Abstract
Exposure to estrogens is a risk factor for breast cancer. Specific estrogen metabolites may initiate breast cancer and other cancers. Genotoxicity may be caused by cytochrome P450 (CYP)–mediated oxidation of catechol estrogens to quinones that react with DNA to form depurinating estrogen-DNA adducts. CYP1B1 favors quinone formation by catalyzing estrogen 4-hydroxylation, whereas NAD(P)H quinone oxidoreductase 1 (NQO1) catalyzes the protective reduction of quinones to catechols. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces CYP1B1 expression through the aryl hydrocarbon receptor (AhR). Resveratrol has anticancer effects in diverse in vitro and in vivo systems and is an AhR antagonist that decreases CYP expression but induces NQO1 expression. The chemopreventive effect of resveratrol on breast cancer initiation was investigated in MCF-10F cells. Its effects on estrogen metabolism and formation of estrogen-DNA adducts were analyzed in culture medium by high-performance liquid chromatography, where as its effects on CYP1B1 and NQO1 were determined by immunoblotting and immunostaining. The antitransformation effects of resveratrol were also examined. TCDD induced expression of CYP1B1 and its redistribution in the nucleus and cytoplasm. Concomitant treatment with resveratrol dose-dependently suppressed TCDD-induced expression of CYP1B1, mainly in the cytoplasm. Resveratrol dose- and time-dependently induced expression of NQO1. NQO1 is mainly in the perinuclear membrane of control cells, but resveratrol induced NQO1 and its intracellular redistribution, which involves nuclear translocation of nuclear factor erythroid 2–related factor 2. Resveratrol decreased estrogen metabolism and blocked formation of DNA adducts in cells treated with TCDD and/or estradiol. Resveratrol also suppressed TCDD and/or estradiol-induced cell transformation. Thus, resveratrol can prevent breast cancer initiation by blocking multiple sites in the estrogen genotoxicity pathway.
Carcinogenesis is a complex multistep process. Prolonged exposure to estrogens is a major known risk factor for breast cancer and other estrogen-mediated cancers (1). Besides the estrogen receptor (ER) pathway (2), reactive estrogen metabolites may also be involved in mutagenesis and breast cancer initiation via an estrogen genotoxicity pathway (3). The geno-toxicity occurs via cytochrome P450 (CYP)–mediated oxidation of estrogens to quinones that react with DNA to form predominantly depurinating DNA adducts. This event may lead to the critical mutations that initiate cancer (3, 4). Two of the estrogen-metabolizing enzymes in this pathway, CYP1B1 and NAD(P)H quinone oxidoreductase 1 (NQO1), are implicated in the etiology of estrogen-mediated tumors by regulating the formation or clearance of genotoxic metabolites (5, 6). CYP1B1 is highly expressed in estrogen-related tissues, such as mammary gland, uterus, and ovary (7), suggesting that CYP1B1 is important in the local control of estrogen metabolism. CYP1B1 is expressed in normal tissues (8, 9) and its overexpression has been implicated in premalignant progression (10). CYP1B1 predominantly catalyzes the 4-hydroxylation of estrone (E1) and estradiol (E2) into 4-OHE1(E2) (refs. 11, 12), favoring quinone formation, whereas the phase II protective enzyme NQO1 catalyzes the reduction of quinones back to CEs (13), which can be methylated by catechol-O-methyltransferase to form 4-OCH3E1(E2). NQO1 is abundantly present in many human tissues (14), and genetic polymorphisms may cause variation in its activity (15, 16). Low NQO1 activity and increased formation of depurinating adducts resulting from unbalanced estrogen metabolism can be critical factors leading to the initiation of breast cancer (17, 18). Therefore, the efficiency of CYP1B1 metabolism of estrogens and the enzymes, including NQO1, that regulate the clearance of quinones are important in the determination of cancer risk. A potential mechanism for the initiation of breast cancer involves altered expression of estrogen-metabolizing enzymes (18). The CYP1 family is inducible by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through high-affinity binding to the aryl hydrocarbon receptor (AhR). The activated AhR translocates into the nucleus, then interacts with the dioxin response element Ah gene battery, responsible for regulating expression of CYP1B1 (19, 20). High CYP1B1 and low NQO1 activity seem to be associated with increased risk of developing breast cancer (21).
Resveratrol (3,5,4′-trihydroxystilbene), a natural antioxidant present in grapes and many other plants (22), has anticarcinogenic effects in diverse in vitro and in vivo systems (23, 24). TCDD induces CYP1B1 expression through the AhR in breast epithelial cells (25, 26). Resveratrol acts as an AhR antagonist that decreases expression of the CYP isoforms catalyzing estrogen metabolism (27). However, it induces NQO1 activity in cultured cells (22) that are capable of metabolically detoxifying carcinogens (28). Induction of NQO1 may be regulated by the antioxidant response element (ARE)/nuclear factor erythroid 2–related factor 2 (Nrf2) pathway (29). Cytosolic transcription factor Nrf2 translocates into the nucleus, where it binds to the ARE to activate transcription of NQO1 mRNA (30, 31). In addition, resveratrol as an antioxidant may reduce semiquinones back to CEs, in turn preventing the formation of depurinating estrogen-DNA adducts (32, 33). Most anticancer studies emphasize resveratrol inhibiting cell proliferation and inducing apoptosis. Neither its preventive role in breast cancer initiation nor its mechanisms of action have been thoroughly characterized. TCDD-induced CYP1B1, which enhances estrogen metabolism and formation of DNA adducts, seems to play a major role in malignant transformation of human breast epithelial cells.4 Low catechol-O-methyltransferase activity and increased formation of estrogen-DNA adducts mediated through the CEs may be contributory factors in the development of breast cancer (34, 35). We hypothesize that resveratrol may prevent estrogen genotoxicity and neoplastic transformation via regulating estrogen-metabolizing enzyme expression and blocking CE and DNA adduct formation (Fig. 1). In this article, we used the human breast epithelial cell line MCF-10F (ER negative and AhR positive), a well-developed cell culture model for studying carcinogenesis through non-ER receptor – mediated pathways (36, 37), to further investigate the chemo-preventive effects of resveratrol on breast cancer initiation. The profile of E2 metabolites and depurinating DNA adducts in resveratrol-treated cells pretreated with TCDD and treated with E2 was analyzed, as well as the effects of resveratrol on the expression pattern of CYP1B1 and NQO1 and the signal transductional mechanism of NQO1 induction. The antitransformation effects of resveratrol were determined by an anchorage-independent growth assay in agar methocel. This study helped us to obtain a deeper understanding of the roles of estrogen-metabolizing enzymes in the genotoxic mechanism of estrogen-initiated cancer.
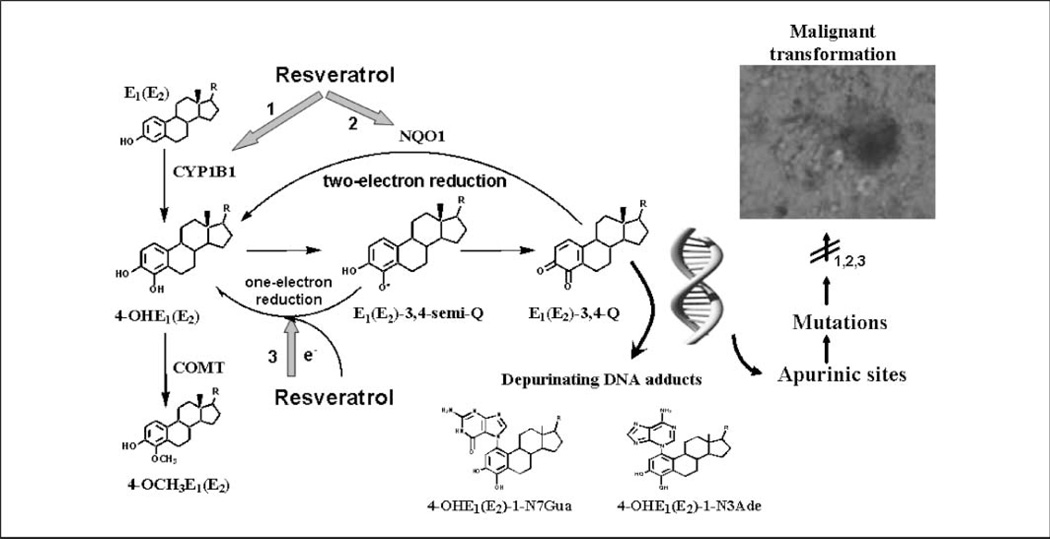
Fig. 1.
Mechanisms by which resveratrol can prevent estrogen-induced breast cancer. 1, resveratrol decreases the formation of 4-OHE1(E2) by inhibiting expression of TCDD-induced CYP1B1. 2, resveratrol induces expression of the protective enzyme NQO1, which catalyzes two-electron reduction of catechol estrogen quinones back to CEs, which can be methylated by catechol-O-methyltransferase (COMT) to form 4-OCH3E1(E2). 3, resveratrol, as an antioxidant, may reduce semiquinones back to CEs, in turn preventing formation of depurinating DNA adducts.
Materials and Methods
Chemicals and cell culture
MCF-10F and MCF-7 cells were obtained from the American Type Culture Collection and cultured in DMEM and Ham's F12 medium as described previously (34). Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. TCDD (>99% pure) was purchased from AccuStandard, Inc. Standards of E2 metabolites and depurinating DNA adducts were prepared as described previously (34). Resveratrol and other chemicals were obtained from Sigma.
Immunoblotting
The expression pattern of E2-metabolizing enzymes (CYP1B1, NQO1) and transcription factor Nrf2 from control and treated MCF-10F cells was analyzed by immunoblotting. Whole-cell lysates were prepared by suspending cells in radioimmunoprecipitation assay buffer with protease inhibitor and lysing by three freeze-thaw cycles. Nuclei and unlysed cellular debris were removed by centrifugation. Protein concentrations were determined by using the BCA protein assay kit (Pierce Biotechnology) and the proteins were separated by SDS-PAGE and transferred to PVD membranes for immunodetection as described previously (34). Dilutions of primary anti-CYP1B1, β-actin (Genetest), NQO1 (Abcam), and Nrf2 (Santa Cruz Biotechnology) antibodies were made in blocking solution (5% nonfat dry milk in PBS). The blots were incubated for 3 h with primary antibody and for 1 h with secondary antibody coupled to horseradish peroxidase at room temperature. After each step, blots were washed with PBST (PBS and 0.1% Tween 20), incubated with enhanced chemiluminescence solution (Amersham Biotech) for 1 min, and visualized with radiographic film. Intensities of the bands were quantified by Alpha DigiDoc 1201 (Alpha Innotech).
Immunocytochemistry
The constitutive cellular localization and TCDD- or resveratrol-induced intracellular redistribution of CYP1B1, NQO1, and Nrf2 in MCF-10F cells were detected by immunocytochemistry. In brief, control and treated cells were grown on coverslips or eight-well chamber slides to 10% confluence (about 2,000 per well) and washed with cold PBS, then fixed with 4% paraformaldehyde and permeabilized with 0.05% Triton X-100 as described previously (38) with the following modifications. The optimal conditions of immunocytochemistry were determined by initial experiments using various concentrations of primary antibodies. The polyclonal antibodies against CYP1B1, NQO1, and Nrf2 were validated by immunoblot analysis. Dilutions (1:2,000 for CYP1B1; 1:1,000 for Nrf2; 1:500 for NQO1) of primary antibodies were made in blocking solution (5% goat serum in bovine serum albumin). For double-labeling immunofluorescence, cells were first incubated with rabbit-derived primary anti-Nrf2 for 2 h, followed by incubation with goat-derived anti-NQO1 for another 2 h. After washing with PBS, the cells were incubated with secondary antibodies conjugated with fluorescence. Immunostaining was evaluated by examination of slides under a confocal laser microscope (Axiovert 135M, Carl Zeiss) and a fluorescent microscope (E600, Nikon). Labeled slides were initially excited at λ = 488 nm and the fluorescing nuclear images (red) were acquired at a ×200 to ×400 magnification. Subsequently, sections were excited at λ = 568 nm to acquire the fluorescing CYP1B1 (or NQO1, Nrf2) image (green). The images were captured by a QImaging digital camera (Burnaby) and Openlab image analysis software (Improvision). Immunosignal was merged with the nuclear signal to determine the intracellular distribution of proteins. The blue color of 4′,6-diamidino-2-phenylindole for the nuclear staining was Cancer converted to red for contrast, whereas the protein signal remained green. All images were acquired under exactly the same conditions. Representative fluorescence images are presented.
Determination of cellular NQO1 activity by high-performance liquid chromatography using E2-3,4-quinone as substrate
NQO1 enzymatic activity was determined by high-performance liquid chromatography (HPLC) with electrochemical detection (ECD). Cellular NQO1 (20 µg) from control and 25 µmol/L resveratrol-treated MCF-10F cells were prepared, as described above. Triplicate enzyme reactions were carried out in a final volume of 100 µL of 0.1 mol/L sodium phosphate (pH 7).We used freshly synthesized E2-3,4-quinone (E2-3,4-Q; ref. 39) as the substrate and NADH (50 µmol/L) as the cofactor in the buffer system containing 0.7 mg/mL bovine serum albumin. E2-3,4-Q alone or E2-3,4-Q plus NADH in the reaction system served as the negative controls. Recombinant NQO1 protein (10 units, Sigma) in the reaction system served as the positive control. Dicumarol (10 µmol/L), an inhibitor of NQO1, was also added into the reaction with resveratrol-treated protein to determine whether the increased NQO1 enzymatic activity can be inhibited by this specific inhibitor. The incubation mixture, except for the E2-3,4-Q substrate, was preincubated for 3 min at room temperature. The reaction was then initiated by adding 5 µL of E2-3,4-Q (100 µmol/L) in acetonitrile and terminated after 27 min at 37°C by adding 100 µL methanol. Following centrifugation to precipitate proteins, the supernatant was passed through a 5,000 molecular weight cutoff filter (Millipore), and 100 µL of each sample were analyzed for the product, 4-OHE2, by HPLC as previously described (34).
HPLC analysis of estrogen metabolites and depurinating DNA adducts
To determine whether resveratrol suppresses E2 metabolism and prevents DNA adduct formation after exposure to TCDD, cells were pretreated with 0.1 to 30 nmol/L TCDD with or without 25 µmol/L resveratrol for 72 h and then incubated with E2 (0.1–10 µmol/L) for 24 h, and the profile of estrogen metabolites and depurinating DNA adducts in culture medium was analyzed by HPLC with ECD. To determine the inhibiting effects of resveratrol on 4-OHE2–induced DNA adduct formation, the cells were treated with increasing concentrations (0.1–10 µmol/L) of 4-OHE2 for 24 h with or without 25 µmol/L resveratrol. Medium was collected and 2 mmol/L ascorbic acid was added to protect E2 metabolites from oxidative degradation. Collected medium was extracted and analyzed by HPLC with ECD. The assay of E2 metabolites and DNA adducts was modified from previously described procedures (34). In brief, media were processed by various concentration methods and the methanol/water mixtures were applied to a Certify II Sep-Pak cartridge. The extracts were subjected to HPLC analysis. The E2 metabolite levels were corrected for recovery and normalized to cell count.
Anchorage-independent transformation assay
To determine whether resveratrol decreased the ability of TCDD and/or E2 to transform MCF-10F cells, control and treated cells (pretreatment with 10 nmol/L TCDD with or without resveratrol) at a density of 1 × 104 per well were cultured in 0.8% methocel soft agar semisolid medium for 21 to 28 d in 24- or 96-well plates precoated with 0.8% agar base medium. Feeding medium was added on the top and changed twice a week. After 24 h postplating, cultures were examined for cell aggregates to ensure that every colony was clonal in origin. Those wells that contained cell aggregates were discarded. Following this incubation period, the colonies formed were analyzed morphologically using a cell stain solution (Chemicon International) under inverted microscopy and photographed. A colony was defined as a cluster of more than 50 cells. The number of colonies was counted, and the results were expressed as colony efficiency − the number of colonies formed per number of cells plated × 100. MCF-7 cells (ER-α–positive breast cancer epithelial cells) were used as positive controls in some anchorage-independent transformation analyses.
Statistical analysis
The statistical significance of the results was determined by Student's t test and ANOVA analysis by using SAS and GraphPad Prism 4.0 software. P < 0.05 was considered significant. All cultures, immunoblottings, and immunostainings were repeated at least three times.
Results
Inhibiting effects of resveratrol on TCDD-induced CYP1B1 expression
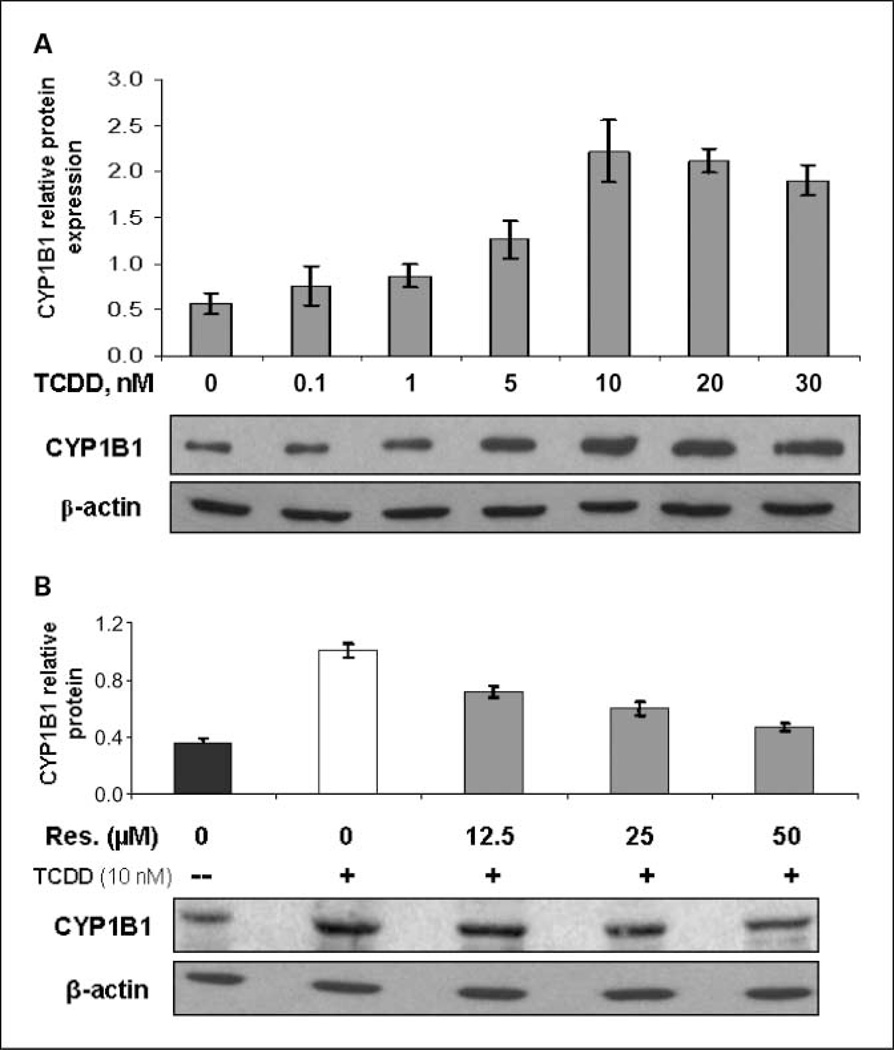
Inclusion of 0.1 to 30 nmol/L TCDD in the culture medium resulted in a concentration-dependent induction of CYP1B1 expression (Fig. 2A). The maximal response in CYP1B1 induction was achieved following treatment with 10 nmol/L TCDD, and the optimal time for maximum induction was 72 h (data not shown). Therefore, 10 nmol/L TCDD was used for subsequent experiments. However, concomitant treatment with 12.5 to 50 µmol/L resveratrol and 10 nmol/L TCDD for 72 h dose dependently suppressed the TCDD-induced expression of CYP1B1 (Fig. 2B). Induction of CYP1B1 by TCDD was almost completely suppressed by 50 µmol/L resveratrol, as assessed by Western blot. Our preliminary data showed that the low constitutive expression of CYP1B1 at the protein level was not affected by 25 µmol/L resveratrol. This is not consistent with a previous study (27) and needs further study.
Fig. 2.
Inhibiting effects of resveratrol on TCDD-induced CYP1B1 expression. The representative immunoblots show that anti-CYP1B1 antibody recognizesa single 60.8-kDa band. Each lane contains30 µg of the cell lysate. Intensity of the bands was quantified and normalized, as described in Materials and Methods (n = 3). A, CYP1B1 expression in MCF-10F cells treated with increasing concentrations of TCDD for 72 h. B, cells were treated with 10 nmol/L TCDD with or without increasing concentrations of resveratrol (0–50 µmol/L) for 72 h.
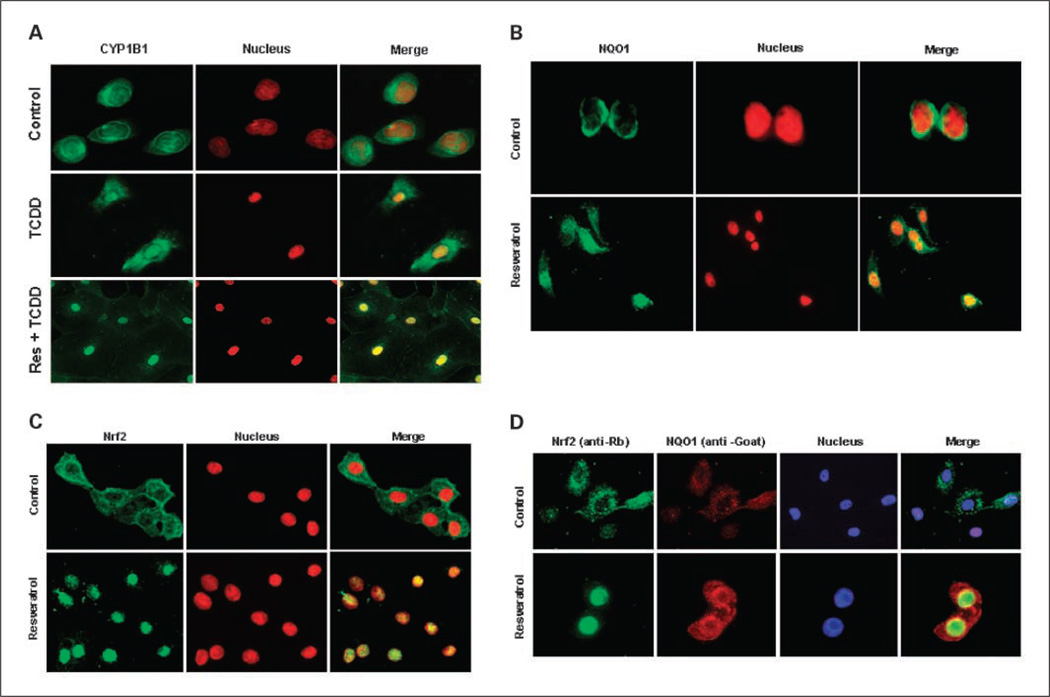
We further determined the inhibiting effects of resveratrol on TCDD-induced CYP1B1 expression by immunofluorescence. The immunostaining pattern of CYP1B1 (FITC-labeled green fluorescence) merged with that of the nucleus (4′,6-diamidino-2-phenylindole–labeled, red) in MCF-10F cells revealed an intracytoplasmic reticulum and distinct strong perinuclear staining that formed a green fluorescent perinuclear ring (Fig. 3A). Almost no CYP1B1 immunostaining is present in the nucleus of MCF-10F cells. In TCDD-treated cells, the merged image showed that the green fluorescence of CYP1B1 colocalized with the red nuclear staining, in addition to cytoplasmic staining, and indicated that TCDD induced the expression of CYP1B1 and its redistribution in the nucleus and cytoplasm. The mechanism by which TCDD induces CYP1B1 expression in the nucleus remains to be elucidated. However, the staining pattern of MCF-10F cells cotreated with resveratrol and TCDD showed that the green fluorescence of CYP1B1 significantly declined in the cytoplasm of TCDD plus resveratrol-cotreated MCF-10F cells, compared with that of control cells (Fig. 3A). The merged image showed that the green fluorescence of CYP1B1 colocalized with the red nuclear staining in TCDD plus resveratrol-cotreated cells and suggested that the redistribution of CYP1B1 into the nucleus by TCDD was not totally compromised by resveratrol cotreatment. These results indicate that resveratrol decreased TCDD-induced CYP1B1 expression mainly in the cytoplasm, with a lesser inhibiting effect in the nucleus (Fig. 3A).
Fig. 3.
Intracellular localization of CYP1B1, NQO1, and Nrf2 in control and treated MCF-10F cellsdetected by immunocytochemistry (green, enzyme or transcription factor; red, nuclear staining). Merged images of nuclei and enzymes allowed visualization of cellular distribution. A, the constitutive cellular localization and effects of TCDD (10 nmol/L) or resveratrol (25 µmol/L) on intracellular redistribution of CYP1B1. B, immunostaining pattern of NQO1 in control or resveratrol-treated MCF-10F cells. C, intracellular localization of Nrf2 in control and resveratrol-treated cells. D, double-labeling immunofluorescence analysis of Nrf2 (green), NQO1 (red), and nucleus (blue).
Induction of NQO1 expression and activity by resveratrol
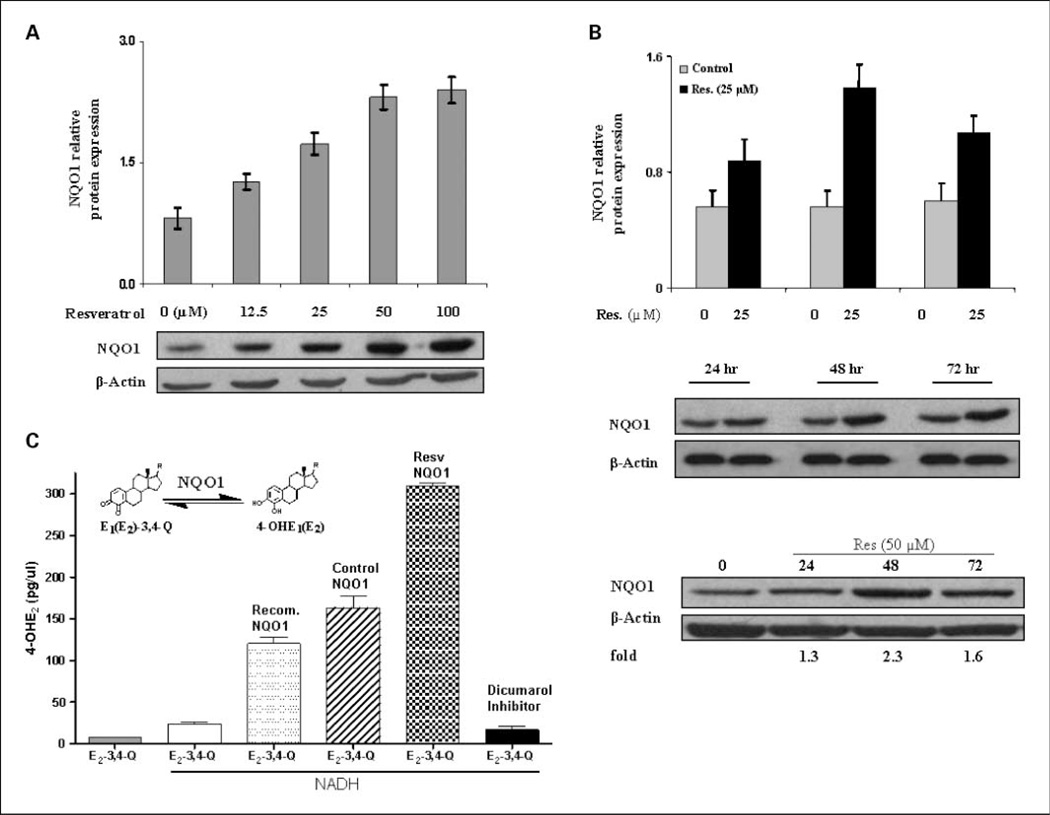
Resveratrol-induced NQO1 expression in MCF-10F cells was determined by immunoblotting. MCF-10F cells were treated with various concentrations of resveratrol (0–100 µmol/L) for 48 h and analyzed for NQO1 protein levels. Densitometric analyses showed that resveratrol dose- and time-dependently induced the expression of NQO1 proteins 2- to 3-fold (Fig. 4A). A time course study using 25 and 50 µmol/L resveratrol for 24 to 72 h showed that induction of NQO1 protein occurred by 24 h, peaked at 48 h, and decreased at 72 h (Fig. 4B). We further investigated resveratrol-induced NQO1 expression by fluorescence-based immunostaining. MCF-10F cells in eight-well chamber slides were treated with resveratrol (25 µmol/L) for 48 h, and control and treated cells were fixed and inmmunostained. Immunostaining was evaluated by examination of slides under a fluorescence microscope and analyzed for NQO1 protein expression (Fig. 3B). The merged image showed that the green fluorescence of NQO1 is mainly in the perinuclear membrane of untreated MCF-10F cells. Resveratrol induced NQO1 expression in both the cytoplasm and the nucleus (Fig. 3B). The green fluorescence of NQO1 increased and colocalized with 4′,6-diamidino-2-phenylindole nuclear staining, in addition to cytoplasmic staining, and indicated that resveratrol induced the expression of NQO1 and its redistribution in the nucleus and cytoplasm.
Fig. 4.
Induction of NQO1 expression and activity by resveratrol. A, NQO1 expression in MCF-10F cells treated with increasing concentrations of resveratrol (0–100 µmol/L) for 48 h. B, cells were treated with 0, 25, or 50 µmol/L resveratrol for 24 to 72 h. Each lane contained 30 µg of cell lysate. Intensity of the bands was quantified by Alpha DigiDoc 1201 and normalized to β-actin. The representative immunoblots (from three replicates) show a single band of NQO1 protein at 30 kDa in MCF-10F cells. C, resveratrol induced enzymatic activity of NQO1 in MCF-10F cells. Freshly made E2-3,4-Q was used as the substrate and NADH as the cofactor. The levels of reaction product, 4-OHE2, in treated cells are significantly different from the untreated cells (P < 0.05, as determined by ANOVA). Recombinant NQO1 protein (10 units) served as the positive control. Dicumarol (10 µmol/L) was used to determine whether resveratrol-induced NQO1 could be inhibited. Gray column, E2-3,4-quinone in buffer; white column, E2-3,4-quinone + NADH; dotted column, E2-3,4-quinone + recombinant NQO1 protein + NADH; striped column, E2-3,4-quinone + cellular protein from nontreated control cells+ NADH; checked column, E2-3,4-quinone + cellular proteins from 25 µmol/L resveratrol-treated cells+ NADH; black column, E2-3,4-quinone + dicumarol + cellular proteins from 25 µmol/L resveratrol-treated cells + NADH.
To further examine the catalytic capacity of induced NQO1, the activity of cellular NQO1 in control and resveratrol-treated cells was determined by an in vitro enzyme assay using HPLC with ECD. Freshly made E2-3,4-Q was used as the substrate and NADH was used as the cofactor in a buffer system. Recombinant NQO1 protein served as the positive control and dicumarol served as the inhibitor of NQO1. The levels of reaction product, 4-OHE2, formed by cellular protein from treated cells are significantly different from the untreated cells (P < 0.05), as determined by ANOVA (Fig. 4C). The levels of 4-OHE2 in reaction mixtures containing E2-3,4-Q alone or E2-3,4-Q plus NADH were very low compared with a reaction mixture containing both substrate and recombinant NQO1 protein. Cellular protein from 25 µmol/L resveratrol-treated cells showed a 2-fold increase in enzymatic activity compared with that from control cells. However, this increased NQO1 enzymatic activity was almost completely inhibited by 10 µmol/L dicumarol. These data suggest that resveratrol enhanced the NQO1 catalytic activity. The increased NQO1 activity in resveratrol-treated cells corresponded to the induction in NQO1 protein level.
Taken together, our results clearly show that resveratrol dose- and time-dependently induced NQO1 protein expression, and the induced cellular NQO1 catalyzes the two-electron reduction of E2-3,4-Q back to 4-OHE2. This is consistent with a previous report that resveratrol both enhances NQO1 catalytic activity and protein expression (22). Furthermore, we showed for the first time nuclear localization of NQO1 in resveratrol-treated MCF-10F cells (Fig. 3B). Although the mechanisms of nucleocytoplasmic transport of induced NQO1 need to be further elucidated, nuclear localization of NQO1 may be very important because resveratrol-induced nuclear NQO1 may directly prevent the formation of quinones in the nucleus, the site of genotoxicity.
Induction of NQO1 by resveratrol may involve nuclear translocation of Nrf2
To determine whether nuclear translocation of Nrf2 is involved in the induction of NQO1, the intracellular localization of Nrf2 in control and resveratrol-treated MCF-10F cells was examined by fluorescence immunocytochemistry. Nrf2 is predominantly in the cytoplasm of nontreated MCF-10F cells (Fig. 3C). Upon treatment with resveratrol, the immunostaining of anti-Nrf2 (green) and 4′,6-diamidino-2-phenylindole (nuclear staining, red) almost completely overlap, strongly suggesting that Nrf2 is localized in the nucleus of resveratrol-treated cells. Therefore, these data indicate that resveratrol induced nuclear translocation of Nrf2 (Fig. 3C). We further determined whether resveratrol-induced NQO1 was accompanied by Nrf2 nuclear translocation by using a double-labeling immunofluorescence analysis. In control cells, Nrf2 remains in the cytoplasm and NQO1 expression is very low. After resveratrol treatment, Nrf2 translocated into the nucleus and the expression of NQO1, which is found in both the cytoplasm and the nucleus (Fig. 3D), was induced. These results suggest that the induction of NQO1 in MCF-10F cells exposed to resveratrol may involve the Nrf2-Keap1-ARE pathway.
Resveratrol decreases estrogen metabolism and prevents formation of depurinating DNA adducts
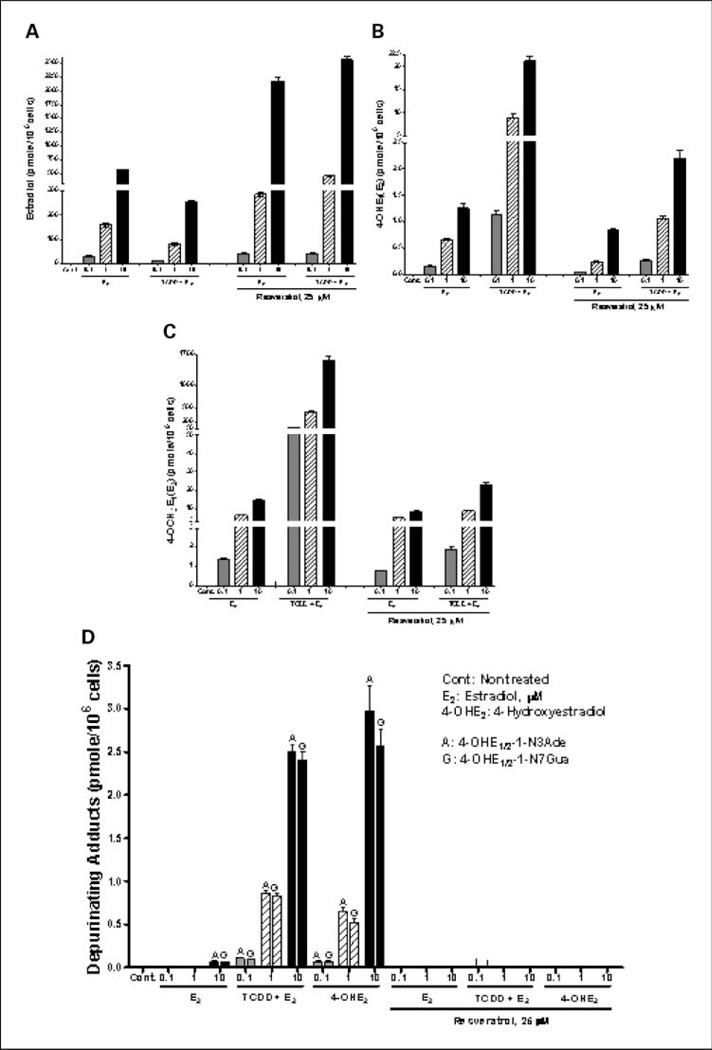
To determine whether resveratrol suppresses estrogen metabolism after exposure of cells to TCDD, MCF-10F cells were (a) pretreated with 10 nmol/L TCDD for 72 h with or without 25 µmol/L resveratrol and then exposed to different concentrations of E2 (0.1–10 µmol/L) for 24 h or (b) treated with E2 (0.1–10 µmol/L) with or without 25 µmol/L resveratrol for 24 h. The middle concentration, 1 µmol/L, is approximately the physiologic concentration of E2 (37). The profile of E2 metabolites [4-OCH3E1(E2), 4-OHE1(E2), and unmetabolized estrogen] in the culture medium of MCF-10F cells pretreated with resveratrol and TCDD and treated with E2 was analyzed by HPLC with ECD. The profile of metabolites was first assessed in control or 10 nmol/L TCDD-pretreated MCF-10F cells subsequently treated with 0.1 to 10 µmol/L E2 for 24 h. In MCF-10F cells treated with E2 alone, metabolism of E2 was very limited. After 24 h, 95% of the E2 recovered was unmetabolized, and the combination of metabolites represented <5% of the total. In contrast, in cells pretreated with TCDD before addition of E2, most of the recovered estrogen was 4-OCH3E1(E2), with a smaller amount of 4-OHE1(E2) (Fig. 5A–C). The level of 4-OHE1(E2) (Fig. 5B) and 4-OCH3E1(E2) (Fig. 5C) increased as E2 concentration increased with both E2 or TCDD plus E2 treatment. The increase in E2 metabolites was lessened by cotreatment with 25 µmol/L resveratrol with E2 or TCDD plus E2 treatment. On the other hand, unmetabolized E2 in resveratrol-treated culture medium is higher than in untreated medium (Fig. 5A). These results indicate that resveratrol decreased estrogen metabolism in MCF-10F cells.
Fig. 5.
Profile of E2 metabolites and depurinating DNA adducts in MCF-10F cells pretreated with resveratrol and TCDD and treated with E2. Levels of (A) unmetabolized E2, (B) 4-OHE1(E2), and (C) 4-OCH3E1(E2) in culture medium pretreated with 10 nmol/L TCDD with or without 25 µmol/L resveratrol for 72 h and then incubated with E2 (0.1–10 µmol/L) for 24 h. D, levels of depurinating DNA adducts in culture medium of cells treated with TCDD and/or increasing concentrations of E2 for 24 h with or without resveratrol. The levels of DNA adducts in resveratrol-treated cells are significantly different from those in the cells not treated with resveratrol (P < 0.05, as determined by ANOVA). The estrogen metabolite and DNA adduct levels were corrected for recovery and normalized to cell numbers. Columns, mean of triplicate cultures from three experiments; bars, SD.
To investigate the implications of the possible effects of resveratrol on E2 metabolism and formation of depurinating DNA adducts, the levels of DNA adducts were determined in control and treated cell culture medium by HPLC with ECD. Enhanced E2 metabolism by TCDD-induced CYP1B1 significantly increased the levels of DNA adducts upon incubation with 0.1 to 10 µmol/L E2 (Fig. 5D). This increase in DNA adducts was dose dependent with increasing concentrations of E2. Resveratrol (25 µmol/L) completely blocked the formation of DNA adducts in both E2- and TCDD plus E2-treated MCF-10F cells. Similar inhibition of DNA adduct formation was also seen in cells treated with 4-OHE2 (0.1–10 µmol/L) plus resveratrol (25 µmol/L) for 24 h (Fig. 5D).
We further determined whether resveratrol prevents DNA adduct formation through blocking the reaction of E2-3,4-Q with DNA by using an in vitro reaction assay. Resveratrol may afford one-electron reduction of E2-3,4-semiquinone back to 4-OHE2 (Fig. 1; refs. 32, 33). These results are consistent with the inhibition of DNA adduct formation (97% inhibition at a ratio of 1:3, 4-OHE2/resveratrol) when lactoperoxidase-activated 4-OHE2 reacted with DNA, but not when E2-3,4-Q reacted with DNA (33).
Antitransformation effects of resveratrol on TCDD- and/or E2-induced transformation
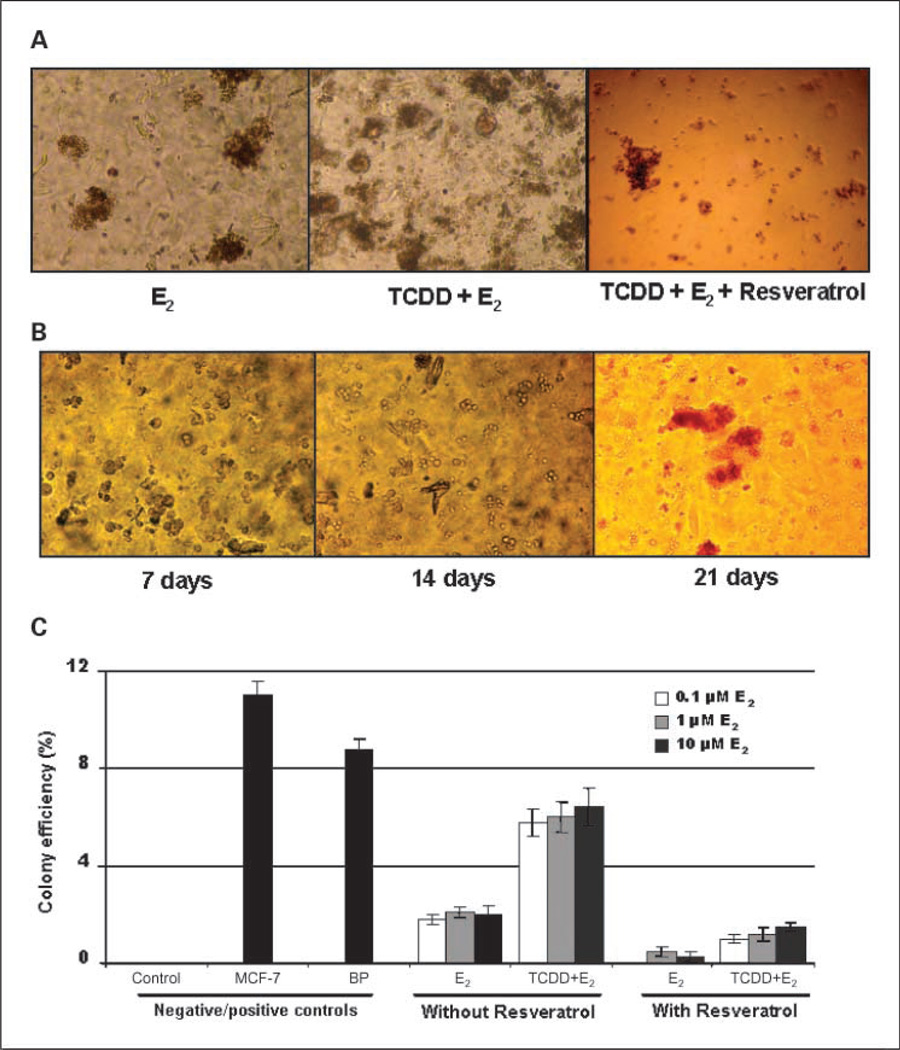
To investigate whether resveratrol decreases the ability of TCDD and/or E2 to transform MCF-10F cells, the inhibiting effects of resveratrol on carcinogen-induced cell transformation were investigated by an anchorage-independent growth assay in agar methocel. After exposure of the cells to TCDD and/or E2, no differences in morphology were observed between control and treated cells. The cells formed flat monolayers without overlapping (data not shown). When control cells were plated in agar methocel, they remained viable for a period of time, but did not form colonies. However, under similar conditions, E2-treated cells formed colonies and pretreatment with TCDD, followed by E2, increased the colony numbers compared with E2 alone (Fig. 6A). The colony size did not differ among the treatments (about 200 µm in diameter). We monitored the colony formation at different periods; after 7 days of incubation, treated cells survived and became enlarged, tending to clump together. After 14 days, small and middle size colonies were formed, and at 21 or 28 days, large colonies can be seen (Fig. 6B). The antitransformation effects of resveratrol were determined by colony efficiency (Fig. 6A and C). The positive control MCF-7 human breast cancer cells (11%) and benzo(a)pyrene-treated MCF-10F cells (9%) had significantly higher colony efficiency than E2-transformed cells (P < 0.05 as determined by Student's t test). MCF-10F cells treated with 0.1 to 10 µmol/L E2 twice a week for 2 weeks formed colonies in agar methocel and the colony efficiency increased to 2% (Fig. 6C). Treatment with TCDD plus E2 increased colony efficiency 3-fold compared with E2 alone, indicating that TCDD enhanced the ability of E2 to transform MCF-10F cells (Fig. 6C). However, when cotreated with resveratrol (50 µmol/L), the colony efficiency was decreased to 0 with 0.1 µmol/L E2 treatment or 0.3% with 1 or 10 µmol/L E2 treatment. Resveratrol also suppressed TCDD plus E2-induced transformation, decreasing colony efficiency from 6.2% to 1.2% (P < 0.01 as determined by Student's t test). Taken together, TCDD enhanced the ability of E2 to transform MCF-10F cells, whereas resveratrol significantly inhibited both E2- and TCDD plus E2-induced transformation.
Fig. 6.
Antitransformation effects of resveratrol on TCDD- and/or E2-induced transformation. Control and treated cells (104 per well) were cultured in methocel agar as described in Materials and Methods. Formed colonies were scored and photographed. A, representative graphs of colony formation in cells treated with TCDD and/or E2 with or without resveratrol. B, the formation of colonies of TCDD plus E2–treated cells at the different incubation periods in methocel agar (7–28 d). C, the results are expressed as colony efficiency (%): The number of colonies formed per number of cells plated × 100. Column, mean of assays from triplicate experiments; bars, SD; P < 0.05.
Discussion
The central role of estrogens in breast tumorigenesis is well known. Numerous lines of evidence suggest that estrogen-mediated cancer is related not only to the ER pathway but also to estrogen genotoxicity resulting from unbalanced estrogen metabolism (3, 4). Estrogens are converted to metabolites, particularly the CE-3,4-quinones, that can react with DNA to form depurinating adducts. These adducts are released from DNA to generate apurinic sites. Error-prone base excision repair of this damage may lead to the mutations that can initiate breast, prostate and other types of cancer (3). Maintaining balanced estrogen metabolism requires interplay between estrogen-activating enzymes, such as CYP1B1, and deactivating enzymes, such as NQO1 (4). Therefore, induction of protective enzymes and/or inhibition of activating enzymes are thought to be potential mechanisms to prevent breast cancer initiation. The chemoprotective effect of resveratrol is considered to be partly due to its free radical scavenging ability (40), as well as its regulating role for phase I activating enzymes and phase II deactivating enzymes. In the present study, we explored how resveratrol acts to regulate the dynamic balance of estrogen metabolism by regulating estrogen-metabolizing enzymes and chemically preventing estrogen metabolite formation in MCF-10F cells.
CYP1B1 is highly expressed in estrogen-related tissues and has been proposed to be an important activating enzyme in controlling estrogen homeostasis. It primarily catalyzes the 4-hydroxylation of E2 with minor catalytic activity for 2-hydroxylation (41). TCDD induces expression of CYP1B1 via the AhR/dioxin response element pathway (12, 19); resveratrol may decrease the levels of reactive estrogen metabolites by suppressing TCDD-induced CYP expression as an AhR antagonist (26). The results reported here show that TCDD induced the expression of CYP1B1 and its redistribution in the nucleus and the cytoplasm (Fig. 3A). Concomitant treatment with resveratrol dose-dependently suppressed TCDD-induced expression of CYP1B1 mainly in the cytoplasm, with less inhibiting effect in the nucleus (Figs. 2B and 3A). Although the mechanism is not understood, we showed for the first time that TCDD elicits the translocation of induced CYP1B1 protein into the nucleus. Activation of estrogens in the nucleus may be very important because unbalanced estrogen metabolism in the nucleus can be a critical factor leading to the initiation of breast cancer. The formation of 4-OHE1(E2) in the nucleus may be prevented at this site of genotoxicity by selectively blocking CYP1B1 or inducing protective enzymes, such as NQO1, in the nucleus using resveratrol as discussed below.
Experiments using transgenic mice with ER-α knocked out (ERKO/Wnt 1 mice) and metabolism in aromatase (CYP19)–overexpressing MCF-7 human breast cancer cells have provided further important evidence for the genotoxic effects of estrogen metabolites, including CE-3,4-quinones, in cancer initiation (3). NQO1 catalyzes the two-electron reduction of quinones to CEs (13, 42), thereby preventing both generation of toxic semiquinone radicals and formation of DNA adducts (3). Thus, increased expression of NQO1 by resveratrol might play a significant role in preventing estrogen-induced carcinogenesis. Resveratrol induced NQO1 expression and activity in MCF-10F cells (Fig. 4). Furthermore, NQO1 is localized in the nucleus in resveratrol-treated MCF-10F cells (Fig. 3B). This may be very important because resveratrol-induced nuclear NQO1 may directly prevent the accumulation of quinones in the nucleus. Therefore, induction of NQO1 by resveratrol suggests that this grape-derived phytochemical is a potential chemopreventive agent against the initiation of breast cancer.
Transcriptional activation of NQO1 depends almost exclusively on intracellular localization of Nrf2 rather than induction of this transcription factor through de novo gene transcription (43, 44). Under normal conditions, Nrf2 remains in the cytoplasm, associated with Keap1, a cytoskeletal protein (45). Antioxidants, in this case resveratrol, modify cysteine thiol groups in Keap1, then the Nrf2/Keap1 dimer dissociates and allows Nrf2 translocation to the nucleus (Fig. 3C), where it could bind to the ARE to activate transcription of NQO1 mRNA (46). Using double-labeling immunofluorescence, we showed that resveratrol-induced NQO1 expression was accompanied by Nrf2 nuclear translocation (Fig. 3D). Nuclear localization of Nrf2 in resveratrol-treated cells revealed that resveratrol may induce NQO1 through an Nrf2-Keap1-ARE pathway, which involves the dissociation of Nrf2 from Keap 1 and facilitates translocation of Nrf2 to the nucleus, where it binds to the ARE to activate the transcription of NQO1 mRNA. Therefore, further elucidating this mechanism may provide new evidence on the regulation of gene expression by resveratrol and other chemopreventive agents.
Induction of CYP1B1 by pretreatment of the cells with TCDD dramatically increased E2 metabolism, with formation of high levels of 4-OHE1(E2) and 4-OCH3E1(E2) (Fig. 5B and C). Little E2 metabolism to 2-OCH3E1(E2) was observed. This response is somewhat different from that in MCF-7 cells treated with TCDD (12, 25), which may reflect different ER-α status and AhR levels in these cell lines. Enhanced estrogen metabolism results in significantly higher levels of depurinating DNA adducts (Fig. 5D). Formation of these adducts and the concomitant apurinic sites in DNA has been shown to induce mutations that are associated with initiation of breast cancer (3, 4). Inclusion of resveratrol decreased estrogen metabolism and eliminated formation of detectable levels of 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua (Fig. 5D). This is the first study to explore the role of resveratrol in the formation of E2 metabolites and depurinating DNA adduct levels in a normal human breast epithelial cell line under conditions in which E2 metabolism has been enhanced by TCDD. Reduced metabolic activation of E2, as well as increased detoxification of reactive estrogen metabolites, is thought to be an important mechanism in breast cancer chemoprevention.
In vitro malignant transformation assays are semiquantitative and measure the morphologic transformation of cell colonies induced by carcinogens. The transformation capabilities of E2 and its metabolites have been shown in MCF-10F and MCF-10A cells (37, 47). We showed again that TCDD plus E2 increased colony efficiency 3-fold compared with E2 alone (Fig. 6C), indicating that TCDD enhanced the ability of E2 to transform MCF-10F cells. However, resveratrol significantly inhibited both E2- and TCDD plus E2-induced transformation (Fig. 6A and C). The colony assay provided evidence consistent with our hypothesis that resveratrol suppresses E2-induced cell transformation by preventing formation of depurinating DNA adducts.
Although prevailing theories for the role of estrogen in mammary gland carcinogenesis have focused on the stimulation of DNA synthesis and breast-cell proliferation by triggering ER-mediated signal transduction (2), evidence also indicates that reactive estrogen metabolites, produced by CYP-catalyzed metabolism of endogenous estrogens, are involved in mutagenesis and breast cancer initiation via an estrogen genotoxicity pathway (3, 4, 37). These two hypotheses are not mutually exclusive and both may contribute significantly to the etiology of estrogen-mediated cancers. Based on these studies, in which resveratrol regulated estrogen-metabolizing enzymes, decreased estrogen metabolism, prevented DNA adduct formation, and suppressed estrogen-induced malignant transformation, we conclude that enhancing estrogen metabolism (in this case, by TCDD-induced CYP1B1) to increase formation of depurinating DNA adducts may play a major role in breast cancer initiation. Resveratrol may act as a potential chemopreventive agent against estrogen-initiated breast cancer by blocking most of the critical steps in the estrogen genotoxicity pathway.
Acknowledgments
Grant support: F. Lu was supported by a fellowship from the University of Nebraska Environmental Toxicology Graduate Program. This study was supported by USPHS grant P01 CA49210 from the National Cancer Institute and Department of Defense grant DAMD17-03-1-0299 from the U.S. Army Breast Cancer Research Program. Core support at the Eppley Institute was provided by grant P30 CA36727 from the National Cancer Institute.
Footnotes
Unpublished data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 2.Dickson RB, Stancel GM. Estrogen receptor-mediated processes in normal and cancer cells. J Natl Cancer Inst Monogr. 2000;27:135–145. doi: 10.1093/oxfordjournals.jncimonographs.a024237. [DOI] [PubMed] [Google Scholar]
- 3.Cavalieri E, Chakravarti D, Guttenplan J, et al. Catechol estrogen quinones as initiators of breast and other human cancers: Implications for biomarkers of susceptibility and cancer prevention. Biochim Biophys Acta-Rev Cancer. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Cavalieri E, Rogan E. Catechol quinones of estrogens in the initiation of breast, prostate and other human cancers. Ann N Y Acad Sci. 2006;1089:286–301. doi: 10.1196/annals.1386.042. [DOI] [PubMed] [Google Scholar]
- 5.Shimada T, Hayes CL, Yamazaki H. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- 6.Talalay P, Dinkova-Kostova AT, Holtzclaw WD. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv Enzyme Regul. 2003;43:121–134. doi: 10.1016/s0065-2571(02)00038-9. [DOI] [PubMed] [Google Scholar]
- 7.Hakkola J, Pasanen M, Pelkonen O, et al. Expression of CYP1B1 in human adult and fetal tissues and differential inducibility of CYP1B1 and CYP1A1 by Ah receptor ligands in human placenta and cultured cells. Carcinogenesis. 1997;18:391–397. doi: 10.1093/carcin/18.2.391. [DOI] [PubMed] [Google Scholar]
- 8.Muskhelishvili L, Thompson PA, Kusewitt DF, Wang C, Kadlubar FF. In situ hybridization and immunohistochemical analysis of cytochrome P450 1B1 expression in human normal tissues. J Histochem Cytochem. 2001;49:229–236. doi: 10.1177/002215540104900210. [DOI] [PubMed] [Google Scholar]
- 9.Tang YM, Chen GF, Thompson PA, Lin DX, Lang NP, Kadlubar FF. Development of an antipeptide antibody that binds to the C-terminal region of human CYP1B1. Drug Metab Dispos. 1999;27:274–280. [PubMed] [Google Scholar]
- 10.Carnell DM, Smith RE, Daley FM. Target validation of cytochrome P450 CYP1B1 in prostate carcinoma with protein expression in associated hyperplastic and premalignant tissue. Int J Radiat Oncol Biol Phys. 2004;58:500–509. doi: 10.1016/j.ijrobp.2003.09.064. [DOI] [PubMed] [Google Scholar]
- 11.Spink DC, Hayes CL, Young NR, et al. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on estrogen metabolism in MCF-7 breast cancer cells: evidence for induction of a novel 17 β-estradiol 4-hydroxylase. J Steroid Biochem Mol Biol. 1994;51:251–258. doi: 10.1016/0960-0760(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 12.Hayes CL, Spink DC, Spink BC. 17β-Estradiol hydroxylation catalyzed by human P450 1B1. Proc Natl Acad Sci U S A. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaikwad NW, Rogan EG, Cavalieri EL. Evidence from ESI-MS for NQO1-catalyzed reduction of estrogen ortho-quinones. Free Radic Biol Med. 2007;43:1289–1298. doi: 10.1016/j.freeradbiomed.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y. NQO1, MPO, and the risk of lung cancer: a HuGE review. Genet Med. 2005;7:463–478. doi: 10.1097/01.gim.0000177530.55043.c1. [DOI] [PubMed] [Google Scholar]
- 15.Chao C, Zhang ZF, Berthiller J, Boffetta P, Hashibe M. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and the risk of lung, bladder, and colorectal cancers: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:979–987. doi: 10.1158/1055-9965.EPI-05-0899. [DOI] [PubMed] [Google Scholar]
- 16.Park SJ, Zhao H, Spitz MR, Grossman HB, Wu X. An association between NQO1 genetic polymorphism and risk of bladder cancer. Mutat Res. 2003;536:131–137. doi: 10.1016/s1383-5718(03)00041-x. [DOI] [PubMed] [Google Scholar]
- 17.Cavalieri EL, Kumar S, Todorovic R, Higginbotham S, Badawi AF, Rogan EG. Imbalance of estrogen homeostasis in kidney and liver of hamsters treated with estradiol: implications for estrogen-induced initiation of renal tumors. Chem Res Toxicol. 2001;14:1041–1050. doi: 10.1021/tx010042g. [DOI] [PubMed] [Google Scholar]
- 18.Rogan EG, Badawi AF, Devanesan PD, et al. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 19.Vidal JD, Vandevoort CA, Marcus CB, Lazarewicz NR, Conley AJ. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces CYP1B1 expression in human luteinized. Arch Biochem Biophys. 2005;439:53–60. doi: 10.1016/j.abb.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Spink BC, Hussain MM, Katz BH, Eisele L, Spink DC. Transient induction of cytochromes P450 1A1 and 1B1 in MCF-7 human breast cancer cells by indirubin. Biochem Pharmacol. 2003;66:2313–2321. doi: 10.1016/j.bcp.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Singh S, Chakravarti D, Edney JA, et al. Relative imbalances in the expression of estrogen-metabolizing enzymes in the breast tissue of women with breast carcinoma. Oncol Rep. 2005;14:1091–1096. [PubMed] [Google Scholar]
- 22.Jang M, Cai L, Udeani GO. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 23.Surh Y-J. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 24.Aziz MH, Kumar R, Ahmad N. Cancer chemoprevention by resveratrol: in vitro and in vivo studies and the underlying mechanisms [review] Int J Oncol. 2003;23:17–28. [PubMed] [Google Scholar]
- 25.Spink DC, Spink BC, Cao JQ, et al. Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis. 1998;19:291–298. doi: 10.1093/carcin/19.2.291. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya Y, Nakajima M, Kyo S, Kanaya T, Inoue M, Yokoi T. Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 2004;64:3119–3125. doi: 10.1158/0008-5472.can-04-0166. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZH, Hurh YJ, Na HK, et al. Resveratrol inhibits TCDD-induced expression of CYP1A1 and CYP1B1 and catechol estrogen-mediated oxidative DNA damage in cultured human mammary epithelial cells. Carcinogenesis. 2004;25:2005–2013. doi: 10.1093/carcin/bgh183. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Cao Z, Zhu H. Upregulation of endogenous antioxidants and phase 2 enzymes by the red wine polyphenol, resveratrol in cultured aortic smooth muscle cells leads to cytoprotection against oxidative and electrophilic stress. Pharmacol Res. 2006;53:6–15. doi: 10.1016/j.phrs.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 29.He X, Chen MG, Lin GX, Ma Q. Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2 × Keap1 × Cul3 complex and recruiting Nrf2 × Maf to the antioxidant response element enhancer. J Biol Chem. 2006;18:23620–23631. doi: 10.1074/jbc.M604120200. [DOI] [PubMed] [Google Scholar]
- 30.Yates MS, Tauchi M, Katsuoka F, et al. Pharmacodynamic characterization of chemopreventive triterpenoidsas exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6:154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 31.Warabi E, Takabe W, Minami T, et al. Shear stress stabilizes NF-E2-related factor 2 and induces antioxidant genes in endothelial cells: role of reactive oxygen/nitrogen species. Free Radic Biol Med. 2007;15:260–269. doi: 10.1016/j.freeradbiomed.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 32.Stivala LA, Savio M, Carafoli F, et al. Specific structural determinants are responsible for the antioxidant activity and the cell cycle effects of resveratrol. J Biol Chem. 2001;276:22586–22594. doi: 10.1074/jbc.M101846200. [DOI] [PubMed] [Google Scholar]
- 33.Zahid M, Gaikwad N, Cavalieri EL, Rogan EG. Inhibition of depurinating estrogen-DNA adducts by natural compounds. Chem Res Toxicol. 2007;20:1947–1953. doi: 10.1021/tx700269s. [DOI] [PubMed] [Google Scholar]
- 34.Lu F, Zahid M, Saeed M, Cavalieri EL, Rogan EG. Estrogen metabolism and formation of estrogen-DNA adducts in estradiol-treated MCF-10F cells. The effects of TCDD induction and catechol-O-methyltransferase inhibition. J Steroid Biochem Mol Biol. 2007;105:150–158. doi: 10.1016/j.jsbmb.2006.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zahid M, Saeed M, Lu F, Gaikwad N, Cavalieri EL, Rogan EG. Inhibition of catechol-O-methyltransferase increases estrogen-DNA adduct formation. Free Radic Biol Med. 2007;43:1534–1540. doi: 10.1016/j.freeradbiomed.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 37.Russo J, Fernandez SV, Russo PA, et al. 17-β-Estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J. 2006;20:1622–1634. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Roy SK. Expression of growth differentiation factor 9 in the oocytes is essential for the development of primordial follicles in the hamster ovary. Endocrinology. 2006;147:1725–1734. doi: 10.1210/en.2005-1208. [DOI] [PubMed] [Google Scholar]
- 39.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem Res Toxicol. 2006;19:164–172. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 40.Fang JG, Lu M, Chen ZH, et al. Antioxidant effects of resveratrol and its analoguesa gainst the free-radical-induced peroxidation of linoleic acid in micelles. Chemistry. 2002;8:4191–4198. doi: 10.1002/1521-3765(20020916)8:18<4191::AID-CHEM4191>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 41.Guengerich FP, Chun YJ, Kim D, Gillam EM, Shimada T. Cytochrome P450 1B1: a target for inhibition in anticarcinogenesis strategies. Mutat Res. 2003;523–4:173–182. doi: 10.1016/s0027-5107(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 42.Ross D, Siegel S. NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol. 2004;382:115–144. doi: 10.1016/S0076-6879(04)82008-1. [DOI] [PubMed] [Google Scholar]
- 43.Numazawa S, Yoshida T. Nrf2-dependent gene expressions: a molecular toxicological aspect. J Toxicol Sci. 2004;29:81–89. doi: 10.2131/jts.29.81. [DOI] [PubMed] [Google Scholar]
- 44.Lee JM, Li J, Johnson DA, et al. Nrf2, a multiorgan protector? FASEB J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 45.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vargas MR, Pehar M, Cassina P, et al. Fibroblast growth factor-1 induces heme oxygenase-1 via nuclear factor erythroid 2-related factor 2 (Nrf2) in spinal cord astrocytes: consequences for motor neuron survival. J Biol Chem. 2005;280:25571–25579. doi: 10.1074/jbc.M501920200. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Lin YC. Transformation of MCF-10A human breast epithelial cells by zeranol and estradiol-17β. Breast J. 2004;10:514–521. doi: 10.1111/j.1075-122X.2004.21410.x. [DOI] [PubMed] [Google Scholar]