Abstract
Telomeres are protective DNA–protein complexes located at the ends of eukaryotic chromosomes, whose length has been shown to predict life-history parameters in various species. Although this suggests that telomere length is subject to natural selection, its evolutionary dynamics crucially depends on its heritability. Using pedigree data for a population of white-throated dippers (Cinclus cinclus), we test whether and how variation in early-life relative telomere length (RTL, measured as the number of telomeric repeats relative to a control gene using qPCR) is transmitted across generations. We disentangle the relative effects of genes and environment and test for sex-specific patterns of inheritance. There was strong and significant resemblance among offspring sharing the same nest and offspring of the same cohort. Furthermore, although offspring resemble their mother, and there is some indication for an effect of inbreeding, additive genetic variance and heritability are close to zero. We find no evidence for a role of either maternal imprinting or Z-linked inheritance in generating these patterns, suggesting they are due to non-genetic maternal and common environment effects instead. We conclude that in this wild bird population, environmental factors are the main drivers of variation in early-life RTL, which will severely bias estimates of heritability when not modelled explicitly.
Keywords: relative telomere length, heritability, maternal effect, sex-linkage, inbreeding, bird
1. Introduction
Telomeres are highly conserved protective DNA–protein complexes based on tandem repeats of a simple sequence of nucleotides. Although telomeric sequences can also be found in the pericentric regions of chromosomes (interstitial telomeres), most interest has gone out to the ends of eukaryotic chromosomes, i.e. the terminal telomeres. Terminal telomeres prevent deterioration of chromosome ends and fusion among chromosomes [1], and their length is shaped by the interplay of pro- and anti-erosion factors [2,3]. Telomere length has been shown to significantly predict life-history parameters in a number of organisms, both when telomeres are measured early in life [4] and during adulthood [5]. For example, telomere length (or their rate of shortening) has been linked to lifespan in humans [6] and several bird species, both in captivity [4] and in natural populations [5,7,8]. More recently, it has been found that telomere length may not only be positively related to individual fitness through its link with lifespan, but also as a mediator of reproductive trade-offs [9]. This further reinforces the idea that telomere length could be subject to directional natural selection. However, whether this also results in an evolutionary response depends on the heritability of telomere length.
Telomere length shows substantial amounts of variation, not only among eukaryote species [10], but also among individuals of the same species and population [5]. Although it is key to obtaining a better understanding of the origin of this individual variation in early-life telomere length, an answer to the question whether variation in telomere length is transmitted from one generation to the next, and if it is, by what mechanism, remains equivocal. Several studies on the mode of transmission and narrow-sense heritability (i.e. the proportion of the phenotypic variance that is attributable to additive genetic effects, h²) of telomere length have been conducted in humans (e.g. [11–14]), as well as in captive and wild populations of other species (e.g. [15–20]). Ranging from 0.18 to 1.23, the majority of these heritability estimates is relatively high, especially considering that heritabilities of traits closely related to fitness are often low [21,22]. However, as they are ratios, heritabilities can be high even if the absolute amount of additive genetic variance is low, if environmentally induced variation is even lower (for example in captivity (e.g. [20], but see [23])). Alternatively, if common environment and parental effects are not accounted for, heritabilities will be overestimated [24]. Indeed, telomere dynamics are known to be modulated by environmental factors, both during development [25–27] and adult life [28,29]. For example, early exposure to steroid hormones triggers an accelerated telomere loss in domestic chickens [30].
Interestingly, a number of studies has found support for sex-specific patterns of inheritance of telomere length (e.g. [13,15–18,31,32]). For example, several of the studies investigating telomere length inheritance in non-human animals (birds and lizards) have found either maternal inheritance [15,16,18] or a much higher son–father than daughter–mother resemblance [17]. Indeed, sex-specific patterns appear to be the rule rather than the exception and suggested mechanisms include parent-specific imprinting, hormonal regulation and sex-chromosome linkage, either acting independently or jointly [13,16,17,32].
Here, we investigate patterns of inheritance of relative telomere length (RTL) in a wild population of white-throated dippers (Cinclus cinclus). We use nestlings from an individual-based long-term study, enabling us to separate phenotypic variation in RTL into variance components attributable to additive genetic, common environment (i.e. nest) and other environmental effects. In addition, we explicitly test for parental and imprinting effects, as well as sex-linked inheritance (Z-linkage).
2. Material and methods
(a). Study system
The white-throated dipper is a medium-sized passerine living along streams and rivers. Since 1987, dippers have been studied at 11 rivers, spanning an area of approximately 400 km² in the proximity of Zurich, Switzerland (8°23′ E/47°25′ N to 8°40′ E/47°10′ N). Here, we use data from the Küsnacht (K), Wehrenbach (W) and Sihl (S) Rivers. Every year, monitoring starts in early February in order to map territories and to find nests. Most pairs are socially monogamous but each year approximately 9% of males are polygynous. Territories are checked regularly between nest building and nestling phase. Nestlings of first broods hatch between early March and April. About 35% of all nestlings are from second broods with hatching dates in early June at the latest. Clutch size is slightly larger for the first compared with the second brood (mean ± s.d.: 4.8 ± 1.0 versus 4.5 ± 0.9), which is also reflected in the number of nestlings (4.4 ± 1.1 versus 3.8 ± 1.2). Incubation takes 16–17 days and offspring fledge at an age of 21–24 days. Both parents feed their offspring but only females incubate. When nestlings are 10–14 days old (min. 7 days, max. 17 days), they are ringed and a small blood sample (max. 30 μl) is collected by puncturing the tarsal vein. Unringed adults (i.e. immigrants) are captured using mist nets and ringed, usually before the breeding season, but at the latest before their offspring are ringed. Fieldwork procedures are licensed by the Swiss Federal Office for the Environment and the Veterinary Office of the Canton of Zurich.
(b). Pedigree reconstruction and inbreeding estimation
Parentage of each brood was determined from behavioural observations, assuming that a nestling's social parents are also its genetic parents. This is a reasonable assumption given the low incidence of extra-pair paternity in this species (2% according to Øigarden et al. [33]; less than 1% in our study population (PJJB 2014, unpublished data)). The identity of the territorial breeding male and female were recorded during territory establishment, incubation and/or feeding of the offspring.
We were able to construct a pedigree spanning 15 generations, covering the cohorts from 1987 to 2012. We calculated Wright's coefficients of inbreeding f [34] for each individual using Pedigree Viewer (available at http://www.personal.une.edu.au/~bkinghor/pedigree.htm). Inbreeding coefficients are relative to the base population, i.e. relative to all birds with unknown parents. In the following analyses, we therefore use only birds that hatched in the study area. Across the study period, 52% of all individuals breeding in one of the three main rivers (n = 662, 1996–2012) settled in their natal river, that is, were philopatric (39% of females and 66% of males). About 20% of the birds not born in their river of breeding originated from another river within the study area, whereas the remaining individuals were of unknown origin.
We selected a total of 177 individuals from the cohorts 2002 to 2011 for RTL measurement, consisting of interconnected groups made up by a sire, a dam and—whenever possible—one female and one male offspring. This study design, which maximizes the number of families rather than the number of individuals per family, will have resulted in a slight reduction in the precision (i.e. the standard error) of our estimate of the (environmental) variance between nests. More importantly however, this also resulted in more independent parent–offspring links, as well as several pedigree links between families, with 16 females and nine males being present in the dataset as both offspring and parent. Furthermore, several individuals have sired offspring with more than one mate (average 1.59 for males and 1.55 for females). Therefore, our dataset also contains pedigree links between grandparents and their grandchildren or between half sibs, etc. On the whole, our study design thereby allows for relatively accurate and precise estimates of additive genetic variance and heritability. Finally, to be able to test for inbreeding effects, we deliberately selected a high proportion of inbred individuals. As a consequence, the mean inbreeding coefficient (±s.d.) of the selected individuals (0.076 ± 0.088; f = 0: N = 64, 0 < f ≤ 0.0625: N = 36, 0.0625 < f ≤ 0.25: N = 62, f > 0.25: N = 15, max. f = 0.3037) was higher than the population-level mean for the same cohorts and rivers (0.023 ± 0.057). Pruning the complete pedigree used for the calculation of inbreeding coefficients (see above) to include only individuals with known RTL, or that provide a pedigree link between two individuals with known RTL (using the R package ‘pedantics’, [35]), resulted in a pedigree containing 380 individuals, 291 maternities, 291 paternities, 221 full sibs, 103 maternal half sibs and 147 paternal half sibs. Mean pedigree depth was 5.0 generations (max. 14 generations).
(c). DNA extraction, storage and sexing
After collection, blood samples were preserved up to several months at approximately 4°C in APS buffer [36]. DNA was extracted using the QIAmp DNA mini kit (BioSprint 96, Qiagen) and then stored in AE buffer at −20°C. Within a few weeks, the DNA concentration was normalized and DNA was stored at −80°C (see [37] for more details). We verified quality and purity with a NanoDrop (Thermo Scientific) spectrophotometer (absorbance ratio A260/280 > 1.7; A260/230 > 1.8) and through gel electrophoresis (more than 10 kbp). Nestling sex was determined by amplifying the CHD-W and CHD-Z genes using modified versions of the P2 and P8 primers [38,39].
(d). Relative telomere length measurements
RTL was assessed using a quantitative real-time amplification (qPCR) procedure [40] adjusted for birds [41]. Because telomeres have never been studied in white-throated dippers before, below we give all necessary details on the measurement methods (following [42]). Although unlike some other methods, qPCR does not discriminate between chromosome ends and interstitial telomeric sequences, qPCR-based telomere measurements have been widely used, including in a number of bird species (e.g. [4,5,15,18,19,41]). We discuss the potential implications of this limitation of the qPCR method in some detail in the Discussion.
RTL was expressed as the ratio of the number of telomeric repeats to the amount of a non-telomeric control gene that is non-variable in copy number in the study species. The amount of telomeric repeats present in the sample (T) and the amount of the control gene (control gene S) are proportional to the number of qPCR amplification cycles needed to reach a threshold fluorescent signal (Cq value) in the exponential growth phase. Here, we used glyceraldehyde-3-phosphate dehydrogenase (GAPDH, GenBank accession no: AF255390) as the control gene. To validate both the expected amplicon size of the control gene (50 bp) and its uniformity in white-throated dippers, 14 randomly chosen samples were run on a standard 1.5% agarose electrophoresis gel in TBE buffer (see electronic supplementary material, figure S1).
Forward and reverse primers for the GAPDH control gene were 5′-AACCAGCCAAGTACGATGACAT-3′ and 5′-CCATCAGCAGCAGCCTTCA-3′, respectively. Primers for amplification of telomeric repeats were 5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ and 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′ (Tel1b and Tel2b, see [41]). qPCRs for both telomeric sequences and GAPDH were performed using 5 ng of DNA with both sets of primers, in a final volume of 10 µl containing 5 µl of Power SYBR© Green PCR Master Mix (Appliedbiosystems, UK). Primer concentrations in the final mix were 100 nM for the telomere assay and 200 nM for the control gene assay. Real-time amplification of telomeric sequences and GAPDH were performed on separate 96-well plates. qPCR conditions for amplification of telomere sequences were 10 min at 95°C, followed by 30 cycles of 1 min at 56°C and 1 min at 95°C. qPCR conditions for GAPDH amplification were 10 min at 95°C, followed by 40 cycles of 1 min at 60°C and 1 min at 95°C.
Samples were randomly assigned to one of six plates. All samples, including both reference samples and dilution series (see below), were analysed in duplicate. The precision of qPCR measurements critically depends on amplification efficiencies [43]. In order to control for variation in the amplification efficiency of the qPCR among plates, serial dilutions (50, 10, 2, 0.4, 0.08 and 0.016 ng) of a reference sample were used to generate a reference curve for each plate. Both a negative control (water) and a melting curve were run for each plate to check for specific amplification of a unique amplicon and for the absence of primer–dimer artefact (electronic supplementary material, figure S2).
Intra-plate mean coefficients of variation for Cq values were 1.35 ± 0.06% for the telomere assay and 0.79 ± 0.04% for the control gene assay (based on duplicates), and inter-plate coefficients of variation based on repeated samples (n = 5) were 1.56% for the telomere assay and 1.35% for the control gene assay (all CV calculated before correction for plate effects). Amplification efficiencies (estimated from the standard curves of serial dilutions) of the qPCR runs were between 98 and 100% for telomeric repeats and between 99 and 100% for the control gene. To take into account both this slight difference in amplification efficiency (E), as well as the non-zero intra- and inter-plate coefficients of variation, we calculated RTL following Pfaffl [44] as
The coefficient of variation for RTL was 12.7%.
(e). Quantitative genetic analyses
We fitted a series of animal models [45] to estimate the absolute and relative amount of additive genetic variance (VA and h2, respectively) underlying telomere length and to test for parental effects, sex-specific inheritance and imprinting. Animal models were fitted using restricted maximum likelihood (REML) in ASReml v. 3.0 [46], except for models including imprinting effects, which were implemented in WOMBAT [47].
RTL was best described by a normal distribution and residuals of the final model did not show any deviations from normality (Shapiro–Wilk test of normality, W = 0.99, p = 0.25). Nestling RTL was modelled as a function of sex and natal population (i.e. river), age at sampling (in days) and (Julian) hatching date. Because within-brood competition might act as a stressor negatively affecting RTL, we fitted brood size, as well as body mass and tarsus length as proxies for body condition, as covariates. We used the residuals of a quadratic regression of tarsus length and body mass against age to account for nestling growth. Additionally, all models included the inbreeding coefficient as a covariate, which in the presence of inbreeding depression in RTL ensures unbiased estimates of additive genetic variance [48,49]. Fixed effects were removed in a stepwise manner, starting with the least significant, as inferred from a conditional Wald F-test. All effects with p ≤ 0.05 were retained in the model, except for the inbreeding coefficient, which was retained irrespective of its significance for reasons mentioned above.
In addition to the fixed effects listed above, we fitted a random additive genetic effect (animal effect), as well as a random nest and year of birth effect. The animal effect estimates the variance in the trait that is due to additive genetic effects (VA), using information on the relatedness and resemblance in telomere length among all individuals in the pedigree. The nest effect estimates the variance among nests (VNEST) that can be attributed to the shared environment of full sibs growing up in the same nest, over and above the variance that is attributable to additive genetic effects. Finally, variation that can be attributed to random environmental variability among years (VYEAR) is accounted for by the year of birth effect. Heritability, the proportion of the phenotypic variance that is explained by additive genetic variance, was calculated as
where VR is the residual variance. Statistical significance of random effects was assessed using likelihood ratio tests, comparing log-likelihoods of models with and without the specific random effect.
To test whether there are general features of the mother or father that affect offspring RTL over and above any additive genetic effects passed on to the offspring (sensu [50]), initial models included maternal and paternal identity as additional random effects. However, both explained little to no additional variation (2.6 × 10−5% and 3.1 × 10−5% of phenotypic variation, respectively). Hence, parental identities were excluded from any further models.
Although the structure of the data did not allow us to unequivocally attribute the increased resemblance among full sibs to properties of the nest or the mother, we were able to directly test for an effect of maternal RTL (as a nestling) on the RTL of her offspring, again over and above the effect of the genes that she passes on to them. To do so, we extended the animal model arrived at above with residual maternal RTL (residuals from a mixed model that corrected RTL for inbreeding coefficient and year of birth) as an additional covariate, as outlined in Lynch & Walsh [51, p. 706]. Similarly, we included residual paternal RTL, as well as both maternal and paternal RTL simultaneously.
Animal models allow for the explicit estimation of sex-linked effects as these follow a different pattern of inheritance than autosomal traits [46,52]. In birds, females are the heterogametic and males the homogametic sex (ZW and ZZ, respectively). In order to quantify Z-linked genetic variance, we used a relatedness matrix that accounts for the specific inheritance of Z-linked genes (e.g. the relatedness between mothers and their daughters is zero) [52] in addition to the usual autosomal relatedness matrix.
Finally, we tested for imprinting effects, which may provide a further source of sex-specific resemblance, using WOMBAT [47]. While in a standard additive genetic model, we express the genetic value of an individual as the sum of the contributions of both maternal and paternal gametes, here we attempted to separate the effects of maternal and paternal gametes [53]. To this end, rather than calculating the (inverse) covariance matrix among individuals, we obtained the inverse variance–covariance matrix among male or female gametes, i.e. the inverse gametic relationship matrices, using code written by Bruce Tier (provided on http://didgeridoo.une.edu.au/womwiki). These were used to fit either a random maternal or paternal imprinting effect, in addition to the animal's additive genetic effect.
In addition to the animal models above, we performed a number of parent–offspring regressions, in which we regressed mean offspring, son or daughter RTL against maternal or paternal RTL. Values that were used for these regressions were residuals taken from a mixed model that accounted for the effects of inbreeding coefficient and year of birth. Not only do these help to visualize our main findings, they also make it possible to directly compare our results to studies on parent–offspring resemblance in RTL in other species.
3. Results
(a). Animal model analyses
RTL was not affected by hatching date (b = 2.5 × 10−4 ± 1.5 × 10−3, F1,102.6 = 0.03, p = 0.87), age at sampling (b = 1.4 × 10−3 ± 1.7 × 10−2, F1,95.4 = 0.01, p = 0.94), sex (male–female length: 1.4 × 10−2 ± 4.1 × 10−2, F1,102.8 = 0.11, p = 0.74) or natal population (F2,93.1 = 1.74, p = 0.18). Similarly, brood size (b = −2.7 × 10−2 ± 2.7 × 10−2, F1,101.5 = 0.97, p = 0.33), age-corrected body mass (b = 4.0 × 10−3 ± 4.6 × 10−3, F1,163.4 = 0.73, p = 0.40) and tarsus length (b = −1.3 × 10−3 ± 1.8 × 10−2, F1,121.3 = 0.01, p = 0.86) did not predict early-life RTL. Although it did not reach statistical significance, inbreeding had a positive effect on RTL (b = 0.53 ± 0.35, F1,64.0 = 2.29, p = 0.14) and was retained in all models to obtain unbiased variance component estimates [48,49].
Nest identity and year of birth explained large and significant proportions of the phenotypic variation (± approximate standard error) (nest identity: 19.6 ± 8.3%, χ² = 8.59, d.f. = 1, p = 0.002; and year of birth: 45.7 ± 13.2%, χ² = 52.06, d.f. = 1, p < 0.001). However, the additive genetic variance was not significantly different from zero (0.007 ± 0.013, χ² = 0.45, d.f. = 1, p = 0.25), and heritability of RTL was estimated to be 3.8 ± 6.9%. For all statistical details, see table 1.
Table 1.
Animal model analysis, explaining variation in nestling RTL with an individual's inbreeding coefficient (f) as well as an additive genetic effect (animal effect), a nest and a year of birth effect (model A, n = 177). Model B (n = 114) additionally includes residual maternal RTL as a covariate. Slopes for covariates, variance components for random effects as well as their proportions are given including approximate standard errors (s.e.) and test statistics (conditional F-test and χ²-test, respectively).
| model A |
model B |
|||||||
|---|---|---|---|---|---|---|---|---|
| covariate | slope ± s.e | test statistic | p-value | slope ± s.e | test statistic | p-value | ||
| f | 0.53 ± 0.35 | F1,64.0 = 2.29 | 0.14 | 0.87 ± 0.43 | F1,46.8 = 4.14 | 0.05 | ||
| maternal RTL | — | — | — | 0.22 ± 0.11 | F1,47.1 = 4.06 | 0.048 | ||
| random effect | variance ± s.e. | proportion ± s.e. | test statistic | p-value | variance ± s.e. | proportion ± s.e. | test statistic | p-value |
|---|---|---|---|---|---|---|---|---|
| animal (VA) | 0.007 ± 0.013 | 0.038 ± 0.069 | χ² = 0.45 | 0.25 | 1 × 10−7 | 6 × 10−7 | χ² = 0 | 0.5 |
| nest (VNEST) | 0.037 ± 0.014 | 0.196 ± 0.083 | χ² = 8.59 | 0.002 | 0.031 ± 0.015 | 0.182 ± 0.092 | χ² = 5.77 | 0.008 |
| year of birth (VYEAR) | 0.086 ± 0.044 | 0.457 ± 0.132 | χ² = 52.06 | <0.001 | 0.075 ± 0.045 | 0.438 ± 0.152 | χ² = 23.71 | <0.001 |
| residual (VR) | 0.058 ± 0.012 | 0.310 ± 0.096 | 0.066 ± 0.012 | 0.380 ± 0.118 |
Including residual maternal RTL resulted in a smaller sample size (n = 114) and in a further reduction of VA (1 × 10−7) and h2 (6 × 10−5%), but did not affect estimates of the other variance components (table 1). Again, the effect of inbreeding was positive and this time did reach statistical significance (b = 0.87 ± 0.43, F1,46.8 = 4.14, p = 0.05). Most importantly, maternal RTL was significantly related to offspring RTL (b = 0.22 ± 0.11, F1,47.1 = 4.06, p = 0.048; table 1). Although the additional inclusion of paternal RTL further reduced the sample size (n = 101), this did not alter the positive point estimate for the effect of the mother's phenotype on offspring phenotype (b = 0.23 ± 0.12, F1,39.4 = 3.35, p = 0.075). However, although there was no effect of the father's phenotype (b = 0.02 ± 0.13, F1,37.9 = 0.04, p = 0.85), the maternal and paternal slopes did not differ significantly from each other based on their overlapping confidence intervals.
Additional analyses attempting to explain these patterns of resemblance by means of sex-linked inheritance revealed a random variance component for Z chromosome-linked variance of 5.3 × 10−8 (χ² = 0, d.f. = 1, p = 0.50), with all other variance component estimates remaining unchanged. Similarly, neither maternal nor paternal imprinting explained any variance (maternal imprinting 1.6 × 10−5, χ² = 0, d.f. = 1, p = 0.50; paternal imprinting: 1.2 × 10−5, χ² = 0, d.f. = 1, p = 0.50).
(b). Parent–offspring regressions
In line with the effects of maternal and paternal RTL in the animal models described above, the slope of the regression of mid-offspring on maternal RTL was significantly positive (b = 0.22 ± 0.10, p = 0.03; n = 59), whereas the slope of the regression of mid-offspring on paternal RTL was small and non-significant (b = 0.04 ± 0.11, p = 0.73; n = 59) (figure 1 and table 2). Again however, the two estimates of the regression slopes were not significantly different from each other (t = 1.21, p = 0.23). Furthermore, single sex offspring–parent regressions revealed that daughter–father and son–father regressions were not significantly different from zero, with estimates close to zero (b = 0.09 ± 0.14, p = 0.50, and b = –0.03 ± 0.12, p = 0.79, respectively). However, single sex offspring–mother regressions showed that resemblance between sons and their mothers (b = 0.27 ± 0.04, p = 0.01) was considerably but not significantly (t = 1.12, p = 0.27) higher than the resemblance between daughters and their mothers (b = 0.11 ± 0.14, p = 0.43).
Figure 1.
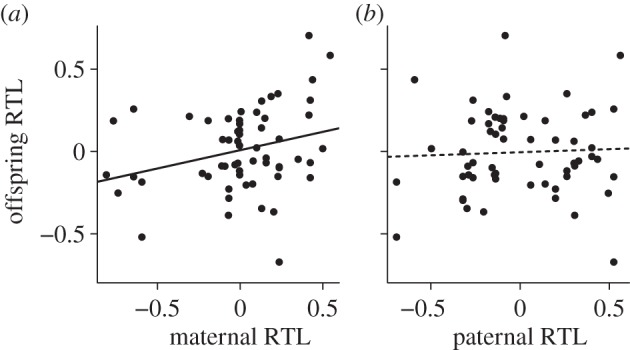
Linear regressions of mid-offspring RTL (mean of nestlings of one nest) against their mother's ((a), n = 59) and father's ((b), n = 59) RTL. RTL values are residuals from a mixed model accounting for an individual's inbreeding coefficient and its year of birth. Whereas offspring resemble their mother (b = 0.22 ± 0.10, p = 0.03), there is no correlation between fathers and their offspring (b = 0.04 ± 0.11, p = 0.73).
Table 2.
Parent–offspring regressions for RTL. Slope including standard error (b ± s.e.), sample size (n) and p-value are given for different regressions of offspring (mid-offspring, son or daughter) on parental RTL. Values of RTL are residuals from a mixed model that accounts for an individual's inbreeding coefficient and its year of birth. All telomere measurements were done on samples taken at the nestling stage.
| parent–offspring combination | n | b ± s.e | p-value |
|---|---|---|---|
| mother–offspring | 59 | 0.22 ± 0.10 | 0.03 |
| mother–son | 58 | 0.27 ± 0.04 | 0.01 |
| mother–daughter | 56 | 0.11 ± 0.14 | 0.43 |
| father–offspring | 59 | 0.04 ± 0.11 | 0.73 |
| father–son | 61 | −0.03 ± 0.12 | 0.79 |
| father–daughter | 57 | 0.09 ± 0.14 | 0.50 |
4. Discussion
Here, we tested whether and how variation in RTL is transmitted across generations. By using pedigree data from a wild population of white-throated dippers, we were able to disentangle the relative effects of genes and the environment on early-life RTL and to test for sex-specific patterns of inheritance.
Previously reported heritability estimates of telomere length range between 0.44 and 0.78 for humans [11–14] and between 0.18 and 1.23 for other vertebrates [15–20]. This suggests that there is a large amount of variation in the degree to which telomere length is heritable, with our estimate being at the lower end of the spectrum. These differences among studies may be the result of biologically interesting variation in either the genetic background of the population or the amount of environmental variation experienced. This might explain differences between wild versus captive populations, between humans and non-human animals, and between wild populations inhabiting environments with varying levels of temporal and spatial heterogeneity. However, below we will briefly explore two alternative sources of variation.
First, some variation may be attributable to differences among statistical methods in their ability to separate genetic and environmental sources of variation [22]. For example, estimates of heritability based on offspring–parent regression might be upwardly biased if non-genetic sources of resemblance between parents and offspring remain unaccounted for [45]. Indeed, based on an offspring–mother regression, we would have obtained a statistically significant and relatively high heritability of 44% (i.e. twice the slope of this regression), even after accounting for the effects of year of birth and inbreeding coefficient. Similarly, not accounting for nest effects in our animal model would have resulted in a heritability of 9.2%, whereas accounting for nest effects reduces the heritability to 3.8% (±6.9%). The latter is in line with a cross-fostering experiment in a wild population of collared flycatchers (Ficedula albicollis), allowing, at least partly, for the separation of additive genetic and common environment effects, which found a heritability of 18% (and not 9% as reported previously (P. Bize 2013, personal communication [19]). Although this value is higher than our estimate in dippers, it is still substantially lower than what has been found in other studies, and due to the experimental design and/or the statistical analyses employed, there still is room for this estimate being inflated [19]. At first sight in contrast to Voillemot et al. [19], another study employing cross-fostering arrived at a heritability of 0.99 [20]. However, as pointed out by the authors, this estimate might also be upwardly biased due to difficulties in separating genetic and non-genetic sources of variation in their breeding design [20].
A second source of variation might be provided by differences among the methods that have been employed to quantify telomere length (reviewed in [42]), and in particular between the qPCR method employed by most studies so far and which measures the total amount of telomeric sequence, and electrophoresis-based methods that quantify the amount of terminal telomeric sequence only [20]. While this complicates their interpretation, several studies have found qPCR-based telomere measures to be biologically meaningful, as exemplified by, for example, their correlation with components of fitness (e.g. [4,5]). Nevertheless, at least in theory the heritability estimate of zero observed in our study may be due to measurement error introduced by variation in interstitial telomeric sequence. However, the strong and significant cohort and nest effects observed, as well as the positive effect of maternal RTL, make this unlikely. Likewise, the strong environmental effects on RTL observed in this system might be shaped by variation in the amount of interstitial, rather than terminal telomeric repeats. However, although their function remains largely unknown, interstitial telomeric repeats are likely to be relatively inert [20,54], making them a less likely target of systematic environmental effects, and more likely to have an additive genetic component.
While we by no means intend to dismiss the limitations of the qPCR method when it comes to measuring variation in telomere length, we at the same time would like to emphasize its major advantage, namely its ability to efficiently screen a large number of individuals. The latter is essential for any quantitative genetic analysis, which typically requires large sample sizes. Furthermore, although several individual-based long-term studies routinely collect DNA to aid, for example, sexing or parentage analysis, the amounts of DNA available will often be insufficient for electrophoresis-based methods. Indeed, to date the only study explicitly estimating a heritability of terminal telomere length in birds is that by Atema et al. [20] for a captive population of zebra finches. While in stark contrast to our findings, they find a heritability estimate close to unity, and no evidence for nest or parental effects, limitations in terms of their sample size and breeding design prevent them from conclusively separating genetic and non-genetic sources of full-sib resemblance in telomere length [20].
Bearing in mind the potential complications when interpreting the currently available heritability estimates, the generality of significant maternal or paternal effects (but see [20]) is striking. For example, a recent study on great reed warblers found significant heritability of RTL of 35–48%, as well as strong brood and maternal effects [15]. Similarly, in kakapos (Strigops habroptilus) and king penguins (Aptenodytes patagonicus), mothers and their offspring resembled each other in telomere length, but fathers and their offspring did not [16,18]. In line with these bird studies, we found a link between maternal and offspring RTL. While our animal models did not provide evidence for a significant effect of maternal identity, or in other words, offspring of the same mother did not resemble each other more than expected on the basis of the genes they shared, this discrepancy can most likely be attributed to three factors. First, generally speaking the estimation of a random effect requires more data than the effect of a fixed covariate, which may explain only a small fraction of the total variance. Second, random maternal effects summarize general features of the mother, and these may comprise both positive and negative effects on RTL. Third, maternal identity tends to be strongly correlated with nest identity, and the separation of their effects requires a large number of females with offspring from multiple nests. In line with the latter, removing the nest effect from the model increased the proportion of variance explained by the maternal effect to 5%. Having said this, the fact that in the absence of maternal identity, the nest identity explained far more variation, suggests that the nest effect is more important than the maternal effect.
One mechanism that could explain the observed patterns of mother–offspring but not father–offspring resemblance is sex-specific gene imprinting. However, we found no evidence of either maternal or paternal imprinting. Similarly, we found no evidence for sex-linkage, specifically Z-linkage. Furthermore, the strong resemblance between mothers and their sons rules out W-linkage. However, it should be noted that, if they do exist in the first place, we could expect the effects of imprinting and Z-linkage to be relatively small, and the double matrix mixed effect models required for their estimation are particularly data-hungry. Hence, our dataset might not provide the statistical power required for their detection. Nevertheless, as of yet few studies have attempted to estimate these effects, and we hope our attempts will aid those with access to larger amounts of data.
We find a major role for environmental effects in shaping variation in RTL. For example, a substantial amount of variation was attributable to environmental differences among birth years (i.e. cohort effects). This is in accordance with previous studies on mammals, lizards and birds [25–27,55–60], which have shown increased telomere shortening in response to suboptimal environmental conditions. For example, Mizutani et al. [60] showed that in black-tailed gulls (Larus crassirostris), the rate of change in telomere length mainly differed with respect to year, and they attributed this to the consequences of El Niño events and the Great Japan Earthquake on food availability.
It remains unclear which environmental variables are responsible for the observed annual variation in early-life RTL in our study population of dippers. We found that birds born in cohorts with relatively high RTL were on average heavier and more likely to produce offspring later in life. However, within cohorts only the correlation between nestling weight and the probability of producing offspring later in life approached statistical significance (p = 0.06). The fact that these associations are observed on the cohort rather than the individual level suggests that these associations are driven by a third, unknown, factor, rather than by a causal relationship between the two.
In addition to the population-level environmental effects acting across years, the micro-environment of the nest was found to be an important determinant of early-life RTL. Indeed, early development is a period characterized by rapid telomere shortening [59], and faster telomere shortening early in life has been demonstrated in the wild, e.g. in jackdaws (Corvus monedula) [8]. Similarly, Andrew et al. [61] showed that 49% of the variation in telomere length could be attributed to (environmental) family effects in a human twin study. This suggests that the main parameters that determine nest microclimate, temperature, humidity and gas composition, may modulate telomere shortening. An additional factor contributing to this large nest effect is parental quality. High-quality parental investment may buffer stressful events during early development and preserve telomeres from adverse stress-related weakening, causing significant nest effects.
Although it only reached statistical significance in the animal model including maternal RTL, we found RTL to be positively related to an individual's inbreeding coefficient. This positive relationship is surprising, as longer telomeres have been associated with increased survival rate and lifespan [4,5], whereas survival and lifespan are typically lower in inbred individuals (for review, see [62]). Interestingly, however, it is in line with findings in laboratory mouse strains [63] and domesticated chicken lines [64]. Furthermore, knowledge about the relationship between telomere length and fitness-related traits such as fecundity is still limited (but see [9]), and the longer telomeres of inbred nestlings may be a by-product of inbreeding effects on factors associated with telomere elongation. Further work focusing on the interplay between telomere length, inbreeding and fitness is warranted.
In summary, we here demonstrate that environmental factors such as the nest and the year of birth are the main drivers influencing early-life RTL in a wild bird population. Furthermore, despite mother–offspring resemblance in early-life RTL, its heritability is very low. These results thereby highlight the importance of including factors of a common environment into analyses that intend to separate genetic and environmental effects on phenotypic variation. On the whole, our study highlights the large amount of interspecific variation in telomere inheritance patterns, and further research should aim to elucidate the underlying causes of this variation.
Acknowledgements
We thank Glauco Camenisch and Thomas Bucher for help in the laboratory, and Pirmin Nietlisbach, Pierre Bize and three anonymous reviewers for helpful comments on previous versions of this manuscript.
Data accessibility
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.b2v37
Funding statement
This work was supported by the CNRS and the Conseil Régional of Alsace.
Authors' contributions
S.R., P.J.J.B., F.C., S.M. and L.F.K. planned and designed the study. J.H. and P.J.J.B. collected samples, S.Z. measured telomere length, P.J.J.B. and E.P. did the statistical analysis, and P.J.J.B., S.R., E.P., F.C., S.M. and L.F.K. interpreted the results and wrote the manuscript.
References
- 1.Blackburn EH. 2000. Telomere states and cell fates. Nature 408, 53–56. ( 10.1038/35040500) [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. 2001. Switching and signaling at the telomere. Cell 106, 661–673. ( 10.1016/S0092-8674(01)00492-5) [DOI] [PubMed] [Google Scholar]
- 3.von Zglinicki T. 2000. Role of oxidative stress in telomere length regulation and replicative senescence. Ann. N. Y. Acad. Sci. 908, 99–110. ( 10.1111/j.1749-6632.2000.tb06639.x) [DOI] [PubMed] [Google Scholar]
- 4.Heidinger BJ, Blount JD, Boner W, Griffiths K, Metcalfe NB, Monaghan P. 2012. Telomere length in early life predicts lifespan. Proc. Natl Acad. Sci. USA 109, 1743–1748. ( 10.1073/pnas.1113306109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bize P, Criscuolo F, Metcalfe NB, Nasir L, Monaghan P. 2009. Telomere dynamics rather than age predict life expectancy in the wild. Proc. R. Soc. B 276, 1679–1683. ( 10.1098/rspb.2008.1817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA. 2003. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361, 393–395. ( 10.1016/S0140-6736(03)12384-7) [DOI] [PubMed] [Google Scholar]
- 7.Haussmann MF, Winkler DW, Vleck CM. 2005. Longer telomeres associated with higher survival in birds. Biol. Lett. 1, 212–214. ( 10.1098/rsbl.2005.0301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salomons HM, Mulder GA, van de Zande L, Haussmann MF, Linskens MHK, Verhulst S. 2009. Telomere shortening and survival in free-living corvids. Proc. R. Soc. B 276, 3157–3165. ( 10.1098/rspb.2009.0517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauch C, Becker PH, Verhulst S. 2013. Telomere length reflects phenotypic quality and costs of reproduction in a long-lived seabird. Proc. R. Soc. B 280, 20122540 ( 10.1098/rspb.2012.2540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes N, Shay JW, Wright WE. 2010. Telomere biology in Metazoa. FEBS Lett. 584, 3741–3751. ( 10.1016/j.febslet.2010.07.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graakjaer J, Pascoe L, Der-Sarkissian H, Thomas G, Kolvraa S, Christensen K, Londoño-Vallejo JA. 2004. The relative lengths of individual telomeres are defined in the zygote and strictly maintained during life. Aging Cell 3, 97–102. ( 10.1111/j.1474-9728.2004.00093.x) [DOI] [PubMed] [Google Scholar]
- 12.Slagboom PE, Droog S, Boomsma DI. 1994. Genetic determination of telomere size in humans: a twin study of three age groups. Am. J. Hum. Genet. 55, 876. [PMC free article] [PubMed] [Google Scholar]
- 13.Broer L, et al. 2013. Meta-analysis of telomere length in 19713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. 21, 1163–1168. ( 10.1038/ejhg.2012.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Njajou OT, et al. 2007. Telomere length is paternally inherited and is associated with parental lifespan. Proc. Natl Acad. Sci. USA 104, 12 135–12 139. ( 10.1073/pnas.0702703104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asghar M, Bensch S, Tarka M, Hansson B, Hasselquist D. 2015. Maternal and genetic factors determine early life telomere length. Proc. R. Soc. B 282, 20142263 ( 10.1098/rspb.2014.2263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn T, Robertson BC, Will M, Eason DK, Elliott GP, Gemmell NJ. 2011. Inheritance of telomere length in a bird. PLoS ONE 6, e17199 ( 10.1371/journal.pone.0017199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsson M, Pauliny A, Wapstra E, Uller T, Schwartz T, Blomqvist D. 2011. Sex differences in sand lizard telomere inheritance: paternal epigenetic effects increases telomere heritability and offspring survival. PLoS ONE 6, e17473 ( 10.1371/journal.pone.0017473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichert S, Rojas ER, Zahn S, Robin JP, Criscuolo F, Massemin S. 2015. Maternal telomere length inheritance in the king penguin. Heredity 114, 10–16. ( 10.1038/hdy.2014.60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voillemot M, Hine K, Zahn S, Criscuolo F, Gustafsson L, Doligez B, Bize P. 2012. Effects of brood size manipulation and common origin on phenotype and telomere length in nestling collared flycatchers. BMC Ecol. 12, 17 ( 10.1186/1472-6785-12-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atema E, Mulder E, Dugdale HL, Briga M, Van Noordwijk AJ, Verhulst S. In press Heritability of telomere length in the zebra finch. J. Ornithol. ( 10.1007/s10336-015-1212-7) [DOI] [Google Scholar]
- 21.Price T, Schluter D. 1991. On the low heritability of life-history traits. Evolution 45, 853–861. ( 10.2307/2409693) [DOI] [PubMed] [Google Scholar]
- 22.Postma E. 2014. Four decades of estimating heritabilities in wild vertebrate populations: improved methods, more data, better estimates. In Quantitative genetics in the wild, (eds A Charmantier, D Garant, LEB Kruuk), pp. 16–33. Oxford, UK: Oxford University Press. [Google Scholar]
- 23.Weigensberg I, Roff DA. 1996. Natural heritabilities: can they be reliably estimated in the laboratory? Evolution 50, 2149–2157. ( 10.2307/2410686) [DOI] [PubMed] [Google Scholar]
- 24.Kruuk LEB, Hadfield JD. 2007. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 20, 1890–1903. ( 10.1111/j.1420-9101.2007.01377.x) [DOI] [PubMed] [Google Scholar]
- 25.Geiger S, Le Vaillant M, Lebard T, Reichert S, Stier A, Le Maho Y, Criscuolo F. 2012. Catching-up but telomere loss: half-opening the black box of growth and ageing trade-off in wild king penguin chicks. Mol. Ecol. 21, 1500–1510. ( 10.1111/j.1365-294X.2011.05331.x) [DOI] [PubMed] [Google Scholar]
- 26.Jennings B, Ozanne S, Dorling M, Hales C. 1999. Early growth determines longevity in male rats and may be related to telomere shortening in the kidney. FEBS Lett. 448, 4–8. ( 10.1016/S0014-5793(99)00336-1) [DOI] [PubMed] [Google Scholar]
- 27.Tarry-Adkins J, Martin-Gronert M, Chen J-H, Cripps R, Ozanne S. 2008. Maternal diet influences DNA damage, aortic telomere length, oxidative stress, and antioxidant defense capacity in rats. FASEB J. 22, 2037–2044. ( 10.1096/fj.07-099523) [DOI] [PubMed] [Google Scholar]
- 28.Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM. 2004. Accelerated telomere shortening in response to life stress. Proc. Natl Acad. Sci. USA 101, 17 312–17 315. ( 10.1073/pnas.0407162101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monaghan P, Haussmann MF. 2006. Do telomere dynamics link lifestyle and lifespan? Trends Ecol. Evol. 21, 47–53. ( 10.1016/j.tree.2005.11.007) [DOI] [PubMed] [Google Scholar]
- 30.Haussmann MF, Longenecker AS, Marchetto NM, Juliano SA, Bowden RM. 2012. Embryonic exposure to corticosterone modifies the juvenile stress response, oxidative stress and telomere length. Proc. R. Soc. B 279, 1447–1456. ( 10.1098/rspb.2011.1913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nawrot TS, Staessen JA, Gardner JP, Aviv A. 2004. Telomere length and possible link to X chromosome. Lancet 363, 507–510. ( 10.1016/S0140-6736(04)15535-9) [DOI] [PubMed] [Google Scholar]
- 32.Nordfjäll K, Larefalk Å, Lindgren P, Holmberg D, Roos G. 2005. Telomere length and heredity: indications of paternal inheritance. Proc. Natl Acad. Sci. USA 102, 16 374–16 378. ( 10.1073/pnas.0501724102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Øigarden T, Borge T, Lifjeld JT. 2010. Extrapair paternity and genetic diversity: the white-throated dipper Cinclus cinclus. J. Avian Biol. 41, 248–257. ( 10.1111/j.1600-048X.2009.04847.x) [DOI] [Google Scholar]
- 34.Wright S. 1969. Evolution and the genetics of populations: vol. 2. The theory of gene frequencies. Chicago, IL: University of Chicago Press. [Google Scholar]
- 35.Morrissey MB, Wilson AJ. 2010. pedantics: an R package for pedigree-based genetic simulation and pedigree manipulation, characterization and viewing. Mol. Ecol. Resour. 10, 711–719. ( 10.1111/j.1755-0998.2009.02817.x) [DOI] [PubMed] [Google Scholar]
- 36.Arctander P. 1988. Comparative studies of avian DNA by restriction fragment length polymorphism analysis: convenient procedures based on blood samples from live birds. J. Ornithol. 129, 205–216. ( 10.1007/BF01647289) [DOI] [Google Scholar]
- 37.Bucher T, Wandeler P, Hegelbach J, Keller L. 2009. Development of microsatellite loci in the European dipper, Cinclus cinclus. Conserv. Genet. Resour. 1, 309–312. ( 10.1007/s12686-009-9071-2) [DOI] [Google Scholar]
- 38.Griffiths R, Double MC, Orr K, Dawson RJ. 1998. A DNA test to sex most birds. Mol. Ecol. 7, 1071–1075. ( 10.1046/j.1365-294x.1998.00389.x) [DOI] [PubMed] [Google Scholar]
- 39.Hoeck P, Bucher T, Wandeler P, Keller L. 2009. Microsatellite primers for the four Galápagos mockingbird species (Mimus parvulus, Mimus macdonaldi, Mimus melanotis and Mimus trifasciatus). Mol. Ecol. Resour. 9, 1538–1541. ( 10.1111/j.1755-0998.2009.02704.x) [DOI] [PubMed] [Google Scholar]
- 40.Cawthon RM. 2002. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 ( 10.1093/nar/30.10.e47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Criscuolo F, Bize P, Nasir L, Metcalfe NB, Foote CG, Griffiths K, Gault EA, Monaghan P. 2009. Real-time quantitative PCR assay for measurement of avian telomeres. J. Avian Biol. 40, 342–347. ( 10.1111/j.1600-048X.2008.04623.x) [DOI] [Google Scholar]
- 42.Nussey DH, et al. 2014. Measuring telomere length and telomere dynamics in evolutionary biology and ecology. Methods Ecol. Evol. 5, 299–310. ( 10.1111/2041-210X.12161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith S, Turbill C, Penn D. 2011. Chasing telomeres, not red herrings, in evolutionary ecology. Heredity 107, 372–373. ( 10.1038/hdy.2011.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruuk LEB. 2004. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. B 359, 873–890. ( 10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilmour AR, Gogel B, Cullis B, Thompson R. 2009. ASReml user guide release 3.0. Hemel Hempstead, UK: VSN International Ltd. [Google Scholar]
- 47.Meyer K. 2007. WOMBAT: a tool for mixed model analyses in quantitative genetics by restricted maximum likelihood (REML). J. Zhejiang Univ. Sci. B 8, 815–821. ( 10.1631/jzus.2007.B0815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Boer I, van Arendonk J. 1992. Prediction of additive and dominance effects in selected or unselected populations with inbreeding. Theor. Appl. Genet. 84, 451–459. ( 10.1007/BF00229506) [DOI] [PubMed] [Google Scholar]
- 49.Hoeschele I, van Raden P. 1991. Rapid inversion of dominance relationship matrices for noninbred populations by including sire by dam subclass effects. J. Dairy Sci. 74, 557–569. ( 10.3168/jds.S0022-0302(91)78203-9) [DOI] [PubMed] [Google Scholar]
- 50.Willham R. 1972. The role of maternal effects in animal breeding: III. Biometrical aspects of maternal effects in animals. J. Anim. Sci. 35, 1288–1293. [DOI] [PubMed] [Google Scholar]
- 51.Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 52.Husby A, Schielzeth H, Forstmeier W, Gustafsson L, Qvarnström A. 2012. Sex chromosome linked genetic variance and the evolution of sexual dimorphism of quantitative traits. Evolution 67, 609–619. ( 10.1111/j.1558-5646.2012.01806.x) [DOI] [PubMed] [Google Scholar]
- 53.Schaeffer LR, Kennedy BW, Gibson JP. 1989. The inverse of the gametic relationship matrix. J. Dairy Sci. 72, 1266–1272. ( 10.3168/jds.S0022-0302(89)79231-6) [DOI] [Google Scholar]
- 54.Rivero MT, Mosquera A, Goyanes V, Slijepcevic P, Fernandez JL. 2004. Differences in repair profiles of interstitial telomeric sites between normal and DNA double-strand break repair deficient Chinese hamster cells. Exp. Cell Res. 295, 161–172. ( 10.1016/j.yexcr.2003.12.031) [DOI] [PubMed] [Google Scholar]
- 55.Ballen C, Healey M, Wilson M, Tobler M, Olsson M. 2012. Predictors of telomere content in dragon lizards. Naturwissenschaften 99, 661–664. ( 10.1007/s00114-012-0941-1) [DOI] [PubMed] [Google Scholar]
- 56.Haussmann MF, Marchetto NM. 2010. Telomeres: linking stress and survival, ecology and evolution. Curr. Zool. 56, 714–727. [Google Scholar]
- 57.Young RC, Kitaysky AS, Haussmann MF, Descamps S, Orben RA, Elliott KH, Gaston AJ. 2013. Age, sex, and telomere dynamics in a long-lived seabird with male-biased parental care. PLoS ONE 8, e0074931 ( 10.1371/journal.pone.0074931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foote C, Gault E, Nasir L, Monaghan P. 2011. Telomere dynamics in relation to early growth conditions in the wild in the lesser black-backed gull. J. Zool. 283, 203–209. ( 10.1111/j.1469-7998.2010.00774.x) [DOI] [Google Scholar]
- 59.Hall ME, Nasir L, Daunt F, Gault EA, Croxall JP, Wanless S, Monaghan P. 2004. Telomere loss in relation to age and early environment in long-lived birds. Proc. R. Soc. Lond. B 271, 1571–1576. ( 10.1098/rspb.2004.2768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizutani Y, Tomita N, Niizuma Y, Yoda K. 2013. Environmental perturbations influence telomere dynamics in long-lived birds in their natural habitat. Biol. Lett. 9, 20130511 ( 10.1098/rsbl.2013.0511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andrew T, et al. 2006. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am. J. Hum. Genet. 78, 480–486. ( 10.1086/500052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller LF, Waller DM. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241. ( 10.1016/S0169-5347(02)02489-8) [DOI] [Google Scholar]
- 63.Manning EL, Crossland J, Dewey MJ, Van Zant G. 2002. Influences of inbreeding and genetics on telomere length in mice. Mammal. Genome 13, 234–238. ( 10.1007/s003350020027) [DOI] [PubMed] [Google Scholar]
- 64.O'Hare TH, Delany ME. 2009. Genetic variation exists for telomeric array organization within and among the genomes of normal, immortalized, and transformed chicken systems. Chromosome Res. 17, 947–964. ( 10.1007/s10577-009-9082-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.b2v37


