Abstract
The circadian system constrains an organism's palette of behaviors to portions of the solar day appropriate to its ecological niche. The central light-entrained clock in the suprachiasmatic nucleus (SCN) of the mammalian circadian system has evolved a complex network of interdependent signaling mechanisms linking multiple distinct oscillators to serve this crucial function. However, studies of the mechanisms controlling SCN development have greatly lagged behind our understanding of its physiological functions. We review advances in the understanding of adult SCN function, what has been described about SCN development to date, and the potential of both current and future studies of SCN development to yield important insights into master clock function, dysfunction, and evolution.
Keywords: circadian, suprachiasmatic, developmental neuroscience, cell fate, transcription factors, clock genes, chronotype, axon pathfinding
From sundials to watches: the evolution of the central clock
From cyanobacteria to humans, a circadian system restricting behavior to specific intervals in the solar day is present. Its ubiquity testifies to the system's utility, allowing organisms to maximize energy efficiency while minimizing environmental risks variant within distinct niche-specific ranges over the solar cycle. In mammals, circadian rhythms regulate crucial homeostatic processes including feeding, metabolism, sleep/wake, hormone secretion, body temperature, waste elimination, and arousal.
In most life forms, molecular clocks control the circadian system on a cellular level. Generally, this takes the form of a negative feedback loop, with a heterodimeric transcriptional complex (in mammals, Bmal1/Clock) driving the expression of genes which feed back to inhibit their own transcription (in mammals, Per and Cry genes). Various sequestration and degradation mechanisms can delay or accelerate this inhibition, letting environmental cues such as food availability, exercise, external temperature, and most importantly light modify the molecular clock's phase and period (reviewed in Buhr and Takahashi, 2013; Yamanaka et al., 2013; Refinetti, 2015).
However, as multicellular organisms arose, some cells became isolated from the primary circadian zeitgeber, or “time-giver”: the sun. In response, specialization began. Many nervous system subdivisions were still accessible to light, and remained photosensitive. In non-mammalian species, this included not just retina, but also the pineal gland (Kappers, 1981; Cahill, 1996) and other deep-brain photoreceptors (von Frisch, 1911; Hartwig and van Venn, 1979; Moore and Whitmore, 2014). The clockworks in these tissues sometimes became synchronous, self-sustaining oscillators, and began to utilize both synaptic and humoral signals such as melatonin to communicate solar time to tissues cut off from direct light input (Steele et al., 2003, 2006; Noche et al., 2011; reviewed in Underwood et al., 2001). In many species, some of these self-sustaining oscillators are essential alone or in combination to coordinate behavioral rhythms. For instance, the eyes are necessary for behavioral rhythmicity in quail, the pineal is essential in many species of sparrows, and both play important roles in pigeons (reviewed in Underwood et al., 2001).
However, in mammals, all of these tissues except the retina lost their photosensitivity, and self-sustaining oscillators in most brain regions outside the suprachiasmatic nucleus (SCN) are short-lived or absent entirely, with a few notable exceptions such as the olfactory bulb and retina (Tosini and Menaker, 1996; Abe et al., 2002; Granados-Fuentes et al., 2004). To fill this void, the SCN evolved to become the mammalian brain's master light-entrained clock. This required a substantial change in the region's gene expression and function; for example, the SCN of lower vertebrates such as frogs express a somewhat different suite of neurotransmitters than in mammals, and the amphibian SCN appears to modulate light-dependent control of skin pigmentation, an ability that it lacks in higher vertebrates (Tuinhof et al., 1994a; Abrahamson and Moore, 2001; Dulcis and Spitzer, 2008). In birds, meanwhile, the medial and/or visual SCN in many ways shares more anatomical and functional characteristics with the mammalian SCN than does the amphibian SCN (reviewed in Underwood et al., 2001; Yoshimura et al., 2001; Cantwell and Cassone, 2006a,b; Ikegami et al., 2009). However, the avian central circadian system remains more distributed, with the retina and pineal playing more important roles in light-entrained circadian function than the medial and visual SCN in many species (reviewed in Underwood et al., 2001; Steele et al., 2003).
In contrast, in mammals the SCN is absolutely essential for behavioral rhythmicity. It has by far the most robust molecular clockwork in the brain (Abe et al., 2002), which is essential for many free-running behavioral rhythms in the absence of environmental light cues (Moore and Eichler, 1972; Stephan and Zucker, 1972; Husse et al., 2014). The SCN is also clock-independently required for entrainment of circadian behavior by light (Moore and Eichler, 1972; Stephan and Zucker, 1972; Husse et al., 2014). Thus, both master pacemaker function and photoentrainment appear to be centralized in the SCN of mammals. Though this evolutionary innovation not shared with other vertebrates, the marked preservation of many of its features across all mammals studied to date suggests that it has been well conserved within our clade (Cassone et al., 1988).
To fulfill its central role in controlling circadian rhythms, the mammalian SCN uses a complex, interdependent hierarchy of signaling molecules to integrate information from assorted afferents and the multiple, robust clocks located in its subdivisions. This diversity implies complicated developmental control of heterogeneous cell fate decisions, formation of assorted connections, and ontogeny of the central clockworks during SCN development. The remainder of this review will focus on the developmental and, to a lesser extent, evolutionary mechanisms that underpin these various aspects of the master clockworks.
Forging the gears: specification and differentiation in the SCN
Neurotransmitter diversity in the adult SCN
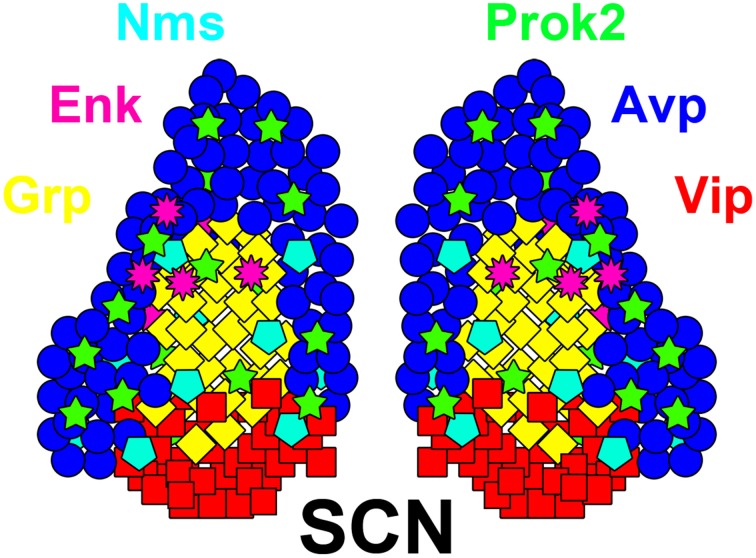
In developmental neuroscience, a given neuron's suite of neurotransmitters is often used as a convenient proxy for cell fate. Highlighting its complexity, the SCN secretes literally hundreds of distinct neuropeptides derived from at least 24 prohormones and numerous other proteins (Southey et al., 2014), along with cytokines, small-molecule neurotransmitters like GABA and nitric oxide, and others. Many of these signals have well-understood circadian roles (reviewed in Slat et al., 2013). For example, vasoactive intestinal peptide (Vip) and gastrin releasing peptide (Grp) maintain synchrony of SCN cellular oscillators and behavioral rhythmicity, and entrain them to light (Piggins et al., 1995; Harmar et al., 2002; Colwell et al., 2003; Aton et al., 2005; Brown et al., 2005; Gamble et al., 2007; An et al., 2011; Maywood et al., 2011). Arginine vasopressin (Avp) also strengthens SCN and behavioral circadian rhythmicity, but unlike Vip and Grp, potentiated phase shifting in animals lacking its receptors suggests Avp signaling acts to buffer and restrain light entrainment of the central clock (Li et al., 2009; Maywood et al., 2011; Yamaguchi et al., 2013). Prokineticin-2 (Prok2) acts primarily as an output that couples the SCN to other brain regions (Cheng et al., 2002; Prosser et al., 2007). More recently, high-throughput peptidomic and circuit mapping approaches have identified novel SCN signals that play important parts in circadian physiology, such as integration of glutamatergic SCN input by the neuropeptide little SAAS and the as-yet-unidentified cue(s) secreted by neuromedin S-(+) neurons that are essential for SCN pacemaker function (Atkins et al., 2010: Lee et al., 2015).
But while our knowledge regarding individual SCN signals continues to swell at an accelerating rate, their interactions are less well-documented, despite the importance of synergy among them for SCN circadian function. For instance, both Grp and Avp partially compensate for loss of Vip's synchronizing effects in slice culture (Brown et al., 2005; Maywood et al., 2011). Furthermore, cannulated neuropeptides have strikingly different effects in vivo when the SCN neuropeptide network is compromised. Under these conditions a single Grp pulse, which normally does not disrupt SCN or behavioral rhythm consolidation even at high doses (Piggins et al., 1995; An et al., 2013), deconsolidates activity rhythms (Bedont et al., 2014). Meanwhile, a Prok2 pulse that normally has negligible circadian effects (Cheng et al., 2002) massively shifts both phase and period of activity rhythms when the network is disrupted (Bedont et al., 2014). The latter may reflect an ancestral function of Prok2 normally overridden by the mammalian SCN network; in crayfish hematopoietic system, the prokineticin-like protein astakine-2 has a surprisingly similar effect on circadian period, though almost surely through a very different mechanism from Prok2 (Negri et al., 2005; Watthanasurorot et al., 2013).
Classically, the panoply of different SCN cell types were neatly divided between a Vip(+) ventrolateral (vlSCN) core and an Avp(+) dorsomedial (dmSCN) shell (Abrahamson and Moore, 2001); however, additional domains such as the central SCN exist in many mammals, and other neuropeptidergic cell types cross domain boundaries (Figure 1; reviewed in Morin et al., 1992; LeSauter and Silver, 1999; Abrahamson and Moore, 2001; Morin, 2007; Bedont et al., 2014; Allen Brain Atlas1). Many cells also express multiple signals, like mouse and rat Vip/Grp(+) SCN neurons, and mouse Vip/Nms-(+) and Avp/Nms-(+) neurons (Okamura et al., 1986; Albers et al., 1991; Lee et al., 2015). There is also extensive patterning of non-neuropeptide SCN gene expression, often along non-core/shell axes. For example, many neuropeptide receptors appear to be expressed in gradients along the anterior/posterior axis in mouse SCN, with Vipr2 and Prokr2 highest in the anterior shell and Avpr1a and Avpr1b highest in the posterior shell (An et al., 2012; Allen Brain Atlas). Thus, though the neuropeptides Vip, Grp, Avp, and Prok2 will be referred to frequently in the following subsections, it should be understood that these neuropeptides almost certainly do not delineate all key neuronal subtypes in the SCN.
Figure 1.
Neuron diversity in the adult SCN. A diagram of the adult SCN, showing a subset of its many neuropeptidergic populations. Neuropeptide colors are indicated by their names, positioned around the SCN. Note that many of these neuropeptides cross or exist outside of the classical core and shell SCN subdomains. Also note that many of these neurotransmitters are at least partially co-expressed, such as in Vip/Grp neurons, though the various probable combinations are not shown here for simplicity. Adapted from Bedont et al. (2014) Graphical Abstract.
Patterning, specification, and neurogenesis in the nascent SCN
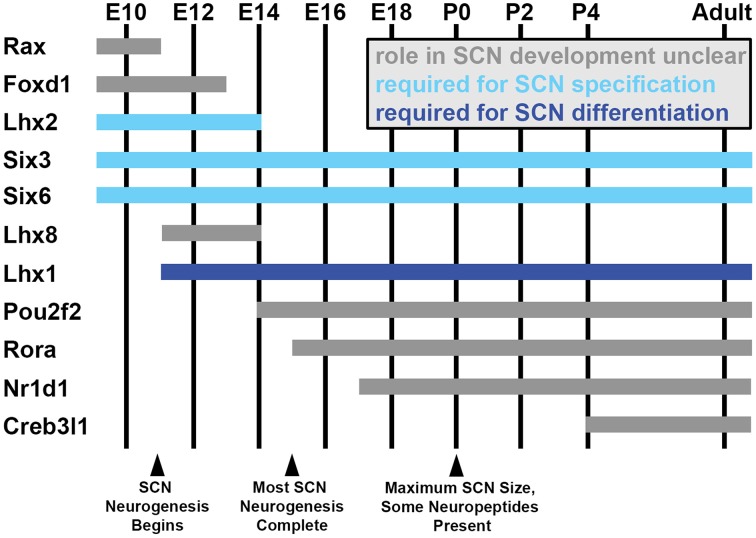
As in much of the rest of the hypothalamus, the study of SCN development has badly lagged understanding of adult SCN diversity, though recent studies have begun to shed light on the topic (reviewed in Bedont et al., 2015). Broadly, the developing anterior hypothalamus (AH) begins as a unitary developmental unit that subsequently divides into two developmentally distinct compartments: a ventral subdivision (vAH) that gives rise to nuclei of the circadian and sleep systems, including the SCN, and a dorsolateral subdivision (dlAH) that gives rise to nuclei of the neuroendocrine system. Very early SCN development shares much in common with the rest of the AH, and later the vAH. For instance, Sonic hedgehog (Shh) signaling from the nearby basal plate is essential for its early patterning and initial specification (Shimogori et al., 2010). Many general AH and vAH transcription factors such as Rax, Foxd1, Nkx2.2, Lhx2, Six3, and Six6 are also expressed in the vAH neuroepithelium prior to the onset of SCN neurogenesis, though expression of many of these factors is lost as the SCN becomes more differentiated from surrounding tissues (Figure 2; Shimogori et al., 2010; VanDunk et al., 2011; Pak et al., 2014; Allen Brain Atlas). Nonetheless, their transient expression is important for early specification of the SCN lineage.
Figure 2.
SCN transcription factor expression during development. Estimated ages of expression for a subset of hypothalamus- and SCN-enriched transcription factors in the developing mouse SCN between embryonic day (E) 10 and adulthood, selected based on interesting expression patterns and/or known developmental functions (indicated by bar color). Note that many general hypothalamic transcription factors (expressed prior to E10) are downregulated as the transcriptional network controlling SCN development ramps up. Not shown in this figure, other transcription factors expressed throughout the SCN during its development in turn become progressively compartmentalized to specific subdomains as the SCN matures (ex: Lhx1, Rorα).
For instance, Lhx2−/− mice fail to down-regulate dlAH markers such as Sim1 and Otp and up-regulate early vAH gene expression such as Vax1 and Lhx1 by embryonic day (E) 12.5, when AH subdivisions are normally clearly defined (Roy et al., 2013). These mice would almost certainly fail to specify an SCN (Figure 2). However, Lhx2 is expressed in cells surrounding the SCN by birth, rather than in the SCN proper, suggesting it may play more of a role in early specification of dlAH and vAH identity than in promoting SCN specification per se (VanDunk et al., 2011).
Unlike Lhx2 and many other early AH and vAH factors that are down-regulated in SCN later in development, Six3 expression remains expressed in the vAH throughout the lifespan, and is required cell-autonomously for initial SCN specification (Figure 2; Shimogori et al., 2010; VanDunk et al., 2011). Its homolog Six6 has a similar, although more restricted, enduring vAH expression pattern, and is also required for initial SCN specification (Figure 2; Shimogori et al., 2010; Clark et al., 2013). Robust but transient Fzd5 receptor expression in mitotic SCN neuroepithelium also suggests that Wnt signaling may regulate SCN specification and/or neurogenesis, but this possibility has so far not been addressed (Shimogori et al., 2010; VanDunk et al., 2011).
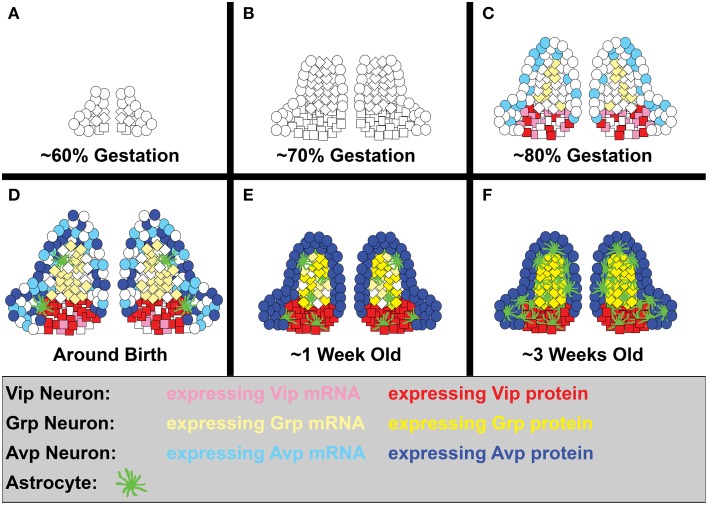
The bulk of SCN neurogenesis occurs between ~60–80% of gestation in rodents, at approximately E9.5-12.5 in hamster, E12-15 in mice, and E13.5-17 in rat (Figures 3A–C; Altman and Bayer, 1978; Crossland and Uchwat, 1982; Davis et al., 1990; Antle et al., 2005; Kabrita and Davis, 2008). Studies in mouse and hamster suggest that genesis of vlSCN neurons consistently peaks earlier than dmSCN neurons in rodents, in contrast to the anteroposterior timing of SCN neurogenesis, which shows much greater species-dependent variation (Davis et al., 1990; Kabrita and Davis, 2008). Correspondingly, cell types enriched in hamster SCN core and central domains such as Vip, Grp, and calbindin (Calb1) are mostly born early, while Avp(+) neurons of the shell can be born at any time during the period of neurogenesis (Antle et al., 2005) (Figures 3A–C). An interesting exception to this trend is a distinct subset of Vip and Calb1 neurons that are born at the end of rat SCN neurogenesis (Abizaid et al., 2004), likely corresponding to the final burst of vlSCN neurogenesis in hamsters (Figures 3C,D; Davis et al., 1990). As in most brain regions, astrogliogenesis follows neurogenesis in the SCN, with the astrocyte marker glial fibrillary acidic protein (Gfap) first detectable shortly before birth, at E15 in hamsters and E20 in rats (Figure 3D; Botchkina and Morin, 1995; Munekawa et al., 2000).
Figure 3.
SCN neurogenesis and differentiation. A simplified diagram showing the development of Vip, Grp, and Avp neurons, as well as astrocytes, in the rodent SCN. While domain boundaries are depicted as being relatively static here to aid in illustrating our points, note that they are actually quite dynamic over the course of development. (A) Early SCN neurogenesis begins at ~60% of gestation (hamster E9.5, mouse E12, and rat E13.5). (B) By ~70% of gestation (hamster E11, mouse E13.5, rat E15), most ventrolateral neurons have already been produced, while dorsomedial neurogenesis is still in full swing. (C) The final major burst of SCN neurogenesis (including a number of ventrolateral neurons) occurs at ~80% of gestation (hamster E12.5, mouse E15, rat E17). Shortly thereafter, many SCN neuropeptide transcripts, but only Vip protein, are detectable (hamster E13, mouse E18-P0). (D) Shortly before and after birth, the first SCN astroglia appear (hamster E15, rat E20), Avp protein expression becomes detectable (P1), and SCN neuron number peaks (hamster P2). (E) By roughly a week into life, a major jump in astrocyte number has occurred (rat P3-P4), cell death brings neuron numbers down to adult levels (hamster P5, mouse P7, rat P6), and Grp protein expression is detectable (hamster P8). (F) By the end of the second major jump in astrocyte number (rat P20-25), the SCN is largely mature. At this stage, the fate of its neurons and the subdomain boundaries they form are relatively stable, though they may remain somewhat malleable under circadian challenge from the environment.
Control of neuronal differentiation in the developing SCN
The mechanism restricting generation of individual SCN cell types to discrete temporal windows is unclear. Given that SCN neuron number peaks at ~P2 in hamsters (Figure 3D; Muller and Torrealba, 1998), there is likely a several-day maturation window for SCN neurons. Thus, one possibility is that extracellular cues could differentially influence the fate of SCN neurons born at different times. For example, although cells of the SCN do not themselves express Shh, at early stages of hypothalamic neurogenesis they are immediately adjacent to cells that do (Shimogori et al., 2010; Alvarez-Bolado et al., 2012). By E14 in the mouse, however, Shh expression is downregulated and moved further posterior from the SCN. Late-born SCN cells are thus exposed to lower Shh levels than early-born cells, possibly influencing their fates (Kabrita and Davis, 2008; Shimogori et al., 2010). Any changes in exposure to Wnt family members during this time would also likely affect Fzd5(+) SCN progenitors (VanDunk et al., 2011). There may even be secreted signals from the maturing SCN itself that influence the fates assumed by late-born neurons (see below). Conversely, SCN progenitor competence to respond to external cues may also change over the course of neurogenesis.
Differential expression of cell-autonomous factors may also play a direct role in birth-date biasing of SCN cell fate, but so far the only known transcription factor involved in any facet of SCN differentiation is LIM homeodomain transcription factor 1 (Lhx1), the earliest selective marker of developing SCN (Figure 2; Shimogori et al., 2010; VanDunk et al., 2011). In Six3-Cre/+;Lhx1lox/lox mice, in which Lhx1 function is disrupted beginning early in hypothalamic neurogenesis, the SCN fails to terminally differentiate (Bedont et al., 2014). This leads to reductions in several neuropeptide lineages important for circadian function, including neurons expressing Vip, Grp, Avp, and Prok2, among others (Bedont et al., 2014). Both bioinformatic and luciferase analysis suggest that Lhx1 directly regulates expression of some of these neuropeptides in the SCN (Bedont et al., 2014; Hatori et al., 2014). Interestingly, induction of Lhx1 expression coincides with the beginning of SCN neurogenesis in mouse (Kabrita and Davis, 2008; Shimogori et al., 2010; VanDunk et al., 2011), and the mostly earlier-born Vip and Grp neurons are more severely depleted than Avp neurons in the Lhx1-deficient SCN (Bedont et al., 2014).
However, it seems unlikely that Lhx1 is necessary for early cell fate decisions or regional patterning in the SCN. Lineage-specific sub-domain markers expressed prior to E16.5 so far seem to be only modestly affected or completely unaffected in Six3-Cre/+;Lhx1lox/lox SCN, and morphological defects in SCN shape and neuron number first appear postnatally in mutant mice (Bedont et al., 2014). Histological data also suggests these morphological defects are shared by RorαCre/Cre;Lhx1lox/lox SCN (though this was not commented on in the original publication), and the Vip lineage is also depleted in this line, which is predicted to delete Lhx1 in SCN beginning at E15, after the generation of Vip(+) neurons is mostly complete (Hatori et al., 2014). While Vip loss appears less severe in RorαCre/Cre;Lhx1lox/lox than Six3-Cre/+;Lhx1lox/lox SCN, interpretation of this phenotype is confounded by the substantially less efficient Lhx1 deletion seen with the RorαCre/Cre driver. It thus seems likely that the less severe phenotypes seen in RorαCre/Cre;Lhx1lox/lox animals result primarily from less efficient Lhx1 deletion, rather than deletion of Lhx1 later in development (Bedont et al., 2014; Hatori et al., 2014). Thus, the sum of these data suggest that Lhx1 primarily acts downstream of other genes that control initial specification of SCN identity.
Instead, Lhx1 likely acts as one of several essential cogs in a complicated transcriptional network controlling SCN terminal differentiation. One probable downstream component is downregulated in RorαCre/Cre;Lhx1lox/lox SCN: the transcription factor Creb3l1 (Hatori et al., 2014). Creb3l1 is a potent, direct regulator of Avp expression, whose circadian expression tracks Avp in adult SCN (Greenwood et al., 2014). Conversely, unlike Vip, both Avp and Creb3l1 lack predicted Lhx1 binding sites in their proximal promoters (Bedont et al., 2014). Thus, Lhx1 likely indirectly controls expression of Creb3l1 and in turn Avp, rather than directly controlling expression as it does for Vip (Hatori et al., 2014).
Interestingly, much like the less severe loss of Avp in the Six3-Cre/+;Lhx1lox/lox SCN relative to Vip, the reduction in Creb3l1 expression in the RorαCre/Cre;Lhx1lox/lox SCN was less severe than for Vip (Bedont et al., 2014; Hatori et al., 2014). Allen Brain Atlas data suggests that Creb3l1 induction may lag behind the earliest Avp expression observed in the SCN during development (Figures 2, 3D). Thus, Creb3l1 may play a more important role in differentiation or maturation of only later-born subpopulations of Avp neurons. Alternatively, it could be required for maintenance, but not induction, of Avp expression. Directly testing Creb3l1 function in the developing SCN is a promising direction going forward, as is testing the contribution of other potential members of the transcriptional network to which Lhx1 and Creb3l1 belong. Candidate factors including Lhx8, Rorα, Rorβ, and Pou2f2, among many others, have been identified as developmentally regulated SCN transcripts by recent screening efforts, but their roles in SCN development have not been examined (or, in the case of Rorα, not examined in detail) (Figure 2; Shimogori et al., 2010; VanDunk et al., 2011).
Lhx1 is also an early, robust SCN marker in amphibians (Dulcis and Spitzer, 2008), despite key differences in gene expression with mammals, such as prominent neuropeptide Y (Npy) expression in frog SCN neurons (Tuinhof et al., 1994b). Frog SCN neurons also express an Lhx5 homolog, which is absent in mammalian SCN but co-expressed with Lhx1 in the mammalian spine, where the two transcription factors jointly regulate terminal differentiation of neuropeptide lineages including Npy(+) cells (Brohl et al., 2008; Shimogori et al., 2010; Allen Brain Atlas). Thus, it would be interesting to assess whether Lhx5 has become re-activated in the human SCN during evolution, as to our knowledge, our species is unique among mammals in expressing Npy locally there (Moore, 1989). Given such tantalizing hints, we suspect that changes in the regional transcriptional network are a major proximal cause of evolutionary changes in the SCN.
Maturation and cell fate plasticity in the developing SCN
The neurotransmitter fate(s) chosen by SCN neurons begin to become clear late in embryonic development. Many SCN neuropeptide mRNAs, including Vip, Avp, Grp, and Prok2, are first detectable by in situ hybridization between E18 and birth in mouse SCN, though generally in numbers lower than seen in adults, suggesting that not all SCN neurons are differentiated at this time (Figure 3C; Shimogori et al., 2010; VanDunk et al., 2011; Bedont et al., 2014; Allen Brain Atlas). In contrast, SCN neuropeptide proteins first become detectable at more variable ages than do their mRNAs. In hamster SCN, Vip is present by E13 (only slightly earlier than Vip mRNA in mouse, adjusted for gestation), while Avp is present by P1, Grp is present by P8, and substance P (Sp) is present by P10 (Figures 3C–E; Romero and Silver, 1990; Botchkina and Morin, 1995; Antle et al., 2005). The first major increase in rat SCN Gfap expression, indicating astrocytic maturation, also occurs during this time at ~P3-P4 (Figure 3E; Munekawa et al., 2000).
Staggered neuropeptide availability may have profound implications for SCN maturation. The earliest synapses form in rat SCN at E19, shortly after Vip first appears in the E13 hamster SCN adjusted for gestation (Moore and Bernstein, 1989). Hamster Vip neurons already begin to extend axons as early as E15, though the scarcity of synapses suggests that any signaling they engage in during neonatal life is likely paracrine (Botchkina and Morin, 1995). This situation changes rapidly with increasing synapse formation between P2-P6, shortly after Avp expression is first detected, followed shortly thereafter by another increase that drives synapse density to adult levels by P10 (Moore and Bernstein, 1989).
SCN cell death begins as synapse formation increases, with substantial death between E21-P6 in rats and P1-P7 in mice, and a sharp drop in neuron number between P2-P5 in hamster (Figures 3D,E; Moore and Bernstein, 1989; Muller and Torrealba, 1998; Ahern et al., 2013). Dying neurons are isolated from neuronal clusters that form during this time in rat SCN, hinting that cell-cell communication regulates SCN neuron survival (Moore and Bernstein, 1989).
It isn't until around this time that SCN adult-like subdomains start to become distinguishable. New transient populations become apparent in the SCN even after birth, with a dorsolateral Calb1 domain appearing as late as P1 in hamster SCN, only to vanish a day after the more ventral adult Calb1 domain appears at P2. The enduring domain then enters a protracted period of maturation, achieving an adult-like appearance by ~P15 (Antle et al., 2005). Many neuropeptide populations follow similar paths and are expressed in gradually shifting, immature patterns until days or even weeks after initial detection (Delville et al., 1994; Botchkina and Morin, 1995; Herzog et al., 2000; Antle et al., 2005). Even stereotypic morphological differences in individual rat core/shell neurons do not appear until around ~P6-P10 (Moore and Bernstein, 1989). Synapse formation, which occurs concurrently with partitioning of the SCN, respects the emerging boundaries; the kinetics of synaptogenesis vary substantially among SCN subdomains, though still within the synaptogenesis window described for the SCN as a whole described by Moore and Bernstein (Laemle et al., 1991).
Against the backdrop of the long SCN maturation period in early postnatal life, non-cell-autonomous regulation of particular SCN lineages has been observed. Rat pups exposed to a pharmacological inhibitor of synthesis for the SCN input serotonin (5HT) from E13-E21 have more Vip and possibly Avp neurons, and increased cellular expression levels of these neuropeptides, in the SCN; however, this effect does not occur if the pups are instead treated with the drug early in post-natal life (Ugrumov et al., 1994; Mirochnik et al., 2005). Paradoxically, similarly increased numbers of Vip and Avp neurons and neuropeptide levels in SCN were observed in monoamine oxidase A null mice, which have constitutively elevated levels of monoamine neurotransmitters including 5HT. Increases in neuropeptide expression levels, but not cell number, could be reversed by 5HT or norepinephrine inhibitors administered post-natally (Vacher et al., 2003). One possible rationalization of these results is that embryonic inhibition of 5HT synthesis causes homeostatic up-regulation in other monoamine pathways to compensate. In another set of studies, mice selectively deficient for PSA-NCAM expressed in the dorsal SCN have three times the usual number of Vip neurons, in an expanded vlSCN (Shen et al., 1997, 1999). Thus, both PSA-NCAM and monoamine neurotransmitters are most likely regulating the differentiation of SCN neuropeptide lineages. However, another more intriguing possibility is that the overall fate of SCN neurons may remain plastic during their maturation, and can be manipulated by these extracellular cues.
Plasticity has been shown in frog SCN, where physiological light levels can dynamically induce dopaminergic marker gene expression in Npy neurons of the frog SCN (Dulcis and Spitzer, 2008). In adult rats, switching of dopamine and somatostatin expression also occurs in other parts of the AH in response to circadian light cycle changes, though such phenomena have not yet been observed in the SCN itself in mammals (Dulcis et al., 2013). However, adult mouse SCN expresses protein markers common to immature and often neurogenic cell populations, including Sox2, transportin-1, doublecortin-like, and possibly doublecortin (Geoghegan and Carter, 2008; Sato et al., 2011; Saaltink et al., 2012; Hoefflin and Carter, 2014) and SCN epigenetic state can be enduringly modified by experience (Azzi et al., 2014). Extreme plasticity of circadian behavioral entrainment is also possible in mice as late as ~P90 (Aton et al., 2004). Thus, it seems likely that SCN cell identity is at least somewhat plastic in response to circadian changes during maturation, and perhaps into adulthood.
SCN astrocyte maturation culminates in a second major jump in Gfap expression between P20-P25 in rats, shortly after the SCN neural network reaches a largely adult state (Figure 3F; Munekawa et al., 2000). Given the well-documented control of neuronal synaptic connectivity by SCN astrocytes and increased neuron-to-neuron apposition in the SCN when astrocyte maturation is disrupted by eye enucleation, a mature astrocyte network may bring relative functional stability to the SCN (Munekawa et al., 2000; Becquet et al., 2008; Girardet et al., 2010). Yet even in adulthood, it is unclear to what extent SCN development is truly fixed when presented with circadian challenge, given the possible contribution of environmentally triggered transdifferentiation and/or induced neurogenesis that have not been closely examined.
Assembling the components: development of SCN connectivity
Connectivity of the adult SCN
The mammalian SCN receives bilateral input of luminance information via the retinohypothalamic tract (RHT) from intrinsically photosensitive ganglion cells of the retina (ipRGCs) that express the photopigment melanopsin (Opn4) (Provencio et al., 2000; Gooley et al., 2001; Hattar et al., 2002, 2006). IpRGC input is essential for SCN light entrainment, but not clock function. Opn4DTA/DTA mice selectively lacking ipRGCs largely phenocopy the circadian effects of Math5−/− mice that fail to develop the entire RHT due to gross retinal malformation—both animals wheel-run under light:dark (LD) conditions on an internal circadian period similar to normal mice, as though they are in constant darkness (DD) (Brzezinski et al., 2005; Guler et al., 2008). ipRGC-derived signals such as synaptically released glutamate and the neuropeptide Pacap mediate this entrainment (Chen et al., 1999; Guido et al., 1999; Hannibal et al., 2008; Kawaguchi et al., 2010; Webb et al., 2013).
The SCN is also intricately interconnected with many other regions of the brain (reviewed in Morin, 2013). One of the most prominent of these afferents comes from the thalamic intergeniculate nucleus (IGL), which sends both indirect photic information and non-photic information such as arousal state to the SCN (Pickard et al., 1987; Hastings et al., 1997; Hattar et al., 2002). The rat SCN also receives prominent inputs from the brainstem raphe nuclei, which commonly converge with IGL innervation on SCN neurons; occasionally these IGL and raphe inputs even form axo-axonic synapses with each other (Guy et al., 1987; Migliarini et al., 2013).
Other major SCN afferents include the olfactory and limbic systems, circumventricular organs, and assorted hypothalamic nuclei, among other regions (Krout et al., 2002). Interestingly, some regions like the lateral septum that project to the mammalian SCN are photosensitive in non-mammalian species (Silver et al., 1988; Li et al., 2004), suggesting that these connections may have routed photic information to our evolutionary ancestors' SCNs. The functions these formerly photic afferents serve in the mammalian circadian network are poorly understood.
SCN efferents innervate many hypothalamic nuclei, including most that reciprocally project to rat and hamster SCN. Prominent targets include the preoptic, paraventricular (PVN), subparaventricular (SPZ), retrochiasmatic, and dorsomedial (DMH) nuclei (Watts and Swanson, 1987; Watts et al., 1987; Morin et al., 1994). There are also major SCN projections to the septum and anterior periventricular thalamus, among other regions (Watts and Swanson, 1987; Watts et al., 1987; Morin et al., 1994). Efferent projections from the SCN to specific hypothalamic nuclei have been implicated in regulating circadian control of distinct subsets of behaviors. These include body temperature for the dorsal SPZ; locomotor activity levels and the sleep/wake cycle for the ventral SPZ; and locomotor activity, sleep, feeding, and stress hormone secretion for the DMH (Lu et al., 2001; Chou et al., 2003; Abrahamson and Moore, 2006).
Reflecting such differences, the afferents and efferents of different SCN subpopulations vary substantially to fulfill their divergent functions within the network. ipRGC innervation is particularly prominent for Vip neurons in the vlSCN, reflecting their key role in light entrainment, though all SCN subdomains receive at least some direct ipRGC input in mammals (Ibata et al., 1989; Tanaka et al., 1993; Muscat et al., 2003; Hattar et al., 2006; Morin et al., 2006; Juarez et al., 2013). Other major afferents, such as IGL-derived Npy and 5HT from the raphe, also preferentially innervate the vlSCN in a closely overlapping pattern, with synaptic terminals seen on Vip neurons (Kiss et al., 1984; Bosler and Beaudet, 1985; Hisano et al., 1987; Abrahamson and Moore, 2001). Conversely, septal neurons project selectively to SCN Avp neurons (Oldfield et al., 1985). Downstream, mammalian SCN target regions are generally innervated by neurons from multiple anatomical subdivisions and/or neuropeptide subtypes, though particular lineages predominate in some areas, such as the prominent Vipergic innervation of PVN (Dai et al., 1997; Abrahamson and Moore, 2001; Leak and Moore, 2001; Kriegsfeld et al., 2004; Zhang et al., 2009; Campos et al., 2014). Intra-SCN connectivity is also enormously complex and heterogeneous, and in some cases controversial (van den Pol and Gorcs, 1986; Daikoku et al., 1992; Ibata et al., 1993; Romijn et al., 1997; Jacomy et al., 1999; Castel and Morris, 2000; Abrahamson and Moore, 2001; LeSauter et al., 2002). In short, correct control of developing SCN connections is likely every bit as important as expression of the proper neuropeptides in establishing adult SCN heterogeneity and function.
Development of SCN afferents
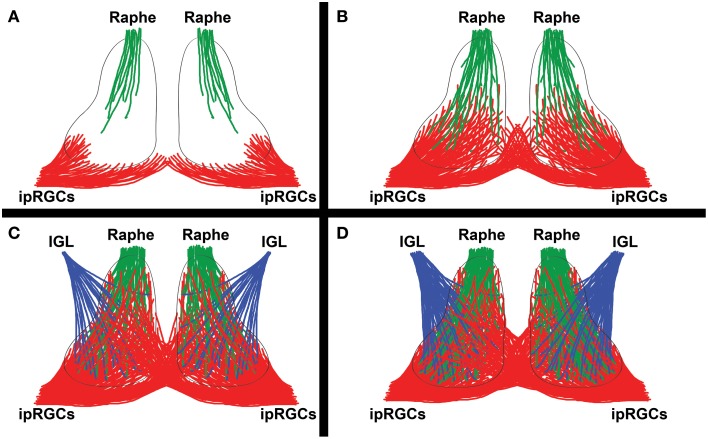
By far the most heavily studied SCN afferent is the ipRGC input. ipRGCs are born in a much wider window than SCN neurons, extending from ~E11-E18 in mouse (McNeill et al., 2011). Opn4 is initially expressed at about 75% of gestation, or ~E15 in mice and ~E18 in rats, roughly concurrent with the end of SCN neurogenesis (Fahrenkrug et al., 2004; McNeill et al., 2011). IpRGC axons are present at the mouse optic chiasm by E17, but for reasons that are unclear they stall here for several days before innervating the SCN, lagging RGC innervation of brain regions involved in image-forming vision (McNeill et al., 2011).
There are substantial species differences in the rate of ipRGC innervation of SCN across mammals. The vlSCN is only gradually innervated around birth in mice and rats, but because of their Opn4 expression, ipRGCs are light responsive and able to activate a subset of vlSCN neurons that they sparsely innervate from P0-P1 onwards (Figure 4A; Speh and Moore, 1993; Hannibal and Fahrenkrug, 2004; Sekaran et al., 2005). Initially, the ipRGCs only innervate the vlSCN ipsilaterally in mouse, but as the projection matures the entire SCN is innervated, with contralateral projections appearing by P4 and becoming commingled with the ipsilateral projections by P7 (Figures 4B,C; McNeill et al., 2011). In rats, the diversity of individual ipRGC-SCN synapses increases dramatically during this time (Lenn et al., 1977). In hamsters this process is slightly delayed but fundamentally similar to the pattern in mice and rats, with initial ipRGC innervation and activation of vlSCN occurring at ~P3 (Speh and Moore, 1993; Kaufman and Menaker, 1994; Duffield et al., 1999). Remarkably, all of these events occur before eye opening in all three species.
Figure 4.
Innervation by major SCN afferents. A simplified diagram showing the development of major SCN afferents from the intrinsically photosensitive retinal ganglion cells (ipRGCs) of the retina, raphe nuclei, and intergeniculate leaflet (IGL). (A) ipRGCs first begin to innervate the vlSCN ipsilaterally at birth (mouse/rat P0-P1) or shortly thereafter (hamster P3). The raphe nuclei first innervate the SCN simultaneously (mouse P0, hamster P3). (B) A few days later, ipRGC innervation of the SCN becomes noticeably denser and broader, with the first contralateral projections appearing in the ventromedial SCN (mouse/rat P4), as raphe innervation also builds. (C) By roughly a week into postnatal SCN development, raphe innervation has become fairly dense (mouse P5, hamster P6), ipRGC contralateral and ipsilateral ipRGC innervation domains are commingled (mouse P7), and the earliest Npy signal from the IGL is detectable (hamster P7, rat P10). Many of the IGL and raphe projections converge, a trend that persists through adulthood. (D) In the following few weeks, more-or-less adult SCN innervation by the raphe (mouse P10), ipRGC (mouse P14), and IGL (rat P20) projections become apparent.
By ~P10 (mouse/rat) or ~P15 (hamster), the ipRGC projection to SCN is mostly mature, with largely adult-like projection density, morphology of individual axon terminals, and light responses (Figure 4C; Moore and Bernstein, 1989; Speh and Moore, 1993; Kaufman and Menaker, 1994; Hannibal and Fahrenkrug, 2004; Sekaran et al., 2005; McNeill et al., 2011), though some maturation continues at least through adolescence (Yellon et al., 1993). During or shortly after the mostly mature P10-P15 range in rodents, rod and cone photoreceptors become mature and the eyes open, rendering Opn4 largely redundant for SCN photoentrainment (Panda et al., 2002; Hannibal and Fahrenkrug, 2004; Sekaran et al., 2005).
The maturation of ipRGC inputs to the SCN in early post-natal life has important implications for SCN development. Multiple NMDA receptor subunits are transiently expressed between birth and eye opening in rat and hamster SCN (Bendova et al., 2009; Duffield et al., 2012), suggesting a period during which SCN development is sensitive to ipRGC-derived glutamate. IpRGC innervation is necessary for certain events in mouse SCN differentiation, such as Calb1 downregulation in dmSCN and Calb2 upregulation in vlSCN during early postnatal life (Ikeda and Allen, 2003), as well as SCN astrocyte maturation (Munekawa et al., 2000). Manipulations of the light cycle during this time also influence SCN development and adult function—rearing in DD decreases adult SCN neuron and glia numbers, while rearing in constant light (LL) decreases adult SCN glia numbers (Cambras et al., 2005). There have also been reports of differences in adult SCN Vip and Avp immunostaining intensity and circadian behavior in rats and mice exposed to postnatal light manipulations; however, the findings have varied substantially, possibly as a result of species-specific differences or variations in experimental protocol (Smith and Canal, 2009; Cambras et al., 2015).
Thus, exceptions to this pattern of gradual postnatal ipRGC innervation of SCN are both developmentally and evolutionarily interesting. For example, in stark contrast to more commonly studied rodents, rabbits are born with an over-abundant ipRGC projection to SCN that is pruned down to adult levels by P19 (Juarez et al., 2013). Though the mechanisms behind this difference are presently unclear, the ultimate cause may relate to the maternal behavior of rabbit dams, which visit the burrow to suckle pups for a much shorter time each day than rodents such as mice and hamsters (Zarrow et al., 1965). Although the SCN-independent food-entrained circadian system is likely most important for anticipating the dam's visit (Caba et al., 2008), the ipRGC projection may have nonetheless evolved to become particularly exuberant in the early postnatal stage, to aid in correctly timing the all-important daily feeding. The impact this very different time-course of ipRGC development may have on development of the rabbit SCN is an intriguing proposition.
After ipRGCs, the most extensively studied afferent projection from a developmental perspective is the ascending raphe projection, which is one of the last raphe efferent projections to form in hamsters. Its inputs to the SCN initially arrive in the SCN at times similar to the ipRGCs, by ~P0 in mouse and ~P3 in hamster, and their maturation process seems to take a similar amount of time (Figures 4B–D; Botchkina and Morin, 1993; Migliarini et al., 2013). Co-transplantation studies suggest that raphe innervation of the rat SCN may require IGL innervation of the SCN to form; if the same is true in normal development, it would explain the frequent convergence of these afferents in the adult SCN (Guy et al., 1987; Ueda et al., 1993, 1995). The SCN is also one of the very few nuclei that requires serotoninergic signaling by raphe neurons in order to receive raphe innervation (Migliarini et al., 2013). As mentioned previously, the raphe projection is also involved in regulating the number of Vip and (possibly) Avp cells in the SCN, a relationship that is likely dependent on the formation of direct synaptic contacts between 5HT terminals and Vip neurons in the SCN (Kiss et al., 1984; Bosler and Beaudet, 1985; Hisano et al., 1987; Mirochnik et al., 2005).
A bit is also known about the development of IGL projections to the SCN. Npy is detectable in the SCN by ~P7 in hamster and ~P10 in rat, suggesting that IGL projections are present by this time (Figure 4C; Romero and Silver, 1990; Takatsuji et al., 1995). This is markedly delayed relative to the early presence of the ipRGC and raphe projections; however, the probable IGL role in raphe innervation of the SCN hints that the IGL projection to SCN may be present much earlier in development than Npy is detectable, a possibility that has not been directly explored (Ueda et al., 1995). In any case, by P20 the IGL projection to SCN is largely mature in rats (Figure 4D; Takatsuji et al., 1995). To our knowledge, the underlying mechanisms and developmental role of this projection have not been studied, beyond the aforementioned transplantation study by Ueda et al. Very little is known about the development of other SCN afferents, aside from the three discussed above.
Development of SCN efferents and paracrine signaling
Virtually nothing is known about the development of SCN efferents. In hamster SCN, one DiI tracing study suggests that efferent growth begins at ~P1, with rapid, largely simultaneous growth toward known SCN target regions (Muller and Torrealba, 1998). This is the only such study we were able to locate.
The surprising lack of data on this topic may stem from the perceived importance of paracrine signals for adult SCN output. The foundation of this long-controversial point is that fetal SCN grafts wrapped in a mesh to block efferent outgrowth are sufficient to rescue circadian activity rhythms in SCN-lesioned hamsters (Silver et al., 1996). However, while both the rat SCN nucleus and its efferents are essential for sleep-wake rhythmicity at P8, SCN efferents but not the SCN nucleus are partially dispensable in younger P1 animals (Gall et al., 2012). Meanwhile, neonatal mouse Cry1−/−;Cry2−/− SCN is surprisingly able to sustain synchronized Per1 and Per2 luciferase (Luc) rhythms for a few days in slice; this ability is lost by ~P7, after which the mutant SCN is arrhythmic as soon as it is explanted (Maywood et al., 2011; Ono et al., 2013). While a neonatal wild-type or Cry1−/−;Cry2−/− SCN can entrain a co-cultured arrhythmic adult Cry1−/−;Cry2−/− SCN in a paracrine fashion, an adult wild-type SCN cannot (Ono et al., 2013). And tellingly, in hamster SCN implantation studies, efficacy is highly correlated with donor age, with ~P7 or older grafts failing to efficiently entrain rhythms (Romero et al., 1993).
In sum, it seems likely that SCN-derived paracrine signaling may indeed be sufficient for behavioral entrainment for a transient period during early postnatal development. However, at least in slice preparations, the SCN neuropeptides that are most highly expressed in adult animals–such as Vip, Grp, and Avp–account for much of the nucleus' paracrine entraining capacity (Maywood et al., 2011). One possible hypothesis rationalizing this seeming paradox is that a modifier of adult neuropeptide signaling is downregulated in early postnatal life, causing a loss of SCN paracrine signaling competence. Possible candidates include regulator(s) of paracrine release or proteolytic enzyme(s) that alter neuropeptide precursors post-transcriptionally. In any case, both the study of SCN paracrine signaling and efferent outgrowth in a developmental context are clearly fruitful areas for future research.
Winding the watch: ontogeny of SCN clockworks
Clock diversity in adult SCN
Mirroring the heterogeneity in SCN gene expression and connectivity, the SCN clockworks are quite complex, with oscillatory clock gene expression varying substantially across different subdomains (reviewed in Honma et al., 2012). For example, a number of Per1Luc and Per2Luc slice studies in both rats and mice show that intrinsic dmSCN rhythms are phase-advanced relative to vlSCN rhythms (Yamaguchi et al., 2003; Foley et al., 2011; Fukuda et al., 2011), likely due in part to shorter intrinsic periods in dmSCN (Noguchi et al., 2004). Conversely, phase-shifts induced by light begin in the vlSCN, before spreading to the dmSCN (Nagano et al., 2003). The phase of subdomain clock gene rhythms also varies across the anteroposterior axis of the mammalian SCN in a manner that, though it depends somewhat upon light cycle, under most conditions is phase-advanced in posterior SCN relative to central and anterior subdomains (Hazlerigg et al., 2005; Sosniyenko et al., 2009; Evans et al., 2011). Variation along this axis phase-locks to the onset and offset of behavioral rhythms, implying behavioral relevance (Inagaki et al., 2007).
Divergent patterns are also noticeable in readouts of SCN activity. There are dramatic differences in the phase of single-unit mouse SCN recordings that strongly correlate with regional variation in Per1GFP rhythms seen across the SCN (Quintero et al., 2003; Schaap et al., 2003). SCN calcium rhythms show a similar pattern, at least insofar as dmSCN is phase advanced relative to vlSCN (Enoki et al., 2012; Brancaccio et al., 2013). Furthermore, cultured rat SCN shows that the circadian rhythm of Avp release is phase-advanced relative to Vip, when the latter is rhythmic (Nakamura et al., 2001). Coding of circadian parameters by SCN neuron activity also varies across compartments; for instance, while population differences in the phase of single-units can be compressed or decompressed by light cycle to encode period, this appears to occur preferentially in the vlSCN, while dmSCN relies more on changes in the period of individual oscillators to encode period (VanderLeest et al., 2007; Brown and Piggins, 2009).
Beyond these subdomains, there is also considerable cell-to-cell variation in the period and amplitude of individual oscillators, and even whether a cell is rhythmic or not. This is true for both clock gene expression (Herzog et al., 2004; Webb et al., 2009) and cellular activity (Nakamura et al., 2001; Herzog et al., 2004; Honma et al., 2004; Webb et al., 2009).
Both cell fate and connectivity play important roles in establishing clock diversity. The period of Bmal1 rhythms varies subdomain-specifically and intrinsically encodes period in mouse SCN neurons, and temporal organization of SCN synchrony seems to arise predominantly from cell-autonomous factors (Myung et al., 2012; Pauls et al., 2014). On the other hand, spatial organization of SCN synchrony is primarily signaling-dependent, likely due to features such as dye coupling of cells within but not between vlSCN and dmSCN, the influence of Vipr2 signaling on Avp neuron period, and the Gαi-protein inhibitor Rgs16's role in driving circadian oscillations of cAMP production in the SCN (Colwell, 2000; Noguchi and Watanabe, 2008; Doi et al., 2011; Pauls et al., 2014). However, these aspects of cell identity seem less important for whether the SCN as a whole is rhythmic or not, despite effects on the parameters of cells that are rhythmic (Webb et al., 2009). The development of this complex clockwork will be the focus of the final section.
Clock ontogeny in developing SCN
It has recently become clear that Per2Luc signal in mouse SCN slices is rhythmic in culture as young as either E13.5 or E15, as found by two different groups (Wreschnig et al., 2014; Landgraf et al., 2015). Regardless of which age of onset is correct, both studies detect Per2 rhythmicity much younger than the earliest detectable SCN clock gene rhythms in previous in vivo studies (Figure 5). One possible explanation is that individual mouse SCN neurons begin to function as isolated oscillators sometime between E13.5-15, which are prematurely aligned to produce rhythms by the synchronizing cue of the culturing process. Supporting this interpretation, early SCN rhythms are unique among multiple tissues studied by Landgraf et al. in strongly phase-locking with culturing time (2014a), consistent with earlier studies showing an elevated dependence of SCN phase on this parameter when cultured from early post-natal mice (Nishide et al., 2008). Alternatively, it is possible that Per2Luc signal amplification simply allows more sensitive detection of rhythmicity, as earlier studies of clock ontogeny in vivo relied upon histological techniques that are less sensitive to very small changes in expression levels. However, a recent in vivo study using more sensitive quantitative real-time PCR methods also found later onsets of rhythmicity more in keeping with the older histological data (Houdek and Sumová, 2014), making it less likely that differences in detection limits are the sole cause of this discrepancy. The data from these Per2Luc studies remains interesting and important in either case, but clarifying their interpretation remains an important future challenge.
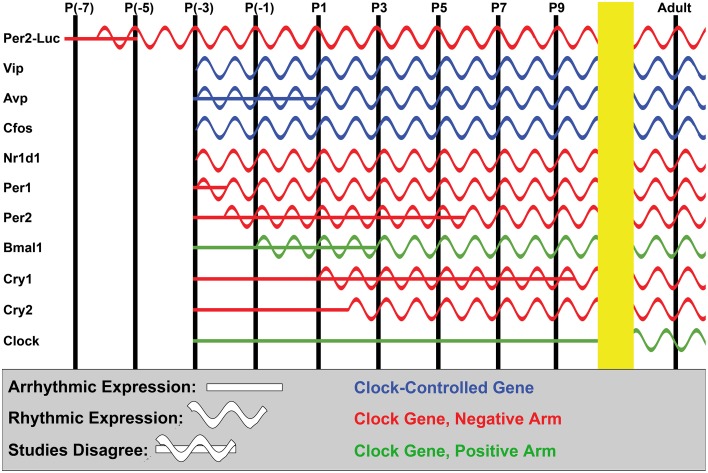
Figure 5.
Ontogeny of the SCN clockworks. A simplified diagram showing the approximate age at which expression and cycling of various cellular clock and clock-controlled genes are known to begin in the SCN. Data from all mouse and rat studies we were able to locate is included in the figure; our P(-x) terminology is used for prenatal time-points to roughly account for the difference in average gestation time in mouse (P0 at ~20 days) and rat (P0 at ~22 days). In vivo studies were surprisingly consistent despite differences in organism and detection methodology among studies, arriving at estimated ages of rhythm ontogeny within a few day window in most cases. Notably late outliers include Per2 (Shimomura et al., 2001) and Cry1 (Ansari et al., 2009). New studies observing Per2-Luc luminescence in slice have arrived at earlier estimates of rhythm induction than in vivo studies of Per2; however, the reason for this is unclear (see text). Finally, note that only induction of expression and rhythmic expression are accurately recapitulated in this figure; relative phase and amplitude of the peaks is incidental.
In vivo, many clock and clock-controlled genes are robustly expressed in the mammalian SCN by ~E13 in hamster, ~E17 in mice, and ~E19 in rat (Figure 5; Shearman et al., 1997; Sladek et al., 2004; Li and Davis, 2005). The SCN clockworks become synchronously rhythmic in a staggered fashion shortly thereafter. Non-core-clock and clock-controlled SCN-enriched gene expression, such as Nr1d1, Vip, Avp, and c-fos mRNA, generally become rhythmic first, by ~E19 in rat and late embryonically in mouse (Figure 5; Huang et al., 2010; Houdek and Sumová, 2014). This corresponds to the onset of rat SCN metabolic rhythms at ~E19, implying that the core clockworks are dispensable for this functional rhythm (Reppert and Schwartz, 1984). Interestingly, reports largely concur that core clock genes are not yet synchronously rhythmic in the developing SCN at this age in vivo (or at least that the rhythms are of quite low amplitude), suggesting that the maternal clock initially drives synchrony of developing pups' SCN clock-controlled gene expression in vivo, perhaps via modulation of Nr1d1 expression (Houdek and Sumová, 2014).
Genes in the negative arm of the core clock become rhythmic next. In some studies, Per1 and Per2 are among the earliest to begin cycling, at ~E17 in mouse and ~E20 in rat (Figure 5; Shearman et al., 1997; Shimomura et al., 2001; Ohta et al., 2002; Kovacikova et al., 2006), while in other studies in mouse and hamster, Cry1 is among the earliest rhythmic clock components (Li and Davis, 2005; Huang et al., 2010). Genes in the positive arm become rhythmic later; Clock is particularly notable in this regard, as it does not begin cycling until sometime after P10 in both mouse and rat (Figure 5; Sladek et al., 2004; Kovacikova et al., 2006; Ansari et al., 2009). Most clock genes are rhythmic by ~90% of gestation across mammals and all become rhythmic within the first several post-natal days, though details vary somewhat among reports (Figure 5; Sladek et al., 2004; Li and Davis, 2005; Kovacikova et al., 2006; Torres-Farfan et al., 2006; Huang et al., 2010; Houdek and Sumová, 2014). SCN neural activity rhythms also appear within this window, at ~E22 in rat SCN (Shibata and Moore, 1987). Combined with the strong correlation between local clock gene expression and adult SCN neural activity (Quintero et al., 2003), it seems that the core clock is required for induction of rhythmic SCN neural activity.
Following their initial emergence, SCN rhythms mature in several ways. Overall expression level of rhythmic genes is divergent in late embryonic mouse SCN, with genes that become rhythmic early increasing and genes that become rhythmic later decreasing in expression (Houdek and Sumová, 2014). Rhythmicity of most core clock and clock-controlled genes in the SCN is initially much weaker than in adults and strengthens throughout late embryonic and early post-natal life, reaching adult or near-adult amplitudes at ~P10 for most genes, excepting Clock (Sladek et al., 2004; Li and Davis, 2005; Kovacikova et al., 2006; Sumová et al., 2006; Ansari et al., 2009; Huang et al., 2010; Houdek and Sumová, 2014; Wreschnig et al., 2014). Rat SCN firing rhythms reach adult amplitudes shortly thereafter, at ~P14 (Shibata and Moore, 1987). The notable exception to increasing rhythmicity with age is Vip, which becomes steadily less rhythmic during post-natal development even as its expression level rises, reaching its diminished adult amplitude by P20 in rat SCN (Ban et al., 1997). Peak and trough phase also changes dramatically for some rhythmic SCN genes during this time, including Cry1, Nr1d1, Avp, and Vip (Ban et al., 1997; Huang et al., 2010; Houdek and Sumová, 2014).
Despite the growing body of research on clock diversity in the adult SCN, to our knowledge there are currently no studies examining the ontogeny of subdomain differences in phase and period of SCN rhythms. However, marked changes in expression patterns of some clock genes within the SCN, such as Rorα, between embryonic development and adulthood suggest substantial shifts in the spatiotemporal clock arrangement (Shimogori et al., 2010; Hatori et al., 2014). These whole-SCN changes during early post-natal life may indicate the existence of fruitful developmental windows to study the emergence of subdomain-specific clocks (Ban et al., 1997; Huang et al., 2010; Houdek and Sumová, 2014).
There is, however, evidence for subdomain-specific clock effects on other aspects of SCN development. For example, there are conflicting reports about SCN cell fate changes in Clock−/− mice. In both studies, Avp neuron number was reduced prior to P6; one of the groups also reported a failure of Vip neurons to increase in number and properly localize after P6, though Vip neuron development before that age seemed normal (Silver et al., 1999; Herzog et al., 2000). There is also a selective increase in Calb1 neurons in Tau mutant hamsters (LeSauter et al., 1999). Thus, the developing clockworks seem to influence SCN cell fate during early post-natal life, and possibly earlier.
Entrainment of developing SCN clocks
As the SCN clock begins ticking, it is faced with a problem: how to align itself with the solar cycle in the absence of light input in the womb. The mother entrains emerging SCN rhythms, but this requires an intact circadian system. SCN lesion of dams at gestational day 7 disrupts within-litter synchrony of SCN metabolic rhythms in E21 rats, melatonin synthesis rhythms in P10 rats, and rest/activity rhythms in P10 hamsters (Reppert and Schwartz, 1986; Davis and Gorski, 1988). Similarly, rhythmic Per1±;Per2± or Cry1±;Per2± mouse pups born to homozygous mutant dams and wild-type studs show reduced within-litter synchrony of activity rhythms (Jud and Albrecht, 2006).
Interestingly, disrupting maternal rhythms shortly before birth by ablating the SCN of pregnant hamster dams at gestational day 14 has a less severe effect on within-litter synchrony of pups than SCN ablation at day 7 (Davis and Gorski, 1988). Similarly, phase shifting a pregnant rat dam at gestational day 18, but not day 20, results in phase shifts in c-fos and AVP rhythms in her developing pups' SCNs (El-Hennamy et al., 2008). Postnatally, fostering blinded rat pups with a phase-reversed dam, or administering a 6-h phase shift, have only very small effects on pup SCN clock gene phase (Ohta et al., 2002), though maternal absence during defined intervals is a much more robust entraining cue (Shimoda et al., 1986; Viswanathan, 1999; Ohta et al., 2003). Thus, excepting conditions of extreme stress such as dam absence, maternal control of synchronization and phase setting of her pups' SCN rhythms rapidly decreases in later embryonic life, as the pup's own rhythms synchronize and begin to oscillate independently of the dam's cues.
But what are these cues? One perennial but controversial candidate is melatonin secreted by the pineal gland. In rat pups born to LL-housed dams, failure of SCN c-fos and Avp rhythms to form by P1 is rescued by E16-E21 melatonin injections (Houdek et al., 2014). Though vehicle injections have a similar effect, melatonin but not vehicle remains able to rescue this defect in pups born to pinealectomized, LL-housed dams (Houdek et al., 2014). Dam pinealectomy at gestational day 7 is also sufficient to disrupt within-litter synchrony of drinking rhythms in rat pups (Bellavia et al., 2006). Melatonin is also present in the milk of mammals, including humans, and exogenous melatonin is sufficient to entrain rat and hamster rhythms both embryonically and in early postnatal life (Davis and Mannion, 1988; Illnerova et al., 1993; Grosse et al., 1996; Viswanathan and Davis, 1997).
On the other hand, propanol-mediated blockade of melatonin secretion of suckling rat dams has no effect on within-litter synchrony of pup melatonin secretion rhythms, though as discussed previously, this study's intervention lagged behind the most important developmental window for maternal entrainment (Rowe and Kennaway, 2002). More tellingly, pineal melatonin secretion has been lost in most laboratory mouse strains, likely due to artificial selective pressures against seasonally regulated reproduction, with no clear effect on the development of SCN rhythmicity (Goto et al., 1989). Given this important counterpoint, while melatonin is very likely one of the maternal entraining cues in mammals, it seems to be largely dispensable, likely due to redundancy with other factors.
Dopamine is a particularly attractive candidate to play a complementary role to melatonin at this stage, given their powerful but diametrically opposed effects on both phase and the cAMP signaling system involved in adult SCN entrainment (Viswanathan and Davis, 1997; Vanecek and Watanabe, 1998; Schurov et al., 2002; An et al., 2011; Atkinson et al., 2011). Dopamine agonists are sufficient to entrain late-embryonic hamsters, and are able to induce c-fos in embryonic rat and hamster SCN with rapid, transient kinetics similar to that induced by light later in life (Weaver et al., 1992, 1995; Viswanathan et al., 1994; Viswanathan and Davis, 1997; Duffield et al., 1999). D1 dopamine receptor (D1r) is transiently expressed in rat SCN, and while D1r is not developmentally regulated in hamster, SCN innervation by dopaminergic afferents is (Weaver et al., 1992; Duffield et al., 1999). Accordingly, rat SCN stops responding to dopamine stimulation at ~P4, as light control of the SCN increases; however, eye enucleation fails to rescue loss of dopamine sensitivity, so different mechanisms control light sensitization and dopamine desensitization in the developing SCN (Weaver and Reppert, 1995).
This handoff between signals controlling SCN entrainment is gradual. As mentioned previously, acute SCN light activation, as measured by c-fos expression, begins between P0 and P1 and reaches adult levels by P10 in mouse and rat, and begins by P4 and matures by P15 in hamster (Kaufman and Menaker, 1994; Leard et al., 1994; Sekaran et al., 2005; Mateju et al., 2009). Acute light induction is observed by P1 for Per1 and P3 for Per2 in rat SCN (Mateju et al., 2009). However, phase-dependent gating of all three genes' responses to light comes significantly later: Per1 induction is gated by P3, Per2 by P5, and c-fos induction is not gated until P10 (Bendova et al., 2004; Mateju et al., 2009). The substantial delay of phase-dependent gating of c-fos induction, relative to light-dependent inducibility, supports the importance of both a fully functional SCN clockwork and neurotransmitter network for controlling SCN activity, as this coincides with the maturation of most clock gene rhythms, arrival of most afferents, and adult-like expression of many cell-specific markers within the SCN.
SCN entrainment by light also developmentally lags behind the ability of light to induce acute responses. Under 8:16 and 16:8 LD cycles, changes are not detectable in rat SCN Per1/2 rhythms until P10 and Cry1 rhythms until P20, while Bmal1 rhythms are still unaffected even at P20 (Kovacikova et al., 2005; Sumová et al., 2006). Similarly, the SCN shell c-fos rhythm fails to respond in an adult pattern to a 16:8 light cycle as late as P10 (Bendova et al., 2004). This starkly contrasts with the marked plasticity of behavior in adolescent and young adult mice later in development, once light entrainment is fully on-line (Aton et al., 2004). Thus, the late developmental onset of full SCN light entrainment may be necessary to prevent maladaptive alterations of the SCN's homeostatic range during the period when its cell fate, connectivity, and clockworks are still extremely plastic, prior to the full maturation of the SCN astrocyte network and/or other stabilizing factors (Munekawa et al., 2000).
Conclusion
In service to the SCN's role as the master light-entrained oscillator, SCN neurons have numerous signals, connections, and clock features that work together, with more advanced functions of the central clock requiring progressively more communication within this heterogeneous network. But for this enormously complex master clock to begin ticking, the gears must be appropriately forged, interlocked, and wound during the development of the organism. Importantly, none of these steps can be accomplished without allowance for the others. Misshapen gears will not interlock. Poorly fitted gears will destroy each other. And a perfectly assembled but unwound clock is useless.
The dramatically differing traits of vlSCNmain and vlSCNmed neurons in the adult rat SCN serve as a perfect exemplar of this point. While vlSCNmain cells are densely innervated by ipRGCs, show Per1 induction by light, and rhythmically express Vip that is often co-expressed with Grp, vlSCNmed cells have little ipRGC innervation, show no Per1 induction by light, and express Vip in a pattern that is neither rhythmic nor co-expressed with Grp (Kawamoto et al., 2003). All of these differences in gene expression, connectivity, and clockworks are consistent within the respective populations and, given this consistency, are presumably important for their respective functions. But to ensure that the appropriate traits co-localize, the development of these various traits must be coordinated.
Thus, although we have separated our discussions of how cell fate, connectivity, and clock ontogeny emerge during SCN development, we also hope to have conveyed the profound integration of these processes. Not coincidentally, this mirrors the current situation in the field. Historically, the study of these topics was limited by a poor understanding of hypothalamic development in general, which prevented genetic tools being pioneered in the cortex and other brain regions from being applied in the SCN. But now, recent large-scale studies mapping hypothalamic gene expression (e.g., Lein et al., 2007; Shimogori et al., 2010; VanDunk et al., 2011) and the burgeoning availability of Cre/lox and other alleles for intersectional genetic approaches in mice have dramatically improved our ability to attack these questions. This directly prompted the ongoing burst of progress in SCN development, particularly the roles of various transcription factors in the process that are discussed in this review and elsewhere (Landgraf et al., 2014).
The interrogation of Lhx1's role in the SCN is a case in point. Constitutive mutants for Lhx1 fail to form anterior head structures entirely (Shawlot and Behringer, 1995), precluding study of the SCN in these animals. It was only by targeting a loxP-flanked Lhx1 allele with Cre drivers whose overlaps with Lhx1 expression are constrained to the developing vAH and retina (Six3-Cre) or SCN and cerebellum (Rorα-Cre) that studies of this gene's role in several aspects of SCN development and function became possible (Zhao et al., 2007; Shimogori et al., 2010; Bedont et al., 2014; Hatori et al., 2014; Allen Brain Atlas).
As such approaches are adopted more broadly in hypothalamic developmental studies, the benefits for understanding the basic biology of the adult SCN will be three-fold. First, developmental mutants that survive to adulthood often present unique and interesting phenotypes difficult to obtain in other ways, such as the loss of multiple neuropeptides in the Lhx1-deficient SCN. Second, by characterizing the spatial and temporal expression of ever more SCN-enriched genes, developmental studies will also steadily improve our ability to intersectionally target genes of interest in the adult SCN. And finally, the study of SCN development in multiple species has given intriguing insights into the evolution of the region, providing tantalizing clues about both the “hows” and “whys” of SCN function.
That said, most work on mammalian SCN development has been conducted in rodent species (predominantly mice, rats, and hamsters), raising some questions about the extent to which this body of work extends to mammals at large, including primates. What is known suggests that the broad strokes of findings in rodents are generalizable within the mammalian clade. The basic structure of the adult SCN and its suite of neuropeptides is largely preserved with mostly modest variations in mammals as diverse as mice, cats, cows, pigs, rhesus macaques, and humans (Sofroniew and Weindl, 1980; Stopa et al., 1984; Cassone et al., 1988; Dai et al., 1997; Abrahamson and Moore, 2001), with the notable exception of the re-emergence of local SCN Npy expression in humans (Moore, 1989). Furthermore, the evidence we discussed earlier suggesting that Lhx1 controls SCN neuropeptide fate in both amphibians and mammals, despite substantial changes in adult SCN neuropeptide composition between these clades, implies that the genetic regulatory networks controlling SCN formation are even better conserved across species than the SCN's adult form (Tuinhof et al., 1994a,b; Abrahamson and Moore, 2001; Brohl et al., 2008; Dulcis and Spitzer, 2008; Bedont et al., 2014).
And perhaps most importantly, the phenomenology of many SCN developmental events is similar in rodents and primates. Sequential induction of SCN neuropeptide protein expression remains the rule in humans, and according to most studies the gestation-adjusted age of initial detection in humans is similar to rodents at ~80% of gestation for Vip and around birth for Avp (Swaab et al., 1990, 1994), though others claim to observe somewhat earlier but similarly sequenced induction (Xu et al., 2003). And much like in rodents, SCN clock ontogeny starts at ~90% of gestation in capuchin monkeys, with Bmal1 and Per2 cycling beginning at P142 (Torres-Farfan et al., 2006). These similarities with the rodent developmental timeline hold despite SCN neurogenesis occurring between E27-E48 (~16–29% of gestation) in rhesus macaques, with the densest production from E30-43, dramatically earlier on a gestationally adjusted basis than in rodents (van Eerdenburg and Rakic, 1994). In sum, the available data suggests that primate SCN development is drawn out over a significantly longer time than, but is mechanistically similar to, rodent SCN development, ultimately producing a functionally similar SCN at correspondent developmental stages. That said, with genetic tools in marmosets coming online in recent years, a direct examination of whether primate SCN developmental mechanisms are fundamentally similar to rodents by intersectional loss-of-function studies is a promising proposition. This is due not only to the topic's profound implications for the basic biology of SCN development and evolution, but also for human health.
Circadian disruption over the course of development in humans is very common. For instance, during adolescence the prevalence of formal delayed sleep phase disorder (DSPD) is estimated at ~7–16% (reviewed in Bartlett et al., 2013), and many teenagers that do not meet this formal diagnosis nonetheless tend toward a “night owl” chronotype. Coupled with social expectations out of phase with biological reality, this circadian change likely contributes to the incidence of psychiatric illness in adolescents, as DSPD and sometimes even the night owl chronotype alone have been linked to depression, obsessive-compulsive disorder, and other mental ailments (Lewy, 2009; Reid et al., 2012; Schubert and Coles, 2013). Early attempts at interventions in a military basic training environment have suggested that simply adapting adolescents' schedules to compensate for their altered chronotype improves sleep quality and learning, while ameliorating mood disturbance (Miller et al., 2012). A better understanding of SCN development may give insight into new approaches and tools for manipulating the circadian system for psychiatric effect during adolescence and at various other developmental stages.
Later in life, low amplitude and late phase of sleep/wake rhythms are predictive of problems such as dementia (Tranah et al., 2011) and progressive degradation of sleep/wake rhythms is apparent in both Alzheimer's and Huntington's disease (Harper et al., 2004; Morton et al., 2005). Age-related circadian dysfunction likely stems at least partially from disruptions in SCN clock output; while core clock gene rhythms remain robust in the SCN throughout the lifespan in rodent models, output-related clock gene expression, electrical activity, and neuropeptide rhythms all decline with age (Roozendaal et al., 1987; Kawakami et al., 1997; Nakamura et al., 2011; Farajnia et al., 2012; Bonaconsa et al., 2014). Similar deficiencies have been noted in the aged human SCN, and many of these deficits are exacerbated in dementia patients, supporting the view that organic decline of the SCN contributes to the late-life sleep/wake fragmentation linked to neurodegeneration (reviewed in Hofman and Swaab, 2006). Understanding how various aspects of SCN output come online during development may give insight into how to restore them later in life, to combat this decline.
Finally, a thorough understanding of SCN cell fate determination is also a prerequisite to differentiate induced pluripotent stem cells (iPSCs) into SCN neurons. Aside from neurodegenerative and psychiatric disease, circadian dysfunction is implicated in a host of metabolic and cardiovascular disorders in humans, as well as cancer and obesity (reviewed in Campos Costa et al., 2013; Froy, 2013; Videnovic et al., 2014; Virag and Lust, 2014; Masri et al., 2015). The ability to differentiate a human SCN from iPSCs will allow the modeling of such patients' central clock in culture, aiding in the identification of abnormalities in SCN function that may contribute to their pathology. iPSC-derived SCN transplants also represent a potential future cure for explicit disorders of the core circadian clock, as such transplants impart their period to the host's circadian behavior in animal studies (Ralph et al., 1990). One such condition is familial advanced sleep phase disorder, a genetic ailment in which clock gene mutations disrupt the patient's internal period (reviewed in Nesbitt and Dijk, 2014). Such an intervention would free these patients from the constant need to rigidly structure their lives, in an effort to shoehorn their misaligned rhythms into following the solar cycle.
In conclusion, we now stand at the edge of a promising new path forward to study SCN development, illuminated by new technologies and following paths laid down in decades of invaluable descriptive studies. As our understanding of the region's developmental program accumulates in the years to come, so too will our understanding of how and why various individual traits of SCN neurons are bound together in the adult network to produce the central circadian clock. And in the process, we stand to gain unique insight not only into how our internal watch is made, but also in how to repair it as time and the environment takes its toll.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Elizabeth Newman for artistic assistance with our figures. This work was supported by National Institutes of Health Grant R01EY020560, and NIH Visual Neuroscience Training Program (5T32EY007143) and NSF Graduate Research Fellowships to JLB.
Glossary
Abbreviations
- 5HT
serotonin (raphe neurotransmitter)
- Avp
arginine vasopressin (SCN neuropeptide)
- Avpr1a/1b
Avp receptors 1a/1b
- Bmal1 (Arntl)
basic helix-loop-helix-PAS domain transcription factor (clock gene, core positive arm)
- Calb1
calbindin, calcium binding protein
- Calb2
calretinin, calcium binding protein
- c-fos
proto-oncogene (induction of gene expression is indicative of neuron activation)
- Clock
basic helix-loop-helix-PAS domain transcription factor (clock gene, core positive arm)
- Creb3l1
cAMP responsive element binding protein 3-like 1 transcription factor
- Cry1/2
cryptochrome 1/2 (clock gene, core negative arm)
- D1r
dopamine receptor 1
- DD
constant darkness
- dlAH
dorsolateral anterior hypothalamus (developmental compartment that gives rise to neuroendocrine hypothalamus)
- DMH
dorsomedial hypothalamus
- dmSCN
dorsomedial shell subdomain of the SCN, classically defined by Avp expression
- E(x)
embryonic day x of gestation
- Foxd1
forkhead box transcription factor D1
- Fzd5
frizzled 5, Wnt receptor
- Gfap
glial fibrillary acid protein (astrocyte marker)
- Grp
gastrin releasing peptide (SCN neuropeptide)
- IGL
intergeniculate leaflet of the thalamus (major SCN afferent)
- ipRGC
intrinsically photosensitive retinal ganglion cells (major SCN afferent)
- LD
light:dark entrainment cycle
- Lhx1/2/8
LIM homeobox transcription factor 1/2/8
- Math5 (Atoh7)
basic helix-loop-helix transcription factor
- Nkx2.2
NK2 homeobox transcription factor 2.2
- Npy
neuropeptide Y (IGL neuropeptide)
- Nr1d1 (Rev-erbα)
nuclear receptor 1 d1 (clock gene, auxiliary negative arm)
- Opn4
melanopsin, ipRGC photopigment
- Otp
orthopedia homeobox transcription factor
- P(x)
post-natal day x of development
- Pacap
pituitary adenylate cyclase-activating peptide (ipRGC neuropeptide)
- Per1/2
period 1/2 (clock gene, core negative arm)
- Pou2f2
Pou class 2 homeobox 2 transcription factor
- Prok2
prokineticin-2 (SCN neuropeptide)
- Prokr2
prokineticin receptor 2
- PVN
paraventricular nucleus of the hypothalamus
- Rax
retinal and anterior neural fold homeobox transcription factor
- RHT
retinohypothalamic tract
- Ror/α/β
retinoic-acid-receptor related orphan nuclear receptor α/β (clock gene, auxiliary positive arm)
- SCN
suprachiasmatic nucleus
- Shh
Sonic hedgehog (lipid-modified polypeptide morphogen)
- Sim1
single-minded homolog 1 basic-helix-loop-helix transcription factor
- Six3/6
Six family homeodomain transcription factor 3/6
- Sox2
sex-determining-region Y box 2 transcription factor
- Sp
substance P, neuropeptide
- SPZ
subparaventricular zone of the hypothalamus
- vAH
ventral anterior hypothalamus (developmental compartment that gives rise to SCN and other nearby nuclei)
- Vax1
ventral anterior homeobox 1
- Vipr2
Vip receptor 2
- Vip
vasoactive intestinal peptide (SCN neuropeptide)
- vlSCN
ventrolateral core subdomain of the SCN, classically defined by Vip expression
- Wnt
wingless family members (lipid-modified glycoprotein morphogens).
Footnotes
1 Allen Brain Atlas (http://www.brain-map.org/). Accessed Sept 1-Dec 31, 2014.
References
- Abe M., Herzog E. D., Yamazaki S., Straume M., Tei H., Sakaki Y., et al. (2002). Circadian rhythms in isolated brain regions. J. Neurosci. 22, 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abizaid A., Mezei G., Sotonyi P., Horvath T. L. (2004). Sex differences in adult suprachiasmatic nucleus neurons emerging late prenatally in rats. Eur. J. Neurosci. 19, 2488–2496. 10.1111/j.0953-816X.2004.03359.x [DOI] [PubMed] [Google Scholar]
- Abrahamson E. E., Moore R. Y. (2001). Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191. [DOI] [PubMed] [Google Scholar]
- Abrahamson E. E., Moore R. Y. (2006). Lesions of suprachiasmatic nucleus efferents selectively affect rest-activity rhythm. Mol. Cell. Endocrinol. 252, 46–56. 10.1016/j.mce.2006.03.036 [DOI] [PubMed] [Google Scholar]
- Ahern T. H., Krug S., Carr A. V., Murray E. K., Fitzpatrick E., Bengston L., et al. (2013). Cell death atlas of the postnatal mouse ventral forebrain and hypothalamus: effects of age and sex. J. Comp. Neurol. 521, 2551–2569. 10.1002/cne.23298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers H. E., Liou S. Y., Stopa E. G., Zoeller R. T. (1991). Interaction of colocalized neuropeptides: functional significance in the circadian timing system. J. Neurosci. 11, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J., Bayer S. A. (1978). Development of the diencephalon in the rat. I. Autoradiographic study of the time of origin and settling patterns of neurons of the hypothalamus. J. Comp. Neurol. 182(4 Pt 2), 945–971 10.1002/cne.901820511 [DOI] [PubMed] [Google Scholar]
- Alvarez-Bolado G., Paul F. A., Blaess S. (2012). Sonic hedgehog lineage in the mouse hypothalamus: from progenitor domains to hypothalamic regions. Neural Dev. 7:4. 10.1186/1749-8104-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Harang R., Meeker K., Granados-Fuentes D., Tsai C. A., Mazuski C., et al. (2013). A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc. Natl. Acad. Sci. U.S.A. 110, E4355–E4361. 10.1073/pnas.1307088110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Irwin R. P., Allen C. N., Tsai C., Herzog E. D. (2011). Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J. Neurophysiol. 105, 2289–2296. 10.1152/jn.00966.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Tsai C., Ronecker J., Bayly A., Herzog E. D. (2012). Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J. Comp. Neurol. 520, 2730–2741. 10.1002/cne.23078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari N., Agathagelidis M., Lee C., Korf H. W., von Gall C. (2009). Differential maturation of circadian rhythms in clock gene proteins in the suprachiasmatic nucleus and the pars tuberalis during mouse ontogeny. Eur. J. Neurosci. 29, 477–489. 10.1111/j.1460-9568.2008.06605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle M. C., LeSauter J., Silver R. (2005). Neurogenesis and ontogeny of specific cell phenotypes within the hamster suprachiasmatic nucleus. Brain Res. Dev. Brain Res. 157, 8–18. 10.1016/j.devbrainres.2005.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins N., Jr., Mitchell J. W., Romanova E. V., Morgan D. J., Cominski T. P., Ecker J. L., et al. (2010). Circadian integration of glutamatergic signals by little SAAS in novel suprachiasmatic circuits. PLoS ONE 9:e12612 10.1371/journal.pone.0012612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S. E., Maywood E. S., Chesham J. E., Wozny C., Colwell C. S., Hastings M. H., et al. (2011). Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J. Biol. Rhythms 26, 210–220. 10.1177/0748730411402810 [DOI] [PubMed] [Google Scholar]
- Aton S. J., Block G. D., Tei H., Yamazaki S., Herzog E. D. (2004). Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J. Biol. Rhythms 19, 198–207. 10.1177/0748730404264156 [DOI] [PubMed] [Google Scholar]
- Aton S. J., Colwell C. S., Harmar A. J., Waschek J., Herzog E. D. (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 8, 476–483. 10.1038/nn1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A., Dallmann R., Casserly A., Rehrauer H., Patrignani A., Maier B., et al. (2014). Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat. Neurosci. 17, 377–382. 10.1038/nn.3651 [DOI] [PubMed] [Google Scholar]
- Ban Y., Shigeyoshi Y., Okamura H. (1997). Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J. Neurosci. 17, 3920–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. J., Biggs S. N., Armstrong S. M. (2013). Circadian rhythm disorders among adolescents: assessment and treatment options. Med. J. Aust. 199, S16–S20. 10.5694/mja13.10912 [DOI] [PubMed] [Google Scholar]
- Becquet D., Girardet C., Guillaumond F., Francois-Bellan A. M., Bosler O. (2008). Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia 56, 294–305. 10.1002/glia.20613 [DOI] [PubMed] [Google Scholar]
- Bedont J. L., LeGates T. A., Slat E. A., Byerly M. S., Wang H., Hu J., et al. (2014). Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. 7, 609–622. 10.1016/j.celrep.2014.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont J. L., Newman E. A., Blackshaw S. (2015). Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip. Rev. Dev. Biol. [Epub ahead of print]. 10.1002/wdev.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia S. L., Carpentieri A. R., Vaque A. M., Macchione A. F., Vermouth N. T. (2006). Pup circadian rhythm entrainment–effect of maternal ganglionectomy or pinealectomy. Physiol. Behav. 89, 342–349. 10.1016/j.physbeh.2006.06.018 [DOI] [PubMed] [Google Scholar]
- Bendova Z., Sumová A., Illnerova H. (2004). Development of circadian rhythmicity and photoperiodic response in subdivisions of the rat suprachiasmatic nucleus. Brain Res. Dev. Brain Res. 148, 105–112. 10.1016/j.devbrainres.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Bendova Z., Sumová A., Mikkelsen J. D. (2009). Circadian and developmental regulation of N-methyl-d-aspartate-receptor 1 mRNA splice variants and N-methyl-d-aspartate-receptor 3 subunit expression within the rat suprachiasmatic nucleus. Neuroscience 159, 599–609. 10.1016/j.neuroscience.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Bonaconsa M., Malpeli G., Montaruli A., Carandente F., Grassi-Zucconi G., Bentivoglio M. (2014). Differential modulation of clock gene expression in the suprachiasmatic nucleus, liver and heart of aged mice. Exp. Gerontol. 55, 70–79. 10.1016/j.exger.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Bosler O., Beaudet A. (1985). VIP neurons as prime synaptic targets for serotonin afferents in rat suprachiasmatic nucleus: a combined radioautographic and immunocytochemical study. J. Neurocytol. 14, 749–763. 10.1007/BF01170826 [DOI] [PubMed] [Google Scholar]
- Botchkina G. I., Morin L. P. (1993). Development of the hamster serotoninergic system: cell groups and diencephalic projections. J. Comp. Neurol. 338, 405–431. 10.1002/cne.903380307 [DOI] [PubMed] [Google Scholar]
- Botchkina G. I., Morin L. P. (1995). Ontogeny of radial glia, astrocytes and vasoactive intestinal peptide immunoreactive neurons in hamster suprachiasmatic nucleus. Brain Res. Dev. Brain Res. 86, 48–56. [DOI] [PubMed] [Google Scholar]
- Brancaccio M., Maywood E. S., Chesham J. E., Loudon A. S., Hastings M. H. (2013). A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78, 714–728. 10.1016/j.neuron.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohl D., Strehle M., Wende H., Hori K., Bormuth I., Nave K. A., et al. (2008). A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev. Biol. 322, 381–393. 10.1016/j.ydbio.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Brown T. M., Hughes A. T., Piggins H. D. (2005). Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J. Neurosci. 25, 11155–11164. 10.1523/JNEUROSCI.3821-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. M., Piggins H. D. (2009). Spatiotemporal heterogeneity in the electrical activity of suprachiasmatic nuclei neurons and their response to photoperiod. J. Biol. Rhythms 24, 44–54. 10.1177/0748730408327918 [DOI] [PubMed] [Google Scholar]
- Brzezinski J. A. IV, Brown N. L., Tanikawa A., Bush R. A., Sieving P. A., Vitaterna M. H., et al. (2005). Loss of circadian photoentrainment and abnormal retinal electrophysiology in Math5 mutant mice. Invest. Ophthalmol. Vis. Sci. 46, 2540–2551. 10.1167/iovs.04-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr E. D., Takahashi J. S. (2013). Molecular components of the mammalian circadian clock. Handb. Exp. Pharmacol. 217, 3–27 10.1007/978-3-642-25950-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caba M., Tovar A., Silver R., Mogado E., Meza E., Zavaleta Y., et al. (2008). Nature's food anticipatory experiment: entrainment of locomotor behavior, suprachiasmatic and dorsomedial hypothalamic nuclei by suckling in rabbit pups. Eur. J. Neurosci. 27, 432–443. 10.1111/j.1460-9568.2008.06017.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill G. M. (1996). Circadian regulation of melatonin production in cultured zebrafish pineal and retina. Brain Res. 708, 177–181. [DOI] [PubMed] [Google Scholar]
- Cambras T., Canal M. M., Cernuda-Cernuda R., Garcia-Fernandez J. M., Diez-Noguera A. (2015). Darkness during early postnatal development is required for normal circadian patterns in the adult rat. Chronobiol. Int. 32, 178–186. 10.3109/07420528.2014.960048 [DOI] [PubMed] [Google Scholar]
- Cambras T., Lopez L., Arias J. L., Diez-Noguera A. (2005). Quantitative changes in neuronal and glial cells in the suprachiasmatic nucleus as a function of the lighting conditions during weaning. Brain Res. Dev. Brain Res. 157, 27–33. 10.1016/j.devbrainres.2005.02.014 [DOI] [PubMed] [Google Scholar]
- Campos Costa I., Nogueira Carvalho H., Fernandes L. (2013). Aging, circadian rhythms and depressive disorders: a review. Am. J. Neurodegener. Dis. 2, 228–246. [PMC free article] [PubMed] [Google Scholar]
- Campos L. M., Cruz-Rizzolo R. J., Watanabe I. S., Pinato L., Nogueira M. I. (2014). Efferent projections of the suprachiasmatic nucleus based on the distribution of vasoactive intestinal peptide (VIP) and arginine vasopressin (AVP) immunoreactive fibers in the hypothalamus of Sapajus apella. J. Chem. Neuroanat. 57–58, 42–53 10.1016/j.jchemneu.2014.03.004 [DOI] [PubMed] [Google Scholar]
- Cantwell E. L., Cassone V. M. (2006a). Chicken suprachiasmatic nuclei: II. Autoradiographic and immunohistochemical analysis. J. Comp. Neurol. 499, 442–457. 10.1002/cne.21124 [DOI] [PubMed] [Google Scholar]
- Cantwell E. L., Cassone V. M. (2006b). Chicken suprachiasmatic nuclei: I. Efferent and afferent connections. J. Comp. Neurol. 496, 97–120. 10.1002/cne.20935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone V. M., Speh J. C., Card J. P., Moore R. Y. (1988). Comparative anatomy of the mammalian hypothalamic suprachiasmatic nucleus. J. Biol. Rhythms 3, 71–91. 10.1177/074873048800300106 [DOI] [PubMed] [Google Scholar]
- Castel M., Morris J. F. (2000). Morphological heterogeneity of the GABAergic network in the suprachiasmatic nucleus, the brain's circadian pacemaker. J. Anat. 196(Pt 1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Buchanan G. F., Ding J. M., Hannibal J., Gillette M. U. (1999). Pituitary adenylyl cyclase-activating peptide: a pivotal modulator of glutamatergic regulation of the suprachiasmatic circadian clock. Proc. Natl. Acad. Sci. U.S.A. 96, 13468–13473. 10.1073/pnas.96.23.13468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M. Y., Bullock C. M., Li C., Lee A. G., Bermak J. C., Belluzzi J., et al. (2002). Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417, 405–410. 10.1038/417405a [DOI] [PubMed] [Google Scholar]
- Chou T. C., Scammell T. E., Gooley J. J., Gaus S. E., Saper C. B., Lu J. (2003). Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms. J. Neurosci. 23, 10691–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. D., Gorman M. R., Hatori M., Meadows J. D., Panda S., Mellon P. L. (2013). Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein Six6. J. Biol. Rhythms 28, 15–25. 10.1177/0748730412468084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell C. S. (2000). Rhythmic coupling among cells in the suprachiasmatic nucleus. J. Neurobiol. 43, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell C. S., Michel S., Itri J., Rodriguez W., Tam J., Lelievre V., et al. (2003). Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285, R939–R949. 10.1152/ajpregu.00200.2003 [DOI] [PubMed] [Google Scholar]
- Crossland W. J., Uchwat C. J. (1982). Neurogenesis in the central visual pathways of the golden hamster. Brain Res. 281, 99–103. 10.1016/0165-3806(82)90117-1 [DOI] [PubMed] [Google Scholar]
- Dai J., Swaab D. F., Buijs R. M. (1997). Distribution of vasopressin and vasoactive intestinal polypeptide (VIP) fibers in the human hypothalamus with special emphasis on suprachiasmatic nucleus efferent projections. J. Comp. Neurol. 383, 397–414. [DOI] [PubMed] [Google Scholar]
- Daikoku S., Hisano S., Kagotani Y. (1992). Neuronal associations in the rat suprachiasmatic nucleus demonstrated by immunoelectron microscopy. J. Comp. Neurol. 325, 559–571. 10.1002/cne.903250408 [DOI] [PubMed] [Google Scholar]
- Davis F. C., Boada R., LeDeaux J. (1990). Neurogenesis of the hamster suprachiasmatic nucleus. Brain Res. 519, 192–199. [DOI] [PubMed] [Google Scholar]
- Davis F. C., Gorski R. A. (1988). Development of hamster circadian rhythms: role of the maternal suprachiasmatic nucleus. J. Comp. Physiol. A 162, 601–610. 10.1007/BF01342635 [DOI] [PubMed] [Google Scholar]
- Davis F. C., Mannion J. (1988). Entrainment of hamster pup circadian rhythms by prenatal melatonin injections to the mother. Am. J. Physiol. 255(3 Pt 2), R439–R448. [DOI] [PubMed] [Google Scholar]
- Delville Y., Mansour K. M., Quan E. W., Yules B. M., Ferris C. F. (1994). Postnatal development of the vasopressinergic system in golden hamsters. Brain Res. Dev. Brain Res. 81, 230–239. 10.1016/0165-3806(94)90309-3 [DOI] [PubMed] [Google Scholar]
- Doi M., Ishida A., Miyake A., Sato M., Komatsu R., Yamazaki F., et al. (2011). Nat. Commun. 2, 237. 10.1038/ncomms1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield G. E., McNulty S., Ebling F. J. (1999). Anatomical and functional characterisation of a dopaminergic system in the suprachiasmatic nucleus of the neonatal Siberian hamster. J. Comp. Neurol. 408, 73–96. [PubMed] [Google Scholar]
- Duffield G. E., Mikkelsen J. D., Ebling F. J. (2012). Conserved expression of the glutamate NMDA receptor 1 subunit splice variants during the development of the Siberian hamster suprachiasmatic nucleus. PLoS ONE 7:e37496. 10.1371/journal.pone.0037496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcis D., Jamshidi P., Leutgeb S., Spitzer N. C. (2013). Neurotransmitter switching in the adult brain regulates behavior. Science 340, 449–453. 10.1126/science.1234152 [DOI] [PubMed] [Google Scholar]
- Dulcis D., Spitzer N. C. (2008). Illumination controls differentiation of dopamine neurons regulating behaviour. Nature 456, 195–201. 10.1038/nature07569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hennamy R., Mateju K., Bendova Z., Sosniyenko S., Sumová A. (2008). Maternal control of the fetal and neonatal rat suprachiasmatic nucleus. J. Biol. Rhythms 23, 435–444. 10.1177/0748730408322635 [DOI] [PubMed] [Google Scholar]
- Enoki R., Kuroda S., Ono D., Hasan M. T., Ueda T., Honma S., et al. (2012). Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 109, 21498–21503. 10.1073/pnas.1214415110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. A., Leise T. L., Castanon-Cervantes O., Davidson A. J. (2011). Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS ONE 6:e15869. 10.1371/journal.pone.0015869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrug J., Nielsen H. S., Hannibal J. (2004). Expression of melanopsin during development of the rat retina. Neuroreport 15, 781–784. 10.1097/00001756-200404090-00008 [DOI] [PubMed] [Google Scholar]
- Farajnia S., Michel S., Deboer T., vanderLeest H. T., Houben T., Rohling J. H., et al. (2012). Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J. Neurosci. 32, 5891–5899. 10.1523/JNEUROSCI.0469-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley N. C., Tong T. Y., Foley D., Lesauter J., Welsh D. K., Silver R. (2011). Characterization of orderly spatiotemporal patterns of clock gene activation in mammalian suprachiasmatic nucleus. Eur. J. Neurosci. 33, 1851–1865. 10.1111/j.1460-9568.2011.07682.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. (2013). Circadian aspects of energy metabolism and aging. Ageing Res. Rev. 12, 931–940. 10.1016/j.arr.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Fukuda H., Tokuda I., Hashimoto S., Hayasaka N. (2011). Quantitative analysis of phase wave of gene expression in the mammalian central circadian clock network. PLoS ONE 6:e23568. 10.1371/journal.pone.0023568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A. J., Todd W. D., Blumberg M. S. (2012). Development of SCN connectivity and the circadian control of arousal: a diminishing role for humoral factors? PLoS ONE 7:e45338. 10.1371/journal.pone.0045338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble K. L., Allen G. C., Zhou T., McMahon D. G. (2007). Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J. Neurosci. 27, 12078–12087. 10.1523/JNEUROSCI.1109-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan D., Carter D. A. (2008). A novel site of adult doublecortin expression: neuropeptide neurons within the suprachiasmatic nucleus circadian clock. BMC Neurosci. 9:2. 10.1186/1471-2202-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardet C., Blanchard M. P., Ferracci G., Leveque C., Moreno M., Francois-Bellan A. M., et al. (2010). Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: contribution of glutamatergic afferents. Eur. J. Neurosci. 31, 359–370. 10.1111/j.1460-9568.2009.07071.x [DOI] [PubMed] [Google Scholar]
- Gooley J. J., Lu J., Chou T. C., Scammell T. E., Saper C. B. (2001). Melanopsin in cells of origin of the retinohypothalamic tract. Nat. Neurosci. 4, 1165. 10.1038/nn768 [DOI] [PubMed] [Google Scholar]
- Goto M., Oshima I., Tomita T., Ebihara S. (1989). Melatonin content of the pineal gland in different mouse strains. J. Pineal Res. 7, 195–204. 10.1111/j.1600-079X.1989.tb00667.x [DOI] [PubMed] [Google Scholar]
- Granados-Fuentes D., Saxena M. T., Prolo L. M., Aton S. J., Herzog E. D. (2004). Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur. J. Neurosci. 19, 898–906. 10.1111/j.0953-816X.2004.03117.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood M., Bordieri L., Greenwood M. P., Rosso Melo M., Colombari D. S., Colombari E., et al. (2014). Transcription factor CREB3L1 regulates vasopressin gene expression in the rat hypothalamus. J. Neurosci. 34, 3810–3820. 10.1523/JNEUROSCI.4343-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse J., Velickovic A., Davis F. C. (1996). Entrainment of Syrian hamster circadian activity rhythms by neonatal melatonin injections. Am. J. Physiol. 270(3 Pt 2), R533–R540. [DOI] [PubMed] [Google Scholar]
- Guido M. E., de Guido L., Goguen D., Robertson H. A., Rusak B. (1999). Differential effects of glutamatergic blockade on circadian and photic regulation of gene expression in the hamster suprachiasmatic nucleus. Brain Res. Mol. Brain Res. 67, 247–257. 10.1016/S0169-328X(99)00074-1 [DOI] [PubMed] [Google Scholar]
- Guler A. D., Ecker J. L., Lall G. S., Haq S., Altimus C. M., Liao H. W., et al. (2008). Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453, 102–105. 10.1038/nature06829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J., Bosler O., Dusticier G., Pelletier G., Calas A. (1987). Morphological correlates of serotonin-neuropeptide Y interactions in the rat suprachiasmatic nucleus: combined radioautographic and immunocytochemical data. Cell Tissue Res. 250, 657–662. 10.1007/BF00218960 [DOI] [PubMed] [Google Scholar]
- Hannibal J., Brabet P., Fahrenkrug J. (2008). Mice lacking the PACAP type I receptor have impaired photic entrainment and negative masking. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R2050–R2058. 10.1152/ajpregu.90563.2008 [DOI] [PubMed] [Google Scholar]
- Hannibal J., Fahrenkrug J. (2004). Melanopsin containing retinal ganglion cells are light responsive from birth. Neuroreport 15, 2317–2320. 10.1097/00001756-200410250-00003 [DOI] [PubMed] [Google Scholar]
- Harmar A. J., Marston H. M., Shen S., Spratt C., West K. M., Sheward W. J., et al. (2002). The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508. 10.1016/S0092-8674(02)00736-5 [DOI] [PubMed] [Google Scholar]
- Harper D. G., Stopa E. G., McKee A. C., Satlin A., Fish D., Volicer L. (2004). Dementia severity and Lewy bodies affect circadian rhythms in Alzheimer disease. Neurobiol. Aging 25, 771–781. 10.1016/j.neurobiolaging.2003.04.009 [DOI] [PubMed] [Google Scholar]
- Hartwig H. G., van Venn T. (1979). Spectral characteristics of visible radiation penetrating into the brain and stimulating extraretinal photoreceptors. J. Comp. Physiol. 130, 277–282 10.1007/BF00614615 [DOI] [Google Scholar]
- Hastings M. H., Duffield G. E., Ebling F. J., Kidd A., Maywood E. S., Schurov I. (1997). Non-photic signalling in the suprachiasmatic nucleus. Biol. Cell 89, 495–503. 10.1016/S0248-4900(98)80005-1 [DOI] [PubMed] [Google Scholar]
- Hatori M., Gill S., Mure L. S., Goulding M., O'Leary D. D., Panda S. (2014). Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. Elife 3:e03357. 10.7554/eLife.03357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S., Kumar M., Park A., Tong P., Tung J., Yau K. W., et al. (2006). Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J. Comp. Neurol. 497, 326–349. 10.1002/cne.20970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S., Liao H. W., Takao M., Berson D. M., Yau K. W. (2002). Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070. 10.1126/science.1069609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlerigg D. G., Ebling F. J., Johnston J. D. (2005). Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr. Biol. 15, R449–R450. 10.1016/j.cub.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Herzog E. D., Aton S. J., Numano R., Sakaki Y., Tei H. (2004). Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythms 19, 35–46. 10.1177/0748730403260776 [DOI] [PubMed] [Google Scholar]
- Herzog E. D., Grace M. S., Harrer C., Williamson J., Shinohara K., Block G. D. (2000). The role of Clock in the developmental expression of neuropeptides in the suprachiasmatic nucleus. J. Comp. Neurol. 424, 86–98. [DOI] [PubMed] [Google Scholar]
- Hisano S., Chikamori-Aoyama M., Katoh S., Maegawa M., Daikoku S. (1987). Immunohistochemical evidence of serotoninergic regulation of vasoactive intestinal polypeptide (VIP) in the rat suprachiasmatic nucleus. Histochemistry 86, 573–578. 10.1007/BF00489549 [DOI] [PubMed] [Google Scholar]
- Hoefflin S., Carter D. A. (2014). Neuronal expression of SOX2 is enriched in specific hypothalamic cell groups. J. Chem. Neuroanat. 61–62C, 153–160. 10.1016/j.jchemneu.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Hofman M. A., Swaab D. F. (2006). Living by the clock: the circadian pacemaker in older people. Ageing Res. Rev. 5, 33–51. 10.1016/j.arr.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Honma S., Nakamura W., Shirakawa T., Honma K. (2004). Diversity in the circadian periods of single neurons of the rat suprachiasmatic nucleus depends on nuclear structure and intrinsic period. Neurosci. Lett. 358, 173–176. 10.1016/j.neulet.2004.01.022 [DOI] [PubMed] [Google Scholar]
- Honma S., Ono D., Suzuki Y., Inagaki N., Yoshikawa T., Nakamura W., et al. (2012). Suprachiasmatic nucleus: cellular clocks and networks. Prog. Brain Res. 199, 129–141. 10.1016/B978-0-444-59427-3.00029-0 [DOI] [PubMed] [Google Scholar]
- Houdek P., Polidarová L., Novakova M., Matějů K., Kubik S., Sumová A. (2014). Melatonin administered during the fetal stage affects circadian clock in the suprachiasmatic nucleus but not in the liver. Dev. Neurobiol. 75, 131–144. 10.1002/dneu.22213 [DOI] [PubMed] [Google Scholar]
- Houdek P., Sumová A. (2014). In vivo initiation of clock gene expression rhythmicity in fetal rat suprachiasmatic nuclei. PLoS One 9:e107360. 10.1371/journal.pone.0107360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Lu C., Chen S., Hua L., Qian R. (2010). Postnatal ontogenesis of clock genes in mouse suprachiasmatic nucleus and heart. Lipids Health Dis. 9:22. 10.1186/1476-511X-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse J., Leliavski A., Tsang A. H., Oster H., Eichele G. (2014). The light-dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J. 28, 4950–4960. 10.1096/fj.14-256594 [DOI] [PubMed] [Google Scholar]
- Ibata Y., Takahashi Y., Okamura H., Kawakami F., Terubayashi H., Kubo T., et al. (1989). Vasoactive intestinal peptide (VIP)-like immunoreactive neurons located in the rat suprachiasmatic nucleus receive a direct retinal projection. Neurosci. Lett. 97, 1–5. [DOI] [PubMed] [Google Scholar]
- Ibata Y., Tanaka M., Ichitani Y., Takahashi Y., Okamura H. (1993). Neuronal interaction between VIP and vasopressin neurones in the rat suprachiasmatic nucleus. Neuroreport 4, 128–130. 10.1097/00001756-199302000-00003 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Allen C. N. (2003). Developmental changes in calbindin-D28k and calretinin expression in the mouse suprachiasmatic nucleus. Eur. J. Neurosci. 17, 1111–1118. 10.1046/j.1460-9568.2003.02515.x [DOI] [PubMed] [Google Scholar]
- Ikegami K., Katou Y., Higashi K., Yoshimura T. (2009). Localization of circadian clock protein BMAL1 in the photoperiodic signal transduction machinery in Japanese quail. J. Comp. Neurol. 517, 397–404. 10.1002/cne.22165 [DOI] [PubMed] [Google Scholar]
- Illnerova H., Buresova M., Presl J. (1993). Melatonin rhythm in human milk. J. Clin. Endocrinol. Metab. 77, 838–841. 10.1210/jc.77.3.838 [DOI] [PubMed] [Google Scholar]
- Inagaki N., Honma S., Ono D., Tanahashi Y., Honma K. (2007). Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc. Natl. Acad. Sci. U.S.A. 104, 7664–7669. 10.1073/pnas.0607713104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H., Burlet A., Bosler O. (1999). Vasoactive intestinal peptide neurons as synaptic targets for vasopressin neurons in the suprachiasmatic nucleus. Double-label immunocytochemical demonstration in the rat. Neuroscience 88, 859–870. 10.1016/S0306-4522(98)00259-0 [DOI] [PubMed] [Google Scholar]
- Juarez C., Morgado E., Meza E., Waliszewski S. M., Aguilar-Roblero R., Caba M. (2013). Development of retinal projections and response to photic input in the suprachiasmatic nucleus of New Zealand White Rabbits. Brain Res. 1499, 21–28. 10.1016/j.brainres.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Jud C., Albrecht U. (2006). Circadian rhythms in murine pups develop in absence of a functional maternal circadian clock. J. Biol. Rhythms 21, 149–154. 10.1177/0748730406286264 [DOI] [PubMed] [Google Scholar]
- Kabrita C. S., Davis F. C. (2008). Development of the mouse suprachiasmatic nucleus: determination of time of cell origin and spatial arrangements within the nucleus. Brain Res. 1195, 20–27. 10.1016/j.brainres.2007.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappers J. A. (1981). A survey of advances in pineal research. in Pineal Gland Boca Raton, ed Reiter R. J. (FL: CRC Press; ), 1–26. [Google Scholar]
- Kaufman C. M., Menaker M. (1994). Ontogeny of light-induced Fos-like immunoreactivity in the hamster suprachiasmatic nucleus. Brain Res. 633, 162–166. [DOI] [PubMed] [Google Scholar]
- Kawaguchi C., Isojima Y., Shintani N., Hatanaka M., Guo X., Okumura N., et al. (2010). PACAP-deficient mice exhibit light parameter-dependent abnormalities on nonvisual photoreception and early activity onset. PLoS ONE 5:e9286. 10.1371/journal.pone.0009286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami F., Okamura H., Tamada Y., Maebayashi Y., Fukui K., Ibata Y. (1997). Loss of day-night differences in VIP mRNA levels in the suprachiasmatic nucleus of aged rats. Neurosci. Lett. 222, 99–102. 10.1016/S0304-3940(97)13355-9 [DOI] [PubMed] [Google Scholar]
- Kawamoto K., Nagano M., Kanda F., Chihara K., Shigeyoshi Y., Okamura H. (2003). Two types of VIP neuronal components in rat suprachiasmatic nucleus. J. Neurosci. Res. 74, 852–857. 10.1002/jnr.10751 [DOI] [PubMed] [Google Scholar]
- Kiss J., Leranth C., Halasz B. (1984). Serotoninergic endings on VIP-neurons in the suprachiasmatic nucleus and on ACTH-neurons in the arcuate nucleus of the rat hypothalamus. A combination of high resolution autoradiography and electron microscopic immunocytochemistry. Neurosci. Lett. 44, 119–124. 10.1016/0304-3940(84)90068-5 [DOI] [PubMed] [Google Scholar]
- Kovacikova Z., Sladek M., Bendova Z., Illnerova H., Sumová A. (2006). Expression of clock and clock-driven genes in the rat suprachiasmatic nucleus during late fetal and early postnatal development. J. Biol. Rhythms 21, 140–148. 10.1177/0748730405285876 [DOI] [PubMed] [Google Scholar]
- Kovacikova Z., Sladek M., Laurinova K., Bendova Z., Illnerova H., Sumová A. (2005). Ontogenesis of photoperiodic entrainment of the molecular core clockwork in the rat suprachiasmatic nucleus. Brain Res. 1064, 83–89. 10.1016/j.brainres.2005.10.022 [DOI] [PubMed] [Google Scholar]
- Kriegsfeld L. J., Leak R. K., Yackulic C. B., LeSauter J., Silver R. (2004). Organization of suprachiasmatic nucleus projections in Syrian hamsters (Mesocricetus auratus): an anterograde and retrograde analysis. J. Comp. Neurol. 468, 361–379. 10.1002/cne.10995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krout K. E., Kawano J., Mettenleiter T. C., Loewy A. D. (2002). CNS inputs to the suprachiasmatic nucleus of the rat. Neuroscience 110, 73–92. 10.1016/S0306-4522(01)00551-6 [DOI] [PubMed] [Google Scholar]
- Laemle L. K., Repke K. B., Hawkes R., Rice F. L. (1991). Synaptogenesis in the rat suprachiasmatic nucleus: a light microscopic immunocytochemical survey. Brain Res. 544, 108–117. 10.1016/0006-8993(91)90891-X [DOI] [PubMed] [Google Scholar]
- Landgraf D., Achten C., Dallmann F., Oster H. (2015). Embryonic development and maternal regulation of murine circadian clock function. Chronobiol. Int. 32, 416–427. 10.3109/07420528.2014.986576 [DOI] [PubMed] [Google Scholar]
- Landgraf D., Koch C. E., Oster H. (2014). Embryonic development of circadian clocks in the mammalian suprachiasmatic nuclei. Front. Neuroanat. 8:143. 10.3389/fnana.2014.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak R. K., Moore R. Y. (2001). Topographic organization of suprachiasmatic nucleus projection neurons. J. Comp. Neurol. 433, 312–334. 10.1002/cne.1142 [DOI] [PubMed] [Google Scholar]
- Leard L. E., Macdonald E. S., Heller H. C., Kilduff T. S. (1994). Ontogeny of photic-induced c-fos mRNA expression in rat suprachiasmatic nuclei. Neuroreport 5, 2683–2687. 10.1097/00001756-199412000-00069 [DOI] [PubMed] [Google Scholar]
- Lee I. T., Chang A. S., Manandhar M., Shan Y., Fan J., Izumo M., et al. (2015). Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85, 1086–1102. 10.1016/j.neuron.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Lenn N. J., Beebe B., Moore R. Y. (1977). Postnatal development of the suprachiasmatic hypothalamic nucleus of the rat. Cell Tissue Res. 178, 463–475. 10.1007/BF00219568 [DOI] [PubMed] [Google Scholar]
- LeSauter J., Kriegsfeld L. J., Hon J., Silver R. (2002). Calbindin-D(28K) cells selectively contact intra-SCN neurons. Neuroscience 111, 575–585. 10.1016/S0306-4522(01)00604-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J., Silver R. (1999). Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J. Neurosci. 19, 5574–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J., Stevens P., Jansen H., Lehman M. N., Silver R. (1999). Calbindin expression in the hamster SCN is influenced by circadian genotype and by photic conditions. Neuroreport 10, 3159–3163. 10.1097/00001756-199910190-00007 [DOI] [PubMed] [Google Scholar]
- Lewy A. J. (2009). Circadian misalignment in mood disturbances. Curr. Psychiatry Rep. 11, 459–465. 10.1007/s11920-009-0070-5 [DOI] [PubMed] [Google Scholar]
- Li H., Ferrari M. B., Kuenzel W. J. (2004). Light-induced reduction of cytoplasmic free calcium in neurons proposed to be encephalic photoreceptors in chick brain. Brain Res. Dev. Brain Res. 153, 153–161. 10.1016/j.devbrainres.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Li J. D., Burton K. J., Zhang C., Hu S. B., Zhou Q. Y. (2009). Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am. J. Physiol. Regul. Integr. Comp. Physiol. 296, R824–R830. 10.1152/ajpregu.90463.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Davis F. C. (2005). Developmental expression of clock genes in the Syrian hamster. Brain Res. Dev. Brain Res. 158, 31–40. 10.1016/j.devbrainres.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Lu J., Zhang Y. H., Chou T. C., Gaus S. E., Elmquist J. K., Shiromani P., et al. (2001). Contrasting effects of ibotenate lesions of the paraventricular nucleus and subparaventricular zone on sleep-wake cycle and temperature regulation. J. Neurosci. 21, 4864–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S., Kinouchi K., Sassone-Corsi P. (2015). Circadian clocks, epigenetics, and cancer. Curr. Opin. Oncol. 27, 50–56. 10.1097/CCO.0000000000000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju K., Bendova Z., El-Hennamy R., Sladek M., Sosniyenko S., Sumová A. (2009). Development of the light sensitivity of the clock genes Period1 and Period2, and immediate-early gene c-fos within the rat suprachiasmatic nucleus. Eur. J. Neurosci. 29, 490–501. 10.1111/j.1460-9568.2009.06609.x [DOI] [PubMed] [Google Scholar]
- Maywood E. S., Chesham J. E., O'Brien J. A., Hastings M. H. (2011). A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. U.S.A. 108, 14306–14311. 10.1073/pnas.1101767108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill D. S., Sheely C. J., Ecker J. L., Badea T. C., Morhardt D., Guido W., et al. (2011). Development of melanopsin-based irradiance detecting circuitry. Neural Dev. 6, 8. 10.1186/1749-8104-6-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliarini S., Pacini G., Pelosi B., Lunardi G., Pasqualetti M. (2013). Lack of brain serotonin affects postnatal development and serotonergic neuronal circuitry formation. Mol. Psychiatry 18, 1106–1118. 10.1038/mp.2012.128 [DOI] [PubMed] [Google Scholar]
- Miller N. L., Tvaryanas A. P., Shattuck L. G. (2012). Accommodating adolescent sleep-wake patterns: the effects of shifting the timing of sleep on training effectiveness. Sleep 35, 1123–1136. 10.5665/sleep.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirochnik V., Bosler O., Tillet Y., Calas A., Ugrumov M. (2005). Long-lasting effects of serotonin deficiency on differentiating peptidergic neurons in the rat suprachiasmatic nucleus. Int. J. Dev. Neurosci. 23, 85–91. 10.1016/j.ijdevneu.2004.07.021 [DOI] [PubMed] [Google Scholar]
- Moore H. A., Whitmore D. (2014). Circadian rhythmicity and light sensitivity of the zebrafish brain. PLoS ONE 9:e86176. 10.1371/journal.pone.0086176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. Y. (1989). The geniculohypothalamic tract in monkey and man. Brain Res. 486, 190–194. 10.1016/0006-8993(89)91294-8 [DOI] [PubMed] [Google Scholar]
- Moore R. Y., Bernstein M. E. (1989). Synaptogenesis in the rat suprachiasmatic nucleus demonstrated by electron microscopy and synapsin I immunoreactivity. J. Neurosci. 9, 2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. Y., Eichler V. B. (1972). Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42, 201–206. 10.1016/0006-8993(72)90054-6 [DOI] [PubMed] [Google Scholar]
- Morin L. P. (2007). SCN organization reconsidered. J. Biol. Rhythms 22, 3–13. 10.1177/0748730406296749 [DOI] [PubMed] [Google Scholar]
- Morin L. P. (2013). Neuroanatomy of the extended circadian rhythm system. Exp. Neurol. 243, 4–20. 10.1016/j.expneurol.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin L. P., Blanchard J., Moore R. Y. (1992). Intergeniculate leaflet and suprachiasmatic nucleus organization and connections in the golden hamster. Vis. Neurosci. 8, 219–230. 10.1017/S095252380000287X [DOI] [PubMed] [Google Scholar]
- Morin L. P., Goodless-Sanchez N., Smale L., Moore R. Y. (1994). Projections of the suprachiasmatic nuclei, subparaventricular zone and retrochiasmatic area in the golden hamster. Neuroscience 61, 391–410. 10.1016/0306-4522(94)90240-2 [DOI] [PubMed] [Google Scholar]
- Morin L. P., Shivers K. Y., Blanchard J. H., Muscat L. (2006). Complex organization of mouse and rat suprachiasmatic nucleus. Neuroscience 137, 1285–1297. 10.1016/j.neuroscience.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Morton A. J., Wood N. I., Hastings M. H., Hurelbrink C., Barker R. A., Maywood E. S. (2005). Disintegration of the sleep-wake cycle and circadian timing in Huntington's disease. J. Neurosci. 25, 157–163. 10.1523/JNEUROSCI.3842-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C., Torrealba F. (1998). Postnatal development of neuron number and connections in the suprachiasmatic nucleus of the hamster. Brain Res. Dev. Brain Res. 110, 203–213. 10.1016/S0165-3806(98)00108-4 [DOI] [PubMed] [Google Scholar]
- Munekawa K., Tamada Y., Iijima N., Hayashi S., Ishihara A., Inoue K., et al. (2000). Development of astroglial elements in the suprachiasmatic nucleus of the rat: with special reference to the involvement of the optic nerve. Exp. Neurol. 166, 44–51. 10.1006/exnr.2000.7490 [DOI] [PubMed] [Google Scholar]
- Muscat L., Huberman A. D., Jordan C. L., Morin L. P. (2003). Crossed and uncrossed retinal projections to the hamster circadian system. J. Comp. Neurol. 466, 513–524. 10.1002/cne.10894 [DOI] [PubMed] [Google Scholar]
- Myung J., Hong S., Hatanaka F., Nakajima Y., De Schutter E., Takumi T. (2012). Period coding of Bmal1 oscillators in the suprachiasmatic nucleus. J. Neurosci. 32, 8900–8918. 10.1523/JNEUROSCI.5586-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Adachi A., Nakahama K., Nakamura T., Tamada M., Meyer-Bernstein E., et al. (2003). An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J. Neurosci. 23, 6141–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. J., Nakamura W., Yamazaki S., Kudo T., Cutler T., Colwell C. S., et al. (2011). Age-related decline in circadian output. J. Neurosci. 31, 10201–10205. 10.1523/JNEUROSCI.0451-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura W., Honma S., Shirakawa T., Honma K. (2001). Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 14, 666–674. 10.1046/j.0953-816x.2001.01684.x [DOI] [PubMed] [Google Scholar]
- Negri L., Lattanzi R., Giannini E., Colucci M. A., Mignogna G., Barra D., et al. (2005). Biological activities of Bv8 analogues. Br. J. Pharmacol. 146, 625–632. 10.1038/sj.bjp.0706376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt A. D., Dijk D. J. (2014). Out of synch with society: an update on delayed sleep phase disorder. Curr. Opin. Pulm. Med. 20, 581–587. 10.1097/MCP.0000000000000095 [DOI] [PubMed] [Google Scholar]
- Nishide S. Y., Honma S., Honma K. (2008). The circadian pacemaker in the cultured suprachiasmatic nucleus from pup mice is highly sensitive to external perturbation. Eur. J. Neurosci. 27, 2686–2690. 10.1111/j.1460-9568.2008.06231.x [DOI] [PubMed] [Google Scholar]
- Noche R. R., Lu P. N., Goldstein-Kral L., Glasgow E., Liang J. O. (2011). Circadian rhythms in the pineal organ persist in zebrafish larvae that lack ventral brain. BMC Neurosci. 12:7. 10.1186/1471-2202-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Watanabe K. (2008). Regional differences in circadian period within the suprachiasmatic nucleus. Brain Res. 1239, 119–126. 10.1016/j.brainres.2008.08.082 [DOI] [PubMed] [Google Scholar]
- Noguchi T., Watanabe K., Ogura A., Yamaoka S. (2004). The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur. J. Neurosci. 20, 3199–3202. 10.1111/j.1460-9568.2004.03784.x [DOI] [PubMed] [Google Scholar]
- Ohta H., Honma S., Abe H., Honma K. (2002). Effects of nursing mothers on rPer1 and rPer2 circadian expressions in the neonatal rat suprachiasmatic nuclei vary with developmental stage. Eur. J. Neurosci. 15, 1953–1960. 10.1046/j.1460-9568.2002.02016.x [DOI] [PubMed] [Google Scholar]
- Ohta H., Honma S., Abe H., Honma K. (2003). Periodic absence of nursing mothers phase-shifts circadian rhythms of clock genes in the suprachiasmatic nucleus of rat pups. Eur. J. Neurosci. 17, 1628–1634. 10.1046/j.1460-9568.2003.02584.x [DOI] [PubMed] [Google Scholar]
- Okamura H. S., Murakami K., Uda T., Sugano Y., Takahashi C., Yanaihara N., et al. (1986). Coexistence of vasoactive intestinal peptide (VIP)-, peptide histidine isoleucine amide (PHI)-, and gastrin releasing peptide (GRP)-like immunoreactivity in neurons of the rat suprachiasmatic nucleus. Biomed. Res. 7, 295–299. [Google Scholar]
- Oldfield B. J., Hou-Yu A., Silverman A. J. (1985). A combined electron microscopic HRP and immunocytochemical study of the limbic projections to rat hypothalamic nuclei containing vasopressin and oxytocin neurons. J. Comp. Neurol. 231, 221–231. 10.1002/cne.902310209 [DOI] [PubMed] [Google Scholar]
- Ono D., Honma S., Honma K. (2013). Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat. Commun. 4, 1666. 10.1038/ncomms2670 [DOI] [PubMed] [Google Scholar]
- Pak T., Yoo S., Miranda-Angulo A. M., Wang H., Blackshaw S. (2014). Rax-CreERT2 knock-in mice: a tool for selective and conditional gene deletion in progenitor cells and radial glia of the retina and hypothalamus. PLoS One 9:e90381. 10.1371/journal.pone.0090381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., Sato T. K., Castrucci A. M., Rollag M. D., DeGrip W. J., Hogenesch J. B., et al. (2002). Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298, 2213–2216. 10.1126/science.1076848 [DOI] [PubMed] [Google Scholar]
- Pauls S., Foley N. C., Foley D. K., LeSauter J., Hastings M. H., Maywood E. S., et al. (2014). Differential contributions of intra-cellular and inter-cellular mechanisms to the spatial and temporal architecture of the suprachiasmatic nucleus circadian circuitry in wild-type, cryptochrome-null and vasoactive intestinal peptide receptor 2-null mutant mice. Eur. J. Neurosci. 40, 2528–2540. 10.1111/ejn.12631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard G. E., Ralph M. R., Menaker M. (1987). The intergeniculate leaflet partially mediates effects of light on circadian rhythms. J. Biol. Rhythms 2, 35–56. 10.1177/074873048700200104 [DOI] [PubMed] [Google Scholar]
- Piggins H. D., Antle M. C., Rusak B. (1995). Neuropeptides phase shift the mammalian circadian pacemaker. J. Neurosci. 15, 5612–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser H. M., Bradley A., Chesham J. E., Ebling F. J., Hastings M. H., Maywood E. S. (2007). Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc. Natl. Acad. Sci. U.S.A. 104, 648–653. 10.1073/pnas.0606884104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I., Rodriguez I. R., Jiang G., Hayes W. P., Moreira E. F., Rollag M. D. (2000). A novel human opsin in the inner retina. J. Neurosci. 20, 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero J. E., Kuhlman S. J., McMahon D. G. (2003). The biological clock nucleus: a multiphasic oscillator network regulated by light. J. Neurosci. 23, 8070–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph M. R., Foster R. G., Davis F. C., Menaker M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978. 10.1126/science.2305266 [DOI] [PubMed] [Google Scholar]
- Refinetti R. (2015). Comparison of light, food, and temperature as environmental synchronizers of the circadian rhythm of activity in mice. J. Physiol. Sci. [Epub ahead of print]. 10.1007/s12576-015-0374-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid K. J., Jaksa A. A., Eisengart J. B., Baron K. G., Lu B., Kane P., et al. (2012). Systematic evaluation of Axis-I DSM diagnoses in delayed sleep phase disorder and evening-type circadian preference. Sleep Med. 13, 1171–1177. 10.1016/j.sleep.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S. M., Schwartz W. J. (1984). The suprachiasmatic nuclei of the fetal rat: characterization of a functional circadian clock using 14C-labeled deoxyglucose. J. Neurosci. 4, 1677–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S. M., Schwartz W. J. (1986). Maternal suprachiasmatic nuclei are necessary for maternal coordination of the developing circadian system. J. Neurosci. 6, 2724–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero M. T., Lehman M. N., Silver R. (1993). Age of donor influences ability of suprachiasmatic nucleus grafts to restore circadian rhythmicity. Brain Res. Dev. Brain Res. 71, 45–52. 10.1016/0165-3806(93)90103-H [DOI] [PubMed] [Google Scholar]
- Romero M. T., Silver R. (1990). Time course of peptidergic expression in fetal suprachiasmatic nucleus transplanted into adult hamster. Brain Res. Dev. Brain Res. 57, 1–6. 10.1016/0165-3806(90)90177-Z [DOI] [PubMed] [Google Scholar]
- Romijn H. J., Sluiter A. A., Pool C. W., Wortel J., Buijs R. M. (1997). Evidence from confocal fluorescence microscopy for a dense, reciprocal innervation between AVP-, somatostatin-, VIP/PHI-, GRP-, and VIP/PHI/GRP-immunoreactive neurons in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 9, 2613–2623. 10.1111/j.1460-9568.1997.tb01691.x [DOI] [PubMed] [Google Scholar]
- Roozendaal B., van Gool W. A., Swaab D. F., Hoogendijk J. E., Mirmiran M. (1987). Changes in vasopressin cells of the rat suprachiasmatic nucleus with aging. Brain Res. 409, 259–264. 10.1016/0006-8993(87)90710-4 [DOI] [PubMed] [Google Scholar]
- Rowe S. A., Kennaway D. J. (2002). Melatonin in rat milk and the likelihood of its role in postnatal maternal entrainment of rhythms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, R797–R804. 10.1152/ajpregu.00228.2001 [DOI] [PubMed] [Google Scholar]
- Roy A., de Melo J., Chaturvedi D., Thein T., Cabrera-Socorro A., Houart C., et al. (2013). LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J. Neurosci. 33, 6877–6884. 10.1523/JNEUROSCI.4216-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaltink D. J., Havik B., Verissimo C. S., Lucassen P. J., Vreugdenhil E. (2012). Doublecortin and doublecortin-like are expressed in overlapping and non-overlapping neuronal cell population: implications for neurogenesis. J. Comp. Neurol. 520, 2805–2823. 10.1002/cne.23144 [DOI] [PubMed] [Google Scholar]
- Sato M., Mizoro Y., Atobe Y., Fujimoto Y., Yamaguchi Y., Fustin J. M., et al. (2011). Transportin 1 in the mouse brain: appearance in regions of neurogenesis, cerebrospinal fluid production/sensing, and circadian clock. J. Comp. Neurol. 519, 1770–1780. 10.1002/cne.22600 [DOI] [PubMed] [Google Scholar]
- Schaap J., Albus H., VanderLeest H. T., Eilers P. H., Detari L., Meijer J. H. (2003). Heterogeneity of rhythmic suprachiasmatic nucleus neurons: implications for circadian waveform and photoperiodic encoding. Proc. Natl. Acad. Sci. U.S.A. 100, 15994–15999. 10.1073/pnas.2436298100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert J. R., Coles M. E. (2013). Obsessive-compulsive symptoms and characteristics in individuals with delayed sleep phase disorder. J. Nerv. Ment. Dis. 201, 877–884. 10.1097/NMD.0b013e3182a5eb13 [DOI] [PubMed] [Google Scholar]
- Schurov I. L., Hepworth T. J., Hastings M. H. (2002). Dopaminergic signalling in the rodent neonatal suprachiasmatic nucleus identifies a role for protein kinase A and mitogen-activated protein kinase in circadian entrainment. Eur. J. Neurosci. 15, 223–232. 10.1046/j.0953-816x.2001.01848.x [DOI] [PubMed] [Google Scholar]
- Sekaran S., Lupi D., Jones S. L., Sheely C. J., Hattar S., Yau K. W., et al. (2005). Melanopsin-dependent photoreception provides earliest light detection in the mammalian retina. Curr. Biol. 15, 1099–1107. 10.1016/j.cub.2005.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawlot W., Behringer R. R. (1995). Requirement for Lim1 in head-organizer function. Nature 374, 425–430. 10.1038/374425a0 [DOI] [PubMed] [Google Scholar]
- Shearman L. P., Zylka M. J., Weaver D. R., Kolakowski L. F., Jr., Reppert S. M. (1997). Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269. 10.1016/S0896-6273(00)80417-1 [DOI] [PubMed] [Google Scholar]
- Shen H., Glass J. D., Seki T., Watanabe M. (1999). Ultrastructural analysis of polysialylated neural cell adhesion molecule in the suprachiasmatic nuclei of the adult mouse. Anat. Rec. 256, 448–457. [DOI] [PubMed] [Google Scholar]
- Shen H., Watanabe M., Tomasiewicz H., Rutishauser U., Magnuson T., Glass J. D. (1997). Role of neural cell adhesion molecule and polysialic acid in mouse circadian clock function. J. Neurosci. 17, 5221–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S., Moore R. Y. (1987). Development of neuronal activity in the rat suprachiasmatic nucleus. Brain Res. 431, 311–315. 10.1016/0165-3806(87)90220-3 [DOI] [PubMed] [Google Scholar]
- Shimoda K., Hanada K., Yamada N., Takahashi K., Takahashi S. (1986). Periodic exposure to mother is potent zeitgeber of rat pups' rhythm. Physiol. Behav. 36, 723–730. [DOI] [PubMed] [Google Scholar]
- Shimogori T., Lee D. A., Miranda-Angulo A., Yang Y., Wang H., Jiang L., et al. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775. 10.1038/nn.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura H., Moriya T., Sudo M., Wakamatsu H., Akiyama M., Miyake Y., et al. (2001). Differential daily expression of Per1 and Per2 mRNA in the suprachiasmatic nucleus of fetal and early postnatal mice. Eur. J. Neurosci. 13, 687–693. 10.1046/j.0953-816x.2000.01438.x [DOI] [PubMed] [Google Scholar]
- Silver R., LeSauter J., Tresco P. A., Lehman M. N. (1996). A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature 382, 810–813. 10.1038/382810a0 [DOI] [PubMed] [Google Scholar]
- Silver R., Sookhoo A. I., LeSauter J., Stevens P., Jansen H. T., Lehman M. N. (1999). Multiple regulatory elements result in regional specificity in circadian rhythms of neuropeptide expression in mouse SCN. Neuroreport 10, 3165–3174. 10.1097/00001756-199910190-00008 [DOI] [PubMed] [Google Scholar]
- Silver R., Witkovsky P., Horvath P., Alones V., Barnstable C. J., Lehman M. N. (1988). Coexpression of opsin- and VIP-like-immunoreactivity in CSF-contacting neurons of the avian brain. Cell Tissue Res. 253, 189–198. 10.1007/BF00221754 [DOI] [PubMed] [Google Scholar]
- Sladek M., Sumová A., Kovacikova Z., Bendova Z., Laurinova K., Illnerova H. (2004). Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 101, 6231–6236. 10.1073/pnas.0401149101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slat E., Freeman G. M., Jr., Herzog E. D. (2013). The clock in the brain: neurons, glia, and networks in daily rhythms. Handb. Exp. Pharmacol. 217, 105–123. 10.1007/978-3-642-25950-0_5 [DOI] [PubMed] [Google Scholar]
- Smith L., Canal M. M. (2009). Expression of circadian neuropeptides in the hypothalamus of adult mice is affected by postnatal light experience. J. Neuroendocrinol. 21, 946–953. 10.1111/j.1365-2826.2009.01914.x [DOI] [PubMed] [Google Scholar]
- Sofroniew M. V., Weindl A. (1980). Identification of parvocellular vasopressin and neurophysin neurons in the suprachiasmatic nucleus of a variety of mammals including primates. J. Comp. Neurol. 193, 659–675. 10.1002/cne.901930305 [DOI] [PubMed] [Google Scholar]
- Sosniyenko S., Hut R. A., Daan S., Sumová A. (2009). Influence of photoperiod duration and light-dark transitions on entrainment of Per1 and Per2 gene and protein expression in subdivisions of the mouse suprachiasmatic nucleus. Eur. J. Neurosci. 30, 1802–1814. 10.1111/j.1460-9568.2009.06945.x [DOI] [PubMed] [Google Scholar]
- Southey B. R., Lee J. E., Zamdborg L., Atkins N., Jr., Mitchell J. W., Li M., et al. (2014). Comparing label-free quantitative peptidomics approaches to characterize diurnal variation of peptides in the rat suprachiasmatic nucleus. Anal. Chem. 86, 443–452. 10.1021/ac4023378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speh J. C., Moore R. Y. (1993). Retinohypothalamic tract development in the hamster and rat. Brain Res. Dev. Brain Res. 76, 171–181. 10.1016/0165-3806(93)90205-O [DOI] [PubMed] [Google Scholar]
- Steele C. T., Tosini G., Siopes T., Underwood H. (2006). Time keeping by the quail's eye: circadian regulation of melatonin production. Gen. Comp. Endocrinol. 145, 232–236. 10.1016/j.ygcen.2005.09.004 [DOI] [PubMed] [Google Scholar]
- Steele C. T., Zivkovic B. D., Siopes T., Underwood H. (2003). Ocular clocks are tightly coupled and act as pacemakers in the circadian system of Japanese quail. Am. J. Physiol. Regul. Integr. Comp. Physiol. 284, R208–R218. 10.1152/ajpregu.00447.2002 [DOI] [PubMed] [Google Scholar]
- Stephan F. K., Zucker I. (1972). Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U.S.A. 69, 1583–1586. 10.1073/pnas.69.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopa E. G., King J. C., Lydic R., Schoene W. C. (1984). Human brain contains vasopressin and vasoactive intestinal polypeptide neuronal subpopulations in the suprachiasmatic region. Brain Res. 297, 159–163. 10.1016/0006-8993(84)90553-5 [DOI] [PubMed] [Google Scholar]
- Sumová A., Bendova Z., Sladek M., Kovacikova Z., El-Hennamy R., Laurinova K., et al. (2006). The rat circadian clockwork and its photoperiodic entrainment during development. Chronobiol. Int. 23, 237–243. 10.1080/07420520500522523 [DOI] [PubMed] [Google Scholar]
- Swaab D. F., Hofman M. A., Honnebier M. B. (1990). Development of vasopressin neurons in the human suprachiasmatic nucleus in relation to birth. Brain Res. Dev. Brain Res. 52, 289–293. [DOI] [PubMed] [Google Scholar]
- Swaab D. F., Zhou J. N., Ehlhart T., Hofman M. A. (1994). Development of vasoactive intestinal polypeptide neurons in the human suprachiasmatic nucleus in relation to birth and sex. Brain Res. Dev. Brain Res. 79, 249–259. 10.1016/0165-3806(94)90129-5 [DOI] [PubMed] [Google Scholar]
- Takatsuji K., Oyamada H., Tohyama M. (1995). Postnatal development of the substance P-, neuropeptide Y- and serotonin-containing fibers in the rat suprachiasmatic nucleus in relation to development of the retino-hypothalamic projection. Brain Res. Dev. Brain Res. 84, 261–270. 10.1016/0165-3806(94)00209-I [DOI] [PubMed] [Google Scholar]
- Tanaka M., Ichitani Y., Okamura H., Tanaka Y., Ibata Y. (1993). The direct retinal projection to VIP neuronal elements in the rat SCN. Brain Res. Bull. 31, 637–640. 10.1016/0361-9230(93)90134-W [DOI] [PubMed] [Google Scholar]
- Torres-Farfan C., Rocco V., Monso C., Valenzuela F. J., Campino C., Germain A., et al. (2006). Maternal melatonin effects on clock gene expression in a nonhuman primate fetus. Endocrinology 147, 4618–4626. 10.1210/en.2006-0628 [DOI] [PubMed] [Google Scholar]
- Tosini G., Menaker M. (1996). Circadian rhythms in cultured mammalian retina. Science 272, 419–421. 10.1126/science.272.5260.419 [DOI] [PubMed] [Google Scholar]
- Tranah G. J., Blackwell T., Stone K. L., Ancoli-Israel S., Paudel M. L., Ensrud K. E., et al. (2011). Circadian activity rhythms and risk of incident dementia and mild cognitive impairment in older women. Ann. Neurol. 70, 722–732. 10.1002/ana.22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinhof R., Artero C., Fasolo A., Franzoni M. F., Ten Donkelaar H. J., Wismans P. G., et al. (1994a). Involvement of retinohypothalamic input, suprachiasmatic nucleus, magnocellular nucleus and locus coeruleus in control of melanotrope cells of Xenopus laevis: a retrograde and anterograde tracing study. Neuroscience 61, 411–420. 10.1016/0306-4522(94)90241-0 [DOI] [PubMed] [Google Scholar]
- Tuinhof R., Gonzalez A., Smeets W. J., Roubos E. W. (1994b). Neuropeptide Y in the developing and adult brain of the South African clawed toad Xenopus laevis. J. Chem. Neuroanat. 7, 271–283. 10.1016/0891-0618(94)90018-3 [DOI] [PubMed] [Google Scholar]
- Ueda S., Matsumoto Y., Azmitia E. C., Kawata M. (1993). Intraocular co-grafts of fetal dorsal raphe nucleus and suprachiasmatic nucleus. Brain Res. 605, 181–186. 10.1016/0006-8993(93)91738-E [DOI] [PubMed] [Google Scholar]
- Ueda S., Matsumoto Y., Nisimura A., Azmitia E. C., Kawata M. (1995). Role of neuropeptide Y projection on the development of serotonergic innervation in the suprachiasmatic nucleus of the rat, shown by triple intraocular grafts. Brain Res. 673, 325–330. 10.1016/0006-8993(94)01454-P [DOI] [PubMed] [Google Scholar]
- Ugrumov M. V., Trembleau A., Calas A. (1994). Altered vasoactive intestinal polypeptide gene expression in the fetal rat suprachiasmatic nucleus following prenatal serotonin deficiency. Int. J. Dev. Neurosci. 12, 143–149. 10.1016/0736-5748(94)90007-8 [DOI] [PubMed] [Google Scholar]
- Underwood H., Steele C. T., Zivkovic B. (2001). Circadian organization and the role of the pineal in birds. Microsc. Res. Tech. 53, 48–62. 10.1002/jemt.1068 [DOI] [PubMed] [Google Scholar]
- Vacher C. M., Fretier P., Creminon C., Seif I., De Maeyer E., Calas A., et al. (2003). Monoaminergic control of vasopressin and VIP expression in the mouse suprachiasmatic nucleus. J. Neurosci. Res. 71, 791–801. 10.1002/jnr.10529 [DOI] [PubMed] [Google Scholar]
- van den Pol A. N., Gorcs T. (1986). Synaptic relationships between neurons containing vasopressin, gastrin-releasing peptide, vasoactive intestinal polypeptide, and glutamate decarboxylase immunoreactivity in the suprachiasmatic nucleus: dual ultrastructural immunocytochemistry with gold-substituted silver peroxidase. J. Comp. Neurol. 252, 507–521. 10.1002/cne.902520407 [DOI] [PubMed] [Google Scholar]
- VanderLeest H. T., Houben T., Michel S., Deboer T., Albus H., Vansteensel M. J., et al. (2007). Seasonal encoding by the circadian pacemaker of the SCN. Curr. Biol. 17, 468–473. 10.1016/j.cub.2007.01.048 [DOI] [PubMed] [Google Scholar]
- VanDunk C., Hunter L. A., Gray P. A. (2011). Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J. Neurosci. 31, 6457–6467. 10.1523/JNEUROSCI.5385-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanecek J., Watanabe K. (1998). Melatonin inhibits the increase of cyclic AMP in rat suprachiasmatic neurons induced by vasoactive intestinal peptide. Neurosci. Lett. 252, 21–24. 10.1016/S0304-3940(98)00530-8 [DOI] [PubMed] [Google Scholar]
- van Eerdenburg F. J., Rakic P. (1994). Early neurogenesis in the anterior hypothalamus of the rhesus monkey. Brain Res. Dev. Brain Res. 79, 290–296. 10.1016/0165-3806(94)90134-1 [DOI] [PubMed] [Google Scholar]
- Videnovic A., Lazar A. S., Barker R. A., Overeem S. (2014). ‘The clocks that time us’-circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 10, 683–693. 10.1038/nrneurol.2014.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag J. A., Lust R. M. (2014). Circadian influences on myocardial infarction. Front. Physiol. 5:422. 10.3389/fphys.2014.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan N. (1999). Maternal entrainment in the circadian activity rhythm of laboratory mouse (C57BL/6J). Physiol. Behav. 68, 157–162. [DOI] [PubMed] [Google Scholar]
- Viswanathan N., Davis F. C. (1997). Single prenatal injections of melatonin or the D1-dopamine receptor agonist SKF 38393 to pregnant hamsters sets the offsprings' circadian rhythms to phases 180 degrees apart. J. Comp. Physiol. A 180, 339–346. 10.1007/s003590050053 [DOI] [PubMed] [Google Scholar]
- Viswanathan N., Weaver D. R., Reppert S. M., Davis F. C. (1994). Entrainment of the fetal hamster circadian pacemaker by prenatal injections of the dopamine agonist SKF 38393. J. Neurosci. 14, 5393–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Frisch K. (1911). Beitrage zur physiologie der pigmentzellen in der fischhaut. Pflü. Arch. Gesammte Physiol. Menschen Tiere 138, 319–387 10.1007/BF01680752 [DOI] [Google Scholar]
- Watthanasurorot A., Saelee N., Phongdara A., Roytrakul S., Jiravanichpaisal P., Soderhall K., et al. (2013). Astakine 2–the dark knight linking melatonin to circadian regulation in crustaceans. PLoS Genet. 9:e1003361. 10.1371/journal.pgen.1003361 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Watts A. G., Swanson L. W. (1987). Efferent projections of the suprachiasmatic nucleus: II. Studies using retrograde transport of fluorescent dyes and simultaneous peptide immunohistochemistry in the rat. J. Comp. Neurol. 258, 230–252. 10.1002/cne.902580205 [DOI] [PubMed] [Google Scholar]
- Watts A. G., Swanson L. W., Sanchez-Watts G. (1987). Efferent projections of the suprachiasmatic nucleus: I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J. Comp. Neurol. 258, 204–229. 10.1002/cne.902580204 [DOI] [PubMed] [Google Scholar]
- Weaver D. R., Reppert S. M. (1995). Definition of the developmental transition from dopaminergic to photic regulation of c-fos gene expression in the rat suprachiasmatic nucleus. Brain Res. Mol. Brain Res. 33, 136–148. 10.1016/0169-328X(95)00117-B [DOI] [PubMed] [Google Scholar]
- Weaver D. R., Rivkees S. A., Reppert S. M. (1992). D1-dopamine receptors activate c-fos expression in the fetal suprachiasmatic nuclei. Proc. Natl. Acad. Sci. U.S.A. 89, 9201–9204. 10.1073/pnas.89.19.9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver D. R., Roca A. L., Reppert S. M. (1995). c-fos and jun-B mRNAs are transiently expressed in fetal rodent suprachiasmatic nucleus following dopaminergic stimulation. Brain Res. Dev. Brain Res. 85, 293–297. 10.1016/0165-3806(95)00016-7 [DOI] [PubMed] [Google Scholar]
- Webb A. B., Angelo N., Huettner J. E., Herzog E. D. (2009). Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 106, 16493–16498. 10.1073/pnas.0902768106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb I. C., Coolen L. M., Lehman M. N. (2013). NMDA and PACAP receptor signaling interact to mediate retinal-induced scn cellular rhythmicity in the absence of light. PLoS ONE 8:e76365. 10.1371/journal.pone.007636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschnig D., Dolatshad H., Davis F. C. (2014). Embryonic development of circadian oscillations in the mouse hypothalamus. J. Biol. Rhythms 29, 299–310. 10.1177/0748730414545086 [DOI] [PubMed] [Google Scholar]
- Xu H., Hu X. Y., Wu L., Zhou J. N. (2003). Neurotensin expressing neurons developed earlier than vasoactive intestinal polypeptide and vasopressin expressing neurons in the human suprachiasmatic nucleus. Neurosci. Lett. 335, 175–178. 10.1016/S0304-3940(02)01184-9 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Isejima H., Matsuo T., Okura R., Yagita K., Kobayashi M., et al. (2003). Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412. 10.1126/science.1089287 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Suzuki T., Mizoro Y., Kori H., Okada K., Chen Y., et al. (2013). Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342, 85–90. 10.1126/science.1238599 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Honma S., Honma K. (2013). Daily exposure to a running wheel entrains circadian rhythms in mice in parallel with development of an increase in spontaneous movement prior to running-wheel access. Am. J. Physiol. Regul. Integr. Comp. Physiol. 305, R1367–R1375. 10.1152/ajpregu.00389.2013 [DOI] [PubMed] [Google Scholar]
- Yellon S. M., Thorn K. J., Buchanan K. L., Kirby M. A. (1993). Retinal input to the suprachiasmatic nucleus before and after puberty in Djungarian hamsters. Brain Res. Bull. 32, 29–33. 10.1016/0361-9230(93)90315-3 [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Yasuo S., Suzuki Y., Makino E., Yokota Y., Ebihara S. (2001). Identification of the suprachiasmatic nucleus in birds. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R1185–R1189. [DOI] [PubMed] [Google Scholar]
- Zarrow M. X., Denenberg V. H., Anderson C. O. (1965). Rabbit: frequency of suckling in the pup. Science 150, 1835–1836. 10.1126/science.150.3705.1835 [DOI] [PubMed] [Google Scholar]
- Zhang C., Truong K. K., Zhou Q. Y. (2009). Efferent projections of prokineticin 2 expressing neurons in the mouse suprachiasmatic nucleus. PLoS ONE 4:e7151. 10.1371/journal.pone.0007151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kwan K. M., Mailloux C. M., Lee W. K., Grinberg A., Wurst W., et al. (2007). LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. U.S.A. 104, 13182–13186. 10.1073/pnas.0705464104 [DOI] [PMC free article] [PubMed] [Google Scholar]