Abstract
Intracellular endosymbiotic bacteria are found in many terrestrial arthropods and have a profound influence on host biology. A basic question about these symbionts is why they infect the hosts that they do, but estimating symbiont incidence (the proportion of potential host species that are actually infected) is complicated by dynamic or low prevalence infections. We develop a maximum-likelihood approach to estimating incidence, and testing hypotheses about its variation. We apply our method to a database of screens for bacterial symbionts, containing more than 3600 distinct arthropod species and more than 150 000 individual arthropods. After accounting for sampling bias, we estimate that 52% (CIs: 48–57) of arthropod species are infected with Wolbachia, 24% (CIs: 20–42) with Rickettsia and 13% (CIs: 13–55) with Cardinium. We then show that these differences stem from the significantly reduced incidence of Rickettsia and Cardinium in most hexapod orders, which might be explained by evolutionary differences in the arthropod immune response. Finally, we test the prediction that symbiont incidence should be higher in speciose host clades. But while some groups do show a trend for more infection in species-rich families, the correlations are generally weak and inconsistent. These results argue against a major role for parasitic symbionts in driving arthropod diversification.
Keywords: Rickettsia, Wolbachia, Cardinium, maximum likelihood, infection
1. Introduction
Terrestrial arthropods carry an array of intracellular endosymbiotic bacteria. These bacteria have a profound influence on their hosts and are thought to affect areas of biology ranging from reproductive mode and resistance to viruses, to effective population size and rate of speciation [1–7]. Some of the bacteria are also remarkable for the breadth of their host range [3,8–10], but relatively little is known about why they infect the hosts that they do.
Several authors have suggested that symbiont infection frequency might vary predictably with host biology [1,3,6,7,9–16]. For example, two distinct arguments predict that symbionts should be more common in host taxa that are species rich. First, some symbionts might cause reproductive isolation in their hosts, thus increasing the number of species in infected groups, relative to uninfected groups [17–19]. Second, if symbionts occasionally switch hosts, and if these switches take place preferentially between closely related host species, then symbionts should be more common in host groups with many closely related species, as these relatives can act as ready sources of infection [20,21].
Hypotheses of this kind are common, but difficult to test in a rigorous comparative framework (though see, e.g. [6,15]). This is partly because symbiont incidence (i.e. the proportion of potential host species that are actually infected) is not easy to measure. While symbionts can be detected with PCR-based screens, infections vary in their prevalence (i.e. the proportion of individuals infected), and so it follows that low prevalence infections will be difficult to detect, that symbiont absence is impossible to prove, and that the number of infected samples might grossly underestimate the number of infected populations [8,22]. Furthermore, the infection status of a population can change rapidly [3,20,23,24], and this makes any single sample a mere snapshot of the ongoing ecological dynamics.
A way to mitigate these problems is to combine data from several populations, and estimate the distribution of prevalences across a group of potential hosts [8]. This distribution might be relatively stable, even when infection in any single species changes rapidly [20,21,25,26], and it allows us to infer the number of unobserved, low prevalence infections, even when few populations were sampled in depth. Such an approach to estimating incidence was pioneered by Hilgenboecker et al. [8] and has since been applied to several bacterial symbionts [8,26,27]. Here, we extend the approach in a full likelihood-based framework; this allows us to place proper confidence intervals on our incidence estimates, and to formally test hypotheses about whether and why incidence varies.
We apply our method to a newly collated database of published screens for three genera of bacterial secondary symbionts: Wolbachia, Rickettsia and Cardinium. Each genus employs a range of transmission strategies but is best known as a reproductive parasite, manipulating the sexual biology of hosts to facilitate vertical transmission via the egg cytoplasm [1,3,4,7]. Most importantly, each genus has been extensively studied, and so our database contains screens of over 150 000 individuals from over 3500 distinct arthropod species.
2. Material and methods
(a). Data collection
We searched the literature for PCR-based screens of Wolbachia, Rickettsia and Cardinium in wild populations of terrestrial arthropods (see electronic supplementary material, S1). For each population, we recorded the host species, the number of individuals screened and the number found to be infected. Our database includes data from 361 source publications, and over 10 000 populations, and is included as electronic supplementary material. Each screened arthropod was classified according to up-to-date taxonomy (electronic supplementary material, figure S1). To estimate the relative species richness of arthropod groups, we used estimates of the number of described species (electronic supplementary material, table S2). These will, of course, be a crude proxy for true species number, but are acceptable for our purposes, given the many difficulties in extrapolation [28].
(b). Model
Following [8], we initially assume that between-population variation in prevalences can be adequately described by a beta distribution, whose parameters are estimated from the screen data. From the best-fit distribution, we calculate the proportion of species infected above a given threshold frequency c, and denote this estimate of symbiont incidence as xc [8]. Most results reported use c = 0.001, and thus we define a population as ‘infected’ if it has a prevalence of greater than 1/1000 individuals. We use this threshold for expedience, but it is clear that the proportion of species in which no single individual is infected will be difficult to estimate with much confidence or precision. Furthermore, the threshold frequency reflects a biologically meaningful distinction between established infections and very low prevalence ‘dead-end’ infections, which are unlikely to persist in the host population [29,30]. Details of the model and numerical methods are found in electronic supplementary material, S2.
3. Results
(a). Estimating symbiont incidence
We begin by estimating symbiont incidence across the terrestrial arthropods as a whole. Figure 1 shows three such estimates for each bacterial genus. The initial estimates (a) were obtained from fitting a simple beta distribution to our complete database of screens. Owing to the shape of the beta distribution, these estimates entail the assumption that no population is completely free from infection (with a prevalence of exactly zero), and no population is completely infected (with a prevalence of exactly one, as with an obligate or primary symbiont). To relax this questionable assumption, we developed a method of fitting a doubly inflated beta distribution [31] which does allow for completely uninfected and completely infected host populations, as well as populations with intermediate prevalence. Comparing the two models on simulated data shows that the doubly inflated distribution is much more accurate when, in reality, a large fraction of populations do not harbour the symbionts (electronic supplementary material, S3 and figure S2). However, for our real data, fitting the doubly inflated distribution had almost no effect on estimates (b), suggesting that the simpler model is reasonably adequate.
Figure 1.
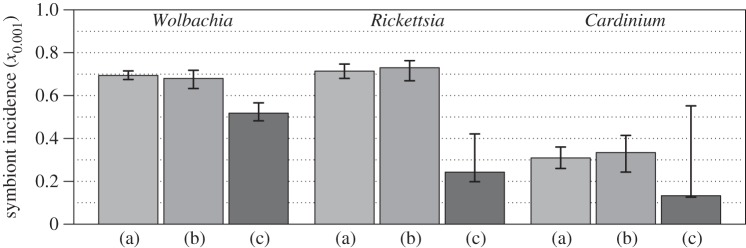
Estimates of symbiont incidence, x0.001 (i.e. the proportion of species infected at a prevalence of greater than 1/1000) in terrestrial arthropods. Estimates obtained from (a) fitting a beta distribution to the complete database; (b) fitting a doubly inflated beta distribution to the complete database, and so allowing for completely uninfected or completely infected species; (c) standardized sampling (i.e. a weighted sum of estimates from the largest arthropod taxa, using the single largest population sample from each sampled species).
More fundamentally, we are interested in the incidence across arthropod species (i.e. the proportion of species infected), and for this purpose, estimates from our complete database will be biased in at least three ways. First, and most obviously, some species are represented by a single population sample, and some by many samples. For example, the vectors of rickettsial disease (Parasitiformes and Siphonaptera) are hugely overrepresented (electronic supplementary material, figure S1). Second, there are clear taxonomic biases in the sampled species. In particular, minor arthropod orders are overrepresented (presumably from studies of symbiont host range; electronic supplementary material, figure S1). Third, and more subtly, sampling might be biased by the concentration of research effort on populations and species that were already known to contain infection [8].
To mitigate and test for these biases, we developed a three-stage process, which we call ‘standardized sampling’. First, we subsampled our data, retaining only the single largest screen from each species. Second, we estimated incidence for each of the major groups of arthropods, and then combined these estimates in a weighted sum, weighting each group by its contribution to total arthropod biodiversity. Third, we tested for differences in symbiont prevalence between multi-individual screens (which are more likely to be carried out on species known to carry infection), and single-individual screens (which are most likely to resemble a quasi-random sample of species). Full details are given in electronic supplementary material, S4, and results obtained with standardized sampling are labelled (c) in figure 1.
These improved estimates are substantially lower than the estimates from the complete database, but they remain remarkably high. We estimate that just over half of terrestrial arthropod species are infected with Wolbachia at a non-negligible frequency (52%, CIs 48–57), around a quarter infected with Rickettsia (24%, CIs 20–42) and around an eighth infected with Cardinium (13%, CIs 13–55). Furthermore, we cannot reject the possibility that Rickettsia and Cardinium incidences are much higher (figure 1). Their large and skewed confidence intervals reflect the underlying distributions of prevalences that we inferred from the data. In particular, for all bacteria, we inferred that most single species were subject to either very high or very low levels of infection at any given time (electronic supplementary material, table S1; [8,26,27]). However, estimates of mean prevalence levels were much lower for Cardinium and Rickettsia (less than 6%) than for Wolbachia (24%; electronic supplementary material, table S1). As such, for Cardinium and Rickettsia, it was difficult to distinguish between low incidence and a high incidence of low prevalence infections; this uncertainty is reflected in the high upper bounds on our estimates.
(b). Variation in incidence between bacteria and major arthropod host groups
We next tested for differences in symbiont incidence between bacteria and between major host groups. Figure 2 compares estimates of incidence for the best-sampled subphyla of arthropods, namely Hexapoda (insects and relatives), and Chelicerata (represented by arachnids in our database), after applying standardized sampling. Results show no significant difference in the incidences of Wolbachia in hexapods (51%) versus chelicerates (61%), and no significant difference in the incidences of the three bacteria in chelicerates (Wolbachia 61%, Rickettsia 51% and Cardinium 60%). As such, the clearest pattern in our data is significantly lower incidences, in hexapod hosts, of Rickettsia (22%) and especially Cardinium (8%) [14]. (We note that results are quite different when standardized sampling is not applied, confirming the benefits of this approach; electronic supplementary material, figure S4.)
Figure 2.
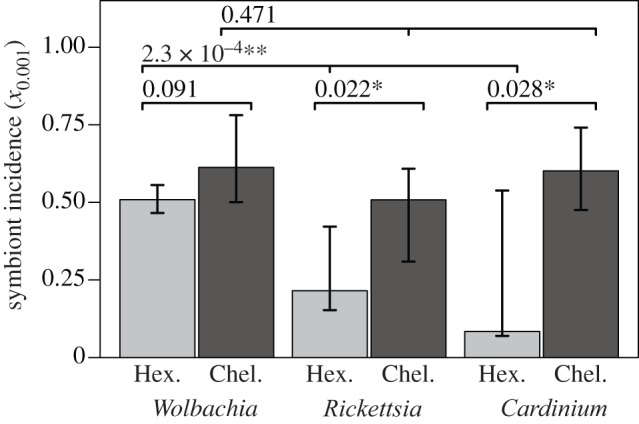
Estimates of symbiont incidence, x0.001 (i.e. the proportion of species infected at a prevalence of greater than 1/1000) in the two major subphyla of Arthropoda. Each pair of bars shows the incidence of a different bacterial genus, and compares estimates for Hexapoda (left-hand bar) and Chelicerata (right-hand bar). Estimates used ‘standardized sampling’ (see main text). P-values above each set of bars are from a likelihood ratio test of heterogeneity in the estimates.
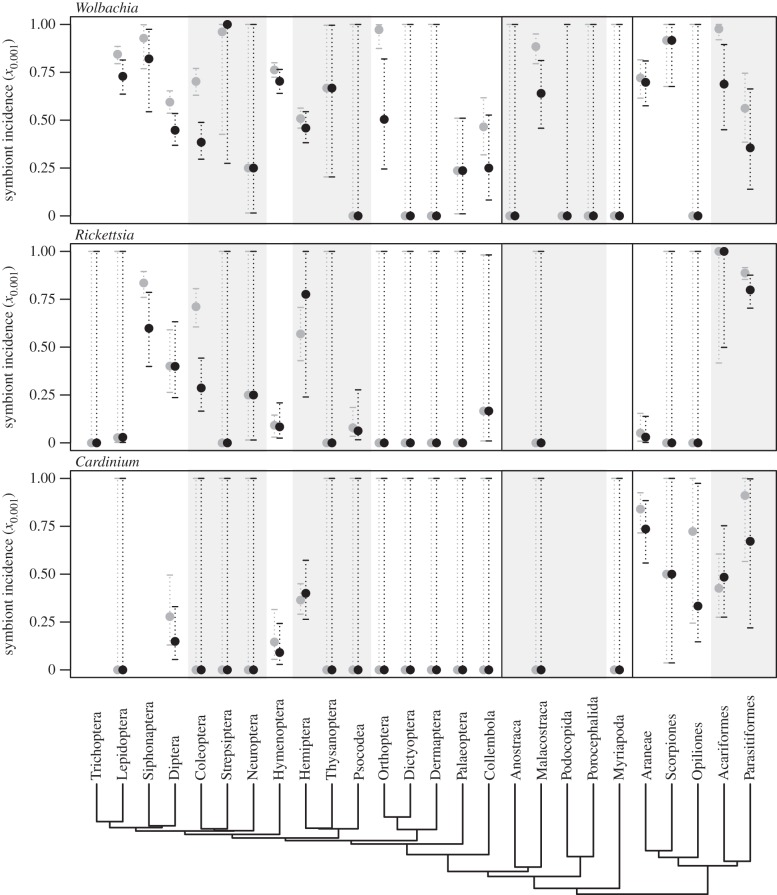
The pattern in figure 2 might be explained in many ways, but one possibility is differences in arthropod innate immunity [32,33]. (WJ Palmer 2014, personal communication.) Comparative genomics shows that chelicerates lack key components of the IMD immune pathway (WJ Palmer 2014, personal communication), which is primarily responsive to Gram-negative bacteria [34], and activated by DAP-type peptidoglycans [35]. Peptidoglycans are not thought to be produced by Wolbachia [36,37], but are produced by Cardinium [36,38], and also by Rickettsia, albeit sometimes at very low levels [39,40]. Therefore, the low incidence in hexapods of Rickettsia and especially Cardinium, might be due to their eliciting an additional immune response, not found in chelicerates, and not induced by Wolbachia in any host group. Suggestive support for this hypothesis comes from within hexapods, where the paraneopteran orders Hemiptera (true bugs), and Psocodea (lice) are also known to lack components of IMD [33]. Figure 3 shows that for Cardinium, the six arthropod groups with the highest estimated incidence (the five sampled chelicerate groups, and Hemiptera) all lack IMD components. This pattern is weakly present in Rickettsia (where it applies the three groups with highest incidence), and is wholly absent in Wolbachia.
Figure 3.
Estimates of symbiont incidence, x0.001 (i.e. the proportion of species infected at a prevalence of greater than 1/1000) for three genera of bacterial endosymbionts, across orders (and some superordinal groups) of terrestrial arthropods. Grey points show estimates from our complete database, and black points show estimates with standardized sampling, in which all sampled species were represented by the single largest population sample. Shading and vertical lines demarcate some major host groups, including Hexapoda (left-hand panel) and Chelicerata (right-hand panel).
Regardless of its cause, figure 3 suggests that closely related groups of host might have similar levels of symbiont incidence. This is borne out in formal tests, where Cardinium and Rickettsia, but not Wolbachia, show weak evidence of phylogenetic signal in their incidence levels (electronic supplementary material, S5.1 and table S4).
(c). Species richness and symbiont incidence
We next tested the prediction that infection levels in a host group will tend to increase with its species richness [18–21]; this is best tested with many taxonomic groups of similar age, and a rough biological similarity [11,13,18], and so we considered arthropod families or genera within major orders. Formally, we regressed estimated incidence in a family against its described species number, using various cut-off frequencies to define an ‘infected’ species (it is unlikely that, say, a speciation event would be caused by a very low prevalence infection; electronic supplementary material, S5.2). Results (electronic supplementary material, table S5 and figure S5), showed no clear pattern. For example, in Coleoptera (beetles), a significant positive relationship between incidence and species richness is found for both Wolbachia and Rickettsia, but Araneae (spiders) show the opposite result. Furthermore, the explanatory power of the model is very low in all cases (only with Wolbachia infections at more than 50% prevalence in Coleoptera does the model yield a pseudo-r2 above 10%).
4. Discussion
We have introduced a maximum-likelihood estimator of symbiont incidence (the proportion of potential host species that are actually infected), and applied our estimator to a large database of PCR screens for Wolbachia, Rickettsia and Cardinium in terrestrial arthropods. We have also introduced methods to account for the most serious sources of sampling bias, including weighting estimates from different groups by their contribution to arthropod diversity. Obviously, other biases will persist (nobody could hope to obtain a truly random sample of all arthropod species) and it remains practically impossible to prove the absence of a symbiont in a given species. Most seriously, our estimates will be reliable only if prevalences in the sampled populations are representative of the species range as a whole. When only a tiny proportion of the species range has been sampled, this is impossible to prove. For although migration between populations and species-specific susceptibilities will act to homogenize prevalences across populations, geographical isolation, habitat variation or intraspecific genetic variation, could lead to large, sustained differences. In the worst-case scenario, prevalences across populations of a given species would be completely uncorrelated with each other. In such a case, our xc would correspond, roughly, to the incidence across populations, while the incidence across species would tend towards 100% (since, with uncorrelated prevalences, it becomes extremely unlikely for all populations of a given species to be free from infection at any given time). Reality must lie somewhere between these two extremes, and so our incidence estimates are probably downwardly biased.
With these caveats, we have estimated that Wolbachia, Rickettsia and Cardinium infect, respectively, around a half, a quarter and an eighth of terrestrial arthropod species (figure 1). These differences mask remarkably similar incidences in chelicerates (figures 2 and 3), and stem from the significantly lower incidences of Rickettsia and especially Cardinium in some hexapod groups [14,25] (electronic supplementary material, table S4). We have speculated that this might reflect evolutionary changes in arthropod immunity, since many of the groups with higher incidences lack components of the IMD pathway [32–34,36,41] (WJ Palmer 2014, personal communication), and IMD is activated by DAP-type peptidoglycan [33], which is produced by Cardinium [36,38] but not Wolbachia [36,37].
Finally, we tested the prediction that incidence levels should be higher in host groups that are more speciose [17–21]. Data from some groups, such as Coleoptera, supported the prediction, but overall, the correlations were inconsistent and generally weak (electronic supplementary material, table S5 and figure S5). There are three possible explanations for these negative results. First, there is limited power, stemming from inadequacies in our data, or the low precision in our incidence estimates. However, several groups did yield significant results—but not consistently in the predicted direction (electronic supplementary material, table S5). Second, a confounding factor might have masked a true underlying correlation. For example, competitive exclusion among symbionts might lead to high incidence of one bacterium being predictably associated with low incidence of another (though see [22,42]), or species richness might correlate with clade age, which might also affect incidence [43]. Alternatively, symbionts might induce speciation without transferring to the new daughter species, or might often drive their hosts extinct [44,45].
Third, and finally, there might be no causal relationship between species number and symbiont incidence. The prediction stems from (i) models of biased host switching, in which symbionts transfer more readily to closely related hosts [20,21] and (ii) suggestions that symbionts might cause host speciation [17–19]. Regarding host shifting, there is strong evidence of between-species transfer in all three bacteria (both phylogenetic [3,7,46,47] and experimental [41,48]), but evidence of bias is indirect, coming from phylogenetic clustering [7,11,13,24,46,47] (strong experimental evidence comes solely from Spiroplasma, an ecologically similar, but phylogenetically distant endosymbiont [49]). The evidence for symbiont-mediated speciation is even sparser. Host reproductive isolation might arise as a passive by-product of host–symbiont coevolution (since any genomic change, whether in host or symbiont, might have negative epistatic fitness effects in a hybrid background [4,19]). But the most plausible route to rapid speciation is through reproductive manipulations that cause isolation, such as cytoplasmic incompatibility, or host parthenogenesis. Not all manipulations have been observed in all host–parasite combinations, and so this might explain the inconsistent results (electronic supplementary material, table S5). Nevertheless, taken together, our results must count as evidence against the claim that symbionts are a major cause of diversification across the arthropods as a whole.
While no consistent effect of species richness has been found, we hope that the methods presented here will prove useful for testing the many further hypotheses about the causes of endosymbiont incidence in nature.
Supplementary Material
Acknowledgements
We are very grateful to J. Beckmann, F. Jiggins, B. Longdon, J. Martinez, W. Palmer, A. Rambaut, J. Thomas, D. Waxman and J. Werren for their help and advice, and to two anonymous reviewers whose comments substantially improved the manuscript.
References
- 1.O'Neill SL, Hoffman A, Werren JH. (eds). 1997. Influential passengers: inherited microorganisms and arthropod reproduction. Oxford, UK: Oxford University Press. [Google Scholar]
- 2.Moran NA. 2006. Symbiosis. Curr. Biol. 16, R866–R871. ( 10.1016/j.cub.2006.09.019) [DOI] [PubMed] [Google Scholar]
- 3.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. ( 10.1038/nrmicro1969) [DOI] [PubMed] [Google Scholar]
- 4.Engelstädter J, Hurst GDD. 2009. The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 40, 127–149. ( 10.1146/annurev.ecolsys.110308.120206) [DOI] [Google Scholar]
- 5.Zchori-Fein E, Bourtzis K. (eds). 2012. Manipulative tenants: bacteria associated with arthropods. Boca Raton, FL: Taylor & Francis. [Google Scholar]
- 6.Henry LM, Peccoud J, Simon J-C, Hadfield JD, Maiden MJC, Ferrari J, Godfray HCJ. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 23, 1713–1717. ( 10.1016/j.cub.2013.07.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinert LA. 2015. The diversity and phylogeny of Rickettsia. In Parasite diversity and diversification: evolutionary ecology meets phylogenetics (eds Morand S, Krasnov B, Littlewood T.), pp. 150–181. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 8.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. ( 10.1111/j.1574-6968.2008.01110.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurst LD. 1991. The incidences and evolution of cytoplasmic male killers. Proc. R. Soc. Lond. B 244, 91–99. ( 10.1098/rspb.1991.0056) [DOI] [Google Scholar]
- 10.Werren JH, Windsor D, Guo L. 1995. Distribution of Wolbachia among Neotropical arthropods. Proc. R. Soc. Lond. B 262, 197–204. ( 10.1098/rspb.1995.0196) [DOI] [Google Scholar]
- 11.Jiggins FM, Randerson JP, Hurst GDD, Majerus MEN. 2002. How can sex ratio distorters reach extreme prevalences? Male-killing Wolbachia are not suppressed and have near-perfect vertical transmission efficiency in Acraea encedon. Evolution 56, 2290–2295. ( 10.1111/j.0014-3820.2002.tb00152.x) [DOI] [PubMed] [Google Scholar]
- 12.Reuter M, Pedersen JS, Keller L. 2004. Loss of Wolbachia infection during colonisation in the invasive Argentine ant Linepithema humile. Heredity 94, 364–369. ( 10.1038/sj.hdy.6800601) [DOI] [PubMed] [Google Scholar]
- 13.Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. 2008. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 17, 557–569. ( 10.1111/j.1365-294X.2007.03608.x) [DOI] [PubMed] [Google Scholar]
- 14.Martin OY, Goodacre SL. 2009. Widespread infections by the bacterial endosymbiont Cardinium in arachnids. J. Arachnol. 37, 106–108. ( 10.1636/SH08-05.1) [DOI] [Google Scholar]
- 15.Toju H, Fukatsu T. 2011. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol. Ecol. 20, 853–868. ( 10.1111/j.1365-294X.2010.04980.x) [DOI] [PubMed] [Google Scholar]
- 16.Russell JA, Funaro CF, Giraldo YM, Goldman-Huertas B, Suh D, Kronauer DJC, Moreau CS, Pierce NE. 2012. A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: broad molecular surveys and a systematic review. PLoS ONE 7, e51027 ( 10.1371/journal.pone.0051027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallin IE. 1927. Symbionticism and the origin of species. Baltimore, MD: Williams & Wilkins Company. [Google Scholar]
- 18.Bordenstein SR. 2003. Symbiosis and the origin of species. In Insect symbiosis (eds Bourtzis K, Miller TA.). New York, NY: CRC Press. [Google Scholar]
- 19.Brucker RM, Bordenstein SR. 2012. Speciation by symbiosis. Trends Ecol. Evol. 27, 443–451. ( 10.1016/j.tree.2012.03.011) [DOI] [PubMed] [Google Scholar]
- 20.Waxman D, Weinert LA, Welch JJ. 2014. Inferring host range dynamics from comparative data: the protozoan parasites of new world monkeys. Am. Nat. 184, 65–74. ( 10.1086/676589) [DOI] [PubMed] [Google Scholar]
- 21.Engelstädter J, Hurst GDD. 2006. The dynamics of parasite incidence across host species. Evol. Ecol. 20, 603–616. ( 10.1007/s10682-006-9120-1) [DOI] [Google Scholar]
- 22.Weinert LA, Tinsley MC, Temperley M, Jiggins FM. 2007. Are we underestimating the diversity and incidence of insect bacterial symbionts? A case study in ladybird beetles. Biol. Lett. 3, 678–681. ( 10.1098/rsbl.2007.0373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA. 2007. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 5, e114 ( 10.1371/journal.pbio.0050114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himler AG, et al. 2011. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332, 254–256. ( 10.1126/science.1199410) [DOI] [PubMed] [Google Scholar]
- 25.Werren JH, Windsor DM. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. B 267, 1277–1285. ( 10.1098/rspb.2000.1139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed MZ, Greyvenstein OFC, Erasmus C, Welch JJ, Greeff JM. 2013. Consistently high incidence of Wolbachia in global fig wasp communities. Ecol. Entomol. 38, 147–154. ( 10.1111/een.12002) [DOI] [Google Scholar]
- 27.Zug R, Hammerstein P. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 7, e38544 ( 10.1371/journal.pone.0038544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. 2011. How many species are there on earth and in the ocean? PLoS Biol. 9, e1001127 ( 10.1371/journal.pbio.1001127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fine PEM. 1978. On the dynamics of symbiote-dependent cytoplasmic incompatibility in culicine mosquitoes. J. Invertebr. Pathol. 31, 10–18. ( 10.1016/0022-2011(78)90102-7) [DOI] [PubMed] [Google Scholar]
- 30.Woolhouse MEJ, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244. ( 10.1016/j.tree.2005.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ospina R, Ferrari SLP. 2010. Inflated beta distributions. Stat. Pap. 51, 111–126. ( 10.1007/s00362-008-0125-4) [DOI] [Google Scholar]
- 32.Waterhouse RM, et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316, 1738–1743. ( 10.1126/science.1139862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerardo NM, et al. 2010. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 11, R21 ( 10.1186/gb-2010-11-2-r21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris HL, Brennan LJ, Keddie BA, Braig HR. 2010. Bacterial symbionts in insects: balancing life and death. Symbiosis 51, 37–53. ( 10.1007/s13199-010-0065-3) [DOI] [Google Scholar]
- 35.Kaneko T, et al. 2004. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649. ( 10.1016/S1074-7613(04)00104-9) [DOI] [PubMed] [Google Scholar]
- 36.Nakamura Y, Gotoh T, Imanishi S, Mita K, Kurtti TJ, Noda H. 2011. Differentially expressed genes in silkworm cell cultures in response to infection by Wolbachia and Cardinium endosymbionts. Insect Mol. Biol. 20, 279–289. ( 10.1111/j.1365-2583.2010.01056.x) [DOI] [PubMed] [Google Scholar]
- 37.Vollmer J, Schiefer A, Schneider T, Jülicher K, Johnston KL, Taylor MJ, Sahl H-G, Hoerauf A, Pfarr K. 2013. Requirement of lipid II biosynthesis for cell division in cell wall-less Wolbachia, endobacteria of arthropods and filarial nematodes. Int. J. Med. Microbiol. 303, 140–149. ( 10.1016/j.ijmm.2013.01.002) [DOI] [PubMed] [Google Scholar]
- 38.Penz T, Schmitz-Esser S, Kelly SE, Cass BN, Müller A, Woyke T, Malfatti SA, Hunter MS, Horn M. 2012. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 8, e1003012 ( 10.1371/journal.pgen.1003012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amano K, Tamura A, Ohashi N, Urakami H, Kaya S, Fukushi K. 1987. Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi. Infect. Immun. 55, 2290–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang H, Winkler HH. 1994. Analysis of the peptidoglycan of Rickettsia prowazekii. J. Bacteriol. 176, 923–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelc RS, McClure JC, Sears KT, Chung A, Rahman MS, Ceraul SM. 2014. Defending the fort: a role for defensin-2 in limiting Rickettsia montanensis infection of Dermacentor variabilis. Insect Mol. Biol. 23, 457–465. ( 10.1111/imb.12094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstadter J, Hurst GD. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6, 27 ( 10.1186/1741-7007-6-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zug R, Koehncke A, Hammerstein P. 2012. Epidemiology in evolutionary time: the case of Wolbachia horizontal transmission between arthropod host species. J. Evol. Biol. 25, 2149–2160. ( 10.1111/j.1420-9101.2012.02601) [DOI] [PubMed] [Google Scholar]
- 44.Anderson RM. 1978. The regulation of host population growth by parasitic species. Parasitology 76, 119–157. ( 10.1017/S0031182000047739) [DOI] [PubMed] [Google Scholar]
- 45.Nice CC, Gompert Z, Forister ML, Fordyce JA. 2009. An unseen foe in arthropod conservation efforts: the case of Wolbachia infections in the Karner blue butterfly. Biol. Conserv. 142, 3137–3146. ( 10.1016/j.biocon.2009.08.020) [DOI] [Google Scholar]
- 46.Russell JA, Goldman-Huertas B, Moreau CS, Baldo L, Stahlhut JK, Werren JH, Pierce NE. 2009. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution 63, 624–640. ( 10.1111/j.1558-5646.2008.00579.x) [DOI] [PubMed] [Google Scholar]
- 47.Perlman SJ, Magnus SA, Copley CR. 2010. Pervasive associations between Cybaeus spiders and the bacterial symbiont Cardinium. J. Invertebr. Pathol. 103, 150–155. ( 10.1016/j.jip.2009.12.009) [DOI] [PubMed] [Google Scholar]
- 48.Carrington LB, Hoffmann AA, Weeks AR. 2010. Monitoring long-term evolutionary changes following Wolbachia introduction into a novel host: the Wolbachia popcorn infection in Drosophila simulans. Proc. R. Soc. B 277, 2059–2068. ( 10.1098/rspb.2010.0166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tinsley MC, Majerus ME. 2007. Small steps or giant leaps for male-killers? Phylogenetic constraints to male-killer host shifts. BMC Evol. Biol. 7, 238 ( 10.1186/1471-2148-7-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.