Abstract
Abscisic acid (ABA)1 is a plant hormone that controls many aspects of plant growth, including seed germination, stomatal aperture size, and cellular drought response. ABA interacts with a unique family of 14 receptor proteins. This interaction leads to the activation of a family of protein kinases, SnRK2s, which in turn phosphorylate substrates involved in many cellular processes. The family of receptors appears functionally redundant. To observe a measurable phenotype, four of the fourteen receptors have to be mutated to create a multilocus loss-of-function quadruple receptor (QR) mutant, which is much less sensitive to ABA than wild-type (WT) plants. Given these phenotypes, we asked whether or not a difference in ABA response between the WT and QR backgrounds would manifest on a phosphorylation level as well. We tested WT and QR mutant ABA response using isotope-assisted quantitative phosphoproteomics to determine what ABA-induced phosphorylation changes occur in WT plants within 5 min of ABA treatment and how that phosphorylation pattern is altered in the QR mutant. We found multiple ABA-induced phosphorylation changes that occur within 5 min of treatment, including three SnRK2 autophosphorylation events and phosphorylation on SnRK2 substrates. The majority of robust ABA-dependent phosphorylation changes observed were partially diminished in the QR mutant, whereas many smaller ABA-dependent phosphorylation changes observed in the WT were not responsive to ABA in the mutant. A single phosphorylation event was increased in response to ABA treatment in both the WT and QR mutant. A portion of the discovery data was validated using selected reaction monitoring-based targeted measurements on a triple quadrupole mass spectrometer. These data suggest that different subsets of phosphorylation events depend upon different subsets of the ABA receptor family to occur. Altogether, these data expand our understanding of the model by which the family of ABA receptors directs rapid phosphoproteomic changes.
Abscisic acid (ABA) is a plant hormone involved in drought response as well as regulation of seed dormancy and germination. The core cellular ABA signaling pathway is mediated by three major events: (i) ABA binding to ABA receptor proteins, i.e. pyrabactin resistance/pyrabactin resistance-like/regulatory component of ABA receptor (PYR/PYL/RCAR) proteins, (ii) inhibition of clade A protein phosphatase 2C (PP2C) activity as a result of ABA:PYR/PYL/RCAR:PP2C ternary complex formation, and (iii) sustained activation via autophosphorylation of PP2C targets, snf-1 related kinase 2s (SnRK2s) (1). Activated SnRK2s phosphorylate multiple substrates, including ion channels such as slow anion channel 1 (SLAC1) (2) and potassium channel in Arabidopsis thaliana 1 (KAT1) (3), transcription factors such as bZIP (4) and bHLH/AKS (5), and flowering time regulators (6). Given that ABA signaling impacts many cellular processes, it is likely that the SnRK2s have additional, undiscovered substrates important for phosphosignaling, genetic regulation, and physiological effects.
Many proteins within the ABA signaling pathway are genetically redundant. There are 14 PYR/PYL/RCARs, nine clade A PP2Cs, of which at least six play a role in ABA signaling (1), and 10 SnRK2s, of which three are known to play roles in ABA signaling (7). This redundancy impeded the discovery of the receptor family until 2009 (8, 9), when a multilocus loss-of-function pyr1/pyl1/pyl2/pyl4 quadruple receptor mutant (QR) was constructed. QR mutants are resistant to ABA-induced inhibition of germination (9), show reduced expression of ABA responsive genes, are deficient in ABA-mediated stomatal closure (10), have a higher stomatal conductance than wild-type (WT) plants, and show dramatically reduced reactive oxygen species-induced calcium influx upon ABA stimulation (11). Loss-of-function sextuple PYR/PYL/RCAR receptor mutants (12), triple SnRK2 mutants (13), and decuple SnRK2 mutants (14) have also been generated and all suffer from stunted growth and ABA insensitivity.
The reliance upon the SnRK2 family of protein kinases to transduce ABA signaling via changes in protein phosphorylation make quantitative mass spectrometry (MS)-based phosphopoteomics an ideal tool to study ABA signaling. The availability of faster and more sensitive mass spectrometers and new techniques for phosphopeptide enrichment has made proteome-wide comprehensive phosphorylation experiments feasible (15). Previous experiments have used a quantitative phosphoproteomic approach to analyze ABA response in loss-of-function snrk2.2/2.3/2.6 mutants (6, 16, 17), however, these focused on long ABA treatments (15 min and longer), even though ABA-responsive phosphorylation occurs as early as 5 min (17). Additionally, none have been performed using the QR mutant, despite multiple phenotypes demonstrating its lack of ABA responsiveness (9 –11).
The two most prevalent quantitative MS techniques for large-scale, untargeted work are in vitro labeling with isobaric tags (i.e. iTRAQ and tandem mass tags) (18, 19) and metabolic labeling with heavy-isotope-labeled inorganic nitrogen or amino acids in the growth medium (i.e. SILAC) (20, 21). Metabolic labeling offers the ability to label proteins as an organism grows, and tissue from isotopic variants is combined immediately after treatment and processed together. Thus, sample processing affects both isotopic variant samples equally, and quantitative measurements are unaffected by inherent downstream processing error. Arabidopsis is uniquely suited for metabolic labeling since its small seed size allows labeling to 95% or greater in a span of approximately 10 days (22, 23).
Untargeted discovery proteomic experiments using MS provide an unequaled means of identifying and quantifying large numbers of phosphorylation sites, but these large-scale untargeted analyses are significantly affected by instrumental variability, which can have a large effect on statistical significance and reproducibility. Analyzing a complex sample multiple times can result in little overlap in peptide identifications between analyses. Low abundance species are not sampled in every analysis, resulting in variable observation and discrepancies in peptide identification between complex samples. One solution is to perform selected reaction monitoring (SRM) (24) assays with a triple quadrupole mass spectrometer. SRM quantification uses ratios comprised of area under elution curves of fragment ions from endogenous phosphopeptides and identical measurements of their respective fragment ions from spiked in heavy-labeled synthetic phosphopeptides. Spiking identical amounts of a synthetic peptide mixture into every sample enables controlled quantification, and mass selection during analysis allows reproducible monitoring of handpicked phosphorylation events. Using a triple quadrupole, SRM allows mass selection at both the precursor and fragment ion levels, improving signal-to-noise levels and enabling detection and quantification of low abundance species. Surprisingly, even with this well-established technique, the set of literature describing targeted MS experiments monitoring protein phosphorylation is noticeably small (25 –30).
To identify early ABA-induced phosphorylation events mediated by the ABA receptors PYR1, PYL1, PYL2, and PYL4, we performed a quantitative phosphoproteomic analysis of WT plants treated with ABA for 5 min. This provided us with the foundation for understanding global ABA-responsive phosphorylation changes at this time point in WT plants. Next, because QR mutants are phenotypically much less sensitive to ABA than WT plants (9 –11), we aimed to determine whether or not this phenotype of QR plants corresponded to a loss of ABA-responsive phosphorylation following treatment. To that end, we used a combination of untargeted high-resolution MS and targeted MS to identify and confirm ABA-dependent phosphorylation events and measure their response to ABA in the QR mutant background. Our results demonstrate that a subset of ABA-responsive phosphorylation depends on a subset of the family of PYR/PYR/RCAR receptors that includes at least one within the group of four mutated in addition to at least one of the nine functional in the QR background, whereas the rest of the ABA-responsive phosphorylation depends on either the receptors knocked out in the QR mutant or at least one of the nine remaining. Altogether, these data suggest that different subsets of phosphorylation events may depend on the functional presence of different subsets of the ABA receptor family for full ABA responsiveness. Further, we have identified putative SnRK2 substrates and found evidence of crosstalk between ABA and other plant stress signaling pathways. This analysis represents comprehensive analysis of early phosphorylation events in ABA signaling and greatly expands the view of ABA-dependent phosphosignaling.
MATERIALS AND METHODS
Arabidopsis Metabolic Labeling and Untargeted MS
Growth conditions, protein extraction, and digestion were all performed as previously described (17). For the WT experiment, two A (14N ABA-treated, 15N mock-treated) replicates were used, and three B (14N mock-treated, 15N ABA-treated) replicates were used. Each A or B experiment consisted of material from two magenta boxes, combined at equal dry, frozen, and ground weights (Fig. S1). For the QR mutant experiments, three A and B replicates were used. When LysC digests were performed, trypsin was replaced with LysC in identical amounts. Offline strong cation exchange fractionation was performed solely on tryptic digests (not the LysC digests) with strong cation exchange chromatography using a method adapted from a previously published protocol (31). Samples were resuspended into buffer A (5 mm KH2PO4, 30%, pH 2.65, 30% C2H3N), loaded onto a 200 × 9.4 mm polySULFOETHYL A column with 5 μm beads with 200Å pores (PolyLC, Inc., Columbia, MD) for 2 min and eluted over 33 min with a gradient to 25% B (5 mm KH2PO4, 350 mm KCl, pH 2.65, 30% C2H3N), after which 100% B was flowed for 5 min, followed by a re-equilibration with 100% A for 20 min. HPLC flow was 3 ml/min. Fractions were collected in 4-min intervals and lyophilized. Fractions were chosen for analysis based on time of elution and intensity of chromatogram, based on 214 nm and 280 nm absorbance. Lyophilized fractions were resuspended in 0.1% TFA, desalted using Waters (Milford, MA) sep-pak columns, and dried down in a vacuum centrifuge. Phosphopeptide enrichment was performed on trypsin digested, strong cation exchange fractionated samples as well as LysC digested samples using 200 μl spin-enrichment TiO2-packed tips (GL Sciences, Inc., Shinjuku, Tokyo) using supplier protocol. This process is detailed in Fig. S1.
Dried phosphopeptides were solubilized into Optima LC/MS-grade 0.1% formic acid in water (Fisher Scientific, Waltham, MA) for injection onto an Linear Trap Quarupole (LTQ) Orbitrap XL using an Agilent 1100 LC system. Solvent A was 0.1% formic acid, and solvent B was 95% acetonitrile, 0.1% formic acid. The sample was loaded directly onto an analytical column of 75 μm inner diameter and 360 μm outer diameter house-packed with ∼13 cm C18 resin (Magic-C18, 200Å, 3 μm, Bruker, Billerica, MA) at 0% B and a flow rate of 0.5 μl/min from 0–45 min, then eluted from 45–235 min at a flow rate of 0.2 μl/min and a gradient to 40% B, then from 235–255 min to 60% B, then from 255–260 min to 100% B. The column was then re-equilibrated by flowing 100% A from 260–278 min, stepping flow rate to 0.5 μl/min at minute 265.
MS1 spectra were collected using the Orbitrap at resolution 100,000 with preview mode enabled. The top five MS2 per MS1 were collected in the LTQ, rejecting +1 and unassigned charge states, using Collision(ally)-Induced Dissociation (CID). With an isolation window of 2.5 m/z, normalized Collision(al) energy (CE) of 35, activation Q of 0.25, activation at 30 ms, and a minimum MS1 signal threshold of 500. Electron Transfer Dissociation (ETD) analysis was performed with identical separation as listed for CID, with a reaction time of 100 ms, precursor charge state-dependent reaction time optimization, and supplemental activation for doubly charged precursor ions. Raw data were converted to .mgf files using default settings in the Trans-Proteomic Pipeline and searched using Mascot v2.2.2 and the Arabidopsis Information Resource (TAIR) protein database (version 9, June 19, 2009) with reverse sequences and common contaminants manually appended (62,522 sequences in total). The enzyme trypsin was specified as the protease and one missed cleavage was allowed. A precursor and fragment ion tolerance of 20 ppm and 0.6 Da, respectively, were used. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation, serine/threonine/tyrosine phosphorylation, and asparagine/glutamine deamidation were set as variable modifications. Mascot output was filtered to 1.0% false discovery rate using an in-house written script, and Census (32, 33) was used to extract ion chromatograms and quantify 14/15N area ratios. Census output was reformatted using an in-house written script, and data were median-normalized per injection. 15N Mascot searches were performed for all .raw files, and phosphopeptide ratios that were not obtained for a given replicate using 14N searching had 15N search-produced ratios added and used in averaging and statistical analysis. Technical replicates were not performed due to high instrument usage from multiple fractions for multiple biological replicates. Phosphopeptides of interest with Mascot score ≤ 30 or potentially ambiguous phosphorylation localization (i.e. multiple phosphoisoforms were identified) were manually validated by raw MS2 spectrum analysis. All data have been provided as supplemental Scaffold (Proteome Software) files, processed Excel files (Tables 2–4), and the raw data have been uploaded to and are publicly available via Chorus (https://chorusproject.org).
To obtain the values highlighted in this manuscript, phosphopeptides of interest had their ratios collapsed into a single data point by averaging all the values for the 14Ntreat/15Ncontrol experiment (A) and the inverse values of the 14Ncontrol/15Ntreat experiment (B). T-tests were performed using the spread of ratios from A experiments and from B experiments prior to inverting the B values.
SRM-MS
Plants were grown and treated as described above, solely in 14N media. Three biological replicates of WT+ABA or Mock and QR+ABA or Mock were performed. Prior to digestion with trypsin/LysC mix, a mixture of heavy labeled internal peptide standards (IS) was added to samples. House-packed, offline TiO2 columns were used for phosphopeptide enrichment, as previously reported (23). Samples were injected onto an AB SCIEX (Framingham, MA) QTRAP 5500 using an Eksigent nanoflex cHiPLC system with a ChromXP C18-CL column (3 μm beads with 120Å pore size packed into a 6 mm × 200 μm column) for trapping and a ChromXP C18-CL column (3 μm beads with 120Å pore size packed into a 15 cm × 75 μm column) for chromatographic separation. Buffer A was 0.1% formic acid in water, and buffer B was 0.1% formic acid in acetonitrile (ANC). Samples were loaded for 1 min in 2% B, and an elution gradient from 2 to 35% B was run for 70 min, upped to 90% B in 2 min., flowed at 90% B for 2 min., dropped to 2% B in 1 min., then flowed at 2% B for 15 min. for a total run time of 90 min. All flow rates were 300 nl/min. Precursor ions were scheduled within 3- or 4-min retention time windows, depending on instrument performance. Every condition save for QR+Mock has at least two injection replicates, and most have 3. Method optimization was performed using MRMPilot software v1.0 (AB SCIEX).
Multiple fragment ions were tested for each peptide using both in silico predictions and acquired MS/MS spectra. The three to five most abundant parent ion-to-fragment ion transitions were selected, and collision energies were optimized to maximize fragment ion intensity. The developed method was pilot tested, and the number of transitions for each peptide was reduced to three through the removal of the lowest quality transitions that performed poorly in complex sample backgrounds. These were monitored during targeted analysis. Quantification was performed using Multiquant software v2.0 (AB SCIEX), and well-behaved (i.e. highest signal intensity, best chromatography) transitions for each peptide were used for quantification. For each peptide, 1–3 transitions were used for quantification with Multiquant's automatic MQ4 function to determine peak area under fragment ion chromatograms.
Further processing occurred in Excel. Injection replicates were averaged per transition, and then transitions were averaged (when more than one was used for quantification) per biological replicate to obtain a ratio for a given peptide for that replicate. Ratios from the control (WT+Mock and QR+Mock) biological replicates (representing average area under chromatograms for endogenous peak:internal standard) were then averaged to get a single value for control condition, and then each WT+ABA or QR+ABA replicate was divided by the respective averaged control value to get a spread of ABA-induced fold changes. This spread was used to create the standard error bars shown in Fig. 4. All reported p values were calculated using the spread of both +ABA and +Mock biological replicate ratios.
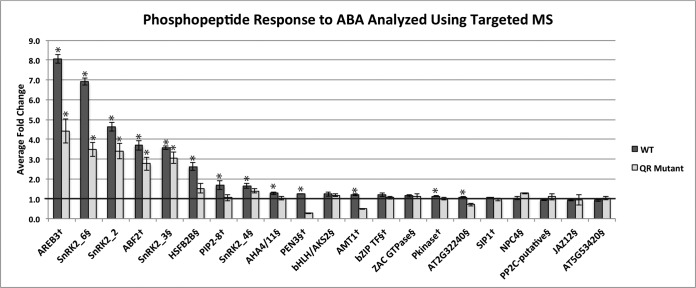
Fig. 4.
Comparison of ABA-responsive phosphorylation changes in WT and pyr1/pyl1/pyl2/pyl4 (QR) mutant analyzed using targeted MS. Phosphopeptides from the dataset presented here are designated with §, and phosphopeptides from previously published discovery work (Kline et al., 2010) (17) are designated with †. Bars are marked (*) to indicate p ≤ .05. See Table S3 for p values.
RESULTS
SnRK2 Protein Kinases and Other Proteins Are Phosphorylated in Response to ABA In Vivo within 5 Min of Treatment
We hypothesized that ABA phosphosignaling may function on a time scale shorter than the majority of plant phosphoproteomic studies have investigated. To test this, untargeted high-resolution MS in conjunction with metabolic labeling was used to examine the effect of 5-min ABA treatment on global phosphorylation in WT plants (Fig. S1). Ratios, calculated using area under extracted ion chromatograms for 14N and 15N versions for every phosphopeptide, were manually examined to find ABA-induced phosphorylation that had:(i) a fold change of greater than 1.15 or less than 0.85,(ii) at least one data point from both reciprocal experiments (see Fig. S1), and (iii) extracted ion chromatograms that exhibited good signal to noise quality. We chose the threshold of 1.15 or 0.85 for fold changes of interest because of the short treatment time, the requirement to see a corresponding fold change in the reciprocal experiment, and because chromatograms were manually examined and validated for each phosphopeptide that passed the aforementioned requirements.
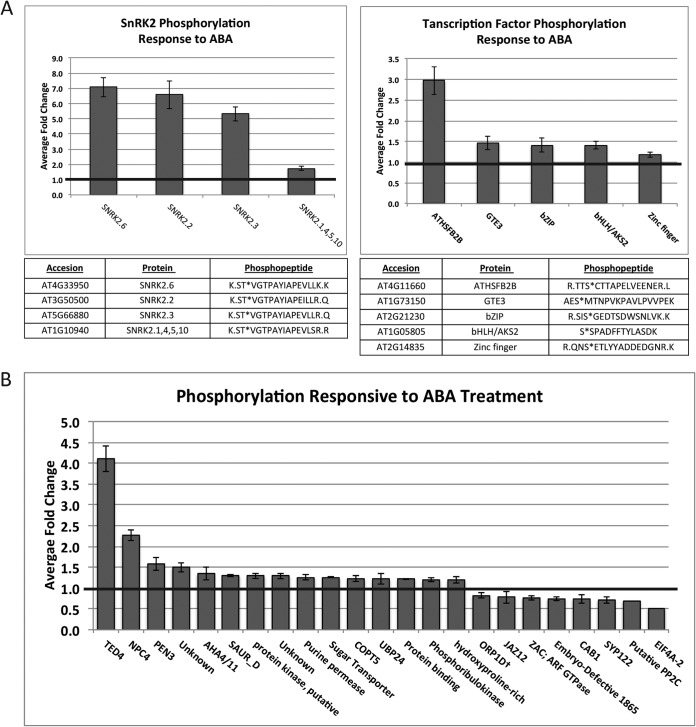
Within 5 min, ABA-dependent phosphorylation increased on residues within the autoactivation loops of SnRK2.2, 3, and 6 (Fig. 1A, Table S1), the three SnRK2 kinases previously demonstrated to be ABA responsive (7). A phosphopeptide that was ambiguous between SnRK2.1, 4, 5, and 10 (due to sequence homology) was also identified and showed a minor ABA-dependent change. We identified ABA-dependent phosphorylation on multiple transcription factors (Fig. 1A, Table S1) including the transcriptional repressor heat shock factor B2b (HSFB2b, AT4G11660), for which the largest ABA-responsive phosphorylation increase of the transcription factors was observed. Additional transcription factors that exhibited ABA-responsive phosphorylation increases include global transcriptional element 3 (GTE3, AT1G73150), a basic leucine zipper (bZIP, AT2G21230), a basic helix-loop-helix/ABA-responsive kinase substrate (bHLH/AKS2, AT1G05805), and a zinc finger (AT2G14835). Interestingly, among these transcription factors, only bHLH/AKS2 has an annotated function. It is one of a family of three ABA-responsive kinase substrates (AKS1, 2, and 3). Upon phosphorylation, AKS1 represses transcription of KAT1, the inward-rectifying K+ channel that positively regulates ABA-induced stomatal closure (5). We identified ABA-induced phosphorylation on an analogous serine on AKS2 occurring within 5 min following treatment. Overall, 3,500 total unique phosphopeptides were sequenced and quantified (Table S2); of these, less than 1% changed after 5 min of ABA treatment: 24 ABA-dependent phosphorylation events increased within this timeframe, and eight decreased (Fig. 1B, Table S1).
Fig. 1.
ABA-responsive phosphorylation changes following 5 min of treatment identified using untargeted MS analyses. Error bars represent standard error. Data displayed in full in Table S1. A, ABA-responsive SnRK2 and transcription factor phosphorylation. (B) Additional ABA-responsive phosphorylation changes.
ABA-Dependent Phosphorylation Increases on Transcription Factors within SnRK2 and MAPK Phosphorylation Motifs
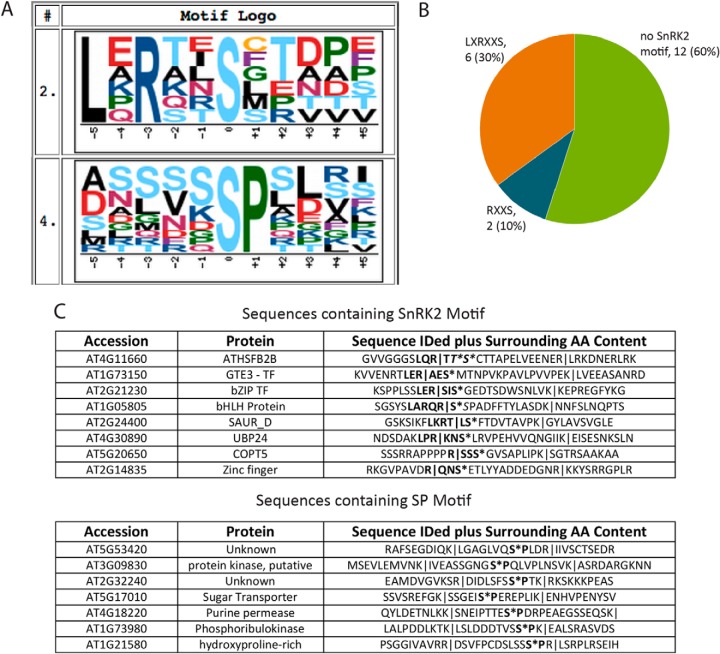
To determine whether the phosphorylation sites that showed rapid changes were occurring on SnRK2 substrates, we analyzed the peptides showing increased ABA-responsive phosphorylation for enriched phosphorylation motifs using Motif-X (34, 35). Using the A. thaliana proteome as a background, the most enriched, highest-scoring sequence motif identified was LXRXXS (Fig. 2A). This sequence has been demonstrated in vitro to be a preferential SnRK2 phosphorylation motif (4, 35), and its enrichment in the dataset suggests that some of these phosphoproteins may be ABA-dependent SnRK2 substrates. The second-highest-scoring motif was SP, which has been computationally predicted to be a target of mitogen-activated protein kinases (MAPKs) (37). The full search results are shown in Fig. S2.
Fig. 2.
Phosphorylation in response to 5 min of ABA treatment occurred on SnRK2 and MAPK motifs. (A) Two enriched motifs were identified, using MOTIF-X, as 10.49- and 3.04-fold enriched (Fig. S2) over the A. thaliana proteome at a significance threshold of 0.005. A window of 11 amino acids, centered on an S residue, with an occurrence threshold of six was used against the A. thaliana proteome as the background. Sequences fed into Motif-X had 10 amino acids from the respective protein sequence before and after the identified sequence added for larger context. (B) Of the phosphopeptides showing increased phosphorylation following ABA treatment, 30% of the increased phosphorylation occurred on the LXRXXS SnRK2 motif and 10% occurred on the RXXS SnRK2 motif. Peptides belonging to SnRK2 were removed form this analysis. (C) Sequences containing identified SnRK2 motif and the MAPK motif. Identified sequences are highlighted with pipes, and motifs are shown in bold.
Of the identified phosphorylation increases, six contain an LXRXXS motif and two contain an RXXS motif, a weaker but still preferential SnRK2 phosphorylation motif (Fig 2B) (4, 36). Interestingly, of the five transcription factors identified, four contain the LXRXXS motif, and one contains the RXXS motif (Fig 2C). Given that phosphorylation occurs concurrent with or following SnRK2 activation and these sequences contain preferential SnRK2 phosphorylation motifs, these transcription factors are likely direct SnRK2 substrates. One of the transcription factors, bHLH/AKS2 (AT1G05805), has previously been shown to exhibit ABA-responsive phosphorylation (5), however, the kinase responsible is not known; our data suggest that the kinase responsible for ABA-responsive bHLH/AKS2 phosphorylation is one of the three SnRK2s we identified as ABA responsive within the same timeframe. Seven of the 20 ABA-responsive phosphorylation increases contain the motif SP (Fig. 2C), a motif likely preferred by MAPKs (37). MAPKs have been implicated in ABA signaling in many studies (38), and these data suggest that their ABA-responsive activity is occurring as early as 5 min after treatment. Interestingly, there is no overlap between the list of phosphopeptides that contains the SnRK2 phosphorylation motifs and the list that contains the MAPK phosphorylation motifs.
ABA-Dependent Phosphorylation in Quadruple ABA Receptor Mutants Exhibits Distinct Patterns
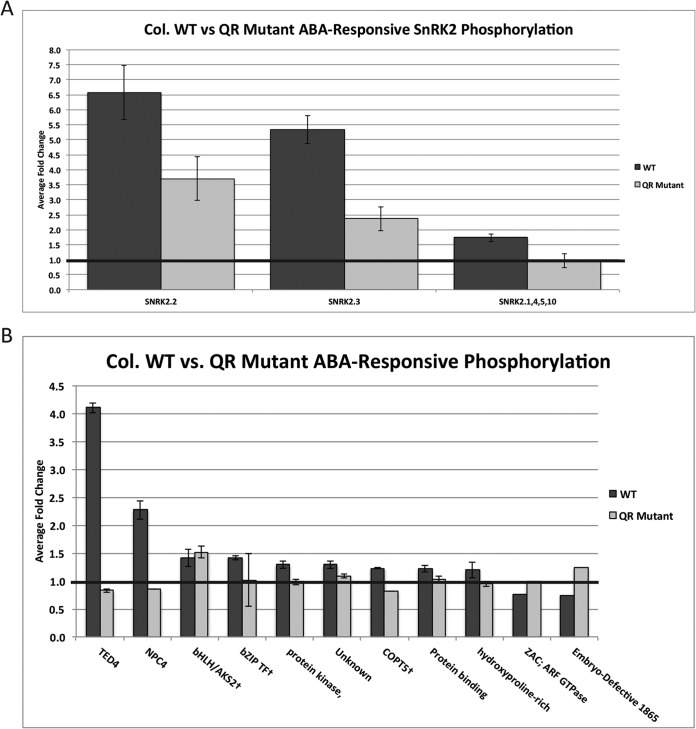
Quadruple ABA receptor (QR) mutants are far less sensitive to ABA than WT plants. This phenotype manifests as a loss of ABA-induced transcriptional changes (12), stomatal closure (10), calcium flux (11), and germination/growth inhibition (9). We predicted that ABA-responsive phosphorylation patterns observed in WT plants would differ in the QR mutant background. To test this, we performed a metabolic labeling experiment with QR mutants to quantify phosphorylation changes induced by ABA (Table S2). We observed that the SnRK2 kinases identified as significantly phosphorylated in response to 5 min of ABA treatment in WT plants are also responsive to ABA in the mutant background, albeit to a lesser degree (Fig. 3A). This indicates that the four mutated receptors only partially account for ABA-responsive SNRK2 phosphorylation. Interestingly, the phosphorylation event within the activation loop of SnRK2.1, 4, 5, and 10 was not ABA responsive in the QR mutant background.
Fig. 3.
Comparison of ABA-responsive phosphorylation changes in WT background to QR mutant background. Error bars represent standard error. (A) ABA-responsive SnRK2 kinase phosphorylation. (B) ABA-responsive phosphorylation changes on other proteins identified and quantified in the QR mutant dataset.
Many of the ABA-responsive phosphorylation changes observed in the WT dataset were identified as unresponsive to ABA in the QR mutant background (Fig. 3B), consistent with the idea that a portion of ABA-dependent phosphorylation relies entirely on at least one of the four knocked out receptors. Surprisingly, ABA-dependent phosphorylation on a single protein, bHLH/AKS2 (AT1G05805), was unaffected in the QR mutant background, indicating that some ABA-responsive phosphorylation events may occur independently of the four knocked out receptors.
In the untargeted studies, we could not compare all of the phosphorylation sites identified in the WT study to the QR mutant study. There was imperfect overlap in phosphopeptide identification between samples, an issue intrinsic to untargeted MS experiments. The nature of data-dependent MS is such that the same pool of low-abundance peptides may not be identified in every analysis, and the effects of this are more apparent when analyzing phosphorylation, which can occur transiently and at low stoichiometries. Thus, we applied a targeted approach using triple quadrupole based MS to reproducibly analyze phosphopeptides identified as ABA responsive and as validation for the changes observed in the untargeted datasets.
Large ABA-Responsive Phosphorylation Changes Are Reproduced in Targeted Analyses
Whereas untargeted MS can be made quantitative by metabolically labeling organisms as they grow prior to analysis, SRM, a type of targeted MS, uses synthetic heavy amino acid-labeled phosphopeptides. Mixtures of synthetic peptides are spiked into extracted protein samples prior to reduction, alkylation, and digestion. Quantification is achieved by comparing the area under extracted ion elution chromatograms of peptide fragment ions for the spiked in heavy isotope-labeled standards to the light, endogenous versions (Fig. S3). Comparing these ratios between samples allows large-scale experiments to be quantitative by focusing on a smaller number of peptides or phosphorylation events in every sample.
The panel of 21 phosphopeptides used for targeted analysis contained those identified from untargeted analyses in this study as well as those in a previously published untargeted study (17). The ABA-responsive fold changes with the largest magnitude in the untargeted study were reproduced in the targeted study (Fig. 4, Table S4). Phosphopeptides corresponding to the activation loop of the SnRK2 kinases showed the same pattern of partially diminished ABA-responsive phosphorylation when in the QR mutant background. The SRM data revealed that ABA-dependent SnRK2.6 phosphorylation is indeed diminished in the mutant background, which could not be determined using the untargeted data due to SnRK2.6 phosphorylation being observed in only one untargeted mutant sample. The ABF (AT1G45249, AT1G49720, AT4G34000, AT3G19290, AT3G56850) and AREB3 (AT5G56850) transcription factors are SnRK2 substrates that are phosphorylated in response to ABA (39, 40). As expected given this relationship, our targeted SRM data indicate their phosphorylation pattern to be similar to that of the SnRK2 kinases in the mutant background. The targeted SRM data also corroborated the ABA-responsive phosphorylation seen on bZIP transcription factor (AT2G21230), which we previously observed in the untargeted data. Additionally, targeted MS confirmed that ABA-responsive bHLH/AKS2 (AT1G05805) phosphorylation occurs fully in both WT and QR mutant background.
Targeted analysis using SRM revealed statistically significant differences that were not previously observed due to inherent limitation of untargeted methods. For example, the transcriptional repressor HSFB2b (AT4G11660) shows diminished ABA-responsive phosphorylation in the QR mutant, similar to the SnRK2 kinases and the known transcription factor substrates. Additionally, we observed an ABA-responsive phosphorylation increase in the WT dataset on a phosphopeptide sequence ambiguous between the plasma membrane ATPases/proton transporters AHA4/11 (AT3G47950, AT5G62670). This phosphopeptide was not observed at all in the mutant untargeted dataset, and the QR response could not be compared with the WT response using the untargeted data. The SRM data revealed that, in the mutant background, an ABA-induced phosphorylation increase on AHA4/11 was not observed, suggesting ABA-induced AHA4/11 phosphorylation depends on at least one of the four mutated ABA receptors. AHA4 has been implicated in growth and salt resistance (41) and little is known about AHA11.
Both Experimental and Biological Variability Contribute to Variation of Small Magnitude Fold Changes in Targeted Analyses
Interestingly, not all the ABA-responsive phosphorylation changes observed in the untargeted WT dataset were reproduced in the targeted dataset. Examples of this include the identified GTPase (AT4G21160), the phospholipase C NPC4 (AT3G03530), and a protein with unknown function accession AT5G53420 (Fig. 4). For the 14 phosphopeptides that we obtained both untargeted and targeted data for in the WT±ABA experiment, we compared the fold changes observed using the two analytical techniques (Fig. S4). Of these, eight had a fold change that was both trending the same direction and of a similar magnitude in the targeted versus the discovery data, and six did not. All of the phosphorylation events that were increased 1.5-fold or greater in the discovery data in response to ABA, with the exception of NPC4 (AT3G03530), were validated by the targeted work, whereas none of the ABA-dependent decreases we monitored were replicated.
Phosphoproteomic experimental workflows involve many steps, making sample processing variability a reasonable concern. Furthermore, the lack of literature surrounding targeted MS protein phosphorylation experiments means widespread quality assurance measures have not been established. Due to this lack of consensus, we tested how much variability we observed in a controlled experiment. Specifically, we assayed the degree of biological variability as well as variability introduced by experimental processing (Fig. S5A). We treated four individual magenta boxes with ABA, then ground, processed, and analyzed the material from the four boxes individually. Observed variability in this experiment was considered to be a result of processing and analysis or biological response variation between samples, given that all were treated and processed individually.
Samples were analyzed using an SRM panel of 55 synthetic peptides, monitoring 106 transitions, including those used in the ABA experiments (Table S4). This panel included 20 of the SRM peptides previously analyzed and 35 from a recently published targeted study from this laboratory (29). A median coefficient of variation of 20% was observed after data were processed identically to the ABA experiment (Fig. S5B). Indeed, many of the small ABA-responsive fold changes fall within this range of variation, suggesting inherent experimental and biological variability may account for discrepancies between discovery and targeted phosphoproteomic data when fold changes are small. The median represents this variability, which may occur at any step following protein extraction or be biological variation inherent to an in vivo system. This result is consistent with the previously published study, which indicated that statistical significance decreases drastically when monitored phosphorylation fold changes are ≤1.5 (29).
DISCUSSION
We used deep phosphoproteomic analyses to determine what ABA-regulated phosphorylation occurs in WT A. thaliana within 5 min of ABA treatment, a time point considerably earlier than has been previously examined in depth. We found ABA-induced phosphorylation changes on multiple proteins, including increased autophosphorylation of the SnRK2 kinases and increased phosphorylation on transcription factors not yet implicated in ABA signaling (Fig. 1). The two most enriched phosphorylation motifs in the set of ABA-responsive phosphorylation were those of the SnRK2 kinases and MAPKs, implicating these two pathways as the earliest activated in response to ABA stimulation (Fig. 2). We then asked if the pattern of observed ABA-responsive phosphorylation was altered in the quadruple ABA receptor mutant pyr1/pyl1/pyl2/pyl4 (QR), a genetic background that is significantly less sensitive to ABA than WT plants (9 –11). Interestingly, the data demonstrated that a subset of phosphorylation events, including the autophosphorylation of the SnRK2 kinases, was lower in magnitude but still present and significantly ABA responsive in the QR mutant background (Fig. 3). This suggests that these events depend on at least one of the four knocked out receptors in addition to at least one of the remaining nine receptors. Another subset of phosphorylation events was unresponsive to ABA in the QR mutant background, suggesting that these events depend on at least one of the four knocked out receptors to occur. A single ABA-induced phosphorylation event on the transcription factor bHLH/AKS2 (AT1G05805) occurred in both the WT and QR mutant background, suggesting its ABA responsiveness depends on at least one of the nine functional receptors present in the QR mutant background. These untargeted phosphoproteomic studies were followed by targeted MS using a panel of 21 synthetic phosphopeptides (Fig. 4). Of the 14 SRM phosphopeptides monitored that were also identified and quantified in the reported untargeted studies, 8/14 (57%) were validated using targeted MS, a subset that included the large ABA-responsive changes (≥1.5) and a portion of the small ones. The remaining phosphopeptides monitored (6/14, 43%), which included small ABA-responsive changes (≤1.5) and ABA-responsive phosphorylation decreases, did not agree with the untargeted results. We tested the variability inherent to our targeted pipeline and found it to be 20%, consistent with the irreproducibility we observed when monitoring low magnitude fold changes in a targeted fashion. Altogether, the data provide comprehensive analysis of early ABA signaling events and expand the view of ABA-regulated phosphorylation signaling.
ABA-Responsive Phosphorylation Occurs within 5 min of Stimulation and Is Likely Downstream of SnRK2 and MAP Kinases
Our analysis of WT plants demonstrated that ABA-induced SnRK2 autophosphorylation occurred as quickly as 5 min following ABA treatment (Fig. 1). The largest ABA-responsive phosphorylation increases were on the three SnRK2 kinases responsible for ABA signaling, corroborating data from a recently published targeted study from this laboratory focusing on crosstalk between signaling pathways (29). We also identified a phosphorylation event ambiguous between SnRK2.1, 4, 5, and 10 that increased in response to ABA treatment. None of these four SnRK2 kinases have yet been implicated in ABA signaling. Phosphoproteomic analyses also identified other ABA-responsive phosphorylation and dephosphorylation within this timeframe; about 1.0% of the quantified phosphoproteome was reproducibly responsive to ABA, and the majority of changes we observed were increased. Two recently published studies used untargeted phosphoproteomic analyses to analyze ABA responsiveness in A. thaliana on timescales ranging from 15–90 min (6, 16). These longer studies found 1.6% and 5.3% of the quantified phosphoproteome were ABA responsive, compared with our 5-min study in which we found 1.0% of the quantified phosphoproteome was ABA responsive. This reduced amount of identified ABA-dependent phosphorylation changes is consistent with the earlier time point following ABA treatment on which our experiments were focused. We compared fold changes for phosphorylation we identified as ABA responsive in WT plants to the same phosphorylation events observed and quantified in the previously published studies (Table S5). Though the quantification methods, time points, and growth conditions were different, our data support some of the smaller fold changes identified in these publications. Our threshold for consideration was 15% increased or decreased across multiple biological replicates, though many changes exceeded 20% or greater. Within the set of increased phosphorylation, the motifs LXRXXS and SP are enriched (Fig. 2.). The first motif is a SnRK2 phosphorylation motif (4, 36), and its enrichment within the identified increased phosphorylation lends confidence to the dataset and is consistent with the SnRK2 activation observed within the same time frame. The second is a predicted MAPK phosphorylation motif (37), and its presence within the increased phosphorylation suggests that ABA-responsive MAPK signaling may be active within 5 min as well. This result is consistent with and may be linked to a recently published targeted MS analysis showing rapid ABA-dependent phosphorylation on a MAP4K (29); however, the MAP4K family is poorly characterized in A. thaliana (42). In S. cerevisiae, the MAP4K Ste20 activates the MAPK cascade responsible for osmotic adaptation in response to osmotic stress (43), but it is unknown if a similar network exists in Arabidopsis. Together, the two lists of phosphopeptides containing the enriched motifs do not overlap and make up the majority of the identified increased phosphorylation, suggesting that the main phosphorylation cascades involved in ABA signaling at this early time point following treatment are the ABA-dependent SnRK2 and MAPK cascades.
ABA-Responsive Phosphorylation Occurs on TFs and Other Substrates Early in ABA Signaling
In addition to the SnRK2 kinases, ABA-dependent phosphorylation also increased on five transcription factors (Fig. 1), HSFB2b (AT4G11660), GTE3 (AT1G73150), a bZIP (AT2G21230), a zinc finger (AT2G14835), and bHLH/AKS2 (AT1G05805). Of these, four contain the SnRK2 phosphorylation motif LXRXXS and one contains a weaker SnRK2 motif, RXXS. Because of the presence of these motifs within the protein sequences and the observation that phosphorylation increases concurrently with or following SnRK2 activation, we consider all of these transcription factors to be candidates of direct SnRK2 kinase phosphorylation. None of these have been previously reported as SnRK2 substrates, and of these, only HSFB2b and bHLH/AKS2 have known function.
HSFB2B (AT4G11660) and HSFB1 (AT4G36990) are heat-inducible transcriptional repressors. Together, they are necessary in A. thaliana for acquired thermotolerance (44) and play a role in pathogen defense signaling (45). Our data indicate that HSFB2b phosphorylation within a SnRK2 motif increases quickly in response to ABA treatment. This is not the first data linking ABA and heat stress response. ABA treatment 1 h before heat shock has been previously demonstrated to increase survival rate post heat shock, and ABI1, a PP2C in the core ABA signaling pathway, is necessary for basal heat shock resistance (46). We predict that at least part of a combined response to ABA and heat shock begins as early as 5 min and involves regulation of heat shock response transcription factors, including HSFB2B. The SnRK2 phosphorylation motif, on which ABA-responsive phosphorylation occurred, is unique to HSFB2B, although HSFB2B contains a DNA binding domain that is highly conserved among both Class A and B heat shock factors.
Though HSFB2b phosphorylation has yet to be functionally linked to ABA signaling, bHLH/AKS2 is known to be phosphorylated in response to ABA at the site we observed in planta (5). Analogous phosphorylation of AKS1 (AT1G51140) by SnRK2.6 was demonstrated in vitro to disrupt AKS1 binding to the promoter of the inward rectifying K+ channel partially responsible for ABA-induced stomatal closure, KAT1. This binding event enhances KAT1 transcription, and phosphorylation prevents AKS1 binding, repressing transcription. Whether AKS2 functions similarly to AKS1 is currently unknown.
Mutant Studies Indicate Dependence of Phosphorylation Events on Subsets of the Family of Receptors
We next asked if the ABA-dependent phosphorylation observed is maintained in a genetic background containing multilocus loss of PYR1/PYL1/PYL2/PYL4 ABA receptor function. Interestingly, of the ABA-responsive phosphorylation observed in the WT background, we observed all three possible differences when examining ABA response in the QR mutant: (1) ABA-dependent phosphorylation that was as responsive in the mutant background as in the WT, (2) ABA-dependent phosphorylation that was unresponsive to ABA in the mutant background, and (3) ABA-dependent phosphorylation that was partially diminished in the mutant background when compared with the WT response (Fig. 3). The distribution was not equal; many of the phosphorylation events that were minimally responsive to ABA in the WT background were completely unresponsive to ABA in the mutant background, and only one event remained fully responsive, that on bHLH/AKS2 (AT1G05805). All of the SnRK2 kinases known to be ABA responsive exhibit the partially sustained phosphorylation in the mutant background, as do well-known SnRK2 kinase substrates. This may be a result of the short time point sampled combined with the lower activity of the SnRK2 kinases. In this case, at longer time points following ABA treatment, ABA-responsive phosphorylation would eventually reach levels observed in the WT. However, this observation could also be the result of a threshold-based response. If this is the case, ABA-responsive SnRK2 autophosphorylation and downstream phosphorylation in the QR mutant background could never reach the levels observed in the WT plants without the presence of the complete set of 14 PYR/PYL/RCAR ABA receptors. Future experiments to classify the nature of ABA-responsive phosphorylation will address this.
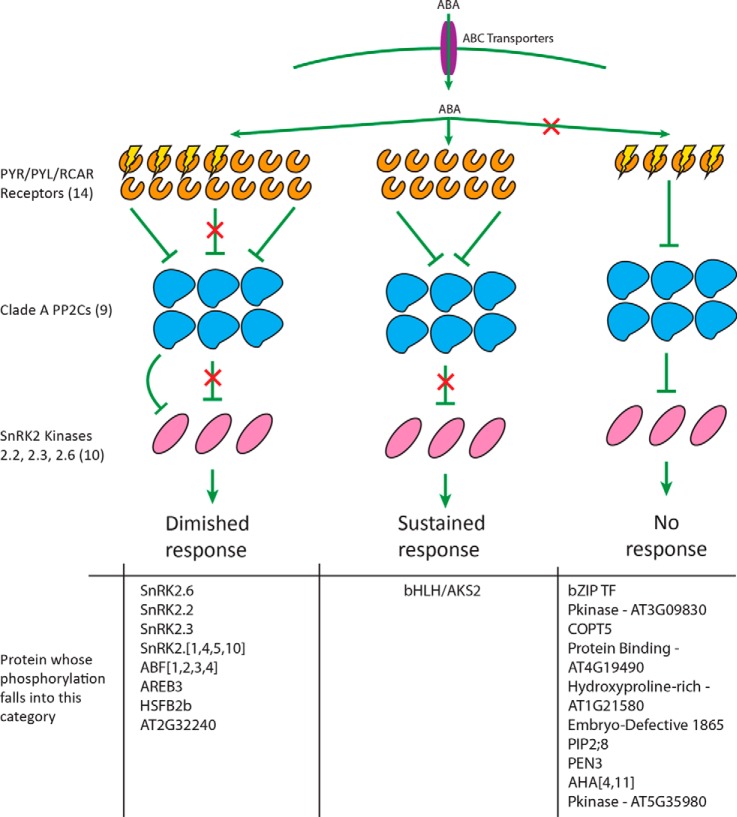
These data suggest that different subsets of ABA-responsive phosphorylation events depend on different subsets of the PYR/PYL/RCAR family of receptors to occur (Fig. 5). Those that are completely unresponsive to ABA in the QR mutant background depend upon the functional presence of at least one of the four mutated receptors. The phosphorylation events that are partially sustained in the QR mutant background depend upon a subset of ABA receptors that includes at least one of the four mutated receptors as well as at least one of the nine remaining in the QR mutant. These data are consistent with previously published data that demonstrate similar responses on a transcriptional level (12). Taken together, these results suggest that diminished ABA-dependent transcriptional response in mutant backgrounds may be the result of lessened ABA-responsive phosphorylation on transcription factors responsible for ABA-stimulated gene regulation. This in turn may be the result of diminished ABA-responsive SnRK2 autophosphorylation in the mutant background. Finally, we identified a phosphorylation event on bHLH/AKS2 that occurred to a similar degree in response to ABA in both the WT and the QR mutant background, a result that we verified with targeted experiments. Thus, this phosphorylation event depends entirely on at least one of the remaining nine receptors present in the QR mutant background to be ABA responsive. Though only one instance of this was identified in these initial experiments, future experiments with other mutants and treatment time points may identify more phosphorylation events that follow an identical pattern.
Fig. 5.
Model of early ABA-responsive phosphorylation. While a subset of ABA-responsive phosphorylation is diminished in the QR mutant, a single event is fully sustained in response to ABA, and another subset is completely unresponsive to ABA in the QR mutant. Proteins displayed had phosphopeptides identified in WT or both WT and QR mutant untargeted datasets shown in this report. Also included are proteins reported in previously published discovery data (Kline et al., 2010), which were followed up with targeted MS reported here. Phosphorylation identified as ABA responsive in untargeted data but not reproduced in targeted analyses is not shown. Brackets indicate ambiguity in protein accession based on the phosphopeptide sequence identified and used for targeted study.
The Largest ABA-Responsive Phosphorylation Changes Were Validated Using SRM-Based MS Analyses
Some phosphorylation observed in the untargeted WT dataset could not be compared with the QR mutant dataset due to intrinsic variability in phosphopeptide identification between samples in untargeted MS experiments. In addition, many of the phosphopeptides were only identified once or twice in the mutant data, preventing statistical analysis. To address this technical limitation, we used SRM-based targeted MS with a triple quadrupole instrument to monitor a panel of 21 phosphopeptides derived from these untargeted experiments and a previously published dataset (17).
The largest magnitude ABA-responsive fold changes (≥1.5) seen in the untargeted work were replicated using targeted assays (Fig. 4, Fig. S4). These included the SnRK2s and transcription factors that are likely ABA-responsive SnRK2 substrates, HSFB2b and bZIP AT2G21320. We also monitored phosphorylation on two peptides from ABFs1/2/3/4 (AT1G45249, AT1G49720, AT4G34000, AT3G19290, AT3G56850), and AREB3 (AT5G56850), known ABA-responsive SnRK2 substrates that were not identified in the discovery experiments performed for this study. The previous discovery work identified increased phosphorylation on ABFs1/2/3/4 and AREB3 only after 30 min of ABA treatment (17), whereas the targeted work demonstrated that these transcription factors are highly phosphorylated within 5 min of treatment. Due to lack of identification at multiple time points in the previous study and in multiple replicates in the discovery data shown here, it is likely that the abundance of ABA-responsive phosphorylation on these transcription factors is too low this early in signaling to monitor without targeted assays, though it is clear that ABA induces their phosphorylation. The phosphorylation pattern on these transcription factors is identical to the SnRK2s, i.e. highly responsive to ABA treatment in WT plants but only partially responsive to ABA in the QR mutant background. Interestingly, though we were unable to observe HSFB2b phosphorylation in the untargeted mutant dataset, our targeted analyses demonstrated a phosphorylation pattern identical to that of both the ABA-responsive SnRK2s and ABF1/2/3/4 and AREB3, providing further evidence that HSFB2b is an ABA-responsive SnRK2 substrate. These experiments demonstrate the ability of targeted assays to monitor phosphorylation events where abundance is too low for untargeted studies to reproducibly detect and quantify.
With respect to the small magnitude ABA-responsive fold changes identified in the untargeted data (≤1.5), some, but not all, were reproduced in the targeted data. The bHLH/AKS2 (AT1G05805) phosphorylation event increased 1.41 and 1.52 fold in the WT and QR mutant background, respectively, in response to ABA in the untargeted experiments (Fig. 3). This increase was repeated using targeted analyses, although lower fold changes were observed (Fig. 4). The bZIP transcription factor (AT2G21230), observed in both approaches, exhibited similar ABA-responsive fold changes of 1.41 and 1.31 in the WT background in the untargeted and targeted experiments, respectively. However, other ABA-dependent phosphorylation changes identified in the untargeted analyses were not reproduced using targeted analysis. For example, the identified ZAC GTPase (AT4G21160) phosphopeptide decreased in abundance upon ABA treatment in WT plants and was unresponsive in the QR mutant plants in the untargeted data. In the targeted dataset, this phosphopeptide appears unchanged upon ABA treatment in both WT and QR mutant plants. Of the 14 phosphorylation events we identified as ABA responsive in the untargeted experiments and monitored using SRM analyses, eight were validated and six did not replicate (Fig. S4). Probing the literature led to a few reports of targeted phosphorylation experiments (25 –30). A single publication from another laboratory describes a similar experimental scheme with human breast cancer tissue samples from high-risk and low-risk patients (30). In this study, a panel of 15 SRM peptides is used to validate an untargeted iTRAQ-based experiment. Of these, 66% (10/15) exhibited good correlation when comparing data points from the discovery and targeted experiments (r2≥0.6, as defined by the authors). This is slightly higher than what we found (57%).
To better understand the basis by which the targeted and untargeted experiments varied, we performed an experiment to determine what degree biological and processing variability affect the changes we can confidently quantify using targeted experiments using a panel of 55 SRM peptides. When treating, processing, and analyzing four plants identically, a median coefficient of variation (CV) of 20% was obtained when analyzing all the ratios of synthetic SRM peptide to endogenous peptide (Fig. S5). This CV value may explain why smaller magnitude fold changes are not reproduced; variation associated with experimental processing and biological variability is 20%, convoluting reproducible determination of smaller magnitude fold changes when performing targeted analyses using the described pipeline. This result is in line with a recent publication from this laboratory, which showed that for fold changes determined by SRM to be ≤1.5, statistical significance greatly decreases (29). Interestingly, when examining the targeted dataset, higher biological CVs were obtained when comparing the mutant data to the WT data (Table S3), suggesting a higher degree of variability in general in the mutant background, although the cause of this remains unclear.
CONCLUSION
Previously, we published a study examining ABA-responsive phosphorylation in WT A. thaliana at 5, 15, and 30 min of treatment (17). This work expands upon this previously published data by more deeply analyzing the 5 min response, both in WT and in the multilocus loss-of-function pyr1/pyl1/pyl2/pyl4 quadruple ABA receptor mutant. We have identified and quantified ABA-responsive phosphorylation changes in WT A. thaliana that occur within 5 min of treatment, representing ∼1.0% of the reproducibly observable phosphoproteome. Two recently published studies also examining ABA-responsive phosphorylation in Arabidopsis found 1.6% and 5.3% of the identified phosphoproteome is affected when examining 15, 30, and 90 min and 30 min of ABA treatment, respectively (6, 16). Considering we are examining an earlier time point after ABA treatment, our data are consistent with these published data. From the WT dataset, we identified multiple transcription factors whose phosphorylation is increased within 5 min of ABA treatment. A subset of these contain the SnRK2 preferential phosphorylation motif LXRXXS, and one contains the weaker RXXS motif, making these transcription factors candidates for direct phosphorylation by the SnRK2 family of protein kinases in response to ABA. Another subset, which does not overlap with the first, contains the MAPK phosphorylation motif SP. These data suggest that the SnRK2 and MAPK phosphorylation cascades are the most activated in the first 5 min following ABA stimulation. Using targeted analyses, the large magnitude ABA-responsive fold changes (≥1.5) and some small fold changes (8/14 total peptides) were validated. Conversely, six out of 13 phosphopeptides we monitored that were responsive to ABA in the untargeted experiments did not replicate using targeted analyses, all of which were small ABA-responsive fold changes in the untargeted experiments (≤1.5). We found that 20% variation in targeted measurements can be attributed to biological and technical variability intrinsic to our targeted workflow. Hopefully, as more laboratories adopt similar workflows, some consensus can be reached in regard to the inherent variability of targeted phosphorylation analyses.
When compared with WT, ABA-responsive phosphorylation in the QR mutants exhibited distinct patterns. The largest ABA-responsive phosphorylation changes, the SnRK2s and transcription factors, showed partially diminished ABA-responsive phosphorylation in the QR mutant. Many of the small ABA-responsive phosphorylation changes were either unresponsive to ABA treatment or reduced to no statistical significance in the QR background. The exception to this was bHLH/AKS2, which maintained full ABA-responsive phosphorylation in the QR mutant background. Taken together, these data add a new layer of understanding to ABA-regulated phosphorylation signaling. Different ABA-induced phosphorylation events depend on different subsets of the family of PYR/PYL/RCAR receptors to occur. A subset of these events, responsive to 5 min of ABA treatment in the WT background, is unresponsive to the same treatment in the QR mutant background. Their ABA-responsive phosphorylation depends on a subset of the ABA receptors that includes at least one of the four mutated in the QR background. Another subset of ABA-responsive phosphorylation partially occurred in the QR mutant background after 5 min of ABA treatment when compared with the WT background, indicating their ABA-responsiveness depends on at least one of the remaining nine ABA receptors present in the QR mutant background in addition to at least one of the four mutated in the QR background. The SnRK2 kinases and some transcription factor phosphorylation events exhibit this partial dependence. Finally, in both the WT and QR mutant background, phosphorylation observed on bHLH/AKS2 increased in response to ABA, indicating its occurrence depends entirely on the functional presence of at least one of the four knocked out receptors. It may be that yet undiscovered ABA-responsive phosphorylation events follow identical patterns, though they may depend on other receptors within the family. Much work by many laboratories has been performed to identify and quantify global phosphorylation changes in response to ABA treatment in A. thaliana, and we hope the data presented here can be used as a starting point for further in planta experiments to be performed within shorter time frames and with multiple permutations of mutants within the family of ABA receptors. Future studies will further elucidate details surrounding each of the PYR/PYL/RCAR receptors and their separate roles within the ABA-regulated phosphorylation network.
Supplementary Material
Acknowledgments
We thank the University of Wisconsin-Madison Biotechnology Center Core Mass Spectrometry Facility and Peptide Synthesis Facility, Greg Barrett-Wilt, Greg Sabat, and Melissa Boersma for technical assistance and consultation. We thank Leigh Anson and Lindsay Traeger for reading and editing the manuscript.
Footnotes
Author contributions: B.B.M., K.E.S., and M.R.S. designed the research; B.B.M. performed research; B.B.M. analyzed the data; B.B.M. and M.R.S. wrote the paper; and K.E.S. developed targeted MS methods.
* This work was supported, by NSF Grants MCB-0929395 (M.R.S.) and DGE-1256259 (K.E.S.), University of Wisconsin-based Morgridge Fellowships (K.E.S. and B.B.M.), and an NHGRI training grant to the Genomic Science Training Program 5T32HG002760 (B.B.M.).
 This article contains supplemental material Tables S1-S5 and Figs. S1-S5.
This article contains supplemental material Tables S1-S5 and Figs. S1-S5.
1 The abbreviations used are:
- ABA
- abscisic acid
- AKS
- ABA-responsive kinase substrate
- bHLH
- basic helix loop helix
- bZIP
- basic leucine zipper
- HSFB
- heat shock factor B
- iTRAQ
- isobaric tag for relative and absolute quantitation
- KAT1
- K (potassium) channel in Arabidopsis thaliana 1
- PP2C
- protein phosphatase 2C
- PYR/PYL/RCAR
- pyrabactin resistance/pyrabactin resistance-like/regulatory component of ABA receptor
- QR mutants
- quadruple ABA receptor mutants
- SLAC1
- slow anion channel 1
- SnRK
- Snf1-related serine/threonine protein kinase
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Cutler S.R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010) Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 [DOI] [PubMed] [Google Scholar]
- 2. Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K. A., Romeis T., Hedrich R. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D. B., Uozumi N. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424, 439–448 [DOI] [PubMed] [Google Scholar]
- 4. Sirichandra C., Davanture M., Turk B. E., Zivy M., Valot B., Leung J., Merlot S. (2010) The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14–3-3 binding site involved in its turnover. PLoS One 5, e13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahashi Y., Ebisu Y., Kinoshita T., Doi M., Okuma E., Murata Y., Shimazaki K. (2013) bHLH transcription factors that facilitate K(+) uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal 6, ra48. [DOI] [PubMed] [Google Scholar]
- 6. Wang P., Xue L., Batelli G., Lee S., Hou Y. J., Van, Oosten M. J., Zhang H., Tao W. A., Zhu J. K. (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110, 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakashima K., Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363 [DOI] [PubMed] [Google Scholar]
- 8. Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E., (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 9. Park S.Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishimura N., Sarkeshik A., Nito K., Park S. Y., Wang A., Carvalho P. C., Lee S., Caddell D. F., Cutler S. R., Chory J., Yates J. R., Schroeder J. I. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y., Chen Z. H., Zhang B., Hills A., Blatt M. R. (2013) PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl- channels through reactive oxygen species-mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol 163, 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonzalez-Guzman M., Pizzio G. A., Antoni R., Vera-Sirera F., Merilo E., Bassel G. W., Fernández M. A., Holdsworth M. J., Perez-Amador M. A., Kollist H., Rodriguez P. L. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fujii H., Zhu J. K. (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujii H., Verslues P. E., Zhu J. K. (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beltran L., Cutillas P.R. (2012) Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids 43, 1009–1024 [DOI] [PubMed] [Google Scholar]
- 16. Umezawa T., Sugiyama N., Takahashi F., Anderson J. C., Ishihama Y., Peck S. C., Shinozaki K. (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal 6, rs8. [DOI] [PubMed] [Google Scholar]
- 17. Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zieske L.R. (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 19. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 20. Everley P. A., Krijgsveld J., Zetter B. R., Gygi S. P. (2004) Quantitative cancer proteomics: Stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Mol. Cell. Proteomics 3, 729–735 [DOI] [PubMed] [Google Scholar]
- 21. Arsova B., Kierszniowska S., Schulze W. X. (2012) The use of heavy nitrogen in quantitative proteomics experiments in plants. Trends Plant Sci. 17, 102–112 [DOI] [PubMed] [Google Scholar]
- 22. Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol. Cell Proteomics 6, 860–881 [DOI] [PubMed] [Google Scholar]
- 23. Minkoff B. B., Burch H. L., Sussman M. R. (2014) A pipeline for 15N metabolic labeling and phosphoproteome analysis in Arabidopsis thaliana. Methods Mol. Biol. 1062, 353–379 [DOI] [PubMed] [Google Scholar]
- 24. Picotti P., Aebersold R. (2012) Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 25. Lam M. P., Scruggs S. B., Kim T. Y., Zong C., Lau E., Wang D., Ryan C. M., Faull K. F., Ping P. (2012) An MRM-based workflow for quantifying cardiac mitochondrial protein phosphorylation in murine and human tissue. J. Proteomics 75, 4602–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mithoe S. C., Boersema P. J., Berke L., Snel B., Heck A. J., Menke F. L. (2012) Targeted quantitative phosphoproteomics approach for the detection of phospho-tyrosine signaling in plants. J. Proteome Res. 11, 438–448 [DOI] [PubMed] [Google Scholar]
- 27. Lam M. P., Lau E., Scruggs S. B., Wang D., Kim T. Y., Liem D. A., Zhang J., Ryan C. M., Faull K. F., Ping P. (2013) Site-specific quantitative analysis of cardiac mitochondrial protein phosphorylation. J. Proteomics 81, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kimura A., Arakawa N., Hirano H. (2014) Mass spectrometric analysis of the phosphorylation levels of the SWI/SNF chromatin remodeling/tumor suppressor proteins ARID1A and Brg1 in ovarian clear cell adenocarcinoma cell lines. J. Proteome Res. 13, 4959–4969 [DOI] [PubMed] [Google Scholar]
- 29. Stecker K. E., Minkoff B. B., Sussman M. R. (2014) Phosphoproteomic analyses reveal early signaling events in the osmotic stress response. Plant Physiol. 165, 1171–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Narumi R., Murakami T., Kuga T., Adachi J., Shiromizu T., Muraoka S., Kume H., Kodera Y., Matsumoto M., Nakayama K., Miyamoto Y., Ishitobi M., Inaji H., Kato K., Tomonaga T. (2012) A strategy for large-scale phosphoproteomics and SRM-based validation of human breast cancer tissue samples. J. Proteome Res. 11, 5311–5322 [DOI] [PubMed] [Google Scholar]
- 31. Villen J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park S. K., Venable J. D., Xu T., Yates J. R., 3rd. (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park S.K., Yates J. R., 3rd (2010) Census for proteome quantification. Curr. Protoc. Bioinformatics Chapter 13, Unit 13 12 11–11 [DOI] [PubMed] [Google Scholar]
- 34. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 35. Chou M.F., Schwartz D. (2011) Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics Chapter 13, Unit 13 15–24 [DOI] [PubMed] [Google Scholar]
- 36. Vlad F., Turk B. E., Peynot P., Leung J., Merlot S. (2008) A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55, 104–117 [DOI] [PubMed] [Google Scholar]
- 37. van Wijk K. J., Friso G., Walther D., Schulze W. X. (2014) Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26, 2367–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Danquah A., de, Zelicourt A., Colcombet J., Hirt H. (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52 [DOI] [PubMed] [Google Scholar]
- 39. Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949 [DOI] [PubMed] [Google Scholar]
- 40. Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vitart V., Baxter I., Doerner P., Harper J. F. (2001) Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J. 27, 191–201 [DOI] [PubMed] [Google Scholar]
- 42. Champion A., Picaud A., Henry Y. (2004) Reassessing the MAP3K and MAP4K relationships. Trends Plant Sci. 9, 123–129 [DOI] [PubMed] [Google Scholar]
- 43. Bahn Y.S. (2008) Master and commander in fungal pathogens: The two-component system and the HOG signaling pathway. Eukaryot. Cell 7, 2017–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikeda M., Mitsuda N., Ohme-Takagi M. (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar M., Busch W., Birke H., Kemmerling B., Nürnberger T., Schöffl F. (2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol. Plant 2, 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larkindale J., Knight M.R. (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128, 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.