Abstract
Reactive oxygen species such as hydrogen peroxide can modify proteins via direct oxidation of their sulfur-containing amino acids, cysteine and methionine. Methionine oxidation, studied here, is a reversible posttranslational modification that is emerging as a mechanism by which proteins perceive oxidative stress and function in redox signaling. Identification of proteins with oxidized methionines is the first prerequisite toward understanding the functional effect of methionine oxidation on proteins and the biological processes in which they are involved. Here, we describe a proteome-wide study of in vivo protein-bound methionine oxidation in plants upon oxidative stress using Arabidopsis thaliana catalase 2 knock-out plants as a model system. We identified over 500 sites of oxidation in about 400 proteins and quantified the differences in oxidation between wild-type and catalase 2 knock-out plants. We show that the activity of two plant-specific glutathione S-transferases, GSTF9 and GSTT23, is significantly reduced upon oxidation. And, by sampling over time, we mapped the dynamics of methionine oxidation and gained new insights into this complex and dynamic landscape of a part of the plant proteome that is sculpted by oxidative stress.
Cellular oxidative stress is the result of an imbalance between oxidizing and reducing cellular agents and enzymes and is considered to be a major damaging factor (1). The identification of those proteins, together with their redox modifications, which are involved in responses to oxidative stress is needed for addressing key questions on understanding how plants perceive and adjust to environmental stress factors. Such adverse environmental conditions indeed influence plant growth and quality; hence, knowing the key protein players might lead to new opportunities to weapon food and feed crops against forecasted climate changes. The latter are expected to trigger sustained exposure of plants to abiotic stress, rendering them vulnerable, thus urging the necessity for sustainable solutions to improve crops. Plant proteomics provides an excellent way to deliver the scientific knowledge that is translatable into useful applications for crop improvement through identification of target proteins affected by the oxidative burden (2 –4).
Importantly, the accumulation of reactive oxygen or nitrogen species upon oxidative stress not only leads to damage of proteins, oligonucleotides, sugars, and lipids but also evokes cellular signaling events (5, 6). Here, hydrogen peroxide is particularly interesting as an oxidative stress signaling molecule (7) because of its relatively long half-life (1 ms) and its size, which enable it to oxidize biomolecules in compartments other than the site of hydrogen peroxide generation and even in neighboring cells (8). However, in plant cells, little is known on how exactly changes in hydrogen peroxide levels can lead to a final oxidative stress response. Perception of and oxidation by hydrogen peroxide (or derived species) have revealed some redox-regulating proteins (9), but for the majority of proteins, it remains unclear whether oxidation leads to signaling events or to damage. To study the in vivo effects of increased hydrogen peroxide levels, Arabidopsis catalase 2 knock-out plants serve as a good model system because these plants have increased intracellular oxidation caused by increased hydrogen peroxide availability, making them a physiologically more relevant model system (10) as compared with the exogenous addition of hydrogen peroxide (11). The catalase 2 enzyme is the major leaf catalase isoform of three encoded in the Arabidopsis genome and as such the leading scavenger of hydrogen peroxide, which is produced by glycolate oxidase in the peroxisomes upon photorespiration (12). Scavenging of hydrogen peroxide is indeed severely reduced in Arabidopsis catalase 2 knock-out plants (cat2-2) as these plants show a loss in catalase enzyme activity to only 10% of that in wild-type plants, illustrating the important contribution of CAT21 to the total leaf catalase activity (13). Furthermore, it became clear that cat2-2 plants are photorespiratory mutants showing a decreased growth relative to wild-type plants in ambient air. This dwarf phenotype becomes already apparent at irradiances of 50–100 μmol m−2 s−1 and is more severe upon increased irradiance (10, 12). The dwarf phenotype is in accordance with an induction of hydrogen peroxide-responsive transcripts (13). However, upon growth under high levels of CO2 or at low light intensities, no phenotypic differences were apparent, and no induction of hydrogen peroxide-responsive transcripts could be detected (13).
Hydrogen peroxide is capable of directly oxidizing the two sulfur-containing amino acids, methionine and cysteine. Oxidation of cysteine in particular has been intensively studied, and it is known that its different oxidation states may impact diverse proteins (14, 15). Methionine oxidation, in contrast, has received less attention, and in plants, only a few examples of functional consequences of this modification have been described (16 –19). Hydrogen peroxide can readily oxidize methionine to methionine sulfoxide (MetO), which can then be further oxidized to methionine sulfone although to a much lesser extent (20, 21). Methionine sulfoxidation can be countered by methionine sulfoxide reductases (MSRs) (22). Upon methionine oxidation, two stereoisomers may form (R-MetO and S-MetO), and accordingly, two families of stereospecific MSRs evolved: MSRA isoforms reduce S-MetO, whereas MSRB isoforms reduce R-MetO (23). Arabidopsis thaliana contains five MSRA genes encoding three cytosolic (MSRA1, MSRA2, and MSRA3), one chloroplastic (MSRA4), and one isoform targeted to the secretory pathway (MSRA5) (24). The nine MSRB proteins present in Arabidopsis are also targeted to different organelles: MSRB1 and -B2 are localized in the chloroplast, B3 is designated to the secretory pathway, whereas the other six MSRB proteins are cytosolic (25, 26). Also other plants such as poplar with five MSRAs and four MSRBs and rice with four MSRAs and three MSRBs (27) express a considerably larger number of MSR enzymes as compared with mammals, which only contain four MSR genes in total (28). In plants, most of the methionine oxidation studies thus far have focused on the different functions of MSRs rather than on the impact and extent of oxidation of protein-bound methionines (29). Methionine oxidation causes conversion of the hydrophobic methionine into the more hydrophilic methionine sulfoxide. This change in physicochemical properties of methionine helps to explain an increase of protein surface hydrophobicity upon methionine oxidation that triggers the exposure of otherwise inaccessible buried regions (30). In plants, a loss of hydrophobic recognition sites upon methionine oxidation was demonstrated for serine phosphorylation of nitrate reductase (31), and a loss of chaperone-like activity upon oxidation of methionine was demonstrated for the small heat shock protein Hsp21 (32).
The lack of antibodies that specifically target oxidized methionine residues has greatly impaired proteome-wide studies of methionine oxidation events in plants (33). Here, we implemented the recently developed COFRADIC proteomics technology that relies on diagonal peptide chromatography and the specificity of MSRs to isolate peptides carrying in vivo oxidized methionines, which are subsequently analyzed by tandem mass spectrometry (34, 35), allowing the identification of the exact sites of methionine oxidation. This enabled us to chart in vivo protein-bound methionine oxidation in A. thaliana following oxidative stress. This pioneering map of methionine oxidation events suggests that under these conditions methionine oxidation is not random but might steer molecular signaling events. Furthermore, we compared differences in methionine oxidation between wild type and catalase 2 mutants and captured the dynamics of oxidation by time-resolved proteome analysis. In addition, a validation study was performed on two members of the plant-specific Phi and Tau glutathione S-transferase (GST) classes, GSTF9 and GSTT23, both identified as targets of in vivo methionine oxidation. Not only do we provide kinetic parameters for both enzymes, but we also show that oxidation negatively affects their activity.
EXPERIMENTAL PROCEDURES
Plant Material, Growth Conditions, and Stress Treatments
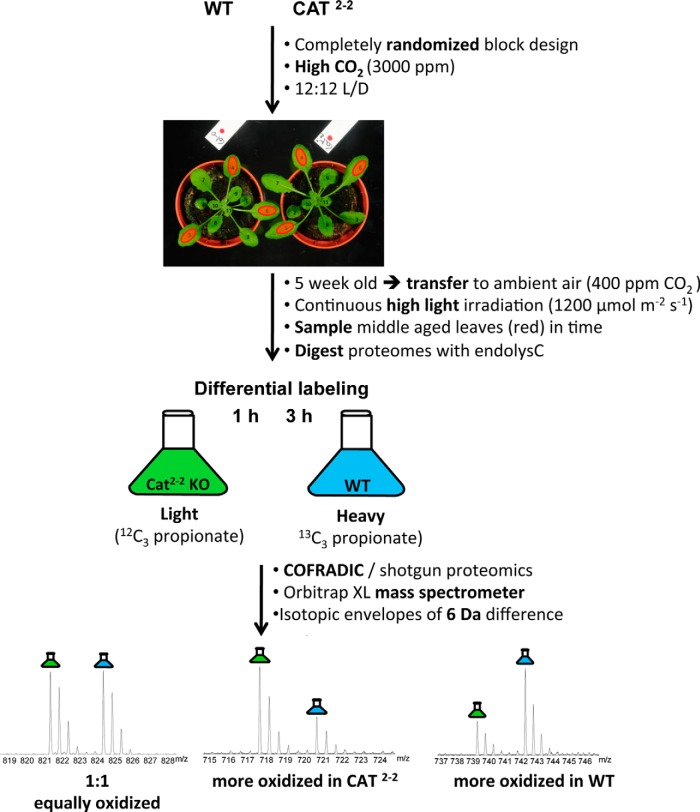
Catalase knock-out (cat2-2) (13) and wild-type (WT) A. thaliana plants, ecotype Columbia-0, were grown under elevated CO2 concentrations (3000 ppm) to impair photorespiration. Individual plants were organized according to a completely randomized block design in a controlled climate chamber (Vötsch Industrietechnik) in a 12:12-h day/night regime (relative humidity of 50%, temperature of 21 °C, and irradiance of 100 μmol m−2 s−1. For high light treatments, 5-week-old plants were transferred to a Sanyo Fitotron plant growth chamber in ambient air conditions (400 ppm CO2, 55% relative humidity, 21 °C) and continuous high light irradiation of 1200 μmol m−2 s−1 for 1 and 3 h. Middle aged leaves (as depicted in Fig. 1) of 15 individual plants per line were sampled, pooled, and frozen in liquid nitrogen.
Fig. 1.
Schematic overview of the experimental workflow. Catalase knock-out (CAT2-2) and WT plants were grown under high CO2 concentrations (3000 ppm) and organized according to a completely randomized block design in a 12:12-h light/dark (L/D) regime. Five-week-old plants were exposed to continuous high light irradiation (1200 μmol m−2 s−1) under ambient air (400 ppm). Middle aged leaves (indicated with red dots) of 15 plants per line were sampled 1 and 3 h after the onset of the high light stress and pooled for further analysis. After digestion with endoproteinase Lys-C and postmetabolic peptide labeling, equal amounts of samples were mixed and applied to COFRADIC-based isolation of MetO peptides. Parallel shotgun proteomics of the same samples allowed correcting for possible differences in overall protein abundance.
Methionine Sulfoxide COFRADIC
Proteins were extracted from 0.5 g of ground tissue by adding 0.5 ml of 250 mm Tris-HCl buffer (pH 8.7) containing 150 mm NaCl, 2 mm EDTA, 1 m guanidinium hydrochloride, 1% CHAPS (Sigma-Aldrich), and the appropriate amount of Complete protease inhibitor mixture (Roche Applied Science). After centrifugation at 16,100 × g for 10 min (twice), protein concentrations were measured using the Bradford assay. By adding the appropriate volumes of extraction buffer, the protein concentration of all samples was adjusted to 1.2 mg/ml using extraction buffer. Then cysteines were reduced and alkylated in 10 mm triscarboxyethylphosphine (Pierce) and 20 mm iodoacetamide (Sigma-Aldrich) upon incubation in the dark for 30 min at 30 °C. Samples were subsequently desalted on a NAPTM-5 column (Amersham Biosciences) in 1 ml of 50 mm triethylammonium bicarbonate buffer (pH 8), heated for 10 min at 95 °C, and cooled on ice before adding sequencing grade endoproteinase Lys-C (Promega) in a ratio of 1:200 (enzyme:substrate, w/w) to digest the samples overnight at 37 °C.
Next, primary α- and ε-amines were labeled using N-hydroxysuccinimide esters of stable isotopic variants of propionate. Peptides originating from cat2-2 proteomes were [12C3]propionated, whereas those from Columbia-0 proteomes were [13C3]propionated by incubation for 1 h at 30 °C to a final concentration of 10 mm N-hydroxysuccinimide ester. After quenching remaining N-hydroxysuccinimide esters in 50 mm glycine for 15 min at 25 °C, partial O-propionylation of serine, threonine, and tyrosine was removed by adding hydroxylamine to a final concentration of 50 mm and incubating for 15 min at 37 °C. The samples were acidified to a pH of 3 with acetic acid, and equal amounts of peptides were mixed. The COFRADIC technology relying on diagonal peptide chromatography and the specificity of MSRA and MSRB3 (Jena Bioscience) to specifically isolate peptides carrying in vivo oxidized methionines was applied to the equivalent of 500 μg of protein material as described (34) after which peptides were analyzed by LC-MS/MS.
Shotgun Proteome Analysis
In parallel, shotgun proteomics was applied on peptides fractionated during the first HPLC runs used for the COFRADIC setup. These fractions were pooled (four fractions per pool), and of each fraction, 10% was kept for LC-MS/MS analysis. These peptide mixtures were vacuum-dried and resolved in 20 μl of 2% acetonitrile of which 2.5 μl was injected for LC-MS/MS analysis as described below.
LC-MS/MS Analysis
The obtained peptide mixtures were introduced into an LC-MS/MS system, the Ultimate 3000 RSLC nano (Dionex, Amsterdam, The Netherlands), in line-connected to an to an LTQ Orbitrap XL (Thermo Fisher Scientific, Bremen, Germany) for analysis. The peptide mixture was first loaded on a trapping column at a flow rate of 10 μl/min (made in house; 100-μm internal diameter × 20 mm, 5-μm beads, C18 Reprosil-HD, Dr. Maisch HPLC GmbH). After flushing from the trapping column, the sample was loaded on a reverse-phase column (made in house; 75-μm internal diameter × 150 mm, 5-μm beads, C18 Reprosil-HD). Peptides were loaded with solvent A (0.1% trifluoroacetic acid and 2% acetonitrile) and separated with a linear gradient applied over 30 min from 2% solvent A′ (0.05% formic acid) to 55% solvent B′ (0.05% formic acid and 80% acetonitrile) at a flow rate of 300 nl/min followed by a wash using solvent B′.
The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the six most abundant peaks in a given MS spectrum. In the LTQ Orbitrap XL, full-scan MS spectra were acquired in the Orbitrap at a target value of 1E6 with a resolution of 60,000. The six most intense ions were then isolated for fragmentation in the linear ion trap with a dynamic exclusion of 60 s. Peptides were fragmented after filling the ion trap at a target value of 1E4 ion counts. From the MS/MS data in each LC run, Mascot generic files were created using the Mascot Distiller software (version 2.3.2.0, Matrix Science). While generating these peak lists, grouping of spectra was allowed with a maximum intermediate retention time of 30 s, and a maximum intermediate scan count of 5 was used where possible. Grouping was done with 0.005-Da precursor tolerance. A peak list was only generated when the MS/MS spectrum contained more than 10 peaks. There was no deisotoping, and the relative signal to noise limit was set at 2. These peak lists were then searched with the Mascot search engine (Matrix Science) using the Mascot Daemon interface (version 2.3, Matrix Science). Spectra were searched against the TAIR database (version 10 containing 27,416 protein-coding genes). Two searches were performed as described (36). In the first search, the quantitation configuration in Mascot was used in which propionate labels were set exclusively to lysines and peptide N termini. The second search left out the propionate label on peptide N termini but considered protein N-terminal acetylation and pyroglutamate formation of glutamine as variable modifications. In both searches, methionine oxidation (to the sulfoxide form) was set as a fixed modification. If an MS/MS spectrum was matched to two different peptides following both searches, the match with the highest score was retained. Mass tolerance on precursor ions was set to ±10 ppm (with Mascot's C13 option set to 1) and on fragment ions to ±0.5 Da. The peptide charge was set to 1+, 2+, 3+, and the instrument setting was on ESI-TRAP. The enzyme was set to endoproteinase Lys-C, allowing for one missed cleavage, and cleavage was also allowed when lysine was followed by proline. Only peptides that were ranked 1 and scored above the threshold score, set at 99% confidence, were withheld. Note that all MS/MS spectra have been uploaded into the PRIDE database (project accession number PXD001286). We calculated a false discovery rate of 2% by using a reverse database of the A. thaliana TAIR database. Searches were performed both against the forward and reverse database, and the false discovery rate was calculated based on the method of Käll et al. (37) using the following formula: false discovery rate = (number of false positives)/(number of true identifications) × 100. Identified peptides were quantified using the Mascot Distiller Toolbox (Matrix Science) in the precursor mode. This software tries to fit an ideal isotopic distribution on the experimental data based on the peptide average amino acid composition. This is followed by extraction of the XIC signal of both peptide components (light and heavy) from the raw data. Ratios are calculated from the area below the light and heavy isotopic envelope of the corresponding peptide (integration method, ‘trapezium”; integration source, “survey”). To calculate this ratio value, a least square fit to the component intensities from the different scans in the XIC peak was created. MS scans used for this ratio calculation are situated in the elution peak of the precursor determined by the Distiller software (XIC threshold, 0.3; XIC smooth, 1; maximum XIC width, 250). To validate the calculated ratio, the standard error on the least square fit has to be below 0.16, and the correlation coefficient of the isotopic envelope should be above 0.97.
Quantitative Data Analysis
Using Huber robust statistics, the median, standard deviation, and 95% confidence intervals were calculated in R for both the methionine sulfoxide data sets as well as for the shotgun data sets. In each setup, the median ratio value was subtracted from the calculated log2 ratio values to rescale the distribution of ratio values around μ = 0. All significant spectra (95% confidence interval, p ≤ 0.05) as well as the peptides denominated “FALSE” by Mascot were manually validated using XCalibur (Thermo Scientific). After recalculation of the Huber limits (95% confidence interval settings) and rescaling, the ratio values of methionine-containing peptides were corrected for protein abundance differences (when available) by subtracting the rescaled log2 values, thus correcting for possible altered protein synthesis/degradation. A final calculation was then done of the 95% confidence interval, which enables identification of the significantly different oxidized proteins after 1 and 3 h of exposure to high light stress.
Recombinant GST Protein Expression and Purification
GatewayTM custom pENTR223 vectors containing coding sequences of full-length GSTF9 (AT2G30860) and GSTT23 (AT1G78320) proteins were recombined into pDEST14 Gateway destination vectors. BL21 StarTM One Shot chemically competent Escherichia coli cells (Invitrogen) were transformed, and GSTF9 and GSTT23 protein production was induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside in growing cultures of successfully transformed E. coli cells reaching an A600 of 0.8. After incubation overnight at 16 °C, cells were centrifuged for 10 min at 2700 × g and resuspended in 20 ml of cold PBS with 1 mm DTT, 1% Triton X-100, and the appropriate amount of Complete protease inhibitor mixture. Sonication for 5 min with 10-s pulses was followed by centrifugation for 10 min at 10,000 × g to remove cellular debris after which the recombinant GST proteins were purified from the supernatant by affinity chromatography using immobilized glutathione columns (GSTrap HP, GE Healthcare). Elution of the affinity-trapped GST proteins was done using a 50 mm Tris-HCl (pH 8) buffer solution containing 10 mm reduced glutathione.
Oxidation of Recombinant GSTF9 and MS Analysis
The reduced glutathione present in the elution buffer was removed by desalting over a 0.7-ml polyacrylamide spin desalting column (Thermo Scientific Pierce) following the manufacturer's instructions. During this step, the GSTF9 protein buffer was exchanged with 10 mm ammonium acetate, pH 7. Then, multiple conditions of hydrogen peroxide-induced protein oxidation were tested (varying concentration of hydrogen peroxide, incubation time, and temperature) to obtain the highest amount of a single oxidized form of GSTF9.
The oxidized GSTF9 protein was first analyzed by LC-MS on a nanoACQUITY UPLC in line-connected to a Q-TOF Premier (Waters). The samples (6 μl) were first loaded on a trapping column (made in house; 100-μm internal diameter × 20 mm, 5-μm beads, Reprosil-pure 300 C4, Dr. Maisch HPLC GmbH) at a flow rate of 10 μl/min in 0.1% TFA and 2% acetonitrile. After 4 min, the trapping column was placed in line with the analytical reverse-phase column (made in-house; 75-mm internal diameter × 150 mm, 3-μm beads, Reprosil-pure 300 C4). The proteins were eluted using a standard linear water-acetonitrile gradient with 0.1% formic acid as ion pairing agent. Optimal conditions of selective oxidation of the GSTF9 proteins were found when 0.03% hydrogen peroxide was used for 135 min at 10 °C, and similar conditions were then used to oxidize GSTT23. To collect oxidized GST9 for further analysis, the Q-TOF Premier mass spectrometer was uncoupled and replaced by a UV detector (214 nm) to trace the single oxidized GSTF9 proteins. This protein fraction was then digested overnight at 37 °C by sequencing grade endoproteinase Lys-C (Wako; 1:50, enzyme:substrate, w/w) and analyzed by LC-MS/MS (Orbitrap Velos, Thermo Fisher Scientific).
GST Enzyme Activity Assays
Reduced glutathione was removed from the recombinant GSTF9 and GSTT23 proteins by dialysis in 100 mm potassium phosphate buffer (pH 6.5) with 0.1% Triton X-100. GST activity was measured in a Spectramax340PC (Molecular Devices) plate reader by following the increase in absorbance at 340 nm due to the conjugation of reduced glutathione with 1-chloro-2,4-dinitrobenzene (CDNB) (38). The reaction took place in a 96-well plate (PolySorb, Nunc, Sanbio) in a total volume of 200 μl containing 2 mm reduced glutathione and 1 mm CDNB as final concentrations. Every 20 s for 8 min the absorbance was measured. From the obtained progress curves, the initial velocities (Vi) in milli-absorbance units/min were calculated. The concentration of CDNB was varied and plotted against the initial velocities divided by the enzyme concentrations according to the formula: Vi/E0 = Vi/(εCDNB·path length per well·E0) = kcat·([S]/(Km + [S])) where E0 is the initial molar GST concentration, ε is the molar extinction coefficient of CDNB, and Vi is the initial velocity. The kinetic parameters (Km and kcat) were calculated after fitting with the Michaelis-Menten equation. The activity of the oxidized GST proteins was determined as described above.
RESULTS
Proteome Analysis of in Vivo Protein-bound Methionine Oxidation
Mapping of oxidation-sensitive methionines in A. thaliana proteins was done by applying the COFRADIC technology (34), which basically consists of three consecutive steps: 1) peptide fractionation based on hydrophobicity (reverse-phase HPLC), 2) treatment of fractionated peptides with recombinant methionine sulfoxide reductases A and B3, and 3) refractionation of the treated fractions by reverse-phase HPLC. The Arabidopsis catalase 2 knock-out line cat2-2 was used as a model system to study the effects on the proteome upon oxidative stress. Because photorespiration is a primary source of hydrogen peroxide in photosynthetic tissue (39), the catalase knock-out plants as well as wild-type plants were grown under elevated CO2 levels (3000 ppm) to block oxygenation of ribulose 1,5-biphosphate and as such hinder photorespiration. By transferring plants after 5 weeks to a continuous high light irradiation regime under ambient air conditions (400 ppm CO2), photorespiratory-dependent generation of hydrogen peroxide was endogenously modulated. Indeed, as these growth conditions boost photorespiration, glycolate is produced that is subsequently converted in the peroxisomes to glyoxylate by glycolate oxidase concomitantly with the release of hydrogen peroxide (12). Thus, here, we assessed primarily the oxidation of protein-bound methionines caused by increased concentrations of hydrogen peroxide originating from the peroxisomes. Middle aged leaves of 5-week-old wild-type and catalase knock-out plants were harvested at 1 and 3 h following the onset of photorespiratory stress. Following protein extraction and proteolytic cleavage (endoproteinase Lys-C), postmetabolic labeling of peptides using either light (12C3) or heavy (13C3) propionate was performed to compare WT versus CAT2-2 proteomes. These “mass tags” on the identified MetO peptides will point to the exact origin of the peptides, whereas the ion signals themselves allow for a quantitative comparison between the genotypes (Fig. 1). To exclude that the observed differences in the degree of methionine oxidation were caused by differences in protein levels, we performed in parallel a quantitative proteomics screen.
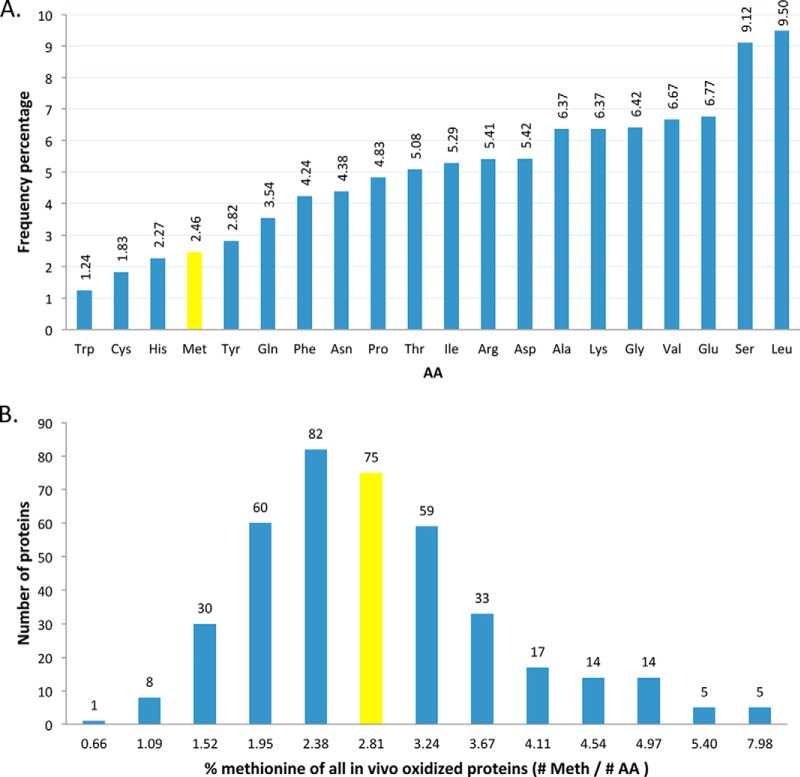
We identified 513 oxidation sites in 403 proteins. Following 1 h of high light exposure, 365 MetO peptides in 296 proteins were found, whereas after 3 h, 354 MetO peptides in 294 proteins were identified (supplemental Fig. 1 and Table 1). There appears to be an overlap of 186 proteins with MetO sites identified at both time points (Table I). Of note is that proteins belonging to the same family show a highly similar sequential context surrounding the MetO sites (supplemental Table 2). Of the 403 proteins carrying an oxidized methionine, 362 proteins were identified with a single MetO peptide, accounting for 90% of the identified proteins. Upon comparison with the overall methionine frequency of 2.46% (Fig. 2A) defined by plotting all annotated A. thaliana proteins (35,386 sequences in TAIR10), about half of the identified oxidized proteins have a lower than average occurrence of protein-bound methionine (Fig. 2B). Furthermore, this number grows when correcting for the fact that the initiator methionine is frequently removed in nascent polypeptides (40), thereby dropping the overall methionine content of Arabidopsis proteins further to 2.21%.
Table I. Overview of the oxidation events identified 1 h and 3 h after the onset of high light stress.
| Methionine oxidation in numbers | 1 h | 3 h | Total unique IDs |
|---|---|---|---|
| MetO peptides | 365 | 354 | 507 |
| Oxidized proteins | 296 | 294 | 403 |
| Proteins significantly more oxidized in CAT2–2 vs WT (≥ 2 fold*) | 24 (7*) | 29 (18*) | 51 |
| Proteins significantly more oxidized in WT vs CAT2–2 (≥ 2 fold*) | 40 (14*) | 18 (8*) | 57 |
| Protein overlap in time (with identical MetO site*) | 187 (175*) | ||
| MetO peptide overlap in time | 206 | ||
| Proteins significant more oxidized over time | 17 | ||
| Proteins significant less oxidized over time | 3 | ||
| Proteins with single oxidation event | 236 | 247 | 362 |
Fig. 2.
Methionine frequency plots across all A. thaliana proteins (A) and within oxidized proteins (B). A, amino acid abundance plot of all annotated A. thaliana proteins. Across all annotated TAIR10 proteins, methionine makes up 2.46% of all amino acids (AA) (yellow bar), and this drops to 2.21% when excluding the initiator methionine. B, methionine (Meth) frequency plot of the identified in vivo oxidized proteins. By plotting the percentage of methionines in all the identified oxidized proteins, a large majority of these proteins were found to contain less than the expected methionine content of 2.46%, the latter falling into the bin highlighted in yellow.
An overview of all MetO-containing proteins identified in our study is available upon request. This overview allows easy tracking of a protein of interest using a query interface, shows the identified peptide(s) mapped onto the amino acid sequence of the protein, and displays differences in methionine oxidation between catalase knock-out and wild-type plants.
Quantification of Methionine Oxidation between Genotypes
Postmetabolic peptide labeling enabled us to assess the influence of the plant genotypic background (cat2-2 versus wild type) on methionine oxidation. 51 proteins were significantly more oxidized in catalase knock-out plants when considering both time points (p ≤ 0.05; 24 proteins after 1 h and 29 proteins after 3 h of high light stress (Table I)). For most of these proteins (18 of 24 after 1 h and all after 3 h of high light stress) only one MetO peptide was identified, although multiple methionines, e.g. up to 69 for the NADH-dependent glutamate synthase 1 (AT5G53460), are present in their sequences (supplemental Table 1). Of note is one protein, At4G23670, that was identified at both time points and found to be oxidized on two sites, methionines 134 and 140. This protein of 151 amino acids is a member of the polyketide cyclase/dehydrase and lipid transport superfamily. It is involved in defense and stress responses as well as in cysteine biosynthesis (41 –43). Moreover, the levels of this protein dropped to about 65% in the catalase mutant compared with wild-type plants, which might hint to increased degradation of this protein upon oxidation. Several proteins were more oxidized in high light-stressed wild-type plants as compared with cat2-2 plants. For instance, upon 3 h of high light stress, all but one of these 18 proteins are oxidized on a single methionine, and all are equally or less abundant in wild-type plants, which could again suggest increased protein degradation caused by methionine oxidation. 14 and eight proteins with at least 2-fold higher oxidation in wild-type plants were identified after 1 and 3 h of high light stress, respectively. In catalase knock-out plants, oxidation events rise from seven to 18 proteins over time (Table I). This increase of more strongly oxidized proteins in the catalase knock-out plant might be explained by elevated hydrogen peroxide production over time, which cannot be eliminated in these plants and as such enhances the overall oxidative stress condition. In contrast, in wild-type plants, the vast majority of proteins with a 2-fold higher degree of oxidation are localized to plastids.
In conclusion, quantification of methionine oxidation revealed 55 MetO peptides in 51 proteins that are significantly more oxidized in the catalase knock-out. Noteworthy is that enhanced oxidation was not restricted to the catalase loss-of-function genotype but was also seen in wild-type plants.
Dynamics of Methionine Oxidation
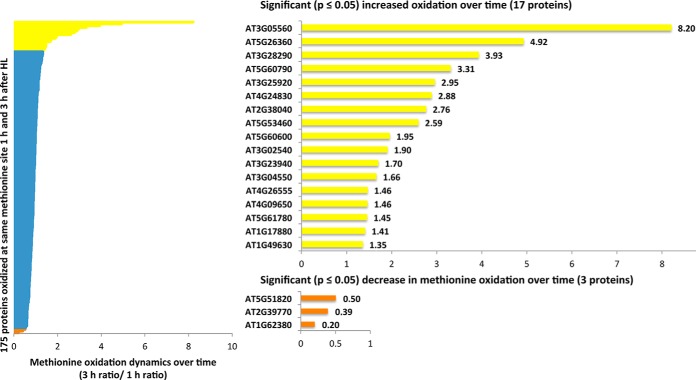
Applying time-resolved analysis by sampling at 1 and 3 h during oxidative stress allowed capturing the dynamics of protein-bound methionine oxidation. The overlap of 186 proteins identified at both time points served as a base for exploring this. More specifically, the dynamics of oxidation of 175 proteins on identical methionine sites could be compared (Fig. 3). 17 proteins displayed significantly higher oxidation (p ≤ 0.05) after 3 h compared with 1 h, whereas only three proteins showed a decrease in methionine oxidation over time. Remarkably, the three proteins whose oxidation is lowered over time show increased protein levels after 3 h of stress treatment. The proteins with a time-dependent increase in oxidation are metabolic enzymes, a chaperone, a transporter, a protease, and a transcription factor among others, and their increased oxidation can be explained by increased concentrations of hydrogen peroxide as the high light stress continues in time in combination with a possible decreased reduction of these MetO sites by methionine sulfoxide reductases.
Fig. 3.
The dynamics of methionine oxidation. The 186 proteins identified at both time points of high light (HL) stress share 175 MetO peptides. Comparing their signals shows the dynamics of methionine oxidation over time; 17 proteins (yellow bars) were significantly (p ≤ 0.05) more oxidized after 3 h of high light stress, whereas only three proteins (orange bars) showed a significant decrease in methionine oxidation.
To investigate a possible role for the primary structure in the oxidation sensitivity of methionines, an iceLogo sequence alignment (44) of neighboring amino acids was generated for the proteins identified at both time points. As a positive input, the 17 proteins with significantly increased oxidation over time at identical methionines were selected. The remaining 158 proteins with MetO peptides identified at both time points were selected as the reference set. Methionine was fixed at position 6 (P6) and the five surrounding amino acids both N- and C-terminal to this site were retrieved (P1–P11). MetO peptides with significantly higher oxidation levels (p ≤ 0.05) after 3 h compared with 1 h revealed a motif of hydrophobic amino acids such as valine, leucine, isoleucine, and methionine in proximity of the oxidation site (Fig. 4).
Fig. 4.
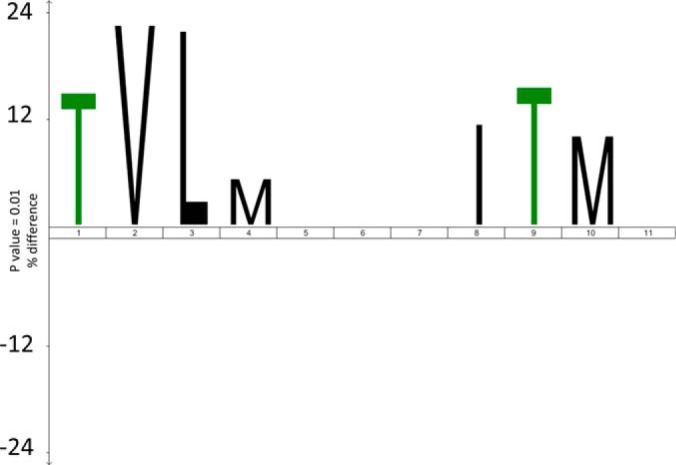
An iceLogo of the amino acid context of the oxidized methionine. The sequence context of proteins with significantly (p ≤ 0.05) increased oxidation over time (n = 17) reveals a preference (p ≤ 0.01) for hydrophobic amino acids surrounding the site of methionine oxidation. As a negative reference set, the remaining 158 proteins with identical MetO peptides identified in both time points were chosen. Methionine was fixed at P6 and the five surrounding amino acids; both N-terminal (P1–P5) and C-terminal amino acids (P7–P11) are shown.
Oxidation Reduces the Activity of Two Plant-specific Glutathione S-Transferases
The A. thaliana genome encodes for 55 GSTs. GSTs are known as stress-responsive proteins and accumulate in biotic and abiotic stress conditions (45, 46). Here, we identified three members of the plant-specific Phi and Tau GST classes as targets of methionine oxidation (GSTs Phi9 at Met-35, Tau21 at Met-48, and Tau23 at Met-47; supplemental Table 1) and decided to evaluate the impact of oxidation on the activity of GSTF9 and GSTT23.
We first determined whether the recombinant GST proteins were enzymatically active by measuring the conjugation of reduced glutathione to CDNB. The increase in absorbance at 340 nm was measured in function of time, and the initial velocities (Vi) were calculated. Recombinant GSTF9 and GSTT23 were found to be active, and at a fixed CDNB concentration of 1 mm, we observed a linear correlation between the Vi and GST in the concentration range of 0.2–4.2 μM and 0.1–2 μm for GSTF9 and GSTT23, respectively (supplemental Fig. 2a). Next, we determined the kinetic parameters of GSTF9 and GSTT23 using 0.4 μm enzyme. CDNB assays were performed with varying concentrations of CDNB, and the data were fitted with a Michaelis-Menten equation (supplemental Fig. 2b). We obtained a kcat/Km specificity constant of 341.32 and 216.45 m−1 s−1 for GSTF9 and GSTT23, respectively (Table II).
Table II. Overview of the kinetic parameters from GSTF9 and GSTT23. The kinetic parameters (Km, kcat and kcat/Km) for the substrate CDNB obtained from the Michaelis-Menten equation.
| kcatCDNB (min−1) | KmCDNB (mm) | kcat/Km (M−1 s-1) | |
|---|---|---|---|
| GSTF9 | 34.20 ± 3.60 | 1.67 ± 0.36 | 341.32 |
| GSTT23 | 23.26 ± 2.55 | 1.79 ± 0.37 | 216.45 |
To examine a possible influence of protein oxidation on the enzymatic activity, we first optimized conditions of in vitro hydrogen peroxide oxidation of GSTF9 to resemble as closely as possible methionine oxidation found in our in planta study. LC-MS measurements were performed on GSTF9 oxidized with varying concentrations of hydrogen peroxide (0.03–0.1%), different incubation times (10 min–10 h), and at different temperatures (10–22 °C). After 135 min of incubation at 10 °C with 0.03% H2O2, a single oxidized species of GSTF9 was most prominently present (supplemental Fig. 3). This oxidized GSTF9 was then enriched by reverse-phase HPLC and digested with endoproteinase Lys-C, and the resulting peptides were analyzed by LC-MS/MS. Oxidation was found on the peptide previously identified to contain a methionine sulfoxide, namely 24GVAFETIPVDLMK36, here again detected with a Met-35 sulfoxide. In fact, two forms of this peptide were found: one with and one without Met-35 oxidized, and both were identified by their MS/MS spectrum (supplemental Fig. 4). A closer look at these MS/MS spectra shows that the mass values of the detected y ions of the oxidized and non-oxidized peptides differ by 16 Da from the y2 ion onward, indicative for sulfoxide formation on Met-35 (supplemental Fig. 4). Taken together, these data show that the in vitro oxidized GSTF9 protein mimics the in vivo oxidized GSTF9 protein and can thus be used to determine possible effects of oxidation on enzyme activity. Both the GSTF9 and GSTT23 were oxidized under the optimized conditions for GSTF9, and their activity was measured in the CDNB assay (Fig. 5). The activity of the in vitro oxidized recombinant GSTF9 and GSTT23 enzymes was significantly (p ≤ 0.01) reduced upon oxidation, and this observation was even more pronounced for higher enzyme concentrations. We observe a loss of about 45% of the total GSTF9 enzyme activity and of about 38% of the activity of the GSTT23 enzyme, which is in agreement with the mixed population of oxidized and non-oxidized enzyme upon our mild conditions of oxidation (and the corresponding degree of oxidation; see supplemental Fig. 3) and thus hints to the fact that oxidation might completely block GST activity.
Fig. 5.
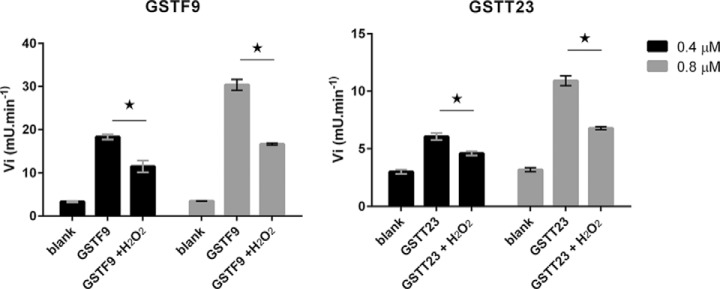
Methionine sulfoxide formation impairs GSTF9 and GSTT23 activities. Enzyme activity measurements in the CDNB assay show a significant (p ≤ 0.01; depicted as *) loss of enzymatic activity for both methionine-oxidized GSTs. Oxidized GSTF9 has a loss of up to ∼45% of the total enzyme activity, whereas the activity of oxidized GSTT23 drops by ∼38%. milli absorption units (mU), milliunits. The error bars represent standard deviation (S.D.).
DISCUSSION
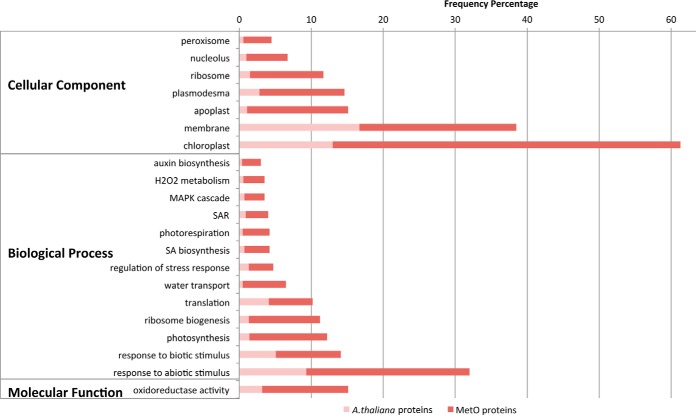
In this study, we identified 403 proteins prone to methionine oxidation under oxidative stress. A gene ontology (GO) enrichment analysis (p ≤ 0.01) performed on these proteins using the complete theoretical proteome of A. thaliana as background revealed some noteworthy trends. The analysis showed that peroxisomal and chloroplast proteins were significantly more abundant in our data set (Fig. 6). Because light irradiation is captured by chloroplasts, excess light could directly affect photosynthetic proteins present in chloroplasts, explaining the prevalence of such proteins when considering the GO terms linked to biological processes. Also, proteins involved in chloroplast relocation (47) were enriched, which might point to the fact that cells try to minimize the captured light by relocating chloroplasts. Higher levels of oxidative stress through the activation of the photorespiratory pathway are illustrated by the enhanced GO classes of proteins involved in photorespiration and hydrogen peroxide metabolism. Nucleolar and ribosomal proteins were also significantly enriched in our data set of oxidized proteins as demonstrated by the enrichment of proteins involved in ribosome biogenesis and translation. Furthermore, proteins known to be localized to plasmodesma and apoplasts were significantly enriched as well as proteins involved in water transport. This suggests two modes of hydrogen peroxide channeling: plasmodesma provide a gate to neighboring cells, whereas hydrogen peroxide could also pass into the apoplast, until now known as a free passage for water and solutes, next to aquaporins, which also transport hydrogen peroxide across membranes (48). Oxidized membrane proteins also appeared to be an enriched GO class. Other significantly enriched GO terms coupled to biological processes include “response to stress” and more specifically response to reactive oxygen species and in particular hydrogen peroxide as well as response to light stimulus. In addition, proteins involved in salt and osmotic stress as well as proteins from the innate immune response and proteins responding to temperature, bacteria, or inorganic substances (chemicals, acids, and metals) were found to be enriched. All together, the enriched GO classes of responses toward abiotic and biotic stimuli point to a general stress response cluster upon oxidation. Noteworthy is that not only executors but also regulators of the cellular response to stress are enriched among the oxidized proteins. Additionally, links to hormone signaling appear as salicylic acid biosynthesis and systemic acquired resistance mediated by salicylic acid signaling are found to be enriched as well as proteins involved in auxin biosynthesis. Proteins linked to the GO term “oxidoreductase activity” include thioredoxin, two MSRs (MSRA2/A3), and the mitochondrial LPD1 and -2 proteins that are part of the glycine decarboxylase complex that releases CO2 during photorespiration. Finally, proteins involved in signal transduction by phosphorylation, more specifically members involved in mitogen-activated protein kinase signaling, were also enriched.
Fig. 6.
Gene ontology enrichment analysis of oxidized proteins. The 403 proteins identified to contain sites of methionine oxidation upon high light stress were submitted to a GO enrichment analysis using AmiGO. The complete proteome of A. thaliana was chosen as a reference set. Significantly enriched GO categories (p < 0.01) discussed in the main text are listed, and the enriched frequencies of methionine-oxidized proteins (dark red) are shown. The frequency of all A. thaliana proteins in these enriched categories is also shown (light red) as part of the total enriched MetO protein frequency bars. SA, salicylic acid; SAR, systemic acquired resistance.
Our compendium of A. thaliana proteins of which methionines get oxidized upon oxidative stress (available upon request) constitutes currently the largest data set of its kind in plants and forms the base for future studies on the functional impact of methionine oxidation. Below we discuss some of these proteins that are particularly interesting given previous findings reported in the literature.
One such protein is the small zinc finger protein methylene blue-sensitive 1 (AT3G02790) that acts as a mediator of singlet oxygen responses in the unicellular green alga Chlamydomonas reinhardtii (49). Upon exposure to high light, this protein translocates to distinct granules in the cytosol, but the mechanism remains unclear. However, in view of the example of the recently reported hypochlorite-responsive transcription factor in E. coli that is activated by methionine oxidation upon oxidative stress (50), oxidation of methionine 62 in methylene blue-sensitive 1 might well be a plausible mechanism to induce this positional shift. Of further note is the nearly 4 times higher degree of oxidation of this protein in light-stressed catalase 2 knock-out plants.
An interesting class of proteins that was oxidized both after 1 and 3 h of high light treatment includes three members of the rotamase family: cyclophilin 38 (AT3G01480), CYP3 (AT2G16600), and CYP4 (AT3G62030). CYP4 was identified to interact with 12-oxo-phytodienoic acid and links 12-oxo-phytodienoic acid signaling to amino acid biosynthesis and cellular redox homeostasis in response to stress (51). Upon interaction, the formation of a complex with O-acetylserine (thiol) lyase B and cysteine synthase is triggered, leading to an increase of thiol metabolites and the buildup of the cellular reduction potential (52). Interestingly, all members of this complex were identified to be oxidized upon high light stress; thus possibly methionine oxidation might regulate a cluster of interacting proteins via a redox modification. We also found that several photorespiratory enzymes were methionine-oxidized, which could be a direct way of sensing elevated hydrogen peroxide levels, in turn directly regulating the pathway involved in methionine oxidation.
So far, studies related to methionine oxidation in plants have focused on the different functions of MSRs. Tarrago et al. (53) explored the possibilities of fishing for MSRB1 substrates via affinity chromatography and identified 24 possible candidate interactors. Of note is that 23 of these were also identified in our data set among which are catalases 2 and 3, confirming oxidation of methionine and demonstrating affinity chromatography as a valuable strategy to look for specific MSR substrates. Recently, glutathione S-transferases F2 and F3 were identified as substrates for MSRB7 where reduction of methionine sulfoxides confers tolerance of A. thaliana to oxidative stress (19). We identified three members of the GST family, GSTF9, -T21, and -T23, of which the Tau family members (GSTT21 and -T23) carried identical sites of methionine oxidation at both time points. Here, we showed that oxidation of GSTF9 and GSTT23 significantly impairs their catalytic activity, and we thus extended the list of GST proteins of which the activity is negatively influenced by methionine oxidation.
To assess the potential specificity of methionine oxidation linked to molecular signaling, different parameters were analyzed such as methionine frequency, the correlation of protein abundance, and the number of methionines with the oxidation degree. Tarrago et al. (53) postulated a positive correlation of methionine oxidation with protein abundance and methionine content as one of the A. thaliana methionine sulfoxide reductases, MSRB1, was found to preferentially interact with proteins with high methionine content. However, we have shown that about half of the oxidized proteins identified in our setup have a lower than average methionine frequency (Fig. 2), meaning that the presence of more methionines in a protein is not a prerequisite for its oxidation. Furthermore, when spectral counts from the shotgun analysis are taken into account as a proxy for protein abundance, an average of 4.8 and 4.9 spectra were linked to proteins identified in the 1- and 3-h high light-stressed samples, respectively. A comparison of these average numbers with the spectral counts of proteins with significantly higher oxidation levels in cat2-2 plants showed that at both time points 76% of these proteins had spectral counts below this average abundance. Thus high protein levels also appear not be prerequisites for protein oxidation. As such, our data argue against random methionine oxidation as an alternative reactive oxygen species scavenging mechanism, leading to indiscriminate protein damage (18).
Further indications for a possible specific regulatory role of methionine oxidation events besides the here illustrated negative effect of methionine oxidation on GST function appeared when comparing the protein sequence surrounding the MetO sites identified in oxidized proteins belonging to the same family (supplemental Table 2). One such example is the rubredoxin protein family (AT3G15640 and AT1G80230), which consists of three proteins with cytochrome c activity and in which the sequence patch surrounding the oxidized methionine is conserved among 43 homologous proteins across the plant kingdom (19 species) (supplemental Fig. 5). The third Arabidopsis gene (AT1G52710) lost this patch and was not picked up in our analysis. Interestingly, both oxidized rubredoxin-like proteins were found to be 4 times more oxidized in the catalase mutant compared with the wild type already 1 h after high light irradiance.
The dynamics of protein-bound methionine oxidation could be captured via considering the overlap of proteins identified at both time points. 17 of the 175 oxidized proteins with identical methionine sites at both time points showed a significantly higher oxidation (p ≤ 0.05) after 3 h. Several of these proteins have previously been linked to stress response such as the RNA-binding protein Tudor-SN (AT5G61780), which stabilizes stress-responsive mRNA molecules encoding for secreted proteins and proved essential for salt stress resistance (54). The basic transcription factor 3 (AT1G17880) homolog in higher plants impairs tolerance of wheat against drought and freezing stresses when silenced and was also found to be differentially regulated by diverse abiotic stresses and hormone treatments (55). Also, the photorespiratory enzyme NADH-dependent glutamate synthase 1 was identified to be more oxidized after 3 h, which might be a consequence of the proximity to the site of hydrogen peroxide production.
The amino acid sequence surroundings of these MetO sites whose oxidation increased over time revealed the oxidized methionine to be embedded in a hydrophobic pocket (Fig. 4). This enriched motif can be explained by either an increased susceptibility of oxidation due to the hydrophobic environment or by decreased reduction by MSRs due to the less preferred substrate recognition of these MetO sites or impaired accessibility. Conversely, proteins of which the levels of methionine oxidation significantly drop over time (three in this case) might have been repaired by the methionine sulfoxide reductases, which are numerously present in plant cells (29). Concerning the latter, a closer look at the expression profiles of the different Arabidopsis MSRs shows a clear up-regulation of MSRB7 and -B9 under elevated reactive oxygen species levels (56, 57). Furthermore, both MSRs cluster together upon transcriptional response toward various perturbations (supplemental Fig. 6A). Also, these two MSR B genes are only weakly expressed in different anatomical parts as well as during stages of development (supplemental Fig. 6, B and C). The vast majority of MSRs present in Arabidopsis together with a sequence preference surrounding the oxidation site thus hint toward specificity of either oxidation or enzymatic reduction. However, which of these two is actually responsible remains to be explored.
Finally, it should be noted that enhanced oxidation was not only detected in cat2-2 plants but also in wild-type plants. This might be the consequence of further oxidation of methionine sulfoxide to sulfone in the cat2-2 plants due to the enhanced hydrogen peroxide production, which we cannot capture with our COFRADIC methodology because the MSR enzymes specifically reduce methionine sulfoxides.
In summary, we have provided the largest data set of methionine oxidation in plants during oxidative stress treatment in which we compare differences in oxidation between Arabidopsis wild-type and catalase 2 knock-out plants, capture the dynamics of oxidation by time-resolved proteome analysis, and pinpoint the exact sites of oxidation. Furthermore, the functional impact of methionine oxidation was proven for two members of the plant-specific GST family.
Supplementary Material
Footnotes
Author contributions: S.J., J.M., F.V., and K.G. designed research; S.J., H.D., K.W., and P.W. performed research; S.J., B.G., P.D., J.M., F.V., and K.G. analyzed data; S.J., B.G., J.M., F.V., and K.G. wrote the paper.
 This article contains supplemental Figs. 1–6 and Tables 1 and 2.
This article contains supplemental Figs. 1–6 and Tables 1 and 2.
1 The abbreviations used are:
- CAT
- catalase
- MetO
- methionine sulfoxide
- MSR
- methionine sulfoxide reductase
- COFRADIC
- combined fractional diagonal chromatography
- WT
- wild-type
- TAIR
- The Arabidopsis Information Resource
- XIC
- extracted ion chromatogram
- CDNB
- 1-chloro-2,4-dinitrobenzene
- GO
- gene ontology.
REFERENCES
- 1. Van Breusegem F., Bailey-Serres J., Mittler R. (2008) Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol. 147, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ischiropoulos H. (2003) Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 305, 776–783 [DOI] [PubMed] [Google Scholar]
- 3. Souza J. M., Daikhin E., Yudkoff M., Raman C. S., Ischiropoulos H. (1999) Factors determining the selectivity of protein tyrosine nitration. Arch. Biochem. Biophys. 371, 169–178 [DOI] [PubMed] [Google Scholar]
- 4. Fries D. M., Paxinou E., Themistocleous M., Swanberg E., Griendling K. K., Salvemini D., Slot J. W., Heijnen H. F., Hazen S. L., Ischiropoulos H. (2003) Expression of inducible nitric-oxide synthase and intracellular protein tyrosine nitration in vascular smooth muscle cells: role of reactive oxygen species. J. Biol. Chem. 278, 22901–22907 [DOI] [PubMed] [Google Scholar]
- 5. Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci. 16, 300–309 [DOI] [PubMed] [Google Scholar]
- 6. Choudhury S., Panda P., Sahoo L., Panda S. K. (2013) Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 8, e23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neill S., Desikan R., Hancock J. (2002) Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5, 388–395 [DOI] [PubMed] [Google Scholar]
- 8. Bienert G. P., Schjoerring J. K., Jahn T. P. (2006) Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 1758, 994–1003 [DOI] [PubMed] [Google Scholar]
- 9. Balsera M., Uberegui E., Schürmann P., Buchanan B. B. (2014) Evolutionary development of redox regulation in chloroplasts. Antioxid. Redox Signal. 21, 1327–1355 [DOI] [PubMed] [Google Scholar]
- 10. Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H. K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58 [DOI] [PubMed] [Google Scholar]
- 11. Claeys H., Van Landeghem S., Dubois M., Maleux K., Inzé D. (2014) What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol. 165, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220 [DOI] [PubMed] [Google Scholar]
- 13. Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657 [DOI] [PubMed] [Google Scholar]
- 14. Couturier J., Chibani K., Jacquot J. P., Rouhier N. (2013) Cysteine-based redox regulation and signaling in plants. Front. Plant Sci. 4, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waszczak C., Akter S., Eeckhout D., Persiau G., Wahni K., Bodra N., Van Molle I., De Smet B., Vertommen D., Gevaert K., De Jaeger G., Van Montagu M., Messens J., Van Breusegem F. (2014) Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 111, 11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drazic A., Winter J. (2014) The physiological role of reversible methionine oxidation. Biochim. Biophys. Acta 1844,1367–1382 [DOI] [PubMed] [Google Scholar]
- 17. Jacques S., Ghesquière B., Van Breusegem F., Gevaert K. (2013) Plant proteins under oxidative attack. Proteomics 13, 932–940 [DOI] [PubMed] [Google Scholar]
- 18. Kim G., Weiss S. J., Levine R. L. (2014) Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta 1840, 901–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S. H., Li C. W., Koh K. W., Chuang H. Y., Chen Y. R., Lin C. S., Chan M. T. (2014) MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J. Exp. Bot. 65, 5049–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Savige W. E., Fontana A. (1977) Interconversion of methionine and methionine sulfoxide. Methods Enzymol. 47, 453–459 [DOI] [PubMed] [Google Scholar]
- 21. Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 [DOI] [PubMed] [Google Scholar]
- 22. Zhang X. H., Weissbach H. (2008) Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol. Rev. Camb. Philos. Soc. 83, 249–257 [DOI] [PubMed] [Google Scholar]
- 23. Boschi-Muller S., Gand A., Branlant G. (2008) The methionine sulfoxide reductases: catalysis and substrate specificities. Arch. Biochem. Biophys. 474, 266–273 [DOI] [PubMed] [Google Scholar]
- 24. Corpas F. J., Chaki M., Fernández-Ocaña A., Valderrama R., Palma J. M., Carreras A., Begara-Morales J. C., Airaki M., del Río L. A., Barroso J. B. (2008) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol. 49, 1711–1722 [DOI] [PubMed] [Google Scholar]
- 25. Vieira Dos Santos C., Cuiné S., Rouhier N., Rey P. (2005) The Arabidopsis plastidic methionine sulfoxide reductase B proteins. Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 138, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Le D. T., Tarrago L., Watanabe Y., Kaya A., Lee B. C., Tran U., Nishiyama R., Fomenko D. E., Gladyshev V. N., Tran L. S. (2013) Diversity of plant methionine sulfoxide reductases B and evolution of a form specific for free methionine sulfoxide. PLoS One 8, e65637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rouhier N., Vieira Dos Santos C., Tarrago L., Rey P. (2006) Plant methionine sulfoxide reductase A and B multigenic families. Photosynth. Res. 89, 247–262 [DOI] [PubMed] [Google Scholar]
- 28. Corpas F. J., Del Río L. A., Barroso J. B. (2008) Post-translational modifications mediated by reactive nitrogen species: nitrosative stress responses or components of signal transduction pathways? Plant Signal. Behav. 3, 301–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tarrago L., Laugier E., Rey P. (2009) Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: gene organization, reduction mechanisms, and physiological roles. Mol. Plant 2, 202–217 [DOI] [PubMed] [Google Scholar]
- 30. Zhao Y., Li H., Gao Z., Gong Y., Xu H. (2006) Effects of flavonoids extracted from Scutellaria baicalensis Georgi on hemin-nitrite-H2O2 induced liver injury. Eur. J. Pharmacol. 536, 192–199 [DOI] [PubMed] [Google Scholar]
- 31. Hardin S. C., Larue C. T., Oh M. H., Jain V., Huber S. C. (2009) Coupling oxidative signals to protein phosphorylation via methionine oxidation in Arabidopsis. Biochem. J. 422, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gustavsson N., Kokke B. P., Anzelius B., Boelens W. C., Sundby C. (2001) Substitution of conserved methionines by leucines in chloroplast small heat shock protein results in loss of redox-response but retained chaperone-like activity. Protein Sci. 10, 1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wehr N. B., Levine R. L. (2012) Wanted and wanting: antibody against methionine sulfoxide. Free Radic. Biol. Med. 53, 1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ghesquière B., Jonckheere V., Colaert N., Van Durme J., Timmerman E., Goethals M., Schymkowitz J., Rousseau F., Vandekerckhove J., Gevaert K. (2011) Redox proteomics of protein-bound methionine oxidation. Mol. Cell. Proteomics 10, M110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghesquière B., Gevaert K. (2014) Proteomics methods to study methionine oxidation. Mass Spectrom. Rev. 33, 147–156 [DOI] [PubMed] [Google Scholar]
- 36. Moruz L., Staes A., Foster J. M., Hatzou M., Timmerman E., Martens L., Käll L. (2012) Chromatographic retention time prediction for posttranslationally modified peptides. Proteomics 12, 1151–1159 [DOI] [PubMed] [Google Scholar]
- 37. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 38. Habig W. H., Pabst M. J., Jakoby W. B. (1974) Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249, 7130–7139 [PubMed] [Google Scholar]
- 39. Noctor G., Veljovic-Jovanovic S., Driscoll S., Novitskaya L., Foyer C. H. (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann. Bot. 89, 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frottin F., Martinez A., Peynot P., Mitra S., Holz R. C., Giglione C., Meinnel T. (2006) The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics 5, 2336–2349 [DOI] [PubMed] [Google Scholar]
- 41. Ascencio-Ibáñez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., Hanley-Bowdoin L. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones A. M., Thomas V., Bennett M. H., Mansfield J., Grant M. (2006) Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 142, 1603–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang Y., Yang B., Harris N. S., Deyholos M. K. (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58, 3591–3607 [DOI] [PubMed] [Google Scholar]
- 44. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 45. Frova C. (2006) Glutathione transferases in the genomics era: new insights and perspectives. Biomol. Eng. 23, 149–169 [DOI] [PubMed] [Google Scholar]
- 46. Dixon D. P., Skipsey M., Edwards R. (2010) Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71, 338–350 [DOI] [PubMed] [Google Scholar]
- 47. Kagawa T., Wada M. (2002) Blue light-induced chloroplast relocation. Plant Cell Physiol. 43, 367–371 [DOI] [PubMed] [Google Scholar]
- 48. Bienert G. P., Chaumont F. (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 1840, 1596–1604 [DOI] [PubMed] [Google Scholar]
- 49. Shao N., Duan G. Y., Bock R. (2013) A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell 25, 4209–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drazic A., Miura H., Peschek J., Le Y., Bach N. C., Kriehuber T., Winter J. (2013) Methionine oxidation activates a transcription factor in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 110, 9493–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kopriva S. (2013) 12-Oxo-phytodienoic acid interaction with cyclophilin CYP20-3 is a benchmark for understanding retrograde signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 110, 9197–9198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park S. W., Li W., Viehhauser A., He B., Kim S., Nilsson A. K., Andersson M. X., Kittle J. D., Ambavaram M. M., Luan S., Esker A. R., Tholl D., Cimini D., Ellerström M., Coaker G., Mitchell T. K., Pereira A., Dietz K. J., Lawrence C. B. (2013) Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. U.S.A. 110, 9559–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tarrago L., Kieffer-Jaquinod S., Lamant T., Marcellin M. N., Garin J. R., Rouhier N., Rey P. (2012) Affinity chromatography: a valuable strategy to isolate substrates of methionine sulfoxide reductases? Antioxid. Redox Signal. 16, 79–84 [DOI] [PubMed] [Google Scholar]
- 54. Frei dit Frey N., Muller P., Jammes F., Kizis D., Leung J., Perrot-Rechenmann C., Bianchi M. W. (2010) The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22, 1575–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kang G., Ma H., Liu G., Han Q., Li C., Guo T. (2013) Silencing of TaBTF3 gene impairs tolerance to freezing and drought stresses in wheat. Mol. Genet. Genomics 288, 591–599 [DOI] [PubMed] [Google Scholar]
- 56. Kim C., Meskauskiene R., Apel K., Laloi C. (2008) No single way to understand singlet oxygen signalling in plants. EMBO Rep. 9, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. (2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.