Abstract
Patients tolerant to a kidney graft display a specific blood cell transcriptional pattern but results from five different studies were inconsistent, raising the question of relevance for future clinical application. To resolve this, we sought to identify a common gene signature, specific functional and cellular components, and discriminating biomarkers for tolerance following kidney transplantation. A meta-analysis of studies identified a robust gene signature involving proliferation of B and CD4 T cells, and inhibition of CD14 monocyte related functions among 96 tolerant samples. This signature was further supported through a cross-validation approach, yielding 92.5% accuracy independent of the study of origin. Experimental validation, performed on new tolerant samples and using a selection of the top-20 biomarkers, returned 91.7% of good classification. Beyond the confirmation of B-cell involvement, our data also indicated participation of other cell subsets in tolerance. Thus, the use of the top 20 biomarkers, mostly centered on B cells, may provide a common and standardized tool towards personalized medicine for the monitoring of tolerant or low-risk patients among kidney allotransplant recipients. These data point to a global preservation of genes favoring the maintenance of a homeostatic and ‘healthy' environment in tolerant patients and may contribute to a better understanding of tolerance maintenance mechanisms.
Keywords: biomarker, gene signature, meta-analysis, renal allograft, tolerance, transplantation
Transplantation is the treatment of choice for end-stage renal disease. Recent advances in immunosuppression (IS) have improved management of acute rejection and graft survival.1 However, due to their toxicity, these drugs have numerous deleterious side effects and only a marginal effect on long-term rejection.2, 3, 4, 5 Tolerance is thus increasingly regarded as an ideal solution.6 This situation, mainly corresponding to incompliant cases, has been observed in renal transplantation7, 8 and current estimates report roughly 100 cases.9 Besides, efforts have been devoted by the European and US transplant community to decipher the regulatory mechanisms and to identify noninvasive biomarkers. These studies could report several gene lists, with evidence converging towards the potential implication of B cells as attested by the identification of numerous related markers,10, 11, 12, 13, 14 a unique differentiation profile,15 and an increasing number of several subtypes in the blood of these patients.13, 14, 16, 17 Although informative, these lists poorly overlapped, raising question about the pertinence of the results for future clinical application. However, as recently exemplified by the identification of rejection markers across multiple transplanted organs,18 comparing and integrating data from several studies by meta-analysis19 is an ideal solution to increase reliability and consistency in the results and conclusions. Hence, when applied to the five existing tolerance-related studies,10, 11, 12, 13, 14 such meta-analysis may also enable to reconcile data.
In this context, the first objective of our work was to define a robust gene signature20 indicative of tolerance. The second objective was to define specific functional and cellular components supported by the signature, as previously defined,20 and that may bring clues for the understanding of peripheral regulatory mechanisms. The third objective was to validate a subset of markers that could be further used in clinics for the stratification of kidney recipients with a low risk of rejection.
Results
Comparison of gene lists from the different data sets failed to identify a robust gene signature of tolerance
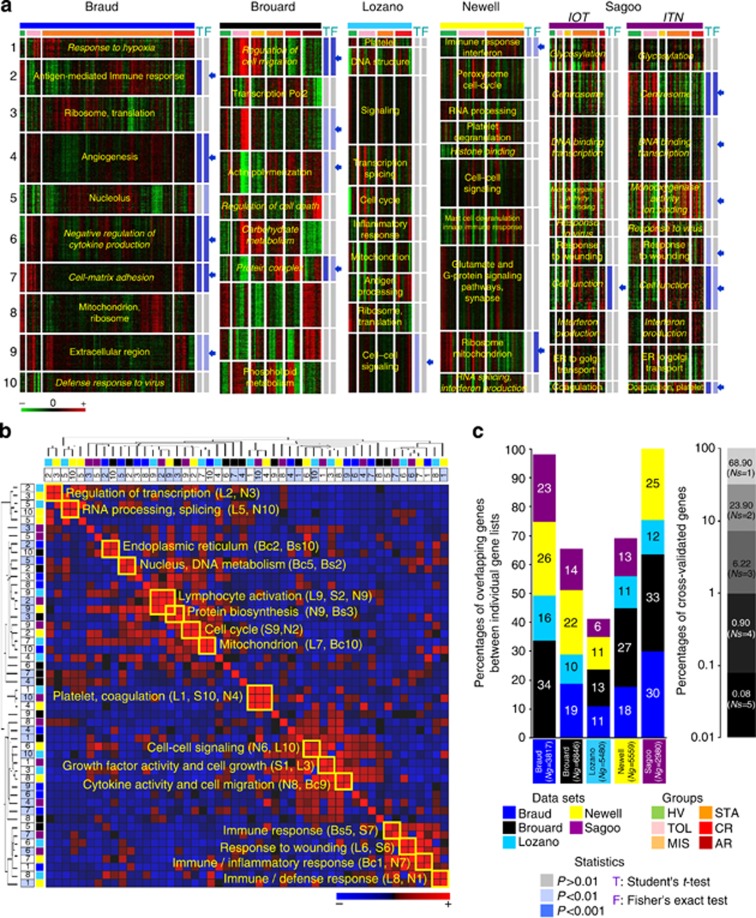
A k-means clustering identified 10 clusters (K1 to K10) in each study (Supplementary Figure SI1 online) with clear functional annotations (Figure 1a). A total of 19 clusters from the 50 were found to be significant (Supplementary Figures SI2 and SI3 online) between tolerant (TOL) and stable (STA), with at least one of the two tests (Student and/or Fisher, P<0.01). These clusters had relevant but limited similarities (Figure 1b, Supplementary Figure SI2 online). From the 19 clusters (Figure 1c), most of the differential genes (81% in average) in one study did not replicate in other studies (Supplementary Figure SI2 online). Only 0.08% (14 genes: APOM, ARHGAP17, AURKB, IGBP1, IL10RA, IL15RA, IL1RL1, INSM1, IRF4, MAOA, MICB, SMAD3, TK1, and YPEL2) was in fact commonly identified across the five studies (Supplementary Figures SI3 and SI4 online). Two of them (TK1 and IRF4) were present either in the 49-gene footprint from Brouard11 or in the top 30 genes from Newell13 (Supplementary Figure SI5 online). These 14 genes did not accurately discriminate TOL from STA (Supplementary Figure SI6 online). Altogether, these data show that comparison of gene lists identified a limited consensus and was not enough to derive a robust meta-signature.
Figure 1.
Comparisons of gene lists from the re-analyzed studies. (a) Individualization of clusters of genes. For each data set (Braud: dark blue bar; Brouard: black bar; Lozano: turquoise blue bar; Newell: yellow bar; Sagoo: purple bar; see legend, ‘data sets'), results from K-means clustering are displayed by a heat-map view using green for gene underexpression, black for gene expression close to the median, and red for gene overexpression (see color scale). Samples are ordered according to the status of origin (see legend, ‘groups'). They correspond to healthy volunteers (HV, green bar), tolerant recipients (TOL, pink bar), stable recipients under minimal immunosuppression (MIS, light orange bar) or classical treatment (STA, dark orange bar), and recipients with chronic (CR, red bar) or acute rejection (AR, brown bar). For each data set, individualized clusters of genes from K-means clustering (K1 to K10) are annotated by a representative term (in yellow) from the Gene Ontology (GO) project. GO terms in italic denote significance of the enrichment but sensitivity to multitesting (false discovery rate adjusted P-values). The propensity of each K cluster to discriminate tolerant (TOL) and stable (STA) recipients was assessed by a Student's t-test (T in blue) applied on its median profile and a Fisher's exact test (F in blue) applied on the contingencies of the related dendrogram (see legend, ‘statistics'). P-values resulting from the tests are indicated on the right of each cluster using shades of blue (see legend, ‘statistics'). The 19 clusters significant at P<0.01 with at least one of the two tests are indicated by arrows. (b) Similarity in gene composition between the 50-K clusters of co-expressed genes. The 50-K clusters from the five re-analyzed studies (10 clusters per study) were compared in terms of overlapping genes. The normalized intersections resulting from pairwise comparisons are presented by a diagonally symmetric matrix ordered by hierarchical clustering (rows and columns) to highlight groups of similar clusters (see trees on top and left). For each cluster, its study of origin (see legend), its number (1–10), and its differential status (light blue square) are given. From the heat-map visualization, a strong overlap between two distinct clusters is portrayed in red and a poor overlap is portrayed in blue (see color scale). Particular similarities are framed on the diagonal along with a representative GO term identified from the functional annotation of the overlap. (c) Intersection of the 19 differential clusters. The 19 clusters (denoted by blue arrows in the top panel) discriminative of TOL and STA groups were compared altogether. The results (left side) are depicted by colored histograms giving for each study (see legend) the percentage of cross-validation of its merged differential clusters (size of the list ‘Ng') with the differential clusters originating from the other data sets. The global cross-validation rate (right side) is indicated by the proportions of differential genes observed in one (Ns=1) to five (Ns=5) data sets. Study from Sagoo comprises two independent cohorts (EU IOT: ‘Indices of Tolerance' US ITN: ‘Immune Tolerance Network') sponsored, respectively.
Meta-analysis by integration of the data sets identified a statistically and functionally relevant gene signature of tolerance
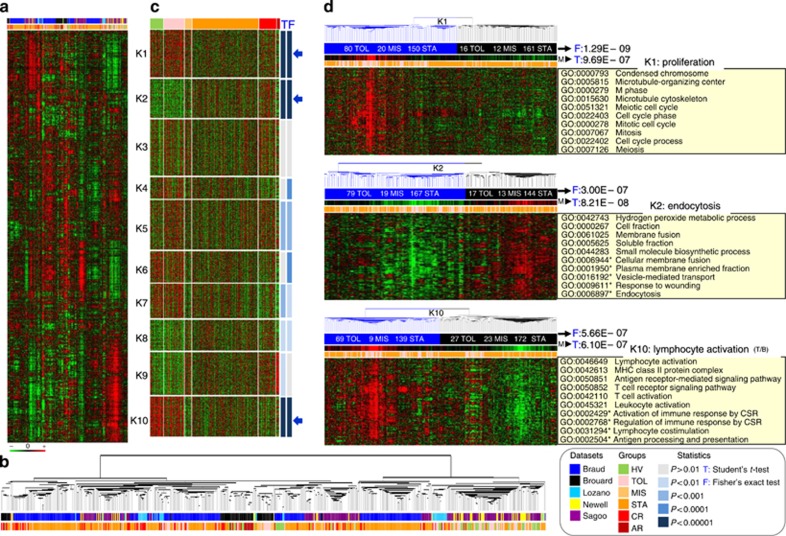
To ensure comparability across studies, the 1846 common genes were retained. This selection is highly skewed towards immune functions (Supplementary Figure SI7 online); however, it covers as high as 65% of the biological functions from the gene ontology. After standardization, data sets were merged and analyzed as a single corpus of data (Figure 2a) comprising 596 samples and available in the Gene Expression Omnibus repository (no. GSE49198). Hierarchical clustering highlighted clear profiles correlated to sample groups (Figure 2a and b). To identify the ones associated with tolerance, we further analyzed the 10-K-means clusters (Figure 2c), K1 (224 genes) linked to proliferation, K2 (183 genes) to endocytosis, and K10 (188 genes) to lymphocyte (B and T) activation and differentiation (Figure 2d, Supplementary Figures SI8 and SI9 online) were found to be the most differential (P<0.00001) between TOL (n=96) and STA (n=343). They thus defined a highly discriminative (P=5.05E−15) 595-gene signature as measured by Fisher's exact test on the contingencies of the sample tree (Figure 2d, Supplementary Figure SI8 online). It comprised eight (APOM, AURKB, IGBP1, IL10RA, IL1RL1, IRF4, SMAD3, and TK1) out of the 14-consensus genes from gene list comparison (Supplementary Figure SI4 online) and also 13 B-cell molecules (AFF3, BLK, BLNK, CD22, CD79B, FCER2, FCRL2, ID3, IGKC, IGLL1, MS4A1, MZB1 and TCL1A) belonging to the top-ranked gene lists (Supplementary Figure SI5 online) from the different studies.10, 11, 12, 13, 14 Altogether, these data show that integrative meta-analysis can identify a robust gene meta-signature of tolerance.
Figure 2.
Meta-analysis of the five studies. (a) Reliability of the meta-matrix. Results from a two-way hierarchical clustering are visualized by a heat map (1846 genes in lines, 596 samples in columns) using the same color code: green for gene underexpression and red for gene overexpression (see color scale). (b) Sample classification. The zoomed tree (bottom) reflects correlation between samples for which the status and the study of origin are indicated (see color legend). (c) Individualization of the meta-clusters. To identify discriminative groups of genes, the meta-data set was partitioned into 10 clusters (from K1 to K10) using a k-means clustering. The results are depicted by a heat map (same color code for gene expression values). Samples are supervised according to their status of origin: healthy volunteer (HV, green), tolerant recipient (TOL, pink), stable recipient under minimum immunosuppression (MIS, light orange) or classical treatment (STA, dark orange), and recipient with chronic (CR, red) or acute (AR, brown) rejection (see legend). The discriminative propensity of each meta-cluster to discriminate tolerant (TOL) from the control group of stable recipients (MIS and STA) was assessed by a Student's t-test (T in blue) applied on its median profile and a Fisher's exact test (F in blue) applied on the contingency of its dendrogram (see legend, ‘statistics'): resulting P-values are indicated on the right of each meta-cluster using shades of blue (see color legend). The three most differential ones (TOL vs. STA/MIS, P<0.00001) with the two tests (namely K1, K2, and K10) are denoted by arrows. (d) Definition of the meta-signature of tolerance. For each of the three most differential (P<0.00001) meta-clusters (K1, K2, and K10) pertaining to the signature, a heat map visualization is depicted. Information provided includes the following: the median profile (M) of the cluster and the P-value from the Student's t-test (T) applied to it; the contingencies (number of TOL, MIS, and STA samples) from the two main branches of the dendrogram (blue and black, respectively); and the P-value from the Fisher's exact test applied to them. For each meta-cluster, the top ten significant Gene Ontology (GO) terms from functional annotation analysis are also given and summarized by a representative term (right side of the panel). Terms with an asterisk indicate significance of the enrichment but sensitivity to multiple testing corrections (FDR-adjusted P-values). CSR, class-switch recombination.
Gene set analysis corroborated functionality and revealed the possible involvement of specific cell populations in tolerance
A screening of the compendium of gene sets retained the 100 best hits for each cluster (K1, K2, and K10; Figure 3, Supplementary Figure SI10 online). Although significant (P<0.00001), these overlaps were partial (mean coverage=8.89±4.82%) but displayed good functional congruence (Figure 3a): the 100 best hits converged to cell proliferation and cell adhesion for K1, inflammatory response for K2 and lymphocyte activation for K10. A similar analysis with a collection of blood transcriptional modules (Supplementary Figure SI11 online)21 significantly (P<0.001) linked K2 genes to pro-inflammatory-related monocyte modules, whereas K10 genes were linked to B-cell and T-cell modules. Particular results (Figure 3b) from different studies22, 23, 24, 25, 26, 27 illustrate the link to T CD4 (panel 1)26 and especially B (panels 2–4 and 6)23, 24, 25, 27 lymphocytes at various differentiation stages. For instance (panels 1 and 4),24, 26 K1 was remarkably enriched (P<1.00E−16) in early proliferating T- and B-cell markers. Conversely, markers of differentiated cell subsets were significantly linked to K10: P=1.01E−13 for T CD4+26 and P=7.55E−09 for B cells.24
Figure 3.
Functional interpretation by Gene Set Analysis (GSA). (a). Functional convergence of the overlapping gene sets. For each differential meta-cluster (K1, K2, and K10), the 100 top overlapping gene sets from the C2_CGP collection (chemical and genetic perturbation sets) of the GSEA Molecular Signature Database (MSigDB v.4) are retained. The functional annotation of these 300 overlapping sets is performed and enrichment P-values for each Gene Ontology (GO) term retained. These values were hierarchically clustered to order and place GO terms (rows) with similar patterns of enrichment across gene sets (columns) in proximity. Results are visualized by a heat map in which each cell represents a specific P-value of enrichment in a specific gene set, using the following color code: blue for poor P-values and yellow for highly significant P-values (see color scale). For each main GO ‘cluster', 10 significant terms are given and summarized by a representative annotation on the right side. (b) Particular similarities with data sets related to immune cell subsets. Overlapping gene sets (see legend) from human (1, 2, 5, and 6) or mice (3 and 4), and related to T- (1) or B-cell development (2, 3 and 4), myeloid-derived dentritic cells (5) or hematopoietic stem cells (6), are detailed by heat map views. Illustration 1 includes human samples corresponding to thymic stromal cultures (TSCs), intrathymic T progenitor (ITTP) cells, ‘double positive' (DP) thymocytes, ‘single positive 4' (SP4) thymocytes, ‘naive' T cells from cord blood (CB4) and ‘naive' T cells from adult blood (AB4). Illustration 2 includes human samples corresponding to bone marrow plasma cells (BMPCs), memory B cells on day 0 of in vitro differentiation (D0 MBCs), activated B cells on day 4 (D4 ActBCs), plasmablasts on day 7 (D7 PBs), and plasma cells on day 10 of culture (D10 PCs). Illustration 3 includes samples from mouse corresponding to plasma cells (plasma), germinal center B cells (GC), naive B cells (naive B), and memory B cells (memory B). Illustration 4 gathers B samples from mouse at the following stages of development: Pre-BI, large Pre-BII, small Pre-BII, immature, and mature. Illustration 5 is on human immature monocyte–derived dendritic cells (MDDCs) untreated (control) or treated with vehicle alone (vehicle) and matured with galectin-1 (GAL-1) or lipopolysaccharide (LPS). Illustration 6 is on human hematopoietic progenitor cells (HPC) corresponding to prothymocytes (Pro-THYM), early pre-proB precursors (ProB), and lymphogranulomacrophagic precursors (Pro-LGM).
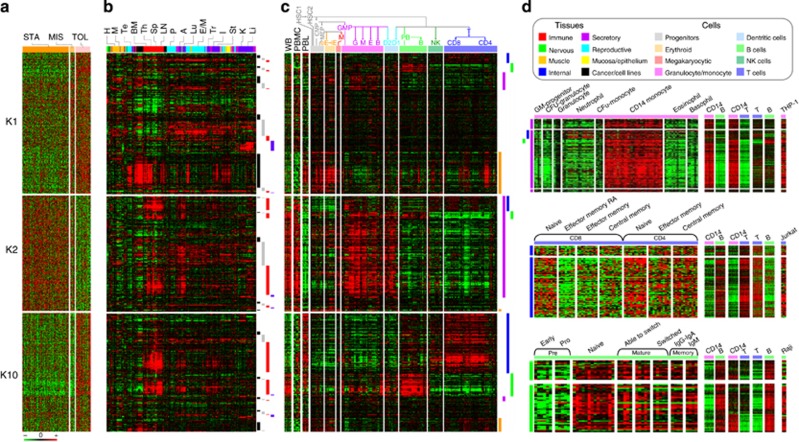
Virtual microdissection analysis revealed the clear participation of B cells, CD4 T lymphocytes, and monocytes in tolerance
The TOL signature (K1, K2, and K10; Figure 4a) was compared with clusters from various tissue (Figure 4b) and blood cell (Figure 4c) samples. This comparison identified K1 as a proliferation cluster (for example, AURKB, CCNB2, CDC20, CHEK1, NEK2, and PLK4) gathering 67% (P=6.31E−13) and 73% (P=1.04E−13), respectively, of the genes from proliferating tissues (for example, testis and skin; Figure 4b) or cells (early blood precursors; Figure 4c). K2 and K10 gathered 90% (P=2.30E−20) of immune tissue markers (for example, bone marrow, thymus, spleen, and lymph nodes; Figure 4b) showing their immunological specificity. K2 contained 82% (P=3.44E−85) of the granulocyte/monocyte lineage markers (Figure 4c) and corresponded to a CD14 monocyte cluster (for example, CD14, CD163, CD68, ITGAM, ITGB2, and PECAM1; Figure 4d). K10 contained 86% (P=5.12E−40) of the T lineage makers (Figure 4c) that were preferentially expressed in naive and differentiated CD4 subsets (for example, CD247, CD28, CD48, CD5, LAT, and MAL; Figure 4d). It also harbored 71% (P=9.31E−10) of the B lineage markers (Figure 4c) expressed in naive and differentiated subsets (for example, BLNK, CD22, CD40, CD79B, FCER2, and MS4A1; Figure 4d). Altogether, these data suggest the involvement of different cell subsets in tolerance, as attested by the specific expansion and differentiation of B and CD4 T cells, and the inhibition of monocyte functions.
Figure 4.
Identification of the cellular component by virtual microdissection analysis (VMDA). (a) Gene meta-signature of tolerance. Gene expression profiles (in rows, same order as (b and c) from the three most differential meta-clusters (K1, K2, and K10) are shown for the following clinical groups (in columns): control group comprising stable recipients under classical treatment (STA, dark orange) or minimal immunosuppression (MIS, light orange) and tolerant recipients (TOL, pink). Results are displayed by a heat map using red for gene overexpression and green for gene underexpression (see color scale). (b and c) Related gene profiles across a large compendium of human tissues (b) and blood cell types (c). A hierarchical clustering was performed on the whole matrix to group genes on the basis of similar profiles across the collection of samples. Results are displayed by expression heat-map views using the same color code. (b) Expression profiles across a large collection of tissues. The 1661 tissue samples (in columns) were ranged according to a hierarchical classification and annotated according to eight main categories (see legend): the immune (red), the nervous (green), the muscle (orange), the internal (dark blue), the secretory (purple), the reproductive (turquoise blue), the mucosa (yellow), and the cancer (black) types of samples. Some particular tissues from these categories are noted on the top: heart (H) and skeletal muscle (M) for the muscle type; testis (Te), prostate (P), and endometrium/myometrium (E/M) for the reproductive type; bone marrow (BM), thymus (Th), spleen (Sp), lymph node (LN) for the immune type; adipose (A), lung (Lu), trachea (Tr), intestine (I), stomach (St), kidney (K), and liver (Li) for the internal type. Four major gene clusters are indicated on the right by colored bars: black bar for overexpression in proliferating tissues; gray bar for ubiquitous overexpression across a large panel of tissue samples; red bar for specific overexpression in immune tissues; blue bar for preferential expression in the liver. (c) Expression profiles across a large compendium of blood cell types. The 681 cell samples (in columns) are ranged according to their respective positions in hematopoiesis (see tree on top) and pertain to eight main categories (see legend): the hematopoietic progenitors (gray) comprising the hematopoietic stem cells (HSC1–2) and the common myeloid (CMP) and myeloid/erythroid (MEP) progenitors; the erythroid lineage (orange) gathering early (eE) to late (lE) erythroid cells; the megakaryocytic category (pink) comprising megakaryocytes (M); the granulocyte/monocyte lineage (purple) comprising granulocyte/monocyte progenitors (GMP), granulocytes (G), monocytes (M), eosinophils (E), and basophils (B); the dentritic category (light blue) comprising dentritic cells (D1–2); the B-cell lineage (light green) gathering pre-B (PB) to mature B cells (B); the natural-killer (NK) lineage (dark green) comprising natural killers (NK); and the T-cell lineage (dark blue) gathering T CD8 (CD8) and T CD4 (CD4) lymphocyte populations. Expression from whole blood (WB), peripheral blood monocytes (PBMC), and peripheral lymphocytes (PL) is also shown on the left side. Four major gene clusters are indicated by colored bars on the right: orange bar for overexpression in proliferating progenitors; purple bar for overexpression in granulocyte/monocyte lineage; blue bar for higher expression in T-cell lineage; green bar for higher expression in B-cell lineage. (d) Tolerance expression profiles specificity of immune cell subtypes. The heat-map view on top details the genes predominantly overexpressed across cell types from the granulocyte/monocyte lineage (purple bar) including: granulocyte/monocyte (GM) progenitors, colony-forming unit (CFU) granulocytes, granulocytes, neutrophils, CFU and CD14 monocytes, eosinophils, and basophils. The heat-map view in middle shows the genes overexpressed in the CD8 and CD4 populations from the T lymphocyte lineage (blue bar) including the following cell subsets: naive, effector memory (TEM and TEMRA), and central memory (CM). The heat-map view on bottom depicts the genes having higher expression in cell subsets from the B-cell lineage (green bar) corresponding to: pre-B (including early and proB), naive B, mature B (gathering samples from B able to switch to fully switched B), and memory B (gathering immunoglobulin IgG and IgA and IgM-secreting B). For more details on the specificity of these gene expression profiles, results from pairwise comparisons (CD14 monocytes vs. B lymphocytes, CD14 monocytes vs. T lymphocytes, and T lymphocytes vs. B lymphocytes) along with expression in specific related cell lines (THP-1, Jurkat, and Raji) are also depicted on the top right of each heat-map view.
Generalization of the results using external data confirmed the biological relevance (functionality and cellularity) of the tolerance gene signature and its reproducibility
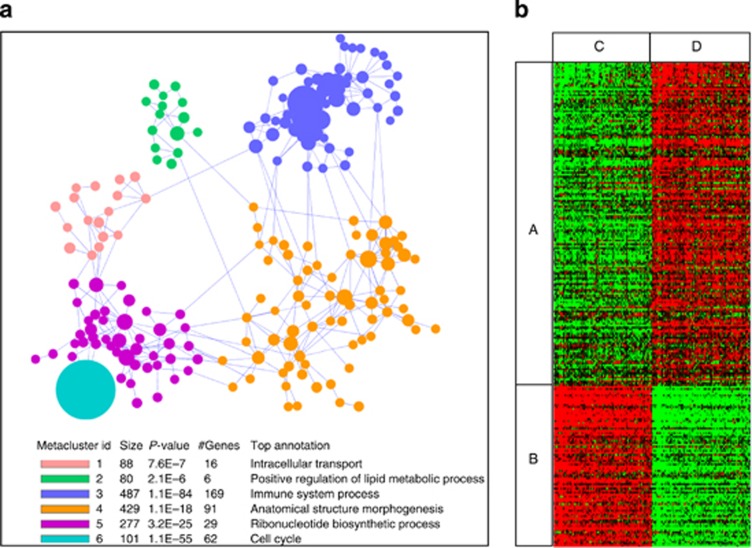
To assess the reproducibility of the signature with external data, similar trends of expression were searched in a collection of data sets using co-expression (Figure 5a) and rank (Figure 5b)-based approaches. A clustering analysis showed that recurrent partners of co-expression fell into six functionally relevant meta-clusters (Figure 5a). The two most significant meta-clusters (M3 and M6) gathered 169 (P=1.1E−84) and 62 (P=1.1E−55) genes that were involved in immune and cell cycle functions. In the second analysis (Figure 5b), 215 distinct pairs of samples significantly (area under curve<0.8; Q-value<10−11) harbored the same pattern in terms of rank differences. A text-mining analysis revealed that 70% (P=3.40E−14) of the matched studies were related to blood (Supplementary Figure SI12 online) or other cell subsets (mononuclear cells: P=3.45E−10; lymphocytes: P=2.88E−07; B cells: P=6.29E−03).
Figure 5.
Validation of the functionality and cellularity of the meta-signature of tolerance. (a) Similar neighbors of co-expression across public data sets. The results (co-expression-based meta-analysis) are visualized by a network representation of the six identified meta-clusters (see color legend and functional annotation) gathering 1462 genes. Nodes from the network are clusters (colored circles on the graph, size proportional to the number of genes they gather) of strongly connected genes (3–101 genes, density ⩾0.5) and edges intercluster density ⩾0.2. (b) Similar gene patterns across public data sets. The results (rank-based meta-analysis) are visualized by a heat map of the 244 top genes (in rows) discriminating tolerant (TOL) from stable (STA) recipients (P<0.005) across 215 sample pairs (in columns) from unique Gene Expression Omnibus (GEO) data sets. (A) Genes upregulated in TOL recipients; (B) genes downregulated in TOL recipients; (C) 215 samples having a ‘stable' (STA)-like pattern. (D) In all, 215 samples with a ‘tolerant' (TOL)-like pattern.
Altogether, these data show that genes from the signature are recurrently associated with the same functionally related neighbors of co-expression and display a TOL-like pattern in numerous blood cell–related studies.
Cross-validation analysis yielded good prediction performances and the use of the top-20 biomarkers enabled the experimental validation of the tolerance gene signature in new samples and new patients
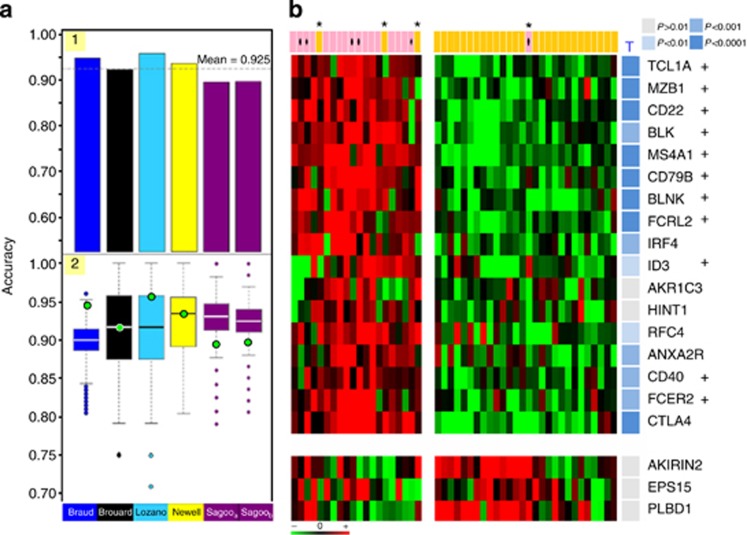
To statistically validate the signature, a cross-validation (Figure 6a) and an experimental validation (Figure 6b) were performed. A full sixfold cross-validation was performed on the set of 1846 genes (Figure 6a). Each initial data set was used as an external validation set, whereas the five others served as training sets for the selection of the top discriminating genes (panel 1). An optimal selection of top-200 genes (Supplementary Figure SI13 online), conserved at 76% across iterations, yielded good mean classification performances (Supplementary Figure SI6 online): 92.5% accuracy, 76.3% sensitivity, 98.2% specificity, 91.1% positive predictive value, and 91.0% negative predictive value. These performances were not compromised regardless of the data set used for test (panel 2).
Figure 6.
Reproducibility of the meta-signature of tolerance. (a) Cross-validation of the meta-signature. In panel 1, the classification performances through the six cross-validation folds are shown. Results are displayed with a histogram in which each bar (one of the six folds) represents the accuracy obtained on one data set used as test, whereas learning is performed on the five others. From left (first fold) to right (sixth fold), tests were data sets from Braud (dark blue), Brouard (black), Lozano (turquoise blue), Newell (yellow), Sagooa (European ‘Indices of Tolerance' (IOT) cohort, purple), and Sagoob (American ‘Immune Tolerance Network' (ITN) cohort, purple). In panel 2, the influence of the origin of a data set on the performances of classification is assessed. The accuracy from the six test data sets (green circle) is compared with the accuracy obtained on comparable test data sets (equal size and same sample composition) constructed by a random selection of tolerant (TOL) and stable (STA) samples from the total pool of samples (whatever the study of origin). Results are depicted by box plots (boxes: interquartile range (IQR); whiskers: 1.5 × IQR) corresponding each to the values obtained after 1000 repeated random selections. Values beyond the range are considered outliers and shown as circles. (b) Experimental validation of the meta-signature. The expression of the 20 top ranked biomarkers discriminating tolerant (TOL) from stable (STA) recipients is assessed in a new collection of 48 samples. The results from real-time PCR are displayed by an expression heat map (red for gene overexpression and green for gene underexpression) showing the patterns from 18 TOL (pink; dot: new cases) and 30 STA (orange). Misclassified samples are denoted by an asterisk. Significance of the individual markers (17 upregulated and 3 downregulated) is assessed by a Student's t-test: resulting P-values are depicted by shades of blue on the right side (see color legend). From the 20 genes, those corresponding to B-cell–related markers are quoted by a cross.
Second, a limited selection of the top-20 markers, mostly centered on B cells and overexpressed in TOL (Supplementary Figure SI14 online), was sufficient to accurately discriminate TOL from STA (Supplementary Figure SI6 online). One of these markers, IRF4, was also identified by gene list comparison (Supplementary Figure SI4 online). Their quantitative measurement by real-time PCR (independent technique) on a new collection of 67 samples (Figure 6b, Supplementary Figure SI15 online) yielded strong discrimination of TOL and STA (P=4.39E−9): 16 markers (80%) were indeed altered (P<0.05) in the same sense. Leave-one-out prediction yielded excellent reproducibility (100% recall) on the 12 new time samples from already analyzed TOL cases and good external validation on the six new TOL cases (83.3% recall). Good classification (91.7%) was achieved (94.4% sensitivity, 90% specificity, 85% positive predictive value, and 96.4% negative predictive value), with one TOL and three STA being misclassified (Supplementary Figure SI16 online).
Altogether, these data show that the signature can be revalidated regardless of the study, the technology, and the sample provided.
A ‘healthy' profile of the top 20 biomarkers in blood from tolerant patients
Of the 20 biomarkers, TOL and HV were closely related as no (0 gene, P<0.001) or minor difference (three genes, P<0.05) could be detected in meta-analysis (Supplementary Figure SI14 online) and RT-PCR (Supplementary Figure SI15 online) sets. This similarity between TOL and HV was reinforced by the observation that only 68 genes out of the 1846 analyzed were differential (P<0.001; not shown). Conversely, 18 of the markers (90%) were also differential between STA and HV, both in the meta-analysis (P<0.001) and in the RT-PCR (P<0.05) sets (Supplementary Figures SI14 and SI15 online). Altogether, these data show that TOL and HV display roughly the same ‘healthy' profile.
Discussion
Five studies were attempted to analyze tolerance through noninvasive blood transcriptomic analyses.10, 11, 12, 13, 14 However, no significant overlap in the expression pattern could emerge.12
In a first attempt to derive a consensus gene signature, we performed comparisons of gene lists generated from the five studies. Although valuable,28 this approach unfortunately led to a limited 14-gene consensus, which is not sufficient to accurately detect tolerant among recipients. This result matches with our previous observations12 as, by comparison, no overlap emerged from the five studies using a per-gene selection (Student's t-test, 5% false discovery rate). Such discrepancies, classically observed,29, 30, 31, 32, 33 raise questions about the reproducibility, validity, and biological significance of microarray outcomes34 when tested on independent data sets.35 Inconsistencies are related to several factors. The different microarray platforms contain probes pertaining to different gene collections, ranging from dedicated10, 11, 14 to all-genome.12, 13 Such a disparity makes comparisons difficult36 and it could reflect the poor percentage of cross-validated genes (<1%). For that reason, the intersection of these platform gene lists is the only genes to be retained for further analysis. Microarray outcomes are also often limited by the small number of samples while analyzing thousands of genes,37, 38 leading to underestimation of variances and reduction of statistic power.39 This can be exacerbated in our situation knowing that blood samples can deeply vary between individuals40, 41, 42 or even the day the sample was taken.42, 43 In addition, the heterogeneity of the clinical outcome of these patients on the long term may add to these problems.7 Moreover, gene lists are also greatly influenced by the subsets of the patients used for their selection, even in the same study,35 and this could reflect the differences between the two cohorts (IOT and ITN) in the study by Sagoo et al..14
In light of this, it has been suggested that more samples are required to reach a decent level of marker stability.44 This was the objective of our second attempt, consisting of the integration of all the initial data sets. Most of the meta-analysis approaches first identify a set of common genes across studies from which they then derive a gene expression signature.36 Because of different data formats and experimental effects,45, 46 direct comparison of raw data is difficult, not straightforward, and sometimes impossible even using standard normalization techniques.47, 48 Standardization of the data obtained from individual research into values derived from a common scale before combination has been successfully applied.49 We adopted this solution to identify a robust signature from the set of 1846 genes.
Our gene signature was defined owing to the meta-analysis of blood transcriptome studies comparing TOL with the more related group of STA patients. This group of patients was chosen as the most appropriate cohort to look at tolerance markers to identify the patients who may benefit from an IS-weaning protocol in the future. Of course, blood may not be the best compartment. However, disease gene profiles showed good concordance between blood and solid tissues.50 In transplantation, gene changes in blood correlate with biopsy-proven rejection.51, 52, 53 Moreover, rare cases of biopsies from tolerant cases did not evidence graft infiltrate,8 suggesting that biopsy may not be informative. Conversely, blood is a promising source of therapeutic molecules,54 and as it flows throughout the body it acts as a pipeline for the immune system and may be a good compartment to analyze complex patterns of recirculation. In tolerance, gene changes observed in blood may thus reflect active processes involving peripheral regulation55 as attested by the characterization of regulatory molecules56 and the possibility to induce tolerance with peripheral lymphocytes in animal models.57
In this context, several cell types (B, CD4 T, and monocytes) may participate in the regulatory mechanisms as attested by related markers. Among all, the confirmation and identification of new B-cell markers appeared thus as one of the interesting findings reinforcing a suspected role for B lymphocytes in tolerance.58 Indeed, B cells are able to drive immune responses59 and there is compelling evidence that they act as multifaceted regulatory cells.60, 61, 62, 63, 64 They may have a crucial role in regulating other cell subtypes reflecting the observed gene changes linked to CD4 T cells and CD14 monocytes. Moreover, although deep depletion of the B-cell compartment is associated with higher incidence rejection,65 its preservation would be a prerequisite to favor the development of tolerance.66 Interestingly, we found that most of the genes differentially expressed in TOL and STA were unchanged between TOL and HV both through the top-20 genes and the whole meta-analysis. Similar results were observed in previous studies both at transcriptional and cell phenotype levels.13, 66 This suggests that, as previously mentioned,67 tolerant patients may harbor a global preservation of their ‘phenotype', especially the B-cell compartment. It may contribute to maintain a physiological cell homeostasis counteracting inflammation and preserving a ‘healthy profile' in these patients. To ascertain that our results are not the only reflection of the absence of treatment, we also look at these markers in patients with chronic rejection and off IS available in the study by Brouard et al.11 As these patients harbor a highly differential profile from TOL, we reasonably exclude an effect of the absence of treatment (Supplementary Figure SI14 online). Even more, these markers, particularly the B-cell ones, were not present in drug-free, operationally tolerant liver recipients.12 Accordingly, operational tolerance in kidney transplants is more often detected in patients who have carried the graft, and thus IS, for a long time.7, 8 In this case, long IS may create a immune restart68, 69 towards a homeostatic equilibrium, as the one observed in healthy volunteers.67
Finally, to assess the reliability of our signature, we first performed a full cross-validation procedure. This analysis yielded good predictions and enabled the validation of B cell–related markers (for example, BLK, BLNK, CD22, CD40, CD79B, CD83, FCER2, FCRL2, and MS4A1) but also genes related to other cell subsets including CD4 T (for example, CD150, CD28, CD52, and NFATC1) and CD14 monocyte (for example, CCR2, CD163, ITGAM, and ITGB2) molecules. For diagnosis purpose, a selection of the top-20 markers from this list, mostly centered on B cells, accurately discriminate tolerant from stable recipients. In a second step, these 20 markers were experimentally revalidated in an independent cohort of new TOL samples, from which six corresponded to new cases. These results showed that our initial findings were not dependent on the technology used or the analyzed set of samples. Both analyses yielded good prediction performances (more than 90%). Hence, our biomarkers could be reliably used to detect tolerance and stratify kidney recipients in clinics. First, they may help for a better follow-up of the tolerant patients. Indeed, several lines of evidence suggest that tolerance is likely not a stable situation for ‘entire life' for most of the studied cases.7 In such situations, immunotherapy could be reinstated before degradation of the graft. Second, these biomarkers may help to monitor kidney-transplanted recipients under classical IS. Such stable cases, presenting a low risk of rejection, would thus be highly eligible for progressive IS weaning. In our meta-analysis, 3% (eight cases) of the STA patients did express this signature and further examination of their clinical status revealed that they were still stable without degradation of renal function, years after the test. This result agreed previous observations ranking from 3.5 to 15%.11, 14, 70, 71 However, only prospective studies of IS weaning in a controlled and randomized setting will enable proof of concept of this hypothesis.
In conclusion, our results indicated the participation of different immune cell subsets in natural operational tolerance. Among them, B cells certainly have a role in the maintenance of tolerance, reinforcing our previous observations. Although the implied regulations are still largely unknown, preservation of this compartment and maintenance of a physiological homeostasis and ‘healthy profile' seem to be necessary for tolerance and may drive current therapies. According to its independent validation and its worldwide origin as collaboration between different teams, we hope that our signature, especially the restricted set of 20 markers, will aid in the identification of ‘low-risk' patients among cohorts of transplanted recipients as recently performed for heart allograft rejection.72
Materials and Methods
Data collection
The data used in this study were published10, 11, 12, 13, 14 and are publicly available. The five microarray data sets and related information on samples were retrieved from Gene Expression Omnibus. They are referred to by the first author of the original publication and include studies from Braud (GSE47755),10 Brouard (GSE47683),11 Lozano (GSE22707),12 Newell (GSE22229),13 and Sagoo (GSE14655)14 (Table 1). Altogether, 596 samples were available (equivalent to 932 distinct hybridizations), gathering 62 samples from HV, 96 samples corresponding to 50 unique operationally TOL (Supplementary Figure SI17 online), 32 samples from recipients under MIS, 311 samples from long-term stable recipients under classical immunosuppressive therapy (STA), 81 samples from patients with CR, and 14 samples from patients undergoing AR. Some of the TOL patients were assessed in more than one study (Supplementary Figure SI17 online): according to extremely good interstudy correlations (Supplementary Figure SI18 online), and as most of these samples were collected at different time-points and processed on different platforms, they were thus analyzed as unique samples. The clinical definition of the patient groups has been described previously.10, 11, 12, 13, 14
Table 1. Summary of the five tolerance-related studies used for the meta-analysis.
| Study | PMID | GEO no. | GPL ID | Description | Total | HV | TOL | MIS | STA | CR | AR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Braud | 17910029 | GSE47755 | GPL8798 | Cancerochip (∼7000 genes)a,c | 250 (528) | 8 (16) | 21 (54) | 0 | 190 (380) | 31 (78) | 0 |
| Brouard | 17873064 | GSE47683 | GPL6271 | Lymphochip (∼18,000 genes)a,c | 67 | 8 | 12 | 10 | 12 | 11 | 14 |
| Lozano | 21827613 | GSE22707 | GPL570 | Affymetrix HG-U133_Plus_2 (∼33,000 genes)b,d | 42 | 6 | 12 | 0 | 12 | 12 | 0 |
| Newell | 20501946 | GSE22229 | GPL570 | Affymetrix HG-U133_Plus_2 (∼33000 genes)b,d | 58 | 12 | 19 | 0 | 27 | 0 | 0 |
| Sagoo IOT | 20501943 | GSE14655 | GPL8136 | RISET 2.0 Agilent custom (∼5000 genes)b,c | 74 (95) | 8 | 10 (13) | 11 (16) | 36 (48) | 9 (10) | 0 |
| Sagoo ITN | 20501943 | GSE14655 | GPL8136 | RISET 2.0 Agilent custom (∼5000 genes)b,c | 105 (142) | 20 | 22 (31) | 11 (14) | 34 (52) | 18 (25) | 0 |
| 596 (932) | 62 (70) | 96 (141) | 32 (40) | 311 (531) | 81 (136) | 14 |
Abbreviations: AR, acute rejection; CR, chronic rejection; GEO, Gene Expression Omnibus; HV, healthy volunteer; MIS, minimally immunosuppressed; STA, stable under classical treatment; TOL, tolerant.
For each study, the numbers of samples in each group—HV, TOL, MIS, STA, CR, AR—are given (brackets: number of hybridizations). Technical information on the microarray platform (GPL ID) is also provided (a: two-channel; b: single-channel; c: dedicated; d: whole genome). Study from Sagoo comprises two independent cohorts (EU IOT: ‘Indices of Tolerance' US ITN: ‘Immune Tolerance Network') sponsored, respectively.
Meta-analysis
To identify a gene signature of tolerance, the comparison was focused on the TOL group (n=96) and patients with stable graft function (n=343) either under standard (STA, n=311) or minimal immunotherapy (MIS, n=32). To this end, we performed two types of meta-analyses. The first captures in each individual data set the clusters of differential genes between the two groups and identifies the overlap as a consensus gene set.12 The second relies on the integration of the different data sets as a single corpus of data73 and identifies, after an analysis similar to the one performed on the individual data sets, the clusters of differentially expressed genes. Details about the procedures can be found online (Supplementary Figure SI19 online) along with MOOSE and PRISMA flowcharts (Supplementary Figure SI20 online).
Reprocessing, integration, and analysis
Data sets were renormalized using a Lowess procedure, log-transformed, and median-centered on genes as previously described.74, 75, 76, 77, 78 Probe annotation was performed using MADGene79 to convert and match the genes across array platforms. Data sets were standardized73 according to the STA group, before their integration and merging on the 1846 consensus genes. The relationship between genes and samples was investigated by hierarchical clustering using the Cluster program.80
Individualization of gene clusters and analysis
Ten clusters per data set (Supplementary Figure SI21 online) were individualized by K-means81 and further analyzed by hierarchical clustering.80 They were functionally interpreted by gene ontology,82 gene set,28 and virtual microdissection83 analyses (Supplementary Figure SI22 online). Their ability to statistically discriminate TOL and STA was evaluated by the Student's t-84 and Fisher's exact85 tests. The discriminative clusters thus defined the ‘signature' as previously shown.20
Independent reproducibility of the results
To confirm the functional and cellular components of the signature, we mined a large collection of 4658 public data sets from Gene Expression Omnibus (Supplementary Figure SI23 online). Two blind analyses were performed, supported by the conservation of co-expressed genes across studies86 and rank-based differences between two biological situations.87
Cross-validation and classification performances
The method was based on the following three parts: a gene selection based on T statistics,88 a sample classifier based on support vector machine,13 the best accurate one in our conditions (Supplementary Figure SI24 online), and a performance evaluation through cross-validation analysis.89 The quality of the gene selection was determined by the classification performances (Supplementary Figure SI25 online) of the predictor over the six data sets.
External validation of the signature by real-time PCR
To confirm our results, we investigated the expression of the top-20 discriminating biomarkers (T statistics selection) by real-time PCR (TLDA cards, Applied Biosystems, Foster City, CA). Experiments were performed using new samples from 19 HV and 48 cases (30 STA, 18 TOL) collected from French transplantation centers (Supplementary Figure SI26 online). The 18 TOL samples (details in Brouard et al.7) correspond to 12 already analyzed cases and 6 new cases (Supplementary Figure SI27 online). The discriminating capacity of the data was then evaluated by a leave-one-out strategy.
Acknowledgments
We thank M Goldman, M Steenman and G Bléas for critical reading and editing the manuscript. We are grateful to the IOT, ITN, and RISET consortia for their collaborative effort in the study of tolerant cohorts. We also acknowledge patients and their families whose trust, support, and cooperation were essential for the collection of the data used in this study. They thank Professors (G Blancho, J Dantal, M Hourmant, C Legendre, C Noel, and J-F Subra) and Doctors (L Braun, D Cantarovitch, R Crochette, A Garnier, B Hurault de Ligny, H Janbon, G Lefrancois, B Le Mauff, H Le Monies De Sagazan, A Meurette, M-C Moal, E Pillebout, M Rabant, A Testa, D Ribes, F Villemain, and J Zuber) for their help in sample collection. This research was supported by INSERM and IHU-CESTI institutes, CENTAURE and ROTRF foundations, Transplantex Labex and ABM agency.
All the authors declared no competing interests.
Footnotes
SUPPLEMENTARY MATERIAL
Figure SI1. Lists the K-clusters from the initial studies.
Figure SI2. Lists cluster pairwise comparisons.
Figure SI3. Lists the 19 differential K-clusters.
Figure SI4. Lists the 14 consensus genes.
Figure SI5. Lists the markers from the original studies.
Figure SI6. Lists the classification performances.
Figure SI7. Lists GO categories for the 1846 genes.
Figure SI8. Lists the 595 gene signature.
Figure SI9. Lists significant terms from GOA.
Figure SI10. Lists significant overlaps from GSA.
Figure SI11. Lists significant overlaps with transcriptional modules.
Figure SI12. Lists significant terms from the text mining analysis.
Figure SI13. Lists the top-200 genes.
Figure SI14. Shows expression of the top-20 markers across samples from the meta-analysis.
Figure SI15. Shows expression of the top-20 markers in the experimental validation set.
Figure SI16. Gives classification performances from the experimental validation set.
Figure SI17. Details numbers of tolerant cases common between studies.
Figure SI18. Details correlation between tolerant samples.
Figure SI19. Gives information and details on procedures.
Figure SI20. Shows MOOSE & PRISMA flowcharts.
Figure SI21. Gives details on the silhouette analysis.
Figure SI22. Lists data sets used for VMDA analyses.
Figure SI23. Lists the 4658 GSE data sets used for rank-based and co-expression-based analyses.
Figure SI24. Details comparison of classifiers.
Figure SI25. Details the calculation of performance metrics.
Figure SI26. Gives clinical data of HV, TOL and STA patients.
Figure SI27. Gives detailed clinical data of the six new tolerant cases.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
Supplementary Material
References
- Pascual M, Theruvath T, Kawai T, et al. Strategies to improve long-term outcomes after renal transplantation. N Engl J Med. 2002;346:580–590. doi: 10.1056/NEJMra011295. [DOI] [PubMed] [Google Scholar]
- Dantal J, Hourmant M, Cantarovich D, et al. Effect of long-term immunosuppression in kidney-graft recipients on cancer incidence: randomised comparison of two cyclosporin regimens. Lancet. 1998;351:623–628. doi: 10.1016/S0140-6736(97)08496-1. [DOI] [PubMed] [Google Scholar]
- Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med. 2007;357:2601–2614. doi: 10.1056/NEJMra064928. [DOI] [PubMed] [Google Scholar]
- Soulillou JP, Giral M. Controlling the incidence of infection and malignancy by modifying immunosuppression. Transplantation. 2001;72:S89–S93. [PubMed] [Google Scholar]
- Wimmer CD, Rentsch M, Crispin A, et al. The janus face of immunosuppression - de novo malignancy after renal transplantation: the experience of the Transplantation Center Munich. Kidney Int. 2007;71:1271–1278. doi: 10.1038/sj.ki.5002154. [DOI] [PubMed] [Google Scholar]
- Lechler RI, Garden OA, Turka LA. The complementary roles of deletion and regulation in transplantation tolerance. Nat Rev Immunol. 2003;3:147–158. doi: 10.1038/nri1002. [DOI] [PubMed] [Google Scholar]
- Brouard S, Pallier A, Renaudin K, et al. The natural history of clinical operational tolerance after kidney transplantation through twenty-seven cases. Am J Transplant. 2012;12:3296–3307. doi: 10.1111/j.1600-6143.2012.04249.x. [DOI] [PubMed] [Google Scholar]
- Roussey-Kesler G, Giral M, Moreau A, et al. Clinical operational tolerance after kidney transplantation. Am J Transplant. 2006;6:736–746. doi: 10.1111/j.1600-6143.2006.01280.x. [DOI] [PubMed] [Google Scholar]
- Orlando G, Hematti P, Stratta RJ, et al. Clinical operational tolerance after renal transplantation: current status and future challenges. Ann Surg. 2010;252:915–928. doi: 10.1097/SLA.0b013e3181f3efb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud C, Baeten D, Giral M, et al. Immunosuppressive drug-free operational immune tolerance in human kidney transplant recipients: Part I. Blood gene expression statistical analysis. J Cell Biochem. 2008;103:1681–1692. doi: 10.1002/jcb.21574. [DOI] [PubMed] [Google Scholar]
- Brouard S, Mansfield E, Braud C, et al. Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci USA. 2007;104:15448–15453. doi: 10.1073/pnas.0705834104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano JJ, Pallier A, Martinez-Llordella M, et al. Comparison of transcriptional and blood cell-phenotypic markers between operationally tolerant liver and kidney recipients. Am J Transplant. 2011;11:1916–1926. doi: 10.1111/j.1600-6143.2011.03638.x. [DOI] [PubMed] [Google Scholar]
- Newell KA, Asare A, Kirk AD, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010;120:1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagoo P, Perucha E, Sawitzki B, et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest. 2010;120:1848–1861. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesneau M, Pallier A, Braza F, et al. Unique B cell differentiation profile in tolerant kidney transplant patients. Am J Transplant. 2014;14:144–155. doi: 10.1111/ajt.12508. [DOI] [PubMed] [Google Scholar]
- Louis S, Braudeau C, Giral M, et al. Contrasting CD25hiCD4+T cells/FOXP3 patterns in chronic rejection and operational drug-free tolerance. Transplantation. 2006;81:398–407. doi: 10.1097/01.tp.0000203166.44968.86. [DOI] [PubMed] [Google Scholar]
- Pallier A, Hillion S, Danger R, et al. Patients with drug-free long-term graft function display increased numbers of peripheral B cells with a memory and inhibitory phenotype. Kidney Int. 2010;78:503–513. doi: 10.1038/ki.2010.162. [DOI] [PubMed] [Google Scholar]
- Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Rovegno F, Mooney D, et al. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene. 2007;401:12–18. doi: 10.1016/j.gene.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Chaussabel D, Quinn C, Shen J, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher JA, Hashimi ST, Levroney EL, et al. Galectin-1-matured human monocyte-derived dendritic cells have enhanced migration through extracellular matrix. J Immunol. 2006;177:216–226. doi: 10.4049/jimmunol.177.1.216. [DOI] [PubMed] [Google Scholar]
- Haddad R, Guardiola P, Izac B, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood. 2004;104:3918–3926. doi: 10.1182/blood-2004-05-1845. [DOI] [PubMed] [Google Scholar]
- Hoffmann R, Seidl T, Neeb M, et al. Changes in gene expression profiles in developing B cells of murine bone marrow. Genome Res. 2002;12:98–111. doi: 10.1101/gr.201501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan M, Caraux A, De VJ, et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009;114:5173–5181. doi: 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Hanspers K, Barker CS, et al. Gene expression profiles during human CD4+ T cell differentiation. Int Immunol. 2004;16:1109–1124. doi: 10.1093/intimm/dxh112. [DOI] [PubMed] [Google Scholar]
- Luckey CJ, Bhattacharya D, Goldrath AW, et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- Miklos GL, Maleszka R. Microarray reality checks in the context of a complex disease. Nat Biotechnol. 2004;22:615–621. doi: 10.1038/nbt965. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Microarrays and molecular research: noise discovery. Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- Moreau Y, Aerts S, Moor BD, et al. Comparison and meta-analysis of microarray data: from the bench to the computer desk. Trends Genet. 2003;19:570–577. doi: 10.1016/j.tig.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Campain A, Yang YH. Comparison study of microarray meta-analysis methods. BMC Bioinformatics. 2010;11:408. doi: 10.1186/1471-2105-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Breitling R. A comparison of meta-analysis methods for detecting differentially expressed genes in microarray experiments. Bioinformatics. 2008;24:374–382. doi: 10.1093/bioinformatics/btm620. [DOI] [PubMed] [Google Scholar]
- Choi JK, Yu U, Kim S, et al. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19:i84–i90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- Cobb JP, Mindrinos MN, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci USA. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady JJ, Wortley GM, Wormstone YM, et al. Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiol Genomics. 2005;22:402–411. doi: 10.1152/physiolgenomics.00080.2005. [DOI] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radich JP, Mao M, Stepaniants S, et al. Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics. 2004;83:980–988. doi: 10.1016/j.ygeno.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Berndt ML, Williams A, et al. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32:e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Barrette TR, Rubin MA, et al. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- Dudley JT, Tibshirani R, Deshpande T, et al. Disease signatures are robust across tissues and experiments. Mol Syst Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz PA, Tsai EJ, Putt ME, et al. Detection of cardiac allograft rejection and response to immunosuppressive therapy with peripheral blood gene expression. Circulation. 2004;110:3815–3821. doi: 10.1161/01.CIR.0000150539.72783.BF. [DOI] [PubMed] [Google Scholar]
- Vasconcellos LM, Schachter AD, Zheng XX, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66:562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- Villeda SA, Plambeck KE, Middeldorp J, et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med. 2014;20:659–663. doi: 10.1038/nm.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuskirk AM, Burlingham WJ, Jankowska-Gan E, et al. Human allograft acceptance is associated with immune regulation. J Clin Invest. 2000;106:145–155. doi: 10.1172/JCI9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger R, Pallier A, Giral M, et al. Upregulation of miR-142-3p in peripheral blood mononuclear cells of operationally tolerant patients with a renal transplant. J Am Soc Nephrol. 2012;23:597–606. doi: 10.1681/ASN.2011060543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino M, Kitazawa Y, Kawasaki M, et al. Differences in lymphocyte gene expression between tolerant and syngeneic liver grafted rats. Liver Transplant. 2004;10:379–391. doi: 10.1002/lt.20082. [DOI] [PubMed] [Google Scholar]
- Kirk AD, Turgeon NA, Iwakoshi NN. B cells and transplantation tolerance. Nat Rev Nephrol. 2010;6:584–593. doi: 10.1038/nrneph.2010.111. [DOI] [PubMed] [Google Scholar]
- LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Jensen PE. Cutting edge: primary B lymphocytes preferentially expand allogeneic FoxP3+ CD4 T cells. J Immunol. 2007;179:2046–2050. doi: 10.4049/jimmunol.179.4.2046. [DOI] [PubMed] [Google Scholar]
- Deng S, Moore DJ, Huang X, et al. Cutting edge: transplant tolerance induced by anti-CD45RB requires B lymphocytes. J Immunol. 2007;178:6028–6032. doi: 10.4049/jimmunol.178.10.6028. [DOI] [PubMed] [Google Scholar]
- Fillatreau S, Sweenie CH, McGeachy MJ, et al. B cells regulate autoimmunity by provision of IL-10. Nat Immunol. 2002;3:944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- Heyzer-Williams LJ, Heyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- Moulin V, Andris F, Thielemans K, et al. B lymphocytes regulate dendritic cell (DC) function in vivo: increased interleukin 12 production by DCs from B cell-deficient mice results in T helper cell type 1 deviation. J Exp Med. 2000;192:475–482. doi: 10.1084/jem.192.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy MR, Watson CJ, Plotnek G, et al. B-cell-depleting induction therapy and acute cellular rejection. N Engl J Med. 2009;360:2683–2685. doi: 10.1056/NEJMc0808481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva HM, Takenaka MC, Moraes-Vieira PM, et al. Preserving the B-cell compartment favors operational tolerance in human renal transplantation. Mol Med. 2012;18:733–743. doi: 10.2119/molmed.2011.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho V, Saitovitch D, Kalil J, et al. Rethinking the multiple roles of B cells in organ transplantation. Curr Opin Organ Transplant. 2013;18:13–21. doi: 10.1097/MOT.0b013e32835c8043. [DOI] [PubMed] [Google Scholar]
- Haynes LD, Jankowska-Gan E, Sheka A, et al. Donor-specific indirect pathway analysis reveals a B-cell-independent signature which reflects outcomes in kidney transplant recipients. Am J Transplant. 2012;12:640–648. doi: 10.1111/j.1600-6143.2011.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes-Vieira PM, Takenaka MC, Silva HM, et al. GATA3 and a dominant regulatory gene expression profile discriminate operational tolerance in human transplantation. Clin Immunol. 2012;142:117–126. doi: 10.1016/j.clim.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Brouard S, Le BA, Dufay A, et al. Identification of a gene expression profile associated with operational tolerance among a selected group of stable kidney transplant patients. Transplant Int. 2011;24:536–547. doi: 10.1111/j.1432-2277.2011.01251.x. [DOI] [PubMed] [Google Scholar]
- Zoller KM, Cho SI, Cohen JJ, et al. Cessation of immunosuppressive therapy after successful transplantation: a national survey. Kidney Int. 1980;18:110–114. doi: 10.1038/ki.1980.116. [DOI] [PubMed] [Google Scholar]
- Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Do KA, Wen S, et al. Merging microarray data, robust feature selection, and predicting prognosis in prostate cancer. Cancer Inform. 2006;2:87–97. [PMC free article] [PubMed] [Google Scholar]
- Baron D, Montfort J, Houlgatte R, et al. Androgen-induced masculinization in rainbow trout results in a marked dysregulation of early gonadal gene expression profiles. BMC Genomics. 2007;8:357. doi: 10.1186/1471-2164-8-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron D, Magot A, Ramstein G, et al. Immune response and mitochondrial metabolism are commonly deregulated in DMD and aging skeletal muscle. PLoS ONE. 2011;6:e26952. doi: 10.1371/journal.pone.0026952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron D, Dubois E, Bihouee A, et al. Meta-analysis of muscle transcriptome data using the MADMuscle database reveals biologically relevant gene patterns. BMC Genomics. 2011;12:113. doi: 10.1186/1471-2164-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamirault G, Meur NL, Roussel JC, et al. Molecular risk stratification in advanced heart failure patients. J Cell Mol Med. 2010;14:1443–1452. doi: 10.1111/j.1582-4934.2009.00913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quille ML, Carat S, Quemener-Redon S, et al. High-throughput analysis of promoter occupancy reveals new targets for Arx, a gene mutated in mental retardation and interneuronopathies. PLoS ONE. 2011;6:e25181. doi: 10.1371/journal.pone.0025181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron D, Bihouee A, Teusan R, et al. MADGene: retrieval and processing of gene identifier lists for the analysis of heterogeneous microarray datasets. Bioinformatics. 2011;27:725–726. doi: 10.1093/bioinformatics/btq710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, et al. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Ross DT, Perou CM, et al. Towards a novel classification of human malignancies based on gene expression patterns. J Pathol. 2001;195:41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- Boorsma A, Foat BC, Vis D, et al. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 2005;33:W592–W595. doi: 10.1093/nar/gki484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein P, Puard V, Chevalier C, et al. Genomic assessment of human cumulus cell marker genes as predictors of oocyte developmental competence: impact of various experimental factors. PLoS ONE. 2012;7:e40449. doi: 10.1371/journal.pone.0040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, et al. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Araki M, Kunimoto R, et al. GEM-TREND: a web tool for gene expression data mining toward relevant network discovery. BMC Genomics. 2009;10:411. doi: 10.1186/1471-2164-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K, et al. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.