Abstract
Genomes of numerous diploid plant and animal species possess traces of interspecific crosses, and many researches consider them as support for homoploid hybrid speciation (HHS), a process by which a new reproductively isolated species arises through hybridization and combination of parts of the parental genomes, but without an increase in ploidy. However, convincing evidence for a creative role of hybridization in the origin of reproductive isolation between hybrid and parental forms is extremely limited. Here, through studying Agrodiaetus butterflies, we provide proof of a previously unknown mode of HHS based on the formation of post-zygotic reproductive isolation via hybridization of chromosomally divergent parental species and subsequent fixation of a novel combination of chromosome fusions/fissions in hybrid descendants. We show that meiotic segregation, operating in the hybrid lineage, resulted in the formation of a new diploid genome, drastically rearranged in terms of chromosome number. We also demonstrate that during the heterozygous stage of the hybrid species formation, recombination was limited between rearranged chromosomes of different parental origin, representing evidence that the reproductive isolation was a direct consequence of hybridization.
Keywords: hybrid speciation, reproductive isolation, chromosome, meiosis, genomic in situ hybridization, amplified fragment length polymorphism
1. Introduction
Speciation is generally regarded as the splitting of a single ancestral lineage in two daughter species. An alternative is hybrid speciation, in which two species hybridize and give rise to a third independent lineage [1–3]. One possibility for hybrid speciation is allopolyploidy, where genome doubling provides instantaneous reproductive isolation from parental species [1,4,5]. Another possibility is homoploid hybrid speciation (HHS), a process by which a new reproductively isolated, sexually reproducing species arises through hybridization and combination of parts of the parental genomes, but without an increase in ploidy [1,4–6]. HHS is generally considered to be relatively rare and difficult [7–9] because, in contrast to polyploidy, the challenge is for hybrid diploid descendants to escape the dissolving effect of gene flow from backcrosses with the parental species [10]. To overcome this difficulty, some models of HHS assume that hybridization could trigger pre-zygotic reproductive isolation between hybrid and parental lineages through formation of novel recombinant phenotypes. These phenotypes allow hybrid species to colonize niches unavailable to parental species [8–14] and/or reject parental species as potential mates [15–18].
A completely different means of escaping the dissolving effect is provided by a chromosomal version of HHS, which represents a mechanism essentially based on post-zygotic reproductive isolation. In the latter version, two species differentiated by several fixed chromosomal rearrangements (CRs) hybridize, and then, following an initial highly heterozygous stage, a novel homozygous chromosome complement is sorted out in the descendants as a result of chromosome segregation [4,19–21]. Each of the CRs fixed in the hybrid is inherited from one of the progenitors but their combination differs from those in the parental species. The hybrid lineage with the novel karyotype is supposed to be post-zygotically protected from gene exchange with the progenitors by (i) a suppressed-recombination mechanism (if CRs are neutral and do not influence fertility of chromosomal heterozygotes) or/and by (ii) a hybrid-sterility mechanism (if CRs are underdominant, i.e. reduce fertility of chromosomal heterozygotes) [22]. Both mechanisms predict a reduced level of genetic recombination between rearranged parental chromosomes during the heterozygous stage: either by definition (i), or because of limited time that underdominant CRs can exist in a heterozygous condition (ii).
Suppressed-recombination results in preservation of parental chromosomal linkage blocks or even whole chromosomes in hybrids. Alternatively, if the linkage blocks of different parental origin are not protected by CRs, recombination leads to homogenization of the derived hybrid genome. Thus, the preservation of intact chromosomal blocks inherited from both progenitors is a signature for post-zygotic isolation caused by CRs. What is more, lack of recombination footprints indicates that the reproductive isolation between hybrid and parental species not only exists currently, but also existed in the past, immediately after the parts of the parental genomes were combined. Therefore, analysis of recombination footprints can provide insights into the early steps of evolution of reproductive isolation, and this is crucially important for inferring conclusions on the creative role of hybridization in HHS.
Revealing cases of chromosomal HHS is difficult as it requires not only demonstration that a hybrid species has a mosaic of the two parental species' genomes but also evidence of a novel karyotype originated from preserved chromosomes of both parental species. Such cases have so far only been reported for hybrid sunflowers where preservation of some parental chromosomal linkage blocks was revealed [1,5,10]. In animals, the genomes of hybrid diploid origin have received little attention as yet, so the factors determining their composition and origin are studied to a lesser extent (but see [23,24]).
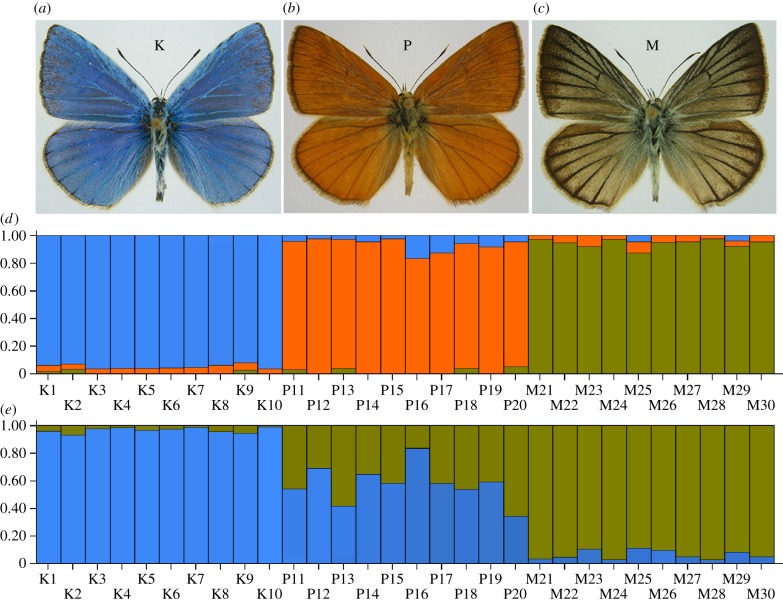
We studied HHS in the blue butterflies of the genus Agrodiaetus (Lepidoptera, Lycaenidae). This genus exhibits a wide diversity of karyotypes, with haploid chromosome numbers (n) of different species ranging from 10 to 134 [25]. This range arose through fusion and fission of chromosomes and is not a consequence of polyploidy [26,27]. The females of Agrodiaetus are brown, whereas the males show considerable variability in wing colour [25]. The colour differences between males are strongly expressed in a group of three closely related sympatric taxa studied here: Agrodiaetus karindus (further in the text abbreviated as K, blue), A. peilei (P, orange) and A. morgani (M, greenish-silver) (figure 1a–c). The opinions on status and identity of these taxa are controversial [28,29]. They are bisexual and true-breeding (not parthenogenetic) taxa [29]. They are endemics of the Zagros Mountains in Iran and were found by Schurian et al. [29] and by us (electronic supplementary material, ESM1) to share identical habitats and periods of flight.
Figure 1.
Male wing colours in K (a), P (b) and M (c) and results of the Bayesian assignment analysis of AFLP markers (d,e). Bar plots showing Bayesian assignment probabilities under assumption of three (d) and two (e) clusters are shown. Each vertical bar corresponds to one individual. The proportion of each bar that is blue, orange and green represents an individual's assignment probability to clusters one, two and three respectively.
The aims of this study were:
(1) using analysis of chromosomal and multiple nuclear molecular markers to demonstrate that M, K and P are distinct, sympatric, reproductively isolated species;
(2) to show that K and M have hybridized in the past; and
(3) to obtain evidence from amplified fragment length polymorphism (AFLP) analysis and genomic in situ hybridization (GISH) experiments that crosses between M and K, differentiated by multiple chromosome fusions/fissions, resulted in a novel fertile species P through the assortment of the parental species' chromosomes.
2. Material and methods
(a). Sample collecting
Adult males (electronic supplementary material, ESM1) were collected with a butterfly net. Testes were removed from the abdomen and placed in 0.5 ml glass vials with fixative (ethanol and glacial acetic acid, 3 : 1) for chromosome analysis. Bodies were placed in 2 ml plastic vials with 100% ethanol for DNA analysis. Wings were stored in glassine envelopes for morphological study. All samples are stored at Zoological Institute, St Petersburg, Russia.
(b). DNA sequencing, alignment, codification, phylogenetic inference and haplotype network
DNA isolation, PCR amplification and sequencing were described previously [25,27,30,31]. COI sequences were aligned using BioEdit software [32]. This resulted in final alignments of 648 bp and 159 specimens. Bayesian phylogenetic trees of COI haplotypes were inferred using beast v. 1.7.1 [33], with the nucleotide substitution model GTR + G + I, as suggested by jModeltest 0.1 [34]. A maximum-parsimony haplotype network was built using tcs v. 1.21 [35] with a 99% parsimony connection limit.
ITS2 sequences were aligned according to secondary structure using the template structure of Neolysandra coelestina (MW99013) [36]. In ITS2, heterogeneous nucleotide positions were identified through dual/multiple peaks present in electropherograms and coded as ‘N’. In evolution of ITS2 sequences, the mono, bi- and mullti-nucleotide insertions/deletions are frequent and contain phylogenetically important information. To account for this, each indel event was coded as a binary character (1/0, presence/absence of the gap independently of its length). Heterogeneous indel positions were identified by a sharp transition in the electropherogram from a clean to garbled sequence, where the transition corresponded to the same position of a homozygous indel in other individuals; they were coded as ‘N’. We cloned ITS2 for five specimens for which we were unable to obtain clear phylogenetic information by using standard sequencing (see electronic supplementary material, ESM2, ESM3 and ESM4). The obtained alignment (499 bp and 49 specimens) and its codification are presented in electronic supplementary material, ESM3 and ESM4. For ITS2, Bayesian trees were inferred using partitioned models: GTR for nucleotide substitutions and standard model for indels as implemented in MrBayes v. 3.2 [37].
(c). Amplified fragment length polymorphism analysis
We used the AFLP Plant Mapping Kit (Applied Biosystems) as previously reported for other butterflies [38]. The manufacturer's protocols were followed, with the following modification: pre-selective amplifications were performed with 25 rather than 20 cycles. We used nine combinations of primers for selective amplification: EcoRI-ACT/MseI-CAT, EcoRI-ACT/MseI-CTC, EcoRI-AAC/MseI-CAC, EcoRI-AGC/MseI-CAC, EcoRI-AGC/MseI-CAT, EcoRI-AGC/MseI-CTT, EcoRI-AAG/MseI-CAA, EcoRI-ACG/MseI-CTC, EcoRI-ACG/MseI-CTT. The forward primer (EcoRI- + 3) from each primer pair was labelled with six FAM fluorophore. The fragments were analysed on a 3500xL analyser (Applied Biosystems). The raw data were visualized, aligned with the internal size standard Gene Scan™ 600 LIZ®, manually controlled and analysed in GeneMapper v. 2.4.2 (Life Technologies). Only fragments between 70 and 500 bp were considered. Thresholds of 100 rfu (relative fluorescence unit) were set to remove instrument noise. The mismatch error rates were scored using aflpscore v. 1.3b software [39] (see details in electronic supplementary material, ESM5). The Bayesian assignment analysis [40] of AFLP markers was performed using structure v. 2.3.4 with a burn-in of 50 000 generations and a Markov chain of 500 000 generations.
(d). Estimation of time to the most recent common ancestor
The time to the most recent common ancestor (TMRCA) of the COI haplotype diversity in P and the clade H1 as a whole was inferred with beast v. 1.7.1 [33] under a coalescent model with constant population size. A dataset consisting of 15 haplotypes (haplogroups H1-H5) was used for the analysis. A lognormal distribution (mean = 0.07, s.d. = 0.91) was used assuming a maximum possible limit of 207 000 years as the 95% HPD of the distribution, trying to allow the maximum exploratory space for MCMC runs. To estimate this prior, we used the maximum COI divergence haplogroup H1 (0.31%) under a rather slow invertebrate mitochondrial substitution rate: 1.5% uncorrected pairwise distance per million years [41]. Since substitution rates are known to overestimate ages for recent lineages still under the coalescence process, we are certain that 207 000 years is a good maximum estimate for the TMRCA of P and the clade H1. The dataset was analysed using the TrN + G model, and a strict molecular clock was applied along the branches. Base frequencies were estimated, six γ rate categories were selected, and a randomly generated initial tree was used. Parameters were estimated using two independent runs of 10 million generations each (with a pre-run burn-in of 100 000 generations) to ensure convergence, checked with the program Tracer v. 1.5 [42]. TMRCA of the COI haplotype diversity in the clearly coalesced node K + M + P was estimated using the whole COI dataset (30 haplotypes), GTR + I + G model of nucleotide substitution, beast v. 1.7.1 software and calibration method as described previously [43].
(e). Statistical approach for distinguishing hybridization and incomplete lineage sorting
Incomplete lineage sorting is a phenomenon when ancestral polymorphisms present in the parental species have not been completely sorted out by genetic drift in the daughter species, resulting in non-monophyletic taxa [44–46]. We applied a parametric statistical approach for distinguishing hybridization from incomplete lineage sorting as implemented in the software jml [47]. The program calculates the minimum distance between sequences of two species and tests whether it is smaller than expected under a simulated scenario that includes incomplete lineage sorting.
First, we performed species-tree analyses for the complete set of COI haplotypes found in K, M and P using the multi-species coalescent methods implemented in *beast [48]. Haplotypes of A. carmon (C) and A. biruni (B) were used as closely and distantly related outgroups, respectively. Thus, the haplotypes were attributed to five different species (K, M, P, C and B) in this analysis. GTR + I + G model of nucleotide substitution was selected as suggested by jModeltest 0.1 [34]. *beast v. 1.7.1 with a strict clock and a linear piecewise demographic model was set for a Markov chain Monte Carlo of 10 million generations sampled every 1000 runs. Two independent runs were performed and convergence was checked using Tracer v. 1.5 [42]. Then, the species-trees from the first million generations were discarded as burn-in in JML, and the remaining 9000 trees were used as input for jml for the simulations and p-value estimations for statistical evidence for hybridization events. The information for the seq-gen command of the jml control file was extracted from jModeltest 0.1 output including nucleotide frequencies, proportion of invariant sites and γ-shape parameter. The relative mutation rate mean of the species-tree posterior distribution was used.
(f). Karyotype analyses
Chromosome preparations were obtained as previously described [25,27,30,31,43], and meiotic karyotypes were studied. The uniformity of lepidopteran chromosomes, the absence of morphological markers such as the localized centromeres and the lack of convenient differential banding techniques make the identification of individual chromosomes difficult [26,49]. Nevertheless, the chromosomes of different size classes (‘large’ and ‘small’) can be relatively easily distinguished not only because of differences in size, but also due to specific architecture of lepidopteran metaphase I spermatocytes with large bivalents located in the centre and small bivalents situated at the periphery [49].
(g). Genomic in situ hybridization and comparative genomic hybridization
Squash preparations of meiosis I metaphase bivalents and meiosis II chromosomes were obtained from unstained testes of adults (electronic supplementary material, ESM1). Suitable metaphase plates were selected using phase-contrast prior to removing the cover glass with the dry ice technique [50]. Whole genomic DNA for GISH and comparative genomic hybridization (CGH) probe construction was isolated from adult males using standard phenol/chloroform nucleic acid extraction protocol. Genomic DNA for GISH was labelled by nick translation in the presence of biotin-16-dUTP using the Nick Translation Kit (Roche) according to the manufacturer's instructions. Genomic DNA for CGH was labelled by nick translation in the presence of biotin-16-dUTP (K probe) and rhodamine-5-dUTP (GeneCraft) (M probe) using the Nick Translation Kit (Roche) according to the manufacturer's instructions. The final probe length (300–1500 bp) was verified on 1% agarose gel. Genomic DNA to be used as the competitor DNA was extracted from adult males as described above and sheared in fragments of 100–1000 bp using DNase (final concentration approx. 3 pg μl−1) in buffer from the Nick Translation Kit for 2–2.5 h at RT. The length of competitor DNA was checked on 1% agarose gel. Females were not used for probe and competitor construction since females in Lepidoptera usually have a ZW sex-chromosome pair, whereas males have a ZZ pair, and hybridization patterns (as well as effects of blocking DNA) of female genomic DNA versus male genomic DNA could differ owing to the presence of the W chromosome in females only [51].
Genomic hybridization was carried out following the described procedure [52]. To determine the amount of blocking DNA needed to completely suppress probe signals, labelled DNA and blocking DNA of K and M were hybridized in preliminary experiments over their own metaphases (self-GISH) in a concentration gradient (1 : 10, 1 : 40, 1 : 60, 1 : 100, 1 : 200). Based on these results, the following proportions of DNA amount were selected for further studies: K-derived probe/M-derived competitor is 1/60–65; M-derived probe/K-derived competitor is 1/12–25. Slides were mounted using glass coverslips and rubber cement. The chromosome slides were incubated for 42–44 h at 37°C. Following hybridization, the slides were washed in 2 × SSC for 3 min at 45°C, then in 50% formamide in 2 × SSC for 10 min at 45°C, twice in 2 × SSC (10 min each) at 45°C, blocked in 1.5% (w/v) BSA/4 × SSC/0.1% Tween-20 for 30 min at 37°C in a humid chamber. Probe was detected with 5 μg ml−1 avidin–Alexa Fluor 488 (Invitrogen). Detection reaction was performed in 1.5% BSA/4 × SSC/0.1% Tween-20 for 1 h at 37°C. Slides were washed three times in 4 × SSC/0.02% Tween-20 (10 min each) at 45° and dehydrated through 70/80/96% ethanol at RT. Chromosome preparations were mounted in a mounting-antifade (ProLong Gold antifade reagent with DAPI, Invitrogen) and covered with glass coverslips. Fluorescence images were taken and analysed with a Leica DM 6000B microscope and Leica DFC 345 FX camera, using Leica Application Suite v. 3.7 software with an Image Overlay module.
3. Results
(a). M, K and P are three distinct sympatric species
We used the approaches based on analysis of genealogical concordance [45] and genotypic clusters [2], and study of chromosomal, nuclear ITS2 and AFLP molecular markers to infer conclusions on the status of K, M and P. According to these approaches, an array of diverse datasets obtained from the taxa under question is used to support or refute the alternative hypotheses of a single or of multiple species within a sample of organisms. The more concordant the various data for each species under question are, and the more multimodal the sample is phenotypically and genotypically in zone of potential or actual hybridization, the more likely a decision of separate species status will be reached [53].
The AFLP analysis of 30 individuals (10K + 10M + 10P) collected in the sympatry zone in Saqqez revealed 1079 bands. A total of 357 markers were fixed, i.e. invariable within each species; 113 markers were non-specific, i.e. were found in all three species and the remaining 244 markers were variable, i.e. specific for one of the taxa studied (K, M or P) or for a combination of two taxa (K + M, K + P or M + P). The Bayesian assignment analysis of AFLP data was performed [42], and three clusters were assumed to determine if K, M and P are three genetically differentiated units. Under this assumption, K, M and P individuals were assigned to their three respective distinct clusters with high probability (figure 1d; electronic supplementary material, table S5.2). In support of monophyly of these three clusters, K possessed 61, M possessed 46 and P possessed five unique fixed AFLP markers.
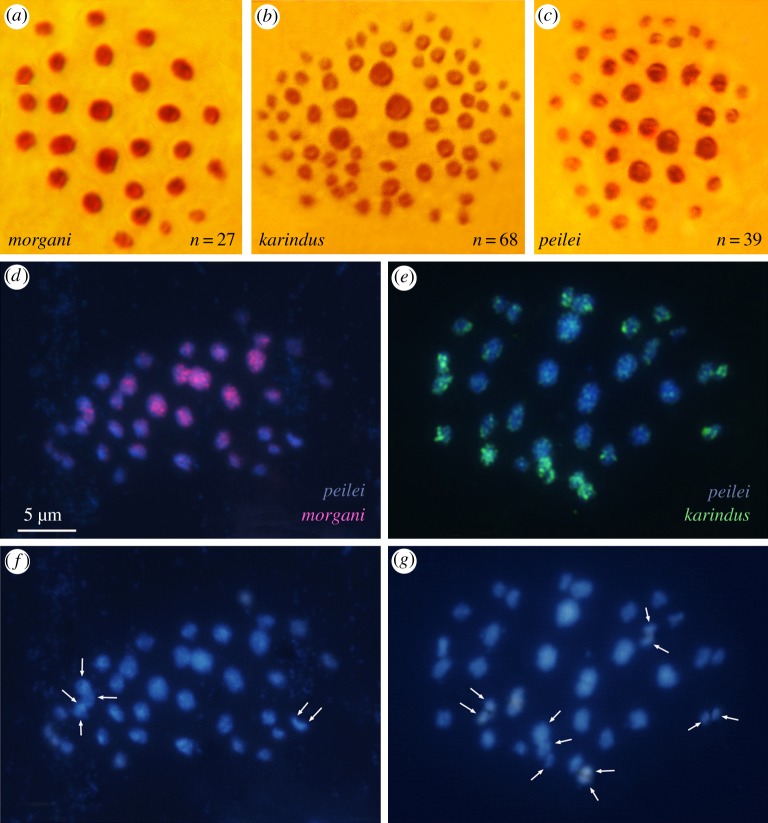
Cytogenetic study of 442 individuals of K, M and P revealed pronounced differences in chromosome number and karyotype structure between these species (figures 2 and 3) and limited within-species variations (electronic supplementary material, ESM1). M had 27 meiotic bivalents in populations from Saqqez, Takab, Zanjan and Chenareh, and 25 bivalents in populations from southeast and north Senandaj (figure 3c); most of these bivalents were large (figure 2a). Few specimens from Chenereh and north Senandaj displayed 26 units (figure 3c). K had 68 bivalents in populations from northwest Iran, and 73 bivalents in populations from central Iran; of these bivalents, five were large and always located in the centre of metaphase plate, and the remaining bivalents were smaller (figures 2b and 3c). P had an intermediate value of chromosome number (mostly 38 or 39 bivalents in meiosis I) and unusual karyotype structure. Whereas in the majority of Agrodiaetus species, all the bivalents have similar sizes or form a gradient size row [49], the bivalents of P could be classified in two groups: one group consisting of 10–11 large bivalents similar in size to the chromosomes of M, and the other group consisting of 27–28 small bivalents similar in size to the majority of bivalents in K (figure 2c). As was found in other Agrodiaetus species [49], the larger bivalents of the first group were always situated in the centre of metaphase plate, whereas the smaller bivalents of the second group were permanently located at the periphery.
Figure 2.
Meiosis I metaphase plates of M, K and P and results of GISH. (a–c) Metaphase plates of M (a), K (b) and P (c). (d) GISH image of M-derived labelled total genomic probe (red signals) hybridized to meiotic chromosomes of P counterstained with DAPI (blue). (e) GISH image of K-derived total genomic probe (green signals) hybridized to meiotic chromosomes of P counterstained with DAPI (blue). (f,g) DAPI images of the meiotic karyotype of P (n = 38). The overlapping bivalents are shown by arrows.
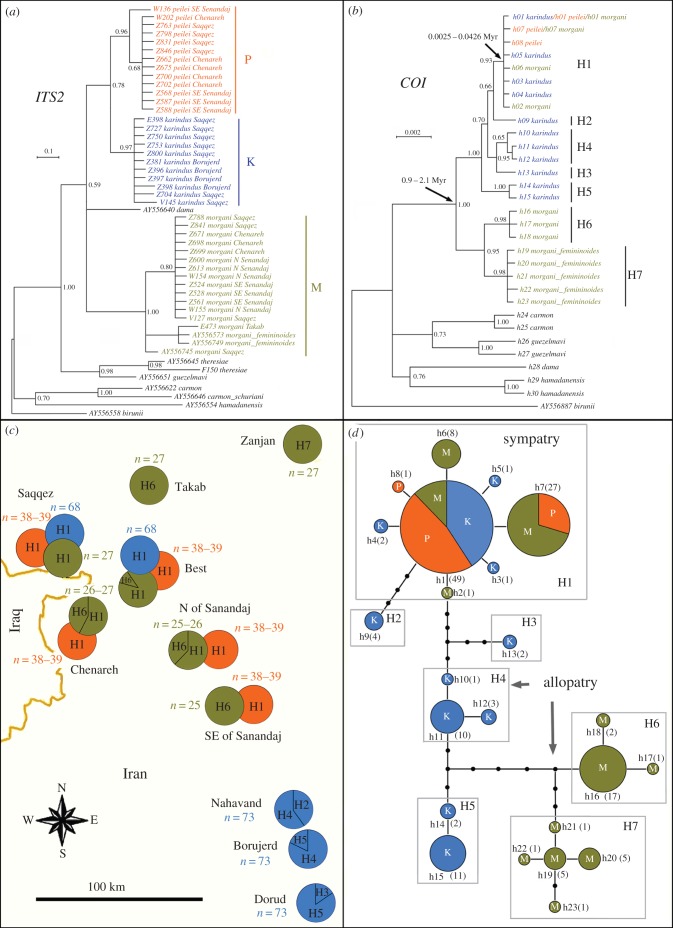
Figure 3.
Bayesian trees for ITS2 and COI and haplotype analysis. (a) Bayesian tree for ITS2. Posterior probabilities (>50%) are shown for each node. (b) Bayesian tree for COI. h1–h30 are COI haplotypes; H1–H7 are haplogroups. Posterior probabilities (>50%) are shown for each node; error intervals covering 95% highest posterior density for age estimation are shown near the arrows. (c) Distribution of haplogroups H1–H7 and haploid chromosome numbers (n) of K (blue), M (greenish) and P (orange) across the Zagros Mountains; overlapping circles indicate zones of sympatry. (d) Most parsimonious COI haplotype network; numbers of studied individuals are given in parentheses.
Analysis of the nuclear molecular marker ITS2 (49 specimens, 50 variable and 20 parsimony informative positions, 13 binary characters) revealed K, M and P as three distinct, but closely related clusters of individuals (figure 3a).
Comparison of the studied AFLP, chromosomal and ITS2 markers revealed a complete concordance between them and wing colours (electronic supplementary material, ESM1). Genotypic cluster analysis [2] of the K + M + P samples from zones of their sympatry demonstrated clear trimodal distribution in wing colours, karyotypes, ITS2 markers and AFLP clusters with no intermediates (electronic supplementary material, ESM1). Thus, both approaches provided strong evidence that K, M and P are three different sympatric species, not simply colour morphs of one species.
(b). Lack of ongoing continuous gene flow between K, M and P
To assess the probability that gene flow occurs between K, M and P, we used cytogenetic analysis of 442 individuals (of which 154 were determined as K, 127 as M and 161 as P according to their wing colours) collected exactly in the localities where K, M and P were sympatric (electronic supplementary material, ESM1). Chromosomal markers represent a powerful tool for detecting ongoing hybridization because they allow a simple discrimination of chromosomally heterozygous F1s and backcrosses from chromosomally homozygous parental species. If parental races are differentiated by multiple fixed chromosome fusions/fissions (and this is exactly our case!), the meiotic karyotype in hybrids will include multiple multivalents that can be easily identified on chromosome preparations. In our cytogenetic analysis, all the individuals were found to be chromosomally homozygous. No individuals were found that might be considered F1s produced from crosses between K, M and P or backcrosses. Thus, we conclude that these taxa are not exchanging genes continuously. Although rare episodes of sporadic hybridization cannot be excluded, the reproductive isolation is strong enough to prevent K, M and P from blending together in places where no temporal and spatial barriers exist between them.
(c). Evidence for mitochondrial introgression in the past
Analysis of the mitochondrial marker COI showed that K and M had complicated genetic structure, each species consisting of several haplotypes clustered in seven divergent haplogroups (figure 3b), with one or two different haplogroups found within each population (figure 3c,d). This pattern most likely reflects a long and complex phylogeographic history of K and M, which could include cases of divergence in allopatry and secondary contact of differentiated populations. Unlike K and M, P was genetically homogenous with respect to COI. All the haplotypes found in P constituted a subset of the haplogroup H1, which also includes specimens of K and M (figure 3b,d). Coalescence-based dating estimated that the TMRCA for the mitochondrial sequence diversity in P was only 2100–36 000 years.
Interestingly, despite pronounced morphological, chromosomal and nuclear DNA differences, and despite the strong COI divergence between allopatric populations of K and M, these species share the same (h1) or very similar (h2–h7) haplotypes in areas of sympatry (figure 3d). This identity can theoretically be explained by interspecific hybridization or alternatively by incomplete lineage sorting [44–46]. In our case, the incomplete lineage sorting scenario seems to be improbable because the lineage K + M + P is old (TMRCA for the mitochondrial sequence diversity in K + M + P was 0.9–2.1 Myr), whereas the clade H1 is very young (TMRCA for the mitochondrial sequence diversity in H1 was 0.0025–0.0426 Myr; figure 3b). Even if the lowest described rate of COI evolution is accepted (1.3% per 1 Myr [41]), the independently evolving descendants of the common ancestor of K + M + P are expected to have accumulated from 7 to 17 nucleotide substitutions in the studied fragment of COI, but not 0–1 as observed in the sympatric populations.
Additionally, the incomplete lineage sorting predicts that differentiated and non-differentiated haplotypes of K and M should be stochastically (i.e. randomly) distributed within different populations. In our case, this distribution was extremely non-random with all the non-differentiated or weakly differentiated haplotypes of K and M concentrated in the zone of their sympatry. The deviation from a random distribution was highly significant (p < 0.001 based on χ2-test; electronic supplementary material, ESM6).
The statistical test for hybridization versus incomplete lineage sorting [44,47] also revealed low probability (p = 0.039) that incomplete lineage sorting could account for the absence of divergence between h1 haplotypes of K and M (electronic supplementary material, ESM7). We conclude therefore that hybridization between K and M that occurred in the past is the most likely explanation for the mitochondrial DNA similarity discovered. Possible phylogeographic scenarios of this process are discussed in electronic supplementary material, ESM8.
(d). Amplified fragment length polymorphism analysis detected a mixed genome in P
The shared mitochondrial haplotypes, unusual colour and intermediate karyotype structure in P indicated a possible hybrid origin of P from K and M. If P is a hybrid species it should possess a genome that is a blend of alleles derived from both K and M. We tested this using a large multilocus genomic dataset, consisting of 1079 AFLP markers, and the Bayesian assignment analysis [12,40] under the assumption that the data represented two separate entities. Individuals from K and M clustered to different groups with high probability, while P was assigned to both groups with moderate probability (figure 1e; electronic supplementary material, table S5.2). This pattern is inconsistent with a bifurcating mode of speciation, where P originated from a single parental species, and suggests that the P genome is a mosaic of the two species. In further support of this hypothesis, 11 AFLP fragments were shared between K and M to the exclusion of P, while P shared 62 unique alleles with K and 59 unique alleles with M. Parsimonious analysis of these data (electronic supplementary material, ESM9) demonstrated that all the bifurcating scenarios of speciation required assumption about multiple homoplasies involved in evolution of AFLP alleles, whereas the scenario of hybrid speciation does not require homoplasies. Thus, the bifurcating scenarios are highly non-parsimonious ones.
(e). Genomic in situ hybridization experiments revealed a hybrid origin and mechanisms of genome evolution in P
Completely independent and even more convincing evidence for a hybrid origin of P from K and M was obtained using GISH experiments. GISH is a molecular cytogenetic technique in which labelled total DNA of parental species is hybridized to the chromosome preparation of a hybrid, enabling highly effective detection of parental chromosomes and/or parental chromosome blocks in the hybrid karyotypes [54]. The great advantage of GISH is the fact it enables direct physical mapping of multiple species-specific DNA sequences on chromosomes and detection of recombination events [55,56], thus representing the most adequate tool for analysis of recombination footprints in hybrid karyotypes. GISH also allows us to discriminate between ‘normal’ divergent, hybrid and introgression models of genome formation even without identification of individual chromosomes (see electronic supplementary material, ESM10).
According to the elaborated experimental design (electronic supplementary material, ESM10), the chromosome preparations of P were used in hybridization with both K-probe/M-competitor and M-probe/K-competitor probe cocktails. In total, chromosome preparations of 20 specimens of P were analysed in two completely independent research trials. In both trials, numerous metaphase I and metaphase II plates and diakinetic nuclei demonstrated the following two highly reproducible features. First, in all cases the strong hybridization signals appeared in both K-probe/M-competitor and M-probe/K-competitor treatments indicating a hybrid origin of the P genome (see electronic supplementary material, ESM11 for more details). Second, the distribution of the signals was very different in these treatments. When the labelled total molecular probe from M hybridized with chromosome preparations of P in the presence of the K-derived competitor, the great majority of hybridization fluorescent signals appeared on the larger chromosomes of P located in the centre of the metaphase plates (figure 2d,f). When the labelled total molecular probe from K hybridized with chromosome preparations of P in the presence of the M-derived competitor, the hybridization signals appeared only on the small chromosomes of P located at the periphery (figure 2e,g). This result can only be explained by hybrid origin of the genome in P, which inherited the majority of its small-sized chromosomes from K and the majority of its large-sized chromosomes from M, and these chromosomes in P were largely non-recombinant (see electronic supplementary material, ESM11 for more details). This conclusion is highly consistent with the fact that the great majority of chromosomes in K are small, and the majority of chromosomes in M are large.
Since the chromosomes in P were anonymous (without specific markers, except for large or small size) and since the K and M probes were hybridized separately to different slides, we cannot exclude that a few chromosomes could hybridize with both K and M probes. However, the number of such chromosomes should be low (if they were present at all) because the separation of chromosomes of these two size classes is generally easy, especially taking into account the specific architecture of lepidopteran spermatocyte metaphase I with large bivalents located in the centre and small bivalents situated at the periphery [49]. Thus, not only chromosome size but also chromosome position can be taken into account for distinguishing chromosomes of these two size classes.
4. Discussion
Our study revealed P to have a mixed nuclear genome obtained partially from K and partially from M, and a mitochondrial genome obtained from K. This mosaic genome is not a result of recent continuous gene flow with K and M since K, M and P were found as three distinct monophyletic phylogenetic lineages differentiated by multiple fixed AFLP, ITS2 and chromosomal alleles, with no signs of ongoing hybridization between them. These facts in combination with complete sympatry provide evidence that K, M and P are different biological species, not intraspecific colour morphs. Thus, P is a hybrid species, not a swarm of hybrid specimens.
The results of GISH analysis clearly indicate that the karyotype of P consists of the chromosomes inherited from M and K. The chromosome sets of the parental species K (n = 68) and M (n = 27) differ by at least 41 fixed chromosome fusions/fissions. Therefore, after the hybridization, the first generations of the hybrid lineage were represented by heterozygotes for multiple CRs, and each of the large chromosomes from M had to conjugate in meiosis with two or more small chromosomes from K. At present the hybrid species P is diploid and chromosomally homozygous. The GISH data does suggest that in P the majority of DNA in the small chromosomes came from K, and the majority of DNA in the large chromosomes came from P; in other words, these chromosomes are largely non-recombinant. Thus, although the GISH method does not allow exclusion of recombinational exchange of small chromosome segments, recombination obviously did not bring about a notable reshuffling of DNA between homologues inherited from the different parental species. Therefore, in P the formation of the genome was mainly realized through meiotic segregation of the parental species' chromosomes with no or limited contribution of chromosome recombination to this process. This reduced recombination is highly consistent with the prediction of chromosomally based HHS thus providing evidence for post-zygotic isolation between P, K and M caused by the chromosome fusions/fissions that differentiate these species.
Schumer et al. [57] have argued that most claims for HHS have been currently published with insufficient evidence that hybridization played a creative role in the speciation process. These authors also proposed a set of three criteria to distinguish ‘true’ HHS from post-speciation introgression and from the formation of hybrid swarms which are not reproductively isolated from their parents. These criteria are (i) demonstration of a mixed genome in a putative hybrid species, (ii) showing reproductive isolation from parental species, and (iii) demonstration that the isolating mechanisms were derived from hybridization. Many HHS studies (e.g. [8,9,11–13,23]) met the first two of these criteria. However, to our knowledge [57,58], until now only studies in sunflowers [1,5,6] and Heliconius butterflies [15,17] also explicitly met the third criterion. In our opinion, the evidence discovered for HHS in P satisfies all three criteria, too. First, both GISH and AFLP markers provide support for the hybrid genome in P. Second, there is reproductive isolation between P, K and M. Lack of F1 hybrids between K, M and P in zones of their sympatry is indirect evidence for pre-zygotic isolation between them. This concurs with the fact that these sympatric species have clear differences in wing colours, i.e. in the characters that represent important signals preventing interspecific mating in butterflies [15,25]. Even more important, GISH analysis revealed preservation in P of intact chromosomal blocks inherited from K and M, and this is a signature for post-zygotic isolation that was caused by CRs and evolved immediately after the parts of the parental genomes were combined. Third, the footprints found of post-zygotic isolation are difficult to interpret in any other way than a direct consequence of chromosomal differences between P and K, and between P and M, and these differences are the result of hybridization and consequent segregation of the parental species' chromosomes. In other words, our case for hybrid speciation combine data on genome-wide patterns of hybridization revealed by GISH with evidence that hybridization per se was a key element in the subsequent development of barriers to gene flow.
Hybrid speciation via chromosome segregation is the oldest model of HHS elaborated in the early years of genetics [19–21]. Despite this, so far it has had insufficient empirical support. This model was confirmed for plants by few laboratory experiments (see [4] for a review), but evidence from nature has been limited to studies in Helianthus species. Its plausibility for animals was even questioned [4] as selfing, which is expected to favour HHS [59], is not so common in animals as in plants [60]. Here we describe a first case of the post-zygotically based, chromosomal HHS for animals.
Additionally, our results represent a previously undescribed form of HHS based on fixation of multiple chromosome fusions/fissions and a new mechanism of genome evolution leading to a change in the diploid number of chromosomes. Interestingly, HHS is frequently defined as ‘hybrid speciation without change in chromosome number’ [3,7,8,11,12,16,17,57]. Our data indicates that this definition is an oversimplification and not always true. The number of chromosomes reflects the number of linkage groups. It affects the rate of meiotic recombination and is therefore an important parameter in genetics and evolution [61]. Chromosome numbers were formerly thought to evolve gradually through step-by-step accumulation of chromosome fusions and fissions between or within species [62], but our results here show the possibility of massive saltatory change in diploid number through hybridization.
The chromosomal version of HHS has been predicted to be far less frequent than speciation via allopolyploidy since homoploid hybrids are expected to suffer from hybrid breakdown while allopolyploids are not [4]. However, recent findings demonstrate that multiple CRs are often not strongly underdominant [62], but contribute to speciation through a suppressed-recombination mechanism [22]. The data obtained here also give indirect evidence that multiple chromosome fusions and fissions did not block fertility in hybrids between K and M. Therefore, one can expect that chromosomally based HHS is more frequent than was supposed, and our data demonstrate that this speciation model was certainly realized in an animal species.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank R. Angus, A. Barabanov, A. Dantchenko, O. Kosterin, N. Pierce, F. Sperling, G. Talavera, A. Vershinina, R. Vila and A. Warren for discussions, comments and help in obtaining material. The work was partially performed using equipment of the ‘Chromas’ Core Facility and Centre for Molecular and Cell Technologies of St Petersburg State University. Postdoctoral fellowship was provided to N.S. from St Petersburg State University.
Data accessibility
The DNA sequences have been deposited in GenBank. All studied butterfly samples are stored at Zoological Institute, St Petersburg, Russia.
Funding statement
The complete financial support for this study was provided by the grant from the Russian Science Foundation no. 14-14-00541 to the Zoological Institute of Russian Academy of Sciences.
Author contributions
V.A.L. designed the study, wrote the manuscript and performed data analysis. V.A.L. and N.A.S. collected the material and performed the field observations. N.A.S. and A.F.S. performed DNA studies. V.A.L. analysed the karyotypes. B.A.A. and V.G.K. performed genomic in situ hybridization. All authors discussed the paper and gave final approval for publication.
Conflict of interests
We have no competing interests.
References
- 1.Rieseberg LH. 1997. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 28, 359–389. ( 10.1146/annurev.ecolsys.28.1.359) [DOI] [Google Scholar]
- 2.Mallet J. 2007. Hybrid speciation. Nature 446, 279–283. ( 10.1038/nature05706) [DOI] [PubMed] [Google Scholar]
- 3.Abbot R, et al. 2013. Hybridization and speciation. J. Evol. Biol. 26, 229–246. ( 10.1111/j.1420-9101.2012.02599.x) [DOI] [PubMed] [Google Scholar]
- 4.Coyne JA, Orr AH. 2004. Speciation. Sunderland, MA: Sinauer. [Google Scholar]
- 5.Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH. 2005. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171, 291–303. ( 10.1534/genetics.105.042242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieseberg LH, Van Fossen C, Desrochers A. 1995. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature 375, 313–316. ( 10.1038/375313a0) [DOI] [Google Scholar]
- 7.Mavárez J, Linares M. 2008. Homoploid hybrid speciation in animals. Mol. Ecol. 17, 4181–4185. ( 10.1111/j.1365-294X.2008.03898.x) [DOI] [PubMed] [Google Scholar]
- 8.Hermansen JS. 2010. Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol. Ecol. 20, 3812–3822. ( 10.1111/j.1365-294X.2011.05183.x) [DOI] [PubMed] [Google Scholar]
- 9.Stemshorn KC, Reed FA, Nolte AW, Tautz D. 2011. Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.). Mol. Ecol. 20, 1475–1491. ( 10.1111/j.1365-294X.2010.04997.x) [DOI] [PubMed] [Google Scholar]
- 10.Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. 2000. The likelihood of homoploid hybrid speciation. Heredity 84, 441–451. ( 10.1046/j.1365-2540.2000.00680.x) [DOI] [PubMed] [Google Scholar]
- 11.Schwarz D, Matta BM, Shakir-Botteri NL, McPheron BA. 2005. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature 436, 546–549. ( 10.1038/nature03800) [DOI] [PubMed] [Google Scholar]
- 12.Gompert Z, Fordyce JA, Forister MA, Shapiro AM, Nice CC. 2006. Homoploid hybrid speciation in an extreme habitat. Science 314, 1923–1925. ( 10.1126/science.1135875) [DOI] [PubMed] [Google Scholar]
- 13.Kuusela J, Zietara MS, Lumme J. 2007. Hybrid origin of Baltic salmon-specific parasite Gyrodactylus salaris: a model for speciation by host switch for hemiclonal organisms. Mol. Ecol. 16, 5234–5245. ( 10.1111/j.1365-294X.2007.03562.x) [DOI] [PubMed] [Google Scholar]
- 14.Gross BL, Rieseberg LH. 2005. The ecological genetics of homoploid hybrid speciation. J. Heredity 96, 241–252. ( 10.1093/jhered/esi026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavarez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M. 2006. Speciation by hybridization in Heliconius butterflies. Nature 441, 868–871. ( 10.1038/nature04738) [DOI] [PubMed] [Google Scholar]
- 16.Melo MM, Salazar C, Jiggins CD, Linares M. 2009. Assortative mating preferences among hybrids offers a route to hybrid speciation. Evolution 63, 1660–1665. ( 10.1111/j.1558-5646.2009.00633.x) [DOI] [PubMed] [Google Scholar]
- 17.Salazar C, Baxter SW, Pardo-Diaz C, Wu G, Surridge A, Linares M, Bermingham E, Jiggins CD. 2010. Genetic evidence for hybrid trait speciation in Heliconius butterflies. PLoS Genet. 6, e1000930 ( 10.1371/journal.pgen.1000930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaral AR, Lovewell G, Coelho MM, Amato G, Rosenbaum HC. 2014. Hybrid speciation in a marine mammal: the Clymene dolphin (Stenella clymene). PLoS ONE 9, e83645 ( 10.1371/journal.pone.0083645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müntzing A. 1930. Outlines to a genetic monograph of the genus Galeopsis with special reference to the nature and inheritance of partial sterility. Hereditas 13, 185–341. ( 10.1111/j.1601-5223.1930.tb02522.x) [DOI] [Google Scholar]
- 20.Stebbins GL. 1957. Self fertilization and population variability in the higher plants. Am. Nat. 91, 337–354. ( 10.1086/281999) [DOI] [Google Scholar]
- 21.Grant V. 1958. The regulation of recombination in plants. Cold Spring Harbor Symp. Quant. Biol. 23, 337–363. ( 10.1101/SQB.1958.023.01.034) [DOI] [PubMed] [Google Scholar]
- 22.Faria R, Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669. ( 10.1016/j.tree.2010.07.008) [DOI] [PubMed] [Google Scholar]
- 23.Nice CC, Gompert Z, Fordyce JA, Forister ML, Lauren K, Lucas LK, Buerkle CA. 2013. Hybrid speciation and independent evolution in lineages of alpine butterflies. Evolution 67, 1055–1068. ( 10.1111/evo.12019) [DOI] [PubMed] [Google Scholar]
- 24.Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukhtanov VA, Kandul NP, Plotkin JB, Dantchenko AV, Haig D, Pierce NE. 2005. Reinforcement of pre-zygotic isolation and karyotype evolution in Agrodiaetus butterflies. Nature 436, 385–389. ( 10.1038/nature03704) [DOI] [PubMed] [Google Scholar]
- 26.Robinson R. 1971. Lepidoptera genetics. Oxford, UK: Pergamon Press. [Google Scholar]
- 27.Kandul NP, Lukhtanov VA, Pierce NE. 2007. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 61, 546–559. ( 10.1111/j.1558-5646.2007.00046.x) [DOI] [PubMed] [Google Scholar]
- 28.Eckweiler W, Häuser H. 1997. An illustrated checklist of Agrodiaetus Hübner, 1822, a subgenus of Polyommatus Latreille, 1804. Nachr. Entomol. Ver. Apollo Suppl. 16, 113–166. [Google Scholar]
- 29.Schurian KG, ten Hagen W, Eckweiler W. 2005. Beitrag zur Biologie und Ökologie von Polyommatus (Agrodiaetus) peilei Bethune-Baker, 1921 (Lepidoptera, Lycaenidae). Nachr. Entomol. Ver. Apollo 26, 197–206. [Google Scholar]
- 30.Lukhtanov VA, Shapoval NA, Dantchenko AV. 2008. Agrodiaetus shahkuhensis sp. n. (Lepidoptera, Lycaenidae), a cryptic species from Iran discovered by using molecular and chromosomal markers. Comp. Cytogen. 2, 99–114. [Google Scholar]
- 31.Lukhtanov VA, Shapoval NA, Dantchenko AV. 2014. Taxonomic position of several enigmatic Polyommatus (Agrodiaetus) species (Lepidoptera, Lycaenidae) from Central and Eastern Iran: insights from molecular and chromosomal data. Comp. Cytogen. 8, 313–322. ( 10.3897/CompCytogen.v8i4.8939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analyses program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 33.Drummond AJ, et al. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. ( 10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 35.Clement M, Posada D, Crandall K. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1660. ( 10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 36.Wiemers M, Keller A, Wolf M. 2009. ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evol. Biol. 9, 300 ( 10.1186/1471-2148-9-300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronquist F, et al. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takami Y, Koshio C, Ishii M, Fujii H, Hidaka T, Shimizu I. 2004. Genetic diversity and structure of urban populations of Pieris butterflies assessed using amplified length polymorphism. Mol. Ecol. 13, 245–258. ( 10.1046/j.1365-294X.2003.02040.x) [DOI] [PubMed] [Google Scholar]
- 39.Whitlock R, Hipperson H, Mannarelli M, Butlin RK, Burke T. 2008. An objective, rapid, and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Mol. Ecol. Resour. 8, 725–735. ( 10.1111/j.1755-0998.2007.02073.x) [DOI] [PubMed] [Google Scholar]
- 40.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quek SP, Stuart JD, Itino T, Pierce NE. 2004. Codiversification in an ant–plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 58, 554–570. ( 10.1111/j.0014-3820.2004.tb01678.x) [DOI] [PubMed] [Google Scholar]
- 42.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. (http://beast.bio.ed.ac.uk/Tracer).
- 43.Dinca V, Lukhtanov VA, Talavera G, Vila R. 2011. Unexpected layers of cryptic diversity in wood white Leptidea butterflies. Nat. Comm. 2, 234 ( 10.1038/ncomms1329) [DOI] [PubMed] [Google Scholar]
- 44.Joly S, McLenachan PA, Lockhart PJ. 2009. A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am. Nat. 174, e54–e70. ( 10.1086/600082) [DOI] [PubMed] [Google Scholar]
- 45.Avise JC. 2004. Molecular markers, natural history and evolution. Sunderland, MA: Sinauer. [Google Scholar]
- 46.Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618. ( 10.1111/mec.12415) [DOI] [PubMed] [Google Scholar]
- 47.Joly S. 2012. JML: Testing hybridization from species trees. Mol. Ecol. Res. 12, 179–184. ( 10.1111/j.1755-0998.2011.03065.x) [DOI] [PubMed] [Google Scholar]
- 48.Heled J, Drummond A. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580. ( 10.1093/molbev/msp274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukhtanov VA, Dantchenko AV. 2002. Principles of highly ordered metaphase I bivalent arrangement in spermatocytes of Agrodiaetus (Lepidoptera). Chromosomes Res. 10, 5–20. ( 10.1023/A:1014249607796) [DOI] [PubMed] [Google Scholar]
- 50.Kuznetsova VG, Maryanska-Nadachowska A, Nokkala S. 2009. Karyotype characterization of planthopper species using AgNOR-, C- and DAPI/CMA3–banding techniques. Comp. Cytogenet. 3, 111–123. ( 10.3897/compcytogen.v3i2.18) [DOI] [Google Scholar]
- 51.Traut W, Sahara K, Marec F. 2008. Sex chromosomes and sex determination in Lepidoptera. Sex. Dev. 1, 332–346. ( 10.1159/000111765) [DOI] [PubMed] [Google Scholar]
- 52.Traut W, Sahara K, Otto TD, Marec F. 1999. Molecular differentiation of sex chromosome probed by comparative genomic hybridization. Chromosoma 108, 173–180. ( 10.1007/s004120050366) [DOI] [PubMed] [Google Scholar]
- 53.Mallet J. 2001. Species, concepts of. In Encyclopedia of biodiversity, vol. 5 (eds Levin S, et al.), pp. 427–440. Oxford, UK: Academic Press, Elsevier. [Google Scholar]
- 54.Markova M, Vyskot B. 2009. New horizons of genomic in situ hybridization. Cytogenet. Genome Res. 126, 368–375. ( 10.1159/000275796) [DOI] [PubMed] [Google Scholar]
- 55.Rampin M, Bi K, Bogart JP, Collares-Pereira MJ. 2012. Identifying parental chromosomes and genomic rearrangements in animal hybrid complexes of species with small genome size using genomic in situ hybridization (GISH). Comp. Cytogenet. 6, 287–300. ( 10.3897/compcytogen.v6i3.3543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bi K, Bogart JP. 2010. Probing the meiotic mechanism of intergenomic exchanges by genomic in situ hybridization on lampbrush chromosomes of unisexual Ambystoma (Amphibia: Caudata). Chromosomes Res. 18, 371–382. ( 10.1007/s10577-010-9121-3) [DOI] [PubMed] [Google Scholar]
- 57.Schumer M, Rosenthal GG, Andolfatto P. 2014. How common is homoploid hybrid speciation? Evolution 68, 1553–1560. ( 10.1111/evo.12399) [DOI] [PubMed] [Google Scholar]
- 58.Lukhtanov VA. 2007. Homoploid hybrid speciation. Science E-letters. See https://www.sciencemag.org/content/314/5807/1923/reply.
- 59.McCarthy EM, Asmussen MA, Anderson WW. 1995. A theoretical assessment of recombinational speciation. Heredity 74, 502–509. ( 10.1038/hdy.1995.71) [DOI] [Google Scholar]
- 60.Jarne P, Charlesworth D. 1993. The evolution of the selfing rate in functionally hermaphrodite plants and animals. Ann. Rev. Ecol. Syst. 24, 441–466. ( 10.1146/annurev.es.24.110193.002301) [DOI] [Google Scholar]
- 61.Dumont BL, Payseur BA. 2011. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 7, e1002116 ( 10.1371/journal.pgen.1002116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lukhtanov VA, Dinca V, Talavera G, Vila R. 2011. Unprecedented within-species chromosome number cline in the wood white butterfly Leptidea sinapis and its significance for karyotype evolution and speciation. BMC Evol. Biol. 11, 109 ( 10.1186/1471-2148-11-109) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequences have been deposited in GenBank. All studied butterfly samples are stored at Zoological Institute, St Petersburg, Russia.