Key Points
BETs promote GATA1 chromatin occupancy and subsequently activate transcription; they are generally not required for repression.
BRD2 and BRD4 are essential for full GATA1 activity whereas BRD3 function overlaps with BRD2.
Abstract
Inhibitors of bromodomain and extraterminal motif proteins (BETs) are being evaluated for the treatment of cancer and other diseases, yet much remains to be learned about how BET proteins function during normal physiology. We used genomic and genetic approaches to examine BET function in a hematopoietic maturation system driven by GATA1, an acetylated transcription factor previously shown to interact with BETs. We found that BRD2, BRD3, and BRD4 were variably recruited to GATA1-regulated genes, with BRD3 binding the greatest number of GATA1-occupied sites. Pharmacologic BET inhibition impaired GATA1-mediated transcriptional activation, but not repression, genome-wide. Mechanistically, BETs promoted chromatin occupancy of GATA1 and subsequently supported transcriptional activation. Using a combination of CRISPR-Cas9-mediated genomic engineering and shRNA approaches, we observed that depletion of either BRD2 or BRD4 alone blunted erythroid gene activation. Surprisingly, depletion of BRD3 only affected erythroid transcription in the context of BRD2 deficiency. Consistent with functional overlap among BET proteins, forced BRD3 expression substantially rescued defects caused by BRD2 deficiency. These results suggest that pharmacologic BET inhibition should be interpreted in the context of distinct steps in transcriptional activation and overlapping functions among BET family members.
Introduction
The mammalian bromodomain and extraterminal motif proteins (BETs) have drawn widespread interest as pharmacologic targets for the treatment of various diseases, including hematologic malignancies and solid tumors.1-4 Within the BET family, BRD2, BRD3, and BRD4 are ubiquitously expressed in mammalian tissues, whereas BRDT is testis-specific. BETs contain 2 tandem bromodomains that mediate association with chromatin by binding to acetylated histones and transcription factors.5-9 BETs function in regulatory complexes that impact messenger RNA (mRNA) production at multiple steps of the transcription cycle, such as modifying and remodeling chromatin and promoting transcription elongation.10-17 Both BRD2 and BRD4 are essential for normal development.18-20 A BRD3 knockout mouse has not been reported.
Promising results obtained with pharmacologic BET inhibitors in animal models of malignancy have sparked clinical trials and intensified efforts to better understand BET function.1,2,4,21 Given the widespread expression and essential functions of BETs, it was initially surprising that BET inhibitors like JQ1 elicit cell- and gene-specific responses. These inhibitors block the acetyl-lysine–binding pockets specifically of BET family bromodomains triggering their release from acetylated lysine residues on histones and transcription factors.16,22,23 JQ1 does not distinguish between BET family members, and the development of additional BET inhibitors with distinct specificities remains an important goal.22,24,25 Functional similarity among BETs is suggested by strict conservation of their bromodomains, association with many of the same regulatory complexes,16 and overlapping genomic-binding profiles.26,27 Additionally, chromosomal translocation of either BRD3 or BRD4 with NUT causes histopathologically indistinguishable carcinoma.28 Despite these shared characteristics, distinct phenotypes result from depletion of individual BET family members.4,18-20,27,29-34 The molecular basis for functional distinctions between BETs remains unclear, and large gaps remain in our understanding of their individual roles.
Erythroid maturation is a developmental process driven in part by the erythroid master transcription factor GATA1, which activates essentially all erythroid-specific genes and silences genes associated with the immature proliferative state.35,36 Mice lacking GATA1 die in utero due to failure to form mature erythroid cells,37 and several types of congenital anemias in humans are associated with GATA1 mutations.38,39 GATA1 is acetylated near its zinc finger DNA-binding domain,40 and mutations of acetylated lysines impair the ability of GATA1 to associate with chromatin in vivo.41 Both BRD3 and BRD4 bind to acetylated GATA1, and chromatin immunoprecipitation (ChIP) studies suggest BRD3 in particular is present at most GATA1-occupied sites.6 Exposure of erythroid cells to BET inhibitors diminishes GATA1 occupancy at a subset of target genes and prevents their activation.6 However, the roles of individual BETs in GATA1 function have not been directly evaluated.
Here, we defined distinct mechanisms through which BETs support GATA1-regulated gene expression and direct erythroid maturation. We characterized the relationship of BETs with GATA1 on a genome-wide scale and demonstrate that BETs facilitate GATA1-mediated transcriptional activation but are largely dispensable for repression. We further found that BETs are required not only for initial GATA1 chromatin occupancy, but also for subsequent transcription activation. GATA1-induced erythroid maturation is highly sensitive to reduced levels of BRD2 or BRD4. Unexpectedly, despite the presence of BRD3 at the great majority of GATA1-occupied sites, BRD3 is not required for normal GATA1-activated transcription. However, BRD3 deficiency exacerbates transcriptional defects associated with BRD2 loss. Moreover, forced expression of BRD3 partially restores defects associated with BRD2 loss, suggesting redundant functions among these 2 BETs. Together, these studies reveal that BETs have overlapping roles, and function at distinct steps of the transcription program controlled by GATA1, which are important considerations when interpreting the functions of chemical BET inhibitors.
Materials and methods
Cell culture
Culture of GATA1-erythroblast (G1E) cells and G1E cells expressing a conditionally active estrogen receptor-GATA1 fusion protein has been described.42 GATA1 was activated in G1E GATA1-ER by addition of 100 nM estradiol for 24 hours (denoted +GATA1). Retroviral creation and infection was performed as described.43 Small hairpin RNAs (shRNAs) were cloned into the vector LMP (Open Biosystems). Supplemental Methods (see supplemental Data available at the Blood Web site) contain hairpin sequences.
ChIP
ChIP was performed as described.44 Antibodies were: GATA1 (sc265-N6; Santa Cruz Biotechnology), BRD2 (A302-583A; Bethyl Laboratories), BRD3 sera,6 BRD4 (A301-985A; Bethyl Laboratories), hemagglutinin (HA; 12CA5). Quantitative polymerase chain reaction (qPCR) was run on ViiA7 System (Life Technologies) using Power SYBR Green (Invitrogen). Supplemental Methods contain primers. High-throughput sequencing was performed on an Illumina Hi-seq2000 as described.36 Reads were mapped to mouse genome assembly mm9. Analysis was performed using Bioconductor,45 bedtools,46 and the Cistrome47 Galaxy48 analysis platform.
mRNA expression analyses
RNA was isolated using TRIzol (Life Technologies). Reverse transcriptase (RT)–qPCR was performed with iScript (Bio-Rad) and Power SYBR Green (Invitrogen). Supplemental Methods contain primers. In microarrays, ERCC RNA Spike-In Mix (Ambion) was added to TRIzol-homogenized samples in proportion to cell number. Mouse Gene 2.0ST arrays (Affymetrix) were hybridized per the manufacturer’s protocols. Analysis was performed using the robust multiarray analysis method and normalized to spike-in controls.49
CRISPR-Cas9-mediated gene knockout
CAS9/green fluorescent protein and guide RNA/mCherry plasmids were transiently cotransfected into G1E cells using an Amaxa II electroporator (Lonza) with program G-016 and Kit R. Single transfected cells were sorted into individual wells in a 96-well plate using a FACSAria II (BD Biosciences), expanded, and screened by DNA sequencing and immunoblot. Supplemental Methods list guide RNAs.
Public data access
ChIP-seq and microarray data are accessible via Gene Expression Omnibus50 number GSE62737.
Results
BRD2, BRD3, and BRD4 recruitment to GATA1-activated genes
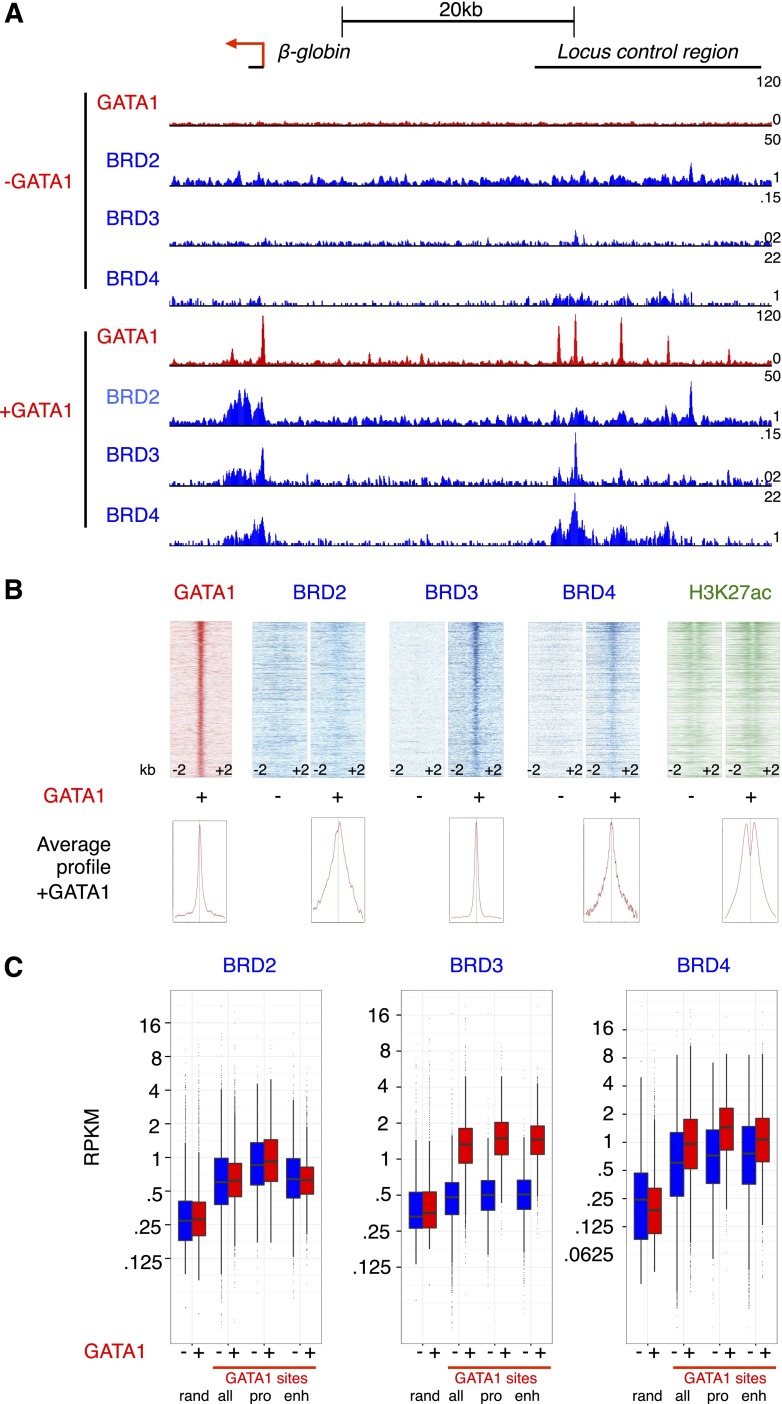
We previously demonstrated that BRD3 and BRD4 directly interact with acetylated GATA1, and BRD3 is recruited to several GATA1-binding sites in a GATA1-dependent manner.6 To comprehensively define the role of BETs in erythroid maturation, we performed chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq) for BRD2, BRD3, and BRD4 in GATA1-null erythroblast (G1E) cells in the absence and presence of an activated estradiol-inducible fusion of GATA1 to the estrogen receptor (GATA1-ER).42 G1E cells recapitulate normal terminal erythroid maturation as verified by gene expression analyses and GATA1 occupancy profiles.35,51 Although we had previously generated a BRD3 ChIP-seq data set,6 we repeated this experiment to improve sequencing depth and compare BET profiles generated on the same platform. BRD2 was included in the present study as well because although early microarray data35 suggested BRD3 and BRD4 were the only BETs expressed in G1E cells, recent genome-wide data36,52 indicated that BRD2 was also expressed. This was confirmed by initial ChIP-qPCR experiments showing JQ1-sensitive BRD2 recruitment to a subset of the GATA1 occupied site (OS) (supplemental Figure 1A). In addition, ChIP-qPCR of exogenous HA-tagged BRD2 produced similar results (supplemental Figure 1B). When extended to a genome-wide scale (ChIP-seq), we found that BRD2, BRD3, and BRD4 occupied GATA1-bound loci, including the β-globin locus Hbb in a GATA1-dependent manner (Figure 1A, supplemental Figure 2). Patterns of BET association with chromatin did not fall into discrete peaks, but instead appear to be broader than proteins that bind directly to DNA. Given these patterns, we focused our analysis on quantitative description of signal intensity at particular sites rather than analysis based on peak calling. In contrast, the sharp binding profiles of GATA1 across the genome readily lent themselves to peak calling using the MACS algorithm. We generated a set of high-confidence GATA1 OSs by intersecting GATA1 peaks called in 2 independent GATA1 ChIP-seq experiments, resulting in 5259 peaks. BRD3 was present at the greatest number of GATA1 OSs, with BRD4 and BRD2 being less frequently associated with GATA1 OS (Figure 1B). We quantified this by calculating ChIP-seq signal density for each BET at GATA1 sites and defining occupancy as a read density >2 standard deviations beyond occupancy at random genomic regions. By this definition, BRD3 was present at 74% of GATA1 sites, BRD4 at 53%, and BRD2 at 27%. For validation, we also examined signals obtained with ectopic HA-tagged forms of BRD2 and BRD3. Although overexpression led to a broadening of the peaks, the results are overall very similar (supplemental Figures 2-3A). Interestingly, when ranked by H3K27ac (Nergiz Dogan, Weisheng Wu, Christapher S. Morrissey, Kuan-Bei Chen, A. Stonestrom, Maria Long, C. A. Keller, Yong Cheng, Deepti Jain, Axel Visel, Len Pennacchio, Mitchell Weiss, G.A.B., and R.C.H., manuscript submitted January 23, 2015), BRD4 occupancy correlated strongly with H3K27ac in the vicinity of GATA1 OS (supplemental Figure 3B). However, the zenith of BRD4 signal localized directly over GATA1 peaks between the twin peaks of maximal H3K27ac, a pattern that likely reflects acetylated nucleosomes flanking GATA1-binding sites. GATA1 OS with high H3K27ac were more likely to be at gene promoters and more likely to be co-occupied by TAL1 (supplemental Figure 3C).
Figure 1.
Reorganization of BET binding following GATA1 complementation. (A) Genome browser tracks showing ChIP-seq signals for the indicated proteins at the mouse β-globin (Hbb) locus. (B) ChIP-seq signal across 4-kb regions centered on GATA1-binding sites ranked from strongest GATA1-binding signal (highest MACS score) to lowest GATA1 signal. (C) Boxplots showing BET reads per kilobase per million (RPKM) at random genomic regions (rand) or GATA1 sites (all, only those at promoters [pro], or only those at candidate enhancers [enh]). Here, we use DNaseI-hypersensitive, H3K4me1-enriched, promoter-excluded sites as enhancers as in Hsiung et al.68
We subsequently divided BET binding at GATA1 sites as binding at candidate enhancers and promoters (Figure 1C). BRD2 signal was higher at GATA1 sites than background sites, but had a similar distribution in the absence and presence of GATA1. In contrast, BRD3 occupancy increased dramatically at both GATA1-bound promoters and enhancers upon GATA1 activation. BRD4 occupancy at GATA1 OS was substantial in the absence of GATA1, and BRD4 signal increased somewhat at these sites following GATA1 activation. These occupancy patterns suggest that BRD3 recruitment is to a large extent influenced by GATA1 whereas BRD2 and BRD4 recruitment is regulated by additional factors.
BETs are required for efficient GATA1-dependent transcriptional activation but not repression
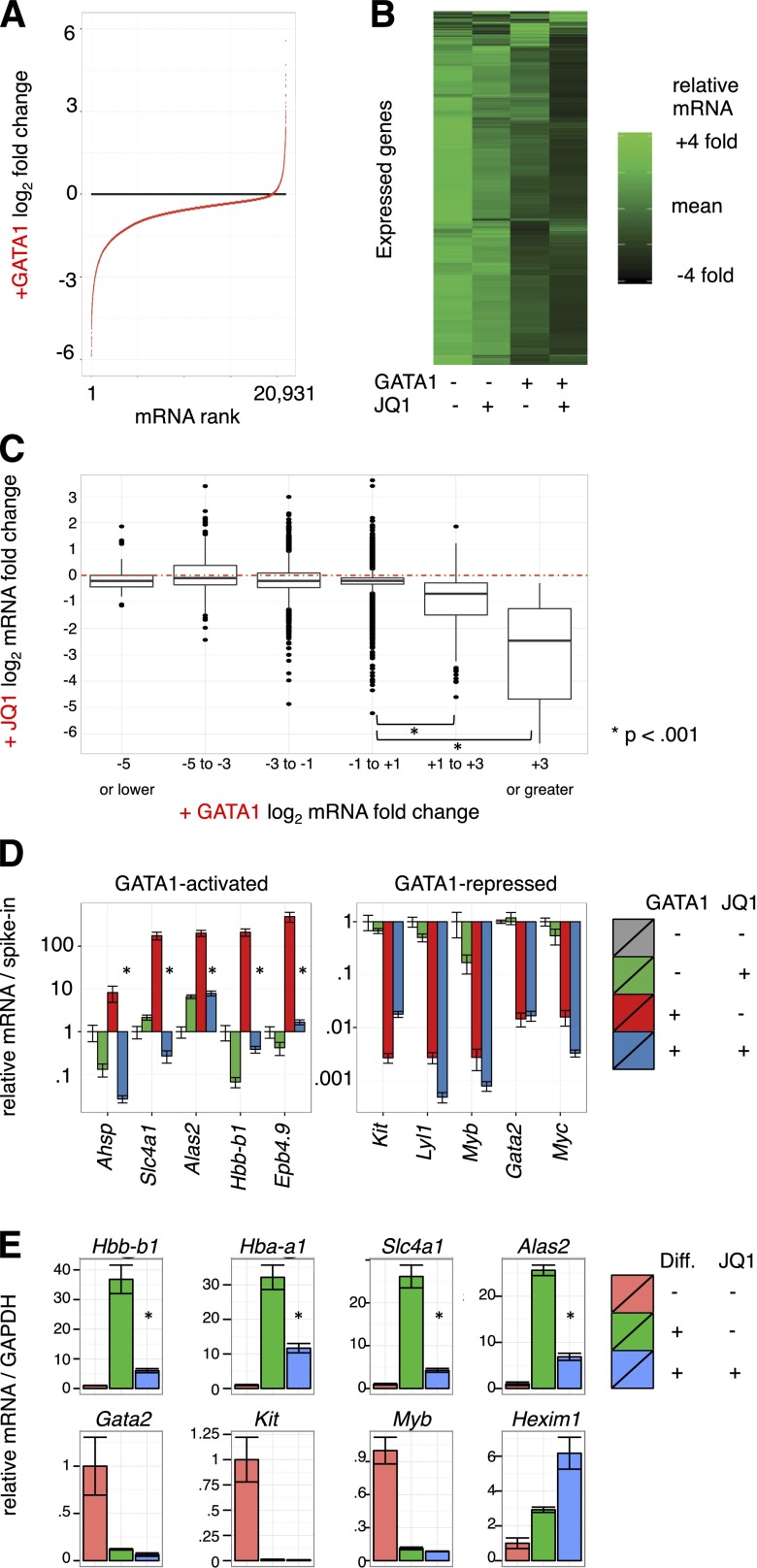
Pharmacologic inhibition of BETs impaired activation of several GATA1 target genes.6 To evaluate the contribution of BETs to GATA1-induced gene expression changes genome-wide, we performed microarray analysis on G1E cells treated with 250 nM JQ1 or dimethylsulfoxide control concurrent with GATA1 activation for 24 hours in biological triplicate. As dramatic alterations in cell size and RNA content occur during erythroid maturation, we added external spike-in RNA controls to each sample in proportion to cell number for normalization.49,53 Focusing first on GATA1’s impact on gene expression, we plotted all transcripts from most repressed to most activated (Figure 2A). Following GATA1 addition, 5094 transcripts decreased whereas only 220 increased using stringent differential expression criteria (twofold change and Bonferroni-corrected, P < .05) (supplemental Figure 4A). The overwhelming predominance of gene repression upon GATA1 induction contrasts with prior studies of GATA1-mediated transcriptome changes that were based on internal standards and concluded that the number of activated and repressed genes are similar.35,51,54,55 This highlights the importance of spike-in controls in transcriptome studies in which global RNA levels change.
Figure 2.
Transcriptome changes driven by GATA1 activation and BET inhibition. (A-C) Microarray expression profiles of G1E cells ± GATA1 induction in the presence or absence of JQ1. Transcript levels were normalized to cell numbers using external RNA spike-in controls. Data represent the mean of 3 biological experiments. (A) Distribution of mRNA changes upon GATA1 activation. (B) Heatmap showing relative expression of each transcript in each condition. (C) Boxplot showing relationship of activation by GATA1 with JQ1 sensitivity. Red dotted line shows no change in mRNA levels with JQ1 treatment. (D) GATA1-activated and -repressed transcript levels as determined by RT-qPCR. Data were plotted relative to untreated G1E cells normalized to cell number by RNA spike-in controls. *P < .01 (2-sample t test). Error bars represent SEM; n = 3. (E) RT-qPCR for erythroid transcripts in primary fetal liver erythroid progenitors induced to differentiate along the erythroid lineage for 24 hours in the presence or absence of 250 nM JQ1. *P < .01 (2-sample t test). Error bars represent SEM; n = 3. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To evaluate the role of BETs in transcriptional regulation by GATA1, we visualized relative mRNA levels of expressed genes as a heatmap arranged by hierarchal clustering (Figure 2B). A preponderance of genes were most highly expressed in untreated G1E cells and declined slightly upon JQ1 treatment. A stronger decline in transcript levels was observed following GATA1 induction, and transcript levels were lowest upon GATA1 activation in the presence of JQ1. We next plotted JQ1 sensitivity at genes activated or repressed by GATA1 by grouping GATA1-responsive genes into bins based on fold activation or repression (Figure 2C, supplemental Figure 4B). Although significant variation was observed across measured genes, consistent with gene selectivity in JQ1 response,2,16,22,29,32 genes most activated by GATA1 were the most sensitive to JQ1. In contrast, transcripts that decreased upon GATA1 induction decreased no more or less on average with concurrent JQ1 treatment. BET inhibition increased mRNA levels at some genes, which could be due to repressive functions of BETs or to indirect action. These results are consistent with BETs functioning principally in GATA1-mediated activation and having little role in repression. We further confirmed this at known GATA-target genes in independent experiments by RT-qPCR (Figure 2D).
We next tested the degree to which BET occupancy determines JQ1 sensitivity. We plotted ChIP-seq read counts at promoters and enhancers against JQ1 response with or without concurrent GATA1 induction (supplemental Figure 5). BET occupancy was not a strong predictor of JQ1 sensitivity overall, however, a weak relationship between JQ1 effects and BRD4 occupancy at promoters was observed (supplemental Figure 2B). This is consistent with observations that not all BET-occupied genes respond to BET inhibitors.22,26,29,32 Both BRD4 and H3K27ac have been used to identify “superenhancers” (SEs),29,56 and BRD4 SEs have been suggested to reside near genes particularly sensitive to JQ1. We defined SEs using the script ROSE29 which aggregates enhancer regions within 12.5 kb (supplemental Figure 6A). Genes adjacent or overlapped by SEs were only minimally more JQ1-sensitive than all expressed genes. In contrast, activation by GATA1 strongly predicted JQ1 responsiveness (supplemental Figure 6B). We further used H3K27ac signal intensity to define SEs, and found that these were even less JQ1-sensitive than those defined using BRD4 signal. Only 1 of 10 genes proximal to the top SEs identified declined by >50% upon BET inhibition (supplemental Figure 6C). These observations do not support categorization of SEs as distinct JQ1-hypersensitive entities.
We sought to verify the transcriptional effects of BET inhibitors in primary erythroid cells as G1E cells are immortalized. We measured gene expression in mouse fetal liver erythroid progenitors differentiated in the presence or absence of JQ1. BET inhibition suppressed surface expression of the erythroid maturation marker TER-119, but had no overt toxic effect (supplemental Figure 7A). Similar to results in G1E cells, activation of erythroid gene expression was impaired by JQ1 whereas gene repression occurred normally (Figure 2E, supplemental Figure 7B). We also noted induction of the transcriptional repressor HEXIM1 upon BET inhibition as has been observed in other cell types.11,57 Together, these results support the role of BETs in GATA1-driven transcriptional activation, and further suggest repression may be largely BET-independent.
Role of BETs in GATA1 occupancy genome-wide
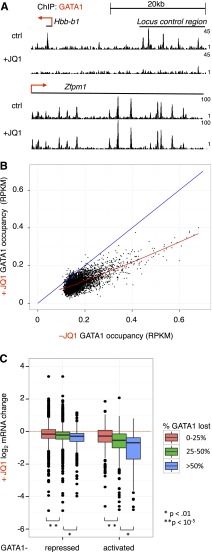
We previously reported that GATA1 must interact with BETs to bind selected OS.6 To evaluate the role of BETs in GATA1 occupancy genome-wide, we performed anti-GATA1 ChIP-seq following 24 hours of GATA1 induction in the absence or presence of JQ1. BET inhibition almost entirely prevented GATA1 binding at some loci (Hbb), while having no measurable effect on binding at others (Zfpm1) (Figure 3A). To quantitatively examine the requirement of BETs for GATA1 occupancy, we compared signal at GATA1 sites (defined in Figure 1) without or with concurrent JQ1 addition (Figure 3B). BET inhibition reduced GATA1 occupancy at 98% of sites (points below blue diagonal), but did so only partially at the great majority of loci. To evaluate the relationship between inhibition of GATA1 occupancy and inhibition of transcription, we plotted fractional maintenance of GATA1 occupancy against transcriptional sensitivity of presumptive target genes within 5 kb of GATA1 sites (Figure 3C). Genes that were expressed at lower levels in response to JQ1 tended to reside near elements at which GATA1 occupancy was sensitive to JQ1 inhibition. GATA1 occupancy was also reduced at a number of sites adjacent to genes whose expression was unaffected by BET inhibition. At these OS, GATA1 binding is either not required for transcription at nearby genes or partial occupancy is sufficient for transcriptional activation. Interestingly, the JQ1 sensitivity of GATA1 occupancy is not predicted by the amounts of BRD2, BRD3, or BRD4 at GATA1 OS (supplemental Figure 8A). This suggests that other GATA1 partners are sufficient for its stable binding to chromatin at some locations. Sites at which GATA1 occupancy was JQ1-sensitive were more likely to be co-occupied by TAL1 (supplemental Table 1), consistent with TAL1 functioning predominantly at GATA1-activated genes.43 JQ1-sensitive GATA1 OSs were similarly present at enhancers and promoters. In independent validation experiments, we observed a similar spectrum of JQ1-mediated impairment of GATA1 occupancy (supplemental Figure 8B). These results suggest that BETs are required for maximal GATA1 occupancy at numerous sites, but that the role of BETs in GATA1-mediated transcription is likely to extend beyond assisting GATA1 in chromatin binding.
Figure 3.
Effects of BET inhibition on GATA1 occupancy genome-wide. (A) Genome browser tracks showing GATA1 binding at the Hbb and Zfpm1 loci in the absence and presence of 250 nM JQ1. Tracks are from 1 biological experiment and representative of 2 with similar results. (B) GATA1 ChIP-seq read density following GATA1 induction for 24 hours in the absence or presence of JQ1. The red line shows a Loess regression; the blue diagonal demarcates no change between control and JQ1 treatment. (C) Boxplot showing relationship between BET dependence of GATA1 occupancy and transcriptional activation. GATA1 peaks are linked to nearest gene within 5 kb. P values reflect results of 2-sample t tests from indicated comparisons.
BETs activate transcription subsequent to establishment of GATA1 occupancy
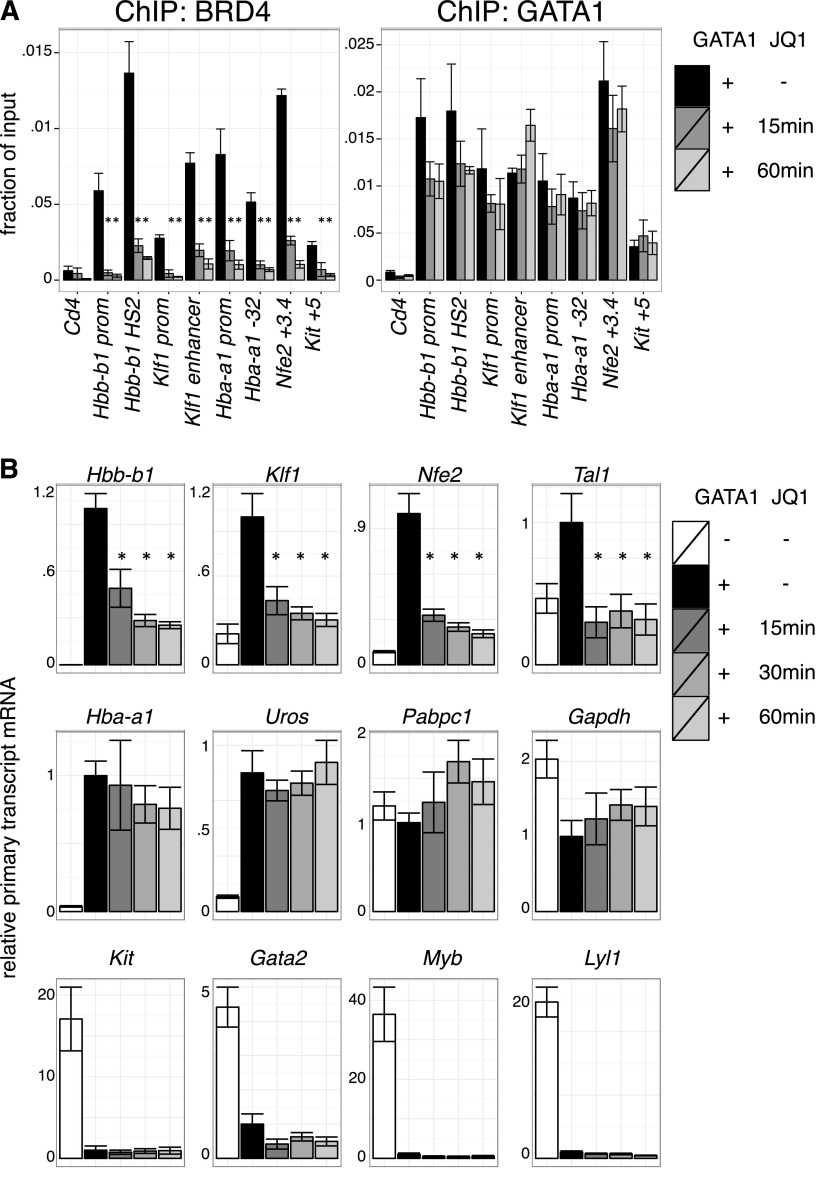
BETs interact with the general transcription machinery and may stimulate transcription directly.12-15,58,59 Based on the results in the previous section, we tested whether BETs act in GATA1-activated transcription subsequent to the establishment of chromatin occupancy. As GATA1 occupancy might be less sensitive to BET inhibition once established, we examined the short-term effects of BET inhibition after establishment of GATA1 occupancy. Indeed, JQ1 treatment of 1 hour removed BETs from all sites examined with relatively little effect on GATA1 occupancy (Figure 4A, supplemental Figure 9A). We next measured primary transcript levels of GATA1-target genes under these conditions. Several GATA1 targets, including Hbb-b1 (β-globin), Klf1, and Nfe2, were immediately repressed upon BET inhibition, suggesting a role for transcription of BETs downstream of GATA1 occupancy (Figure 4B, supplemental Figure 9B). In contrast, transcription of other genes like Hba-a1 (α-globin) and Uros (involved in heme synthesis) were unperturbed by short-term JQ1 treatment despite their sensitivity to long-term JQ1 exposure and proximity to BET-bound regulatory elements. At these genes, BETs might function predominantly by assisting GATA1 occupancy or by secondary mechanisms. As expected, genes repressed by GATA1, such as Gata2 and Kit, remained inactive upon JQ1 treatment (Figure 4B). We conclude that at a subset of genes BETs facilitate GATA1 function at a step subsequent to binding to chromatin.
Figure 4.
Transcriptional requirement of BET proteins after establishment of GATA1 occupancy. (A) ChIP for BRD4 and GATA1 in G1E GATA1-ER cells treated with 250 nM JQ1 for up to 60 minutes following GATA1 induction. Cd4 served as negative control. Error bars represent SEM; n = 3. *P < .05 that indicated JQ1-treated sample mRNA is lower than untreated (2-sample t test). (B) Primary transcript RT-qPCR of indicated transcripts following JQ1 treatment in GATA1-induced cells. Error bars represent SEM; n = 4. *P < .05 comparing JQ1 treated and control samples (2-sample t test).
Partially overlapping function among BETs
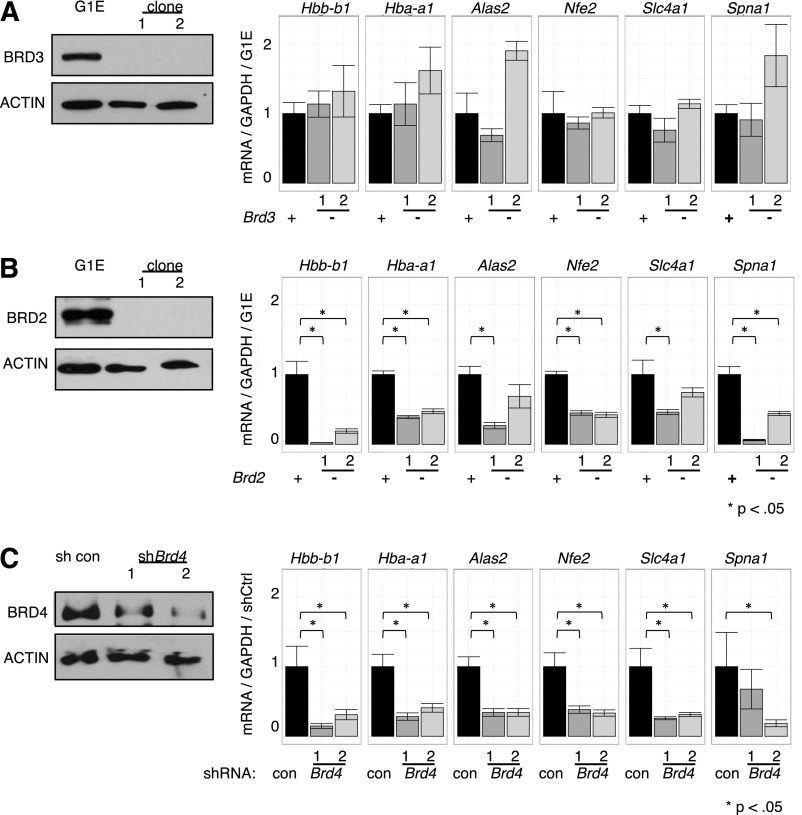
Current knowledge of BET function is largely built on studies using inhibitors that do not distinguish between individual BETs.1,4,22,27,29,60 To dissect the roles of individual BETs in GATA1-driven erythropoiesis, we used a loss-of-function approach combining CRISPR-Cas9-engineered gene disruption61 and shRNA-mediated knockdown. As BRD3 occupies nearly all GATA1 OSs, we had speculated that it was the most relevant BET in GATA1-mediated transcription. Surprisingly, cells engineered to produce no detectable BRD3 expressed all examined GATA1-target genes at essentially normal levels upon GATA1 induction (Figure 5A), suggesting that BRD3 is not essential. In contrast, in BRD2-deficient cells, GATA1 failed to induce several of its archetypical target genes to normal levels (Figure 5B). However, the effects of BRD2 depletion were less pronounced than those observed with JQ1 treatment implicating additional BETs in GATA1-driven erythroid gene expression. Attempts at functional deletion of BRD4 failed, perhaps due to its requirement for cell growth.18 However, transient shRNA-mediated depletion of BRD4 significantly decreased GATA1-induced gene expression (Figure 5C) supporting its importance in this process. Although no growth defects were observed upon BRD3 ablation, cells deficient in BRD2 or BRD4 proliferated more slowly, indicating that these genes are required for normal G1E cell proliferation (data not shown). These results suggest BRD2 and BRD4 are individually required for normal GATA1-mediated transcriptional activation.
Figure 5.
Functions of individual BETs in GATA1-activated transcription. (A-C) Left, Western blots with antibodies against indicated BET proteins. Right, Relative transcript levels following GATA1 activation in cells depleted of (A) BRD3, (B) BRD2, or (C) BRD4. BRD4 reduction was achieved by shRNA-mediated Brd4 knockdown. P-value comparisons are the results of 2-sample t tests.
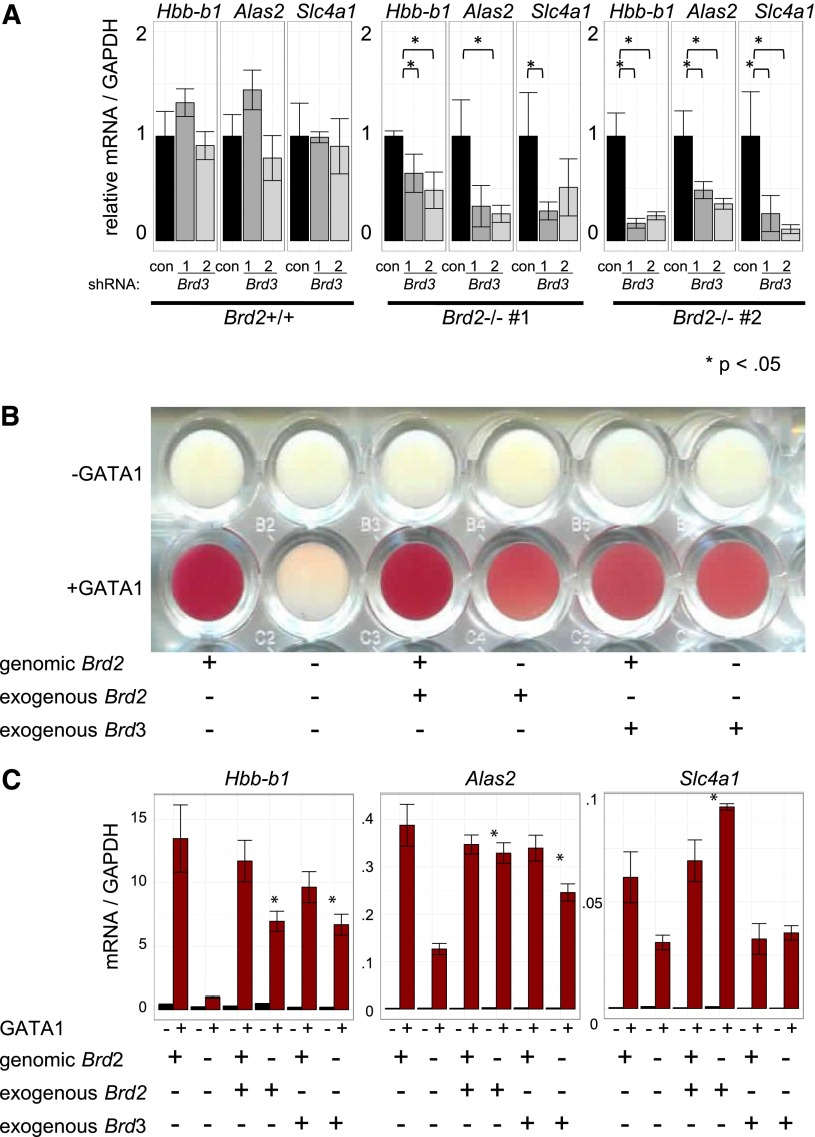
Given the direct physical association of BRD3 with acetylated GATA1 and their genome-wide colocalization, the indifference of GATA1-activated transcription to BRD3 depletion was unexpected. We therefore tested whether other BETs might compensate for BRD3 loss. To this end, we used shRNAs to deplete BRD3 in BRD2-replete and BRD2-deficient cells (Figure 6A, supplemental Figure 10A). As expected, BRD3 knockdown on its own had no significant impact on gene activation. However, BRD3 knockdown exacerbated the consequences of BRD2 deficiency on GATA1-activated gene expression. Hence, BRD2 and BRD3 may assist GATA1 in an at least a partially overlapping manner. Additionally, BRD3 knockdown impaired the growth of BRD2-deficient, but not of BRD2-replete, cells (data not shown). To further test the idea of functional overlap between BRD2 and BRD3, we examined erythroid maturation as reflected in hemoglobinization (red coloring) following GATA1 activation (Figure 6B). BRD2-deficient cells failed to hemoglobinize. Retroviral BRD2 expression restored this defect, confirming specificity of BRD2 gene targeting. Remarkably, overexpression of BRD3 also restored hemoglobinization in BRD2-deficient cells, indicating substantial functional overlap between BRD2 and BRD3 in this system. Consistent with phenotypic results, BRD3 overexpression also rescued BRD2 deficiency at most, but not all, genes examined (Figure 6C, supplemental Figure 10B). Interestingly, Hexim1 mRNA increased in BRD2-deficient cells. This was reversed by either BRD2 or BRD3 expression, suggesting these BETs contribute to Hexim1 repression. In sum, although BET proteins serve unique and essential functions during erythroid maturation, BRD2 and BRD3 can functionally compensate for each other.
Figure 6.
BRD3 function revealed in BRD2-deficient cells. (A) shRNA-mediated Brd3 knockdown in BRD2 replete vs deficient cells. Error bars represent SEM; n = 3. P-value comparisons are the results of 2-sample t tests. (B-C) BRD2 replete and deficient G1E cells in the presence and absence of GATA1 with or without retroviral BRD2 or BRD3 expression. (B) Photograph of cell pellets in wells of a 96-well plate. One representative experiment is shown of 3 with similar results. (C) mRNA levels. Error bars represent SEM; n = 3. *P < .05 that sample expressing exogenous Brd2 or Brd3 has higher mRNA levels than control Brd2-deficient cells.
Discussion
The advance of BET inhibitors into human clinical trials is a strong incentive to better understand the role of BETs in normal physiology. The present study reveals that GATA1 induces recruitment of different BETs to GATA1-occupied sites during erythroid maturation. BET function is critical for GATA1-mediated gene activation, but not repression, by both facilitating GATA1 occupancy and subsequently activating transcription. Despite the association of BRD3 with GATA1 at nearly all genomic-binding sites, loss-of-function experiments uncover the more essential roles of BRD2 and BRD4 in GATA1-driven transcription. Notably, BRD2 and BRD3 are able to at least partially substitute for each other.
Despite the similarity of their bromodomains, individual BETs have dynamic and distinct occupancy patterns during erythroid maturation. BRD3 is recruited to nearly all GATA1 sites whereas BRD4 occupies approximately one-half. BET-binding patterns are likely determined by association not only with acetylated GATA1 but also histone acetylation, which increases at many sites upon GATA1 activation.62 In addition, other acetylated transcription factors might contribute to BET recruitment. BET occupancy is maximal between the highest points of histone acetylation, suggesting acetylated transcription factors in nucleosome-free regions might generally be major contributors to BET recruitment.
The mere presence of BETs at a given gene does not predict JQ1 response. Moreover, even the amounts of BETs as defined by read counts at promoters or enhancers were poor indicators of JQ1 sensitivity. However, the assignment of enhancers to target genes by genomic distance is unreliable, potentially obscuring possible relationships between enhancer BET binding and JQ1 response.4 Significantly, the most JQ1-sensitive genes were those activated by GATA1, consistent with the notion that BETs function generally in inducible transcription,22,63,64 and activation by GATA1 predicted JQ1 sensitivity better than BET occupancy at superenhancers.
Linking ChIP-seq and transcriptome data sets requires reliable quantification of transcripts. Cell-state transitions such as maturation can dramatically alter both total RNA and mRNA content, limiting reliability of normalization with internal standards.49,53 Using spike-in controls, we measured a greatly reduced ratio of GATA1 activated to repressed genes compared with previous estimates.35,36,51,55 This enabled us to more accurately gauge the role of BETs during GATA1-induced changes in transcription, and established BETs chiefly as transcriptional coactivators. We also found that GATA1-dependent repression is largely unaffected by BET inhibition. This includes genes that are adjacent to JQ1-sensitive GATA1 peaks. Decreases in mRNA levels during GATA1-induced maturation in the presence of JQ1 may be the compound effect of repression by GATA1 and by BET inhibition.
Despite the strong effects of JQ1 on GATA1-dependent erythroid differentiation, no evidence suggests that BET inhibition causes anemia in animals or humans. Whether BET inhibitors have deleterious effects on erythropoiesis in patients will depend both on pharmacokinetic factors and how well laboratory models predict physiology. Importantly, mammals efficiently compensate for disease-induced ineffective erythropoiesis and traumatic blood loss via powerful feedback mechanisms. GATA1 is also essential for megakaryocyte development and platelet formation, which might also be affected. Dose-limiting toxicity could result from functional inhibition of any transcription factor that utilizes BETs.27,33,65
Surprisingly, despite a high degree of co-occupancy with GATA1, BRD3 was dispensable for GATA1-induced transcription. Instead, BRD2 and BRD4 were required for induction. A requirement for BRD3 was only uncovered in the context of BRD2 depletion, indicating at least partially overlapping functions among these proteins. Interestingly, Brd3 mRNA is approximately fourfold less abundant than Brd2 mRNA according to RNA-seq data sets in G1E cells.52 Together with the ability of Brd3 overexpression to rescue BRD2 deficiency, it is possible that phenotypic differences between these proteins may be at least partially due to different expression levels. This implies that the combined amounts of BRD2 and BRD3 may be functionally as relevant as the levels of the individual proteins or any potentially unique features of them. Many reports attribute the effects of BET inhibitors to BRD4 based on experiments in which individual BETs are depleted.32,58,66,67 It is important to consider that because of the functional overlap of BRD2 and BRD3, it is possible that the combined contribution of these molecules has been underestimated. Future dissection of the mechanisms through which BETs act distinctly or compensate for each other will be critical when considering the development of BET inhibitors directed against specific members of this family.
Acknowledgments
The authors thank Christopher Vakoc and Chris Hsiung for helpful discussion, Ryan Wychowanec and Hetty Rodriguez for technical assistance, and James Bradner for JQ1.
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK054937 (G.A.B.), R56-DK065806 (R.C.H., G.A.B), and T32-DK007780 (A.J.S.), National Cancer Institute grant F30-CA189553 (S.C.H.), and a generous donation by the DiGaetano family (G.A.B., P.E.).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.J.S. and G.A.B. conceived the study, designed experiments, and wrote the manuscript; A.J.S., B.M.G., P.E., R.C.H., and G.A.B. analyzed data; A.J.S., S.C.H., K.S.J., P.H., C.A.K, S.K., and A.E.C. performed experiments; and all authors edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerd A. Blobel, Children’s Hospital of Philadelphia, Abramson Research Center 316H, 3615 Civic Center, Philadelphia, PA 19104-4318; e-mail: blobel@email.chop.edu.
References
- 1.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12(7):465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367(7):647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 3.Prinjha RK, Witherington J, Lee K. Place your BETs: the therapeutic potential of bromodomains. Trends Pharmacol Sci. 2012;33(3):146–153. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54(5):728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100(15):8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamonica JM, Deng W, Kadauke S, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci USA. 2011;108(22):E159–E168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishiyama A, Dey A, Miyazaki J, Ozato K. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol Biol Cell. 2006;17(2):814–823. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S-Y, Lee A-Y, Lai H-T, Zhang H, Chiang C-M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49(5):843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B, Yang X-D, Zhou M-M, Ozato K, Chen L-F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itzen F, Greifenberg AK, Bösken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42(12):7577–7590. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem. 2012;287(43):36609–36616. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel MC, Debrosse M, Smith M, et al. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol. 2013;33(12):2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Yik JH, Chen R, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 15.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30(1):51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S-Y, Chiang C-M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 18.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22(11):3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without type 2 diabetes. Biochem J. 2009;425(1):71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238(4):908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicodème E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picaud S, Da Costa D, Thanasopoulou A, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 2013;73(11):3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 26.Anders L, Guenther MG, Qi J, et al. Genome-wide localization of small molecules. Nat Biotechnol. 2014;32(1):92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 2012;7:247–265. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 29.Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Ma Q, Wong K, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155(7):1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapuy B, McKeown MR, Lin CY, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagarajan S, Hossan T, Alawi M, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Reports. 2014;8(2):460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190(7):3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104(10):3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Wu W, Kumar SA, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19(12):2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93(22):12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols KE, Crispino JD, Poncz M, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24(3):266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell AE, Wilkinson-White L, Mackay JP, Matthews JM, Blobel GA. Analysis of disease-causing GATA1 mutations in murine gene complementation systems. Blood. 2013;121(26):5218–5227. doi: 10.1182/blood-2013-03-488080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19(5):3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108(12):3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17(3):1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripic T, Deng W, Cheng Y, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113(10):2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letting DL, Chen Y-Y, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci USA. 2004;101(2):476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu T, Ortiz JA, Taing L, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lovén J, Orlando DA, Sigova AA, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu M, Riva L, Xie H, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36(4):682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paralkar VR, Mishra T, Luan J, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, Holstege FC. Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep. 2003;4(4):387–393. doi: 10.1038/sj.embor.embor798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu W, Cheng Y, Keller CA, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21(10):1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doré LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119(16):3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaidos A, Caputo V, Gouvedenou K, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123(5):697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 58.Anand P, Brown JD, Lin CY, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devaiah BN, Lewis BA, Cherman N, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA. 2012;109(18):6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mele DA, Salmeron A, Ghosh S, Huang H-R, Bryant BM, Lora JM. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210(11):2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23(4):1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng S, Zhang L, Tang Y, et al. BET inhibitor JQ1 blocks inflammation and bone destruction. J Dent Res. 2014;93(7):657–662. doi: 10.1177/0022034514534261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan YM, Kirkham P, Barnes PJ, Adcock IM. Brd4 is essential for IL-1β-induced inflammation in human airway epithelial cells. PLoS ONE. 2014;9(4):e95051. doi: 10.1371/journal.pone.0095051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang Y, Gholamin S, Schubert S, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20(7):732–740. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahai V, Kumar K, Knab LM, et al. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther. 2014;13(7):1907–1917. doi: 10.1158/1535-7163.MCT-13-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knoechel B, Roderick JE, Williamson KE, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46(4):364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsiung CC-S, Morrissey CS, Udugama M, et al. Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res. 2014;25(2):213–225. doi: 10.1101/gr.180646.114. [DOI] [PMC free article] [PubMed] [Google Scholar]