Abstract
The relationship between protein three-dimensional structure and function is essential in determining mechanism. Unfortunately, most techniques do not provide a direct measurement of this relationship. Structural data are usually limited to static pictures and function must be inferred. Conversely, functional assays usually provide little information on structural conformation. We developed a single-molecule technique combining optical tweezers and fluorescence microscopy that allows for both measurements simultaneously. Here, we present measurements of UvrD, a DNA repair helicase, that directly and unambiguously reveal the connection between its structure and function. Our data reveal that UvrD exhibits two distinct types of unwinding activity regulated by its stoichiometry. Furthermore, two UvrD conformational states, termed ‘closed’ and ‘open’, correlate with movement toward or away from the DNA fork.
Helicases are vectorial enzymes that play a critical role in genome maintenance, hydrolyzing NTPs to translocate along nucleic acids and separate the duplex strands. UvrD (DNA helicase II) is a prototypical Superfamily 1 (SF 1) helicase involved primarily in nucleotide excision repair and methyl-directed mismatch repair in E. coli (1, 2). While a UvrD monomer is known to translocate along single-stranded DNA (ssDNA) in a 3’-5’ direction (3-6), studies indicate that highly processive duplex DNA unwinding requires at least a dimer (4, 7-11). UvrD and structurally related SF 1 homologs (including Rep and PcrA) consist of four domains (12-16): the RecA-like motor core domains 1A and 2A and the accessory domains 1B and 2B (Fig. 1). Previous studies have shown that the 2B domain of UvrD (14, 16) and homologs (12, 13) can exhibit two orientations—‘open’ and ‘closed’ relative to the other domains (Fig. 1)—believed to regulate activity (6, 9, 14, 16). Despite detailed structural and biochemical data (4, 9-11, 14, 17), a lack of direct evidence linking UvrD conformational state to its function has prevented a more complete understanding of its mechanism. Here, we measured simultaneously the unwinding activity and conformation of UvrD helicase using an instrument combining high-resolution optical traps and single-molecule confocal microscopy (fig. S1 (18, 19)). Through these direct measurements, we demonstrate the link between UvrD oligomeric state and processivity, and conformational state and unwinding vs. rezipping activity.
Fig. 1. Experimental layout.
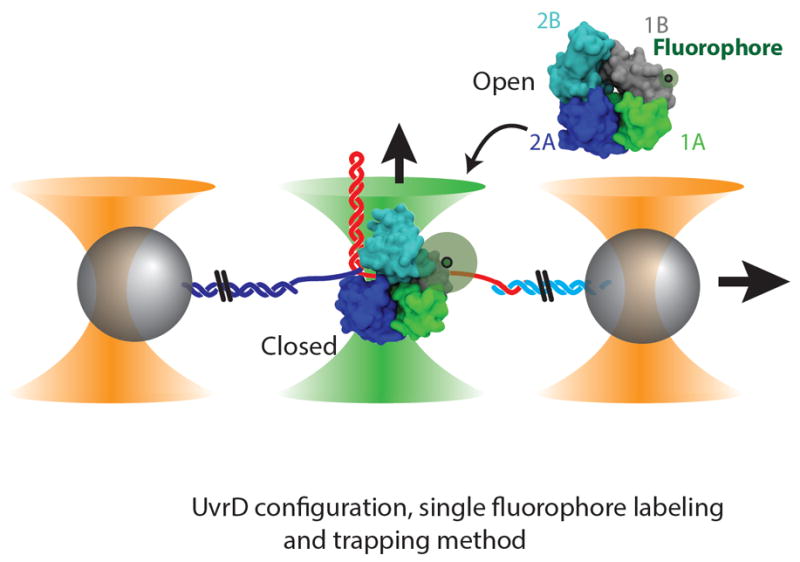
Structure of UvrD monomer in the ‘open’ (upper right, PDB ID 3LFU, (16)) and ‘closed’ (lower left, PDB ID 2IS2, (14)) conformations with single fluorophore location. Two microspheres (gray) in dual optical traps (orange cones) are tethered together by a DNA hairpin. One or more UvrD monomers bind ssDNA and unwind the hairpin in the presence of ATP. A confocal microscope (green cone) detects the configuration of the same fluorescently labeled UvrD unwinding complex.
Helicase activity was monitored by detecting the unwinding of a DNA hairpin with an optical trap, as described previously (20, 21). The trap maintained a constant tension (4–15 pN; 10 pN for data shown unless otherwise specified (19)), and tracked the number of hairpin base pairs unwound. The composition and conformation of the same UvrD unwinding complexes, site-specifically labeled with one or two fluorophores (with labeling efficiencies ranging from 71-85% (19)), were detected with the confocal microscope (Fig. 1).
We observed two distinct types of DNA duplex unwinding activity, which we termed ‘frustrated’ and ‘long-distance’. During ‘frustrated’ activity (Figs. 2A, 3B and 3C, lower panels), UvrD exhibited repetitive, bi-directional motion on DNA, during which <20 of available hairpin base pairs unwound and rezipped (13.6±1.8 bp, mean±SEM unless otherwise noted). This bi-directional activity is distinct from the repetitive ssDNA translocation observed previously with UvrD (6) and its SF 1 homologs (22, 23). Reversals in direction occurred frequently (mean unwinding and rezipping durations 0.25±0.01 s and 0.23±0.02 s respectively), both mid-hairpin and after complete hairpin rezipping, and typically repeated many times before UvrD dissociation (9.2±1.2 repetitions for 7.8±1.3 s). In contrast, during ‘long-distance’ activity (Fig. 2B, lower panel) UvrD systematically unwound >20 bp (38.9±5.6 bp on average). UvrD motion was far less repetitive, although reversals in direction did occur (mean unwinding and rezipping durations 2.82±0.30 s and 1.38±0.13 s, respectively) at mid-hairpin (e.g., at 33 s, Fig. 2B), at the end of the hairpin upon complete unwinding (89 bp, fig. S2), and after full hairpin rezipping. The mean rezipping speed was ATP dependent and nearly the same as for unwinding (fig. S3), strongly suggesting active translocation of UvrD. An alternate in which rezipping results from backsliding (14, 21) would predict nearly instantaneous rezipping events, inconsistent with the data. Since UvrD is a strict 3’-5’ ssDNA translocase (4) with tight ATP-coupling (24), rezipping must correspond to 3’-5’ translocation away from the ss-dsDNA junction, allowing the duplex to base pair in its wake. Thus, reversals in direction are likely the result of switching ssDNA strands, as first proposed by Dessinges et al. (25).
Fig. 2. Effect of UvrD oligomeric state on DNA unwinding activity.
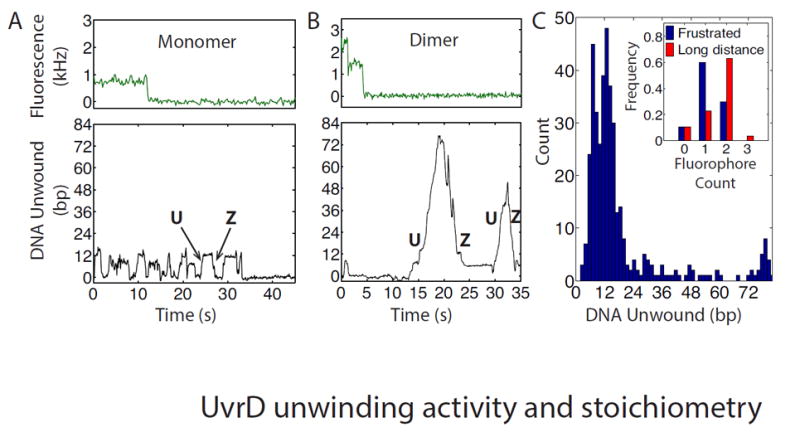
Representative time traces of unwinding activity for a UvrD monomer (A) and dimer (B), respectively. Upper panels: Fluorescence photobleaching from a monomer (A) and a dimer (B) (240 and 120 ms per point respectively). Lower panels: Simultaneous measurements of hairpin unwinding and rezipping. The UvrD monomer displays ‘frustrated’ unwinding (A) (15 bp unwound) whereas the dimer displays long-distance unwinding (72 bp) (B). (C) Histogram of the maximum number of base pairs unwound per unwinding attempt (N = 401) showing frustrated (<20 bp) and long-distance (>20 bp) unwinding. Inset: Distribution of fluorophore count for frustrated (blue) or long-distance (red) unwinding attempts.
Fig. 3. Effect of ‘open’ vs. ‘closed’ UvrD conformation on unwinding activity.
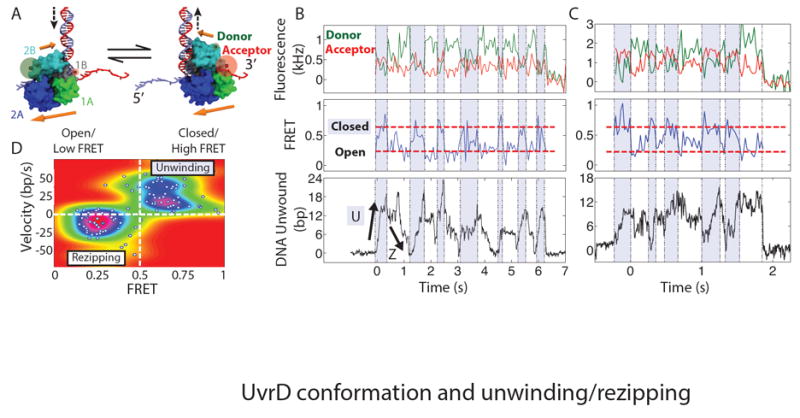
(A) Location of donor and acceptor fluorophores for FRET measurement and model of UvrD conformational switching. Upper (lower) orange arrows denote 2B (1A-2A) domain orientation. See text and (19) for details on the model. (B) and (C) Representative time traces of monomeric UvrD conformation and activity. Upper panels: donor (green) and acceptor (red) fluorescence intensity. Middle panels: corresponding FRET efficiency showing UvrD reversibly switching between ‘open’ (low FRET) and ‘closed’ (high FRET, dashed red lines) conformations. Fluorescence and FRET data are integrated to 60 and 50 ms per point for (B) and (C) respectively. Shaded and unshaded areas denote low and high FRET intervals, respectively. Lower panels: simultaneous measurements of unwinding and rezipping of the DNA hairpin. Fluorescence intensity and ‘frustrated’ unwinding activity are consistent with monomeric UvrD-DNA complexes (which dissociate at t = 6.2 and 1.8 s). (D) Correlation between UvrD activity and conformation. The mean FRET efficiency and mean UvrD velocity determined over each time interval are plotted (white points; N = 109 intervals, 15 molecules). The colormap represents the probability distribution of FRET state and velocity (19).
Our claim that frustrated and long-distance unwinding are distinct activities of UvrD is supported by measurements of the number of consecutive base pairs unwound per unwinding attempt (Fig. 2C (19)) which show a dramatic decrease past ~20 bp. Moreover, the two distinct activities correlate with the number of UvrD present on the DNA hairpin, measured by counting singly-labeled UvrD (19). When we observed frustrated unwinding activity, we usually detected only a single fluorophore, indicating a single UvrD loaded (Fig. 2A, top panel). In contrast, when we observed long-distance unwinding, we were more likely to detect two fluorophores, indicating two UvrD (Fig. 2B, top panel). These trends were corroborated over many UvrD-DNA complexes (Fig. 2C, inset). These observations are consistent with previous reports that UvrD dimers are required for long-distance unwinding (4, 7-9), but also demonstrate that monomers are competent to unwind a limited amount of DNA under tension (fig. S4 (19)).
The ability of a single UvrD to unwind and rezip DNA reversibly many times and in succession suggests that it can switch strands without dissociating from its DNA substrate. Previous studies showed that beyond the 1A-2A motor domains contacting ssDNA, the 2B domain can contact the junction duplex (6, 14). We thus used our instrument to detect the unwinding activity of individual UvrD monomers via optical trapping simultaneously with conformational changes of the 2B domain via single-molecule FRET (26). UvrD was labeled with donor and acceptor fluorophores such that high/low FRET efficiency revealed the ‘closed’/‘open’ conformation, respectively (16) (Fig. 3A (19)).
Figure 3B and 3C show two example data traces of UvrD monomer conformation and unwinding activity measured simultaneously. The bottom panels show DNA duplex unwinding and rezipping as detected by the optical trap. The top panels show highly dynamic donor (green) and acceptor (red) fluorescence signals during activity. In the middle panels, the corresponding FRET efficiency fluctuates between high and low values (0.7±0.2 and 0.3±0.01, respectively for all molecules), consistent with the ‘closed’ and ‘open’ conformations, respectively (16). Dividing the unwinding and fluorescence data traces into time intervals determined by FRET state (i.e. conformation) reveals a correlation between unwinding vs. rezipping activity and 2B orientation. When a monomer is in the closed conformation (Fig. 3B and 3C, blue shaded intervals), the DNA duplex unwinds, while upon switching to the open conformation (Fig. 3B and 3C, unshaded intervals) the duplex rezips. Previously proposed models (9, 14, 16) suggested that the open vs. closed conformations of UvrD correspond to moving vs. stalled states; however, our direct observations demonstrate that these conformations instead correlate with UvrD rezipping vs. unwinding activity. Plotting the average speed vs. FRET efficiency for many individual FRET-determined time intervals (Fig. 3D) corroborates our finding that unwinding (positive velocity) and rezipping (negative velocity) correspond to high and low FRET states, respectively. Dividing the data traces into time intervals by unwinding speed instead yields similar results (fig. S5).
A simple model explains the correlation between 2B domain orientation and UvrD unwinding/rezipping activity (Fig. 3A, movie S1 (19)). In the ‘closed’ conformation, the 1A-2A motor domains are oriented such that 3’-5’ translocation occurs toward the duplex, in the unwinding direction (14). In the transition from ‘closed’ to ‘open’, the 2B and 1A-2A domains rotate ~150° relative to each other (16), consistent with our FRET efficiencies. We propose that an unwinding-to-rezipping reversal is initiated by the 1A-2A domains disengaging from the ssDNA strand, while 2B anchors UvrD at the DNA junction preventing its complete dissociation. A switch from ‘closed’ to ‘open’ along with rotation of 2B about the duplex then reorients the 1A-2A domains to bind the opposing ssDNA strand in the proper orientation. 3’-5’ translocation is now directed away from the fork junction, allowing the duplex to rezip. These steps are reversible, leading to robust back-and-forth switching between unwinding and rezipping activities. Our model is compatible with previous studies of UvrD (6) and its SF 1 homologs (19, 22, 23).
Although our model provides a mechanism for monomer reversals, it does not explain why these occur in the first place or why they occur less frequently during dimer unwinding. Additional measurements varying duplex stability indicate that reversals occur whenever UvrD encounters an energetic barrier to unwinding (19). UvrD dimers (and higher order assemblies) may be more likely to overcome this barrier, leading to long-distance unwinding. Our data, together with past mutational studies (6, 27), point to strand-switching being the primary inhibitor of long-distance unwinding. For dimers, direct contact between helicases may inhibit strand-switching by the lagging UvrD either by applying force to the leading UvrD, stabilizing it at the duplex junction, or by preventing 2B interactions with the duplex DNA.
It is plausible that these two levels of unwinding activity play biological roles. UvrD is involved in multiple, distinct DNA maintenance processes that require different levels of processivity. During nucleotide excision repair, it is estimated that only ~15 bp of DNA are unwound (28). The strand-switching model above may provide a mechanism by which UvrD can unwind a small number of base pairs yet remain engaged with the DNA near the site of damage. In contrast, methyl-directed mismatch repair can require over 1000 bp of DNA to be unwound (29). UvrD conformation and stoichiometry may be critical in enabling and regulating these disparate functions. Interactions of UvrD with accessory proteins, such as MutL (2), may also influence its conformation, stoichiometry and activities.
Supplementary Material
Acknowledgments
We thank members of the Chemla (including Zhi Qi), Ha (including Kyung-Suk Lee) and Lohman laboratories (including Eric Tomko) for scientific discussion and assistance in the experimental design. Funding was provided by National Science Foundation grants MCB-0952442 (CAREER to Y.R.C) and PHY-1430124 (Center for the Physics of Living Cells to Y.R.C. and T.H), National Institutes of Health grants R21 RR025341 (to Y.R.C. and T.H), R01 GM065367 (to T.H.), and R01 GM045948 (to T.M.L.) and the Alfred P. Sloan Research Fellowship (to Y.R.C). T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
References and Notes
- 1.Husain I, Van Houten B, Thomas D, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci U S A. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 3.Matson SW. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3’ to 5’ direction. J Biol Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 4.Fischer C, Maluf N, Lohman T. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Tomko E, Fischer C, Niedziela-Majka A, Lohman T. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomko E, et al. 5’-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maluf N, Fischer C, Lohman T. A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 8.Maluf N, Ali J, Lohman T. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J Biol Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 9.Lohman T, Tomko E, Wu C. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 10.Yokota H, Chujo Y, Harada Y. Single-molecule imaging of the oligomer formation of the nonhexameric Escherichia coli UvrD helicase. Biophys J. 2013;104:924–933. doi: 10.1016/j.bpj.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K, Balci H, Jia H, Lohman T, Ha T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat Comm. 2013;4:1878. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korolev S, Hsieh J, Gauss G, Lohman T, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 13.Velankar S, Soultanas P, Dillingham M, Subramanya H, Wigley D. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 16.Jia H, et al. Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J Mol Biol. 2011;411:633–648. doi: 10.1016/j.jmb.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mechanic L, Hall M, Matson S. Escherichia coli DNA helicase II is active as a monomer. J Biol Chem. 1999;274:12488–12498. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- 18.Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See supplementary materials on Science Online.
- 20.Dumont S, et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Z, Pugh R, Spies M, Chemla Y. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. eLife. 2013;2 doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 23.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomko E, Fischer C, Lohman T. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J Mol Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci U S A. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brendza K, et al. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T, Erie D. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 30.Ali J, Maluf N, Lohman T. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J Mol Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- 31.Min TL, Mears PJ, Golding I, Chemla YR. Chemotactic adaptation kinetics of individual Escherichia coli cells. Proc Natl Acad Sci U S A. 2012;109:9869–9874. doi: 10.1073/pnas.1120218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 33.Landry MP, McCall PM, Qi Z, Chemla YR. Characterization of photoactivated singlet oxygen damage in single-molecule optical trap experiments. Biophys J. 2009;97:2128–2136. doi: 10.1016/j.bpj.2009.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 35.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 36.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci U S A. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parzen E. On estimation of a probability density function and mode. Ann Math Stat. 1962;33:1065–1076. [Google Scholar]
- 38.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huguet J, et al. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc Natl Acad Sci U S A. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owczarzy R, Moreira BG, You Y, Behlke MA, Walder JA. Predicting stability of DNA duplexes in solution containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 41.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


