Key Points
TKI resistance can be caused by the action of TKIs on MSCs.
Inhibition of the IL-7R/Janus kinase pathway diminishes TKI resistance in MSC milieu.
Abstract
Tyrosine kinase inhibitors (TKIs) are used as a frontline therapy for BCR-ABL+ acute lymphoblastic leukemia (ALL). However, resistance to TKI therapy arises rapidly, and its underlying molecular mechanisms are poorly understood. In this study, we identified a novel cascade of events initiated by TKIs and traversing through mesenchymal stem cells (MSCs) to leukemic cells, leading to resistance. MSCs exposed to TKIs acquired a new functional status with the expression of genes encoding for chemo-attractants, adhesion molecules, and prosurvival growth factors, and this priming enabled leukemic cells to form clusters underneath the MSCs. This cluster formation was associated with the protection of ALL cells from therapy as leukemic cells switched from BCR-ABL signaling to IL-7R/Janus kinase signaling to survive in the MSC milieu. Our findings illustrate a novel perspective in the evolution of TKI resistance and provide insights for advancing the treatment of BCR-ABL+ ALL.
Introduction
In Philadelphia chromosome–positive acute lymphoblastic leukemia (ALL), which is mediated by the BCR-ABL fusion oncoprotein, resistance to the ABL kinase inhibitors can arise from both BCR-ABL–independent and BCR-ABL–dependent mechanisms.1,2 The BCR-ABL–independent mechanisms consist of extra-chromosomal abnormalities, disruptions in drug intake and efflux, and activation of alternative signaling pathways.2,3 The BCR-ABL–dependent mechanisms, including mutations in the ABL kinase domain (such as T315I) and amplification of the BCR-ABL gene,4 usually develop following an initial response to tyrosine kinase inhibitor (TKI) treatment.5 Overcoming BCR-ABL–independent resistance to TKIs is expected to eliminate leukemic cells early in the disease course and to greatly reduce the occurrence of BCR-ABL–dependent resistance. Recent studies showed that the bone marrow milieu, which includes mesenchymal stem cells (MSCs), may play an essential role in the activation of an alternative survival signaling pathway in leukemic cells that protects leukemic cells from chemotherapy.6-10 However, the origin of this resistance in the complex leukemic microenvironment has not been identified. In this study, we used a p190 BCR-ABL–transformed mouse B-cell ALL model to investigate the cascade of events causing the resistance of BCR-ABL+ ALL cells to TKIs.
Study design
Animal studies
All mouse experiments were reviewed and approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. For details of leukemic cell transplantation, bioluminescence imaging, and TKI dosage, see supplemental Methods, available on the Blood Web site.
Viral vectors, transduction, and cell culture
Details of the viral vector construction, virus transduction, and conditions used for culturing MSCs and leukemic cells are described in supplemental Methods.
Microscopy
Phase contrast and mCherry fluorescence images of cultured cells were taken using an Axio Observer.Z1 microscope, an AxioCam MRm camera, and the AxioVision software (Zeiss, Jena, Germany). Total number of leukemic cell clusters (defined as more than 10 leukemic cells) underneath MSCs was obtained from images taken from 10 different fields (×10 objective).
Gene expression microarray analysis
Gene expression profiling analysis was performed as described previously.11 Details of the analysis are provided in supplemental Methods.
Results and discussion
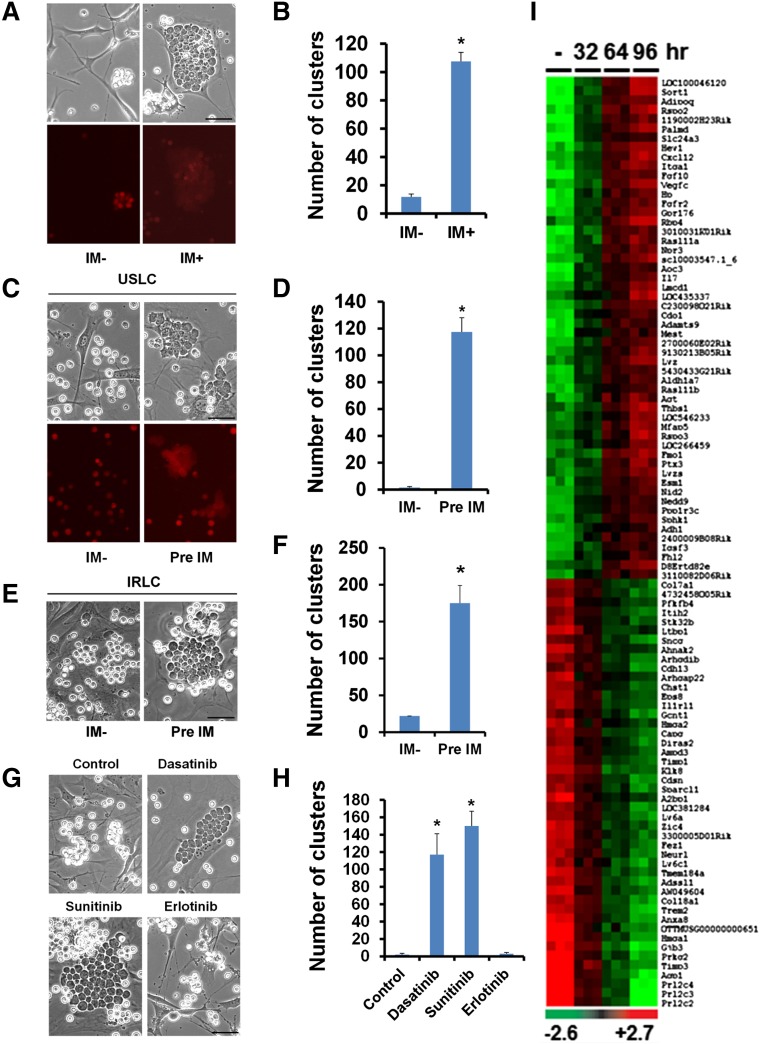
In cocultures of the mouse primary MSC line OP9 (supplemental Figure 1) and mouse ALL cells (also referred to as unselected leukemic cells [USLCs]) (supplemental Figure 2A-B), we observed that the ALL cells closely clustered underneath the OP9 cells in the presence of the BCR-ABL prototype inhibitor imatinib (IM),12,13 whereas the number of cell clusters was significantly reduced in the absence of IM (Figure 1A-B). ALL cell cluster formations were associated with the protection of leukemic cells from IM-induced apoptosis (supplemental Figure 3A-B). We detected reduced phosphorylation levels of platelet-derived growth factor receptor α and β in the IM-exposed OP9 cells, suggesting that IM targets are indeed inhibited by IM treatment (supplemental Figure 4). Although IM treatment reduced the proliferation of OP9 cells (supplemental Figure 5), the treatment did not alter the viability (supplemental Figure 6A) or differentiation (data not shown) and did not induce senescence (supplemental Figure 6B) of the OP9 cells.
Figure 1.
IM-induced alterations in OP9 cells promote the interaction between OP9 cells and leukemic cells. (A) Microscopic visualization of cocultured OP9 cells and mCherry-labeled leukemic cells treated with vehicle (IM−) or IM for 4 days (top: phase contrast; bottom: mCherry fluorescence). (B) Quantification of leukemic cell clusters in (A). (C) Microscopic visualization of leukemic cell clusters formed within 2 hours after seeding of mouse BCR-ABL+ ALL cells onto OP9 cells pretreated with the vehicle (IM−) or IM for 4 days (top: phase contrast; bottom: mCherry fluorescence). (D) Quantification of leukemic cell clusters in (C). (E) Microscopic visualization of leukemic cell clusters formed within 2 hours after seeding of IRLCs onto OP9 cells pretreated with the vehicle (IM−) or IM for 4 days. (F) Quantification of leukemic cell clusters in (E). (G) Microscopic visualization of leukemic cell clusters formed within 2 hours after seeding of leukemic cells onto OP9 cells pretreated with a vehicle (control), dasatinib, sunitinib, or erlotinib for 4 days. Note that, like IM, dasatinib and sunitinib are TKIs, whereas erlotinib is an epidermal growth factor receptor inhibitor. (H) Quantification of leukemic cell clusters in (G). (I) Heat map showing the top 100 differentially expressed genes in OP9 cells treated with a vehicle (−) or IM for 32, 64, or 96 hours. Scale bars, 50 µm. Data are shown as the mean ± SEM. *P < .05 as determined by Student t test.
To determine which cells (OP9 cells or ALL cells) initiated the cluster formation in the presence of IM, we seeded USLCs onto OP9 cells that had been pretreated with IM for 4 days. We observed that the OP9 cells were small and slender in the absence of IM but became enlarged and polygonal when treated with IM (supplemental Figure 7). Notably, ALL cells seeded onto the IM-pretreated OP9 cells formed clusters robustly in as little as 2 hours. In contrast, very few clusters (2 vs 118) were observed within 2 hours after seeding the ALL cells onto the untreated OP9 cells (Figure 1C-D and supplemental Figure 8). Similar results were obtained with BCR-ABL+ B-cell ALL cells from human patients cultured with IM-pretreated OP9 cells (supplemental Figure 9A-B). These data suggest that IM causes alterations in OP9 cells that in turn induce cluster formation that contributes to the survival of leukemic cells. We found that mouse primary MSCs pretreated with IM also showed significantly increased leukemic cell cluster formation compared with untreated MSCs, albeit at a lower frequency than that observed with OP9 cells (supplemental Figure 10A-B).
We established an IM-resistant ALL cell line (IRLC) by continuously culturing USLCs in the presence of IM and interleukin (IL)-7 for 4 to 5 weeks. Like USLCs, the IRLCs that were seeded onto the IM-pretreated OP9 cells formed significantly more clusters than did the IRLCs that were seeded onto the untreated OP9 cells (Figure 1E-F). These findings indicate that the IM pretreatment of the OP9 cells, not the IM pretreatment of the ALL cells, induced ALL cell cluster formation. As with IM, pretreatment of OP9 cells with two other TKIs, dasatinib and sunitinib, induced leukemic cell cluster formation (Figure 1G-H), but the conventional chemotherapeutic agents cyclophosphamide, cytarabine, l-asparaginase, and doxorubicin could neither induce leukemic cell cluster formation nor disrupt existing clusters (supplemental Figure 11A-B).
To identify IM-induced molecular changes in OP9 cells, we performed whole-genome gene expression profiling of OP9 cells with and without IM treatment. We found that 791 genes were differentially expressed in OP9 cells under these two conditions (Figure 1I). Analysis of the top 50 differentially regulated genes through Ingenuity Pathway Analysis revealed the functional association of these genes with chemo-attraction (CXCL12), cell adhesion (ITGA1), and pro-survival growth factor signaling (IL-7). Real-time reverse transcription-polymerase chain reaction analysis validated the upregulation of the top 10 genes identified in the array analysis, including IL-7 (supplemental Figure 12A-D). We also found that mouse primary MSCs treated with IM expressed elevated levels of CXCL12 and IL-7 (supplemental Figure 10C).
IL-7 signaling is critical in normal B-cell development and is mediated primarily through IL-7R. To evaluate the role of IL-7R in ALL cell resistance to IM, progenitor B cells were obtained from the bone marrow of IL-7Rfl/fl mice and transformed with BCR-ABL. IL-7R deletion was then induced with self-excising Cre recombinase (Figure 2A). Real-time reverse transcription-polymerase chain reaction and flow cytometry analysis confirmed the depletion of IL-7R expression in Cre-transduced cells (Figure 2B and supplemental Figure 13). Although ALL cells with IL-7R deletion formed clusters when seeded onto IM-pretreated OP9 cells or primary MSCs, they could not survive beyond 24 hours in the presence of IM, suggesting that IL-7R was required for the survival of the ALL cells, even when they were in clusters (Figure 2C and supplemental Figure 14A-C). Consistently, treatment with IL-7–neutralizing antibody also caused death of the clustered ALL cells in the coculture with IM-pretreated OP9 cells (Figure 2D). These findings suggest that when BCR-ABL signaling is inhibited by IM, leukemic cells rely on alternative survival signals delivered through IL-7/IL-7R and that blocking of IL-7/IL-7R signaling sensitizes leukemic cells to IM. IRLCs, which were established by continuous culture in the presence of IM and IL-7, also showed drastically reduced proliferation, profoundly increased apoptosis, and markedly diminished S-phase cell numbers upon withdrawal of IL-7 (supplemental Figure 15A-D). To evaluate the role of IL-7R in the development of drug resistance in vivo, we transplanted luciferase-labeled IL-7Rfl/fl and IL-7R knockout (KO) ALL cells into nonobese diabetic-severe combined immunodeficiency (NOD-SCID) mice and then treated the mice with dasatinib. We found that, upon dasatinib treatment, leukemia progressed much more slowly and survival was longer in the mice transplanted with IL-7R–deleted ALL cells than in the mice transplanted with IL-7Rfl/fl ALL cells (Figure 2E-F).
Figure 2.
Inhibition of IL-7R/JAK signaling diminishes IM resistance of BCR-ABL+ ALL cells. (A) Schematic presentation of the generation of IL-7R–deficient ALL cells. (B) Relative IL-7R and Cre-recombinase gene expression in IL-7R KO ALL cells compared with that in IL-7Rfl/fl ALL cells. Note that Cre is undetectable in KO cells owing to the use of self-excising Cre. (C) Microscopic visualization of IL-7Rfl/fl and IL-7R–deficient ALL cell clusters 24 hours after seeding of leukemic cells onto OP9 cells pretreated with the vehicle (IM−) or IM for 4 days (left). Quantification of the ALL cell clusters (middle). Representative bioluminescence images of IL-7Rfl/fl and IL-7R–deficient (IL-7R KO) ALL cells 24 hours after seeding onto the vehicle-pretreated (IM−) or IM-pretreated OP9 cells (right). (D) Microscopic visualization of IL-7Rfl/fl ALL cell clusters after treatment with control IgG or IL-7–neutralizing antibody for 48 hours. OP9 cells had been treated with the vehicle (IM−) or IM for 4 days before the leukemic cells were seeded and antibodies (6 µg/ml) were added (left). Quantification of the ALL cell clusters (middle). Bioluminescence images of IL-7Rfl/fl ALL cells cultured with vehicle- (IM−) or IM-pretreated OP9 cells and treated with control IgG or IL-7 neutralizing antibody for 48 hours (right). (E) Bioluminescence imaging of non-irradiated NOD-SCID mice transplanted with IL-7Rfl/fl or IL-7R–deficient ALL cells (4 million cells per mouse) and treated with dasatinib from day 2 to day 8 posttransplantation. Images from day 5 and day 13 are shown (n = 5 per group). (F) Kaplan-Meier survival analysis of leukemic mice transplanted with IL-7Rfl/fl or IL-7R–deficient ALL cells and treated with vehicle (n = 3) or dasatinib (n = 5). The arrows indicate 7-day treatment from day 2 to day 8 posttransplantation. P values were determined by log-rank test. (G) Bioluminescence imaging of non-irradiated NOD-SCID mice transplanted with ALL cells (3 million cells per mouse) and treated with dasatinib alone or in combination with tofacitinib from posttransplantation day 4 onwards. Images from day 1 and day 29 are shown (n = 6 per group). Note that combinatorial treatment with dasatinib and tofacitinib markedly reduced BCR-ABL+ ALL progression. (H) Morphology of peripheral blood smears from leukemic mice at day 30 posttransplantation. Note that there are fewer leukemic blasts in the mice with combinatorial treatment than in the mice with dasatinib treatment alone. Insets, magnified view of blasts. (I) Percentage of leukemic cell blasts in total white blood cells from the peripheral blood smears in (H). (J) Kaplan-Meier survival analysis of leukemic mice treated with vehicle or dasatinib or with the combination of dasatinib and tofacitinib. Arrow indicates the beginning of the treatment at 4 days posttransplantation. P values were determined by log-rank test. (K) Schematic illustration of IM-induced alterations in MSCs and MSC-mediated IM resistance of leukemic cells. In the absence of IM, the growth of BCR-ABL+ ALL cells is driven by BCR-ABL signaling and does not require support from MSCs. In the presence of IM, MSCs undergo morphologic and functional changes and produce a multitude of supporting molecules, including IL-7, thus activating IL-7R/JAK and other growth factor signaling pathways in ALL cells, whereas BCR-ABL signaling is blocked by IM. As a result, growth of the BCR-ABL+ ALL cells switches from being dependent on BCR-ABL signaling to being dependent on growth factor signaling. Note that JAK1 and JAK3, but not JAK2, are likely involved in the IL-7/IL-7R pathway. Scale bars, 50 µm. Data are shown as the mean ± SEM. *P < .05 by Student t test. ND, not detectable.
IL-7R signaling is mediated through Janus kinase 1 (JAK1) and JAK3, which are associated with the IL-7Rα chain and the common γ chain, respectively.14 Our western blot analysis showed elevated levels of phosphorylated JAK1 (pJAK1), markedly reduced pJAK2, and unaltered pJAK3 in IRLCs compared with USLCs (supplemental Figure 16A-B). The reduced pJAK2 levels in IRLCs might have been due to the inhibition of BCR-ABL activity by IM since BCR-ABL was reported to phosphorylate JAK2.15-18 These findings also suggest that JAK2 is dispensable for IL-7 signaling,19 and that JAK1 and JAK3 are likely to mediate IL-7 signaling in IRLCs. To test the effect of JAK signaling intervention on overcoming IM resistance, we treated IRLCs with tofacitinib (a pan-JAK inhibitor),20 TG101209 (a JAK2 and JAK3 inhibitor),21 or ruxolitinib (a JAK2 and JAK1 inhibitor)22 and found that the proliferation of IRLCs was significantly reduced by each of these 3 inhibitors (supplemental Figure 17A-C). Furthermore, treatment of cocultured OP9 and USLCs with IM and TG101209 enhanced apoptosis, disrupted leukemic cell clusters, and eliminated IM-induced OP9 cell-mediated protection of leukemic cells (supplemental Figure 17D-E). To test the effect of combinatorial inhibition of BCR-ABL and JAK signaling on leukemia progression in vivo, NOD-SCID mice transplanted with BCR-ABL+ ALL cells were treated with dasatinib and/or tofacitinib. Our results showed lower bioluminescence signal intensity, reduced peripheral blood leukemic blast counts, and prolonged survival in mice that received the combinatorial treatment compared with those treated with dasatinib alone (Figure 2G-J and supplemental Figure 18), suggesting a therapeutic benefit of inhibiting both the conventional BCR-ABL signaling and the unconventional JAK signaling in BCR-ABL+ ALL cells.
In this study, we suggest that the cascade of events that lead to the development of resistance to TKIs is triggered by TKI therapy itself. At the initial stages of the development of this resistance, MSCs exposed to TKIs become less proliferative and undergo dramatic morphologic and molecular changes, leading to the elevated production of chemo-attractants, adhesion molecules, and pro-survival factors. Stepwise actions of these molecular cues culminate in the initiation of stable interactions, ie, ALL cell cluster formation beneath the MSCs and the emergence of MSC-mediated IM resistance (Figure 2K). Cluster formation and the associated IM resistance suggest that when BCR-ABL signaling is inhibited, BCR-ABL+ ALL cells can adapt to the growth factor–driven signaling provided by MSCs for normal progenitor B-cell survival. Whereas BCR-ABL mediates its oncogenic effect through the activation of JAK2 and other signal transducers, IL-7/IL-7R–induced signaling is mediated through the activation of JAK1 and JAK3.14,23 In the resistant state of leukemic cells, although JAK2 is inactivated upon BCR-ABL inhibition by IM, JAK1 and JAK3 remain active through IL-7 signaling. This compensatory survival support provided by activated JAK1 and JAK3 may be vital to the evolution of drug resistance in the MSC milieu.24 Our results demonstrate that inhibition of IL-7/IL-7R or JAK (JAK1 and JAK3) signaling can effectively abolish ALL cell cluster formation, sensitize ALL cells to IM, and hence diminish the resistance to TKIs. In similar lines, concurrent treatment with JAK2 and BCR-ABL inhibitors demonstrated enhanced eradication of stem cells in chronic myeloid leukemia.25 In conclusion, our findings provide mechanistic insights into the cascade of events leading to the emergence of TKI resistance and could advance therapeutic modalities in BCR-ABL+ ALL.
Acknowledgments
The authors thank K. Dorshkind (The University of California, Los Angeles, Jonsson Comprehensive Cancer Center) for help with pro–B-cell culturing; H. Pelicano and K. Harutyunyan for discussions; K. Ramirez, K. Acklin, D. Bonilla, R. Jewell, and D. Dwyer for help with flow cytometry sorting; and C. Robertson (University of Houston) for proofreading the manuscript.
This study was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases grant 1R21AI103652-01A1 (X.S.), an American Cancer Society Research Scholar grant (X.S.), The University of Texas MD Anderson Cancer Center Institutional Research grant (X.S.), a Leukemia SPORE Developmental Research Award (X.S.) and the MD Anderson Cancer Center Support grant from the National Institutes of Health, National Cancer Institute (CA016672).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.M., X.L., and X.S. planned the study, designed the experiments, analyzed the data, and wrote the manuscript; S.M. performed most of the experiments; X.L., H.M., J.Z., J.L., H.W., Y.Y., B.S., A.N.P., and K.C.-D. performed some experiments and collected data; K.L., Y.L., and J.-S.L. contributed to gene expression profiling experiments; Y.G., R.B.A., Z.C., H.S., and S.A.M. participated in designing and instructing the study; M.K., M.A., X.H., and C.F.H.-G. provided the human samples; B.F. and X.G. provided chemical inhibitors; and X.S. provided overall supervision of the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiaoping Sun, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 37, Houston, TX 77030; e-mail: xsun@mdanderson.org.
References
- 1.Liu-Dumlao T, Kantarjian H, Thomas DA, O’Brien S, Ravandi F. Philadelphia-positive acute lymphoblastic leukemia: current treatment options. Curr Oncol Rep. 2012;14(5):387–394. doi: 10.1007/s11912-012-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee HJ, Thompson JE, Wang ES, Wetzler M. Philadelphia chromosome-positive acute lymphoblastic leukemia: current treatment and future perspectives. Cancer. 2011;117(8):1583–1594. doi: 10.1002/cncr.25690. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer H, Lange T, Wystub S, et al. Prevalence and dynamics of bcr-abl kinase domain mutations during imatinib treatment differ in patients with newly diagnosed and recurrent bcr-abl positive acute lymphoblastic leukemia. Leukemia. 2012;26(7):1475–1481. doi: 10.1038/leu.2012.5. [DOI] [PubMed] [Google Scholar]
- 4.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 5.Soverini S, De Benedittis C, Papayannidis C, et al. Drug resistance and BCR-ABL kinase domain mutations in Philadelphia chromosome-positive acute lymphoblastic leukemia from the imatinib to the second-generation tyrosine kinase inhibitor era: the main changes are in the type of mutations, but not in the frequency of mutation involvement. Cancer. 2014;120(7):1002–1009. doi: 10.1002/cncr.28522. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Li M, McDonald T, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-β-catenin signaling. Blood. 2013;121(10):1824–1838. doi: 10.1182/blood-2012-02-412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, O’Leary H, Fortney J, Gibson LF. Ph+/VE-cadherin+ identifies a stem cell like population of acute lymphoblastic leukemia sustained by bone marrow niche cells. Blood. 2007;110(9):3334–3344. doi: 10.1182/blood-2007-01-068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Mallampati S, Sun B, et al. Wnt pathway contributes to the protection by bone marrow stromal cells of acute lymphoblastic leukemia cells and is a potential therapeutic target. Cancer Lett. 2013;333(1):9–17. doi: 10.1016/j.canlet.2012.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt T, Kharabi Masouleh B, Loges S, et al. Loss or inhibition of stromal-derived PlGF prolongs survival of mice with imatinib-resistant Bcr-Abl1(+) leukemia. Cancer Cell. 2011;19(6):740–753. doi: 10.1016/j.ccr.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Tabe Y, Jin L, Iwabuchi K, et al. Role of stromal microenvironment in nonpharmacological resistance of CML to imatinib through Lyn/CXCR4 interactions in lipid rafts. Leukemia. 2012;26(5):883–892. doi: 10.1038/leu.2011.291. [DOI] [PubMed] [Google Scholar]
- 11.Mallampati S, Sun B, Lu Y, et al. Integrated genetic approaches identify the molecular mechanisms of Sox4 in early B-cell development: intricate roles for RAG1/2 and CK1ε. Blood. 2014;123(26):4064–4076. doi: 10.1182/blood-2013-12-543801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344(14):1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 13.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 14.Rochman Y, Kashyap M, Robinson GW, et al. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc Natl Acad Sci USA. 2010;107(45):19455–19460. doi: 10.1073/pnas.1008271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson-Rawls J, Liu J, Laneuville P, Arlinghaus RB. P210 Bcr-Abl interacts with the interleukin-3 beta c subunit and constitutively activates Jak2. Leukemia. 1997;11(suppl 3):428–431. [PubMed] [Google Scholar]
- 16.Warsch W, Walz C, Sexl V. JAK of all trades: JAK2-STAT5 as novel therapeutic targets in BCR-ABL1+ chronic myeloid leukemia. Blood. 2013;122(13):2167–2175. doi: 10.1182/blood-2013-02-485573. [DOI] [PubMed] [Google Scholar]
- 17.Xie S, Wang Y, Liu J, et al. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 2001;20(43):6188–6195. doi: 10.1038/sj.onc.1204834. [DOI] [PubMed] [Google Scholar]
- 18.Tao W, Leng X, Chakraborty SN, Ma H, Arlinghaus RB. c-Abl activates janus kinase 2 in normal hematopoietic cells. J Biol Chem. 2014;289(31):21463–21472. doi: 10.1074/jbc.M114.554501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parganas E, Wang D, Stravopodis D, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93(3):385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 20.Flanagan ME, Blumenkopf TA, Brissette WH, et al. Discovery of CP-690,550: a potent and selective Janus kinase (JAK) inhibitor for the treatment of autoimmune diseases and organ transplant rejection. J Med Chem. 2010;53(24):8468–8484. doi: 10.1021/jm1004286. [DOI] [PubMed] [Google Scholar]
- 21.Pardanani A, Hood J, Lasho T, et al. TG101209, a small molecule JAK2-selective kinase inhibitor potently inhibits myeloproliferative disorder-associated JAK2V617F and MPLW515L/K mutations. Leukemia. 2007;21(8):1658–1668. doi: 10.1038/sj.leu.2404750. [DOI] [PubMed] [Google Scholar]
- 22.Quintás-Cardama A, Vaddi K, Liu P, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010;115(15):3109–3117. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilaria RL, Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271(49):31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 24.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21(18):2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallipoli P, Cook A, Rhodes S, et al. JAK2/STAT5 inhibition by nilotinib with ruxolitinib contributes to the elimination of CML CD34+ cells in vitro and in vivo. Blood. 2014;124(9):1492–1501. doi: 10.1182/blood-2013-12-545640. [DOI] [PMC free article] [PubMed] [Google Scholar]