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. USA. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitohy B., Hammarstrom S., Danielsson A., Hammarstrom M.L. Basal lymphoid aggregates in ulcerative colitis colon: a site for regulatory T cell action. Clin. Exp. Immunol. 2008;151:326–333. doi: 10.1111/j.1365-2249.2007.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- Zhou L., et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasan S., et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- McDonald K.G., Leach M.R., Huang C., Wang C., Newberry R.D. Aging impacts isolated lymphoid follicle development and function. Immun. Ageing. 2011;8:1. doi: 10.1186/1742-4933-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr R.H., et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owyang A.M., et al. Interleukin 25 regulates type 2 cytokine-dependent immunity and limits chronic inflammation in the gastrointestinal tract. J. Exp. Med. 2006;203:843–849. doi: 10.1084/jem.20051496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K.G., McDonough J.S., Newberry R.D. Adaptive immune responses are dispensable for isolated lymphoid follicle formation: antigen-naive, lymphotoxin-sufficient B lymphocytes drive the formation of mature isolated lymphoid follicles. J. Immunol. 2005;174:5720–5728. doi: 10.4049/jimmunol.174.9.5720. [DOI] [PubMed] [Google Scholar]
- Bain C.C., Mowat A.M. CD200 receptor and macrophage function in the intestine. Immunobiology. 2012;217:643–651. doi: 10.1016/j.imbio.2011.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.