Abstract
In mitochondria, carbamoyl-phosphate synthetase 1 activity produces carbamoyl phosphate for urea synthesis, and deficiency results in hyperammonemia. Cytoplasmic carbamoyl-phosphate synthetase 2, however, is part of a tri-functional enzyme encoded by CAD; no human disease has been attributed to this gene. The tri-functional enzyme contains carbamoyl-phosphate synthetase 2 (CPS2), aspartate transcarbamylase (ATCase) and dihydroorotase (DHOase) activities, which comprise the first three of six reactions required for de novo pyrimidine biosynthesis. Here we characterize an individual who is compound heterozygous for mutations in different domains of CAD. One mutation, c.1843-1G>A, results in an in-frame deletion of exon 13. The other, c.6071G>A, causes a missense mutation (p.Arg2024Gln) in a highly conserved residue that is essential for carbamoyl-phosphate binding. Metabolic flux studies showed impaired aspartate incorporation into RNA and DNA through the de novo synthesis pathway. In addition, CTP, UTP and nearly all UDP-activated sugars that serve as donors for glycosylation were decreased. Uridine supplementation rescued these abnormalities, suggesting a potential therapy for this new glycosylation disorder.
Introduction
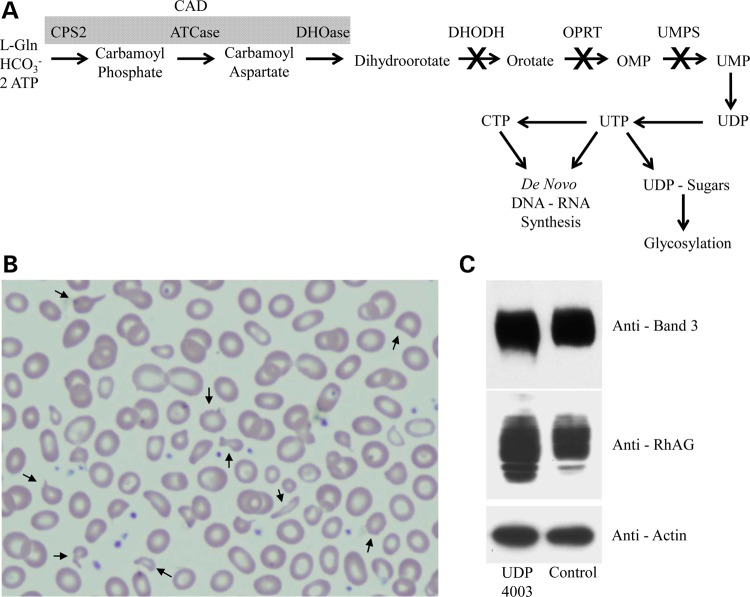
Six enzymatic reactions are required for de novo pyrimidine biosynthesis, an essential step in the synthesis of nucleotides including uridine triphosphate (UTP) and cytosine triphosphate (CTP) (Fig. 1A). The pyrimidines are mainly utilized for de novo DNA and RNA synthesis, but UTP is also used to make UDP nucleotide sugars for glycosylation (1,2).
Figure 1.
Schematic of the de novo pyrimidine biosynthesis pathway. UDP4003 Pedigree and characterization of CDAII/HEMPAS like phenotype. (A) Diagram showing the de novo pyrimidine biosynthesis pathway with known disorders highlighted with an (X) over the arrow. (B) Peripheral blood smear showing abnormal erythrocytes with characteristics of varying sizes and abnormal shapes (anisopoikilocytosis), spiked cell membrane (acanthocytes) and fragmented with pointed ends (schistocytes) resulting in a dyserythropoietic anemia phenotype. (C) Western blot analysis of RBC proteins, RhAG and Band-3, showing slower migration consistent with abnormal glycosylation.
The first three of six enzymatic steps are carried out by CAD, which is synthesized as a single large polypeptide with three highly conserved enzymatic activities: carbamoyl-phosphate synthetase 2 (CPS2), aspartate transcarbamylase (ATCase) and dihydroorotase (DHOase) (3–6). The fourth step occurs within the inner mitochondrial membrane where dihydroorotate dehydrogenase (DHODH) catalyzes dihydroorotate into orotate (7). Uridine monophosphate synthetase (UMPS) is a dual functioning protein with N-terminal orotate phosphoribosyltransferase activity and C-terminal OMP decarboxylase activity (8,9). UMP then serves as a substrate for subsequent steps that lead to the synthesis of UDP and ultimately UTP. These substrates can also be derived directly from uridine via the pyrimidine salvage pathway (10).
Extremely rare human genetic disorders occur in the last three steps of the highly regulated, de novo pathway. Miller syndrome (also known as Genee–Weidemann or postaxial acrofacial dysostosis syndrome) (MIM-263750) was the first Mendelian disorder solved via whole exome sequencing; causal mutations were identified in DHODH (MIM-126064) (11). Miller syndrome is characterized by postnatal growth deficiency, micrognathia, ear and eye anomalies, skeletal defects and gastrointestinal problems such as intestinal malrotation (11–13). Interestingly, individuals with fetal exposure to methotrexate, a known inhibitor of purine biosynthesis, also have malformations commonly seen in Miller syndrome (11). Likewise, treating mice with leflunomide, a specific inhibitor of DHODH activity, recapitulates the highly penetrant limb and craniofacial malformations present in Miller syndrome (13). Co-administration of uridine with leflunomide reverses this phenotype, but it is unknown if administering uridine can reverse the any of the phenotypes in Miller syndrome.
Mutations in UMPS (MIM-613891) cause hereditary orotic aciduria (MIM-258900), an autosomal recessive disorder characterized by growth retardation, varying degrees of intellectual disabilities, megaloblastic anemia and urinary excretion of orotic acid (orotic acid crystalluria) (14). Importantly, providing exogenous uridine reduces urinary orotic acid and improves clinical and hematological abnormalities (15). A bovine model termed ‘DUMPS', or Deficiency of UMP Synthase, containing a homozygous premature stop codon (p.Arg405*), is stillborn or dies shortly after birth (16).
Here we identify a 4-year-old boy, enrolled in the National Institutes of Health Undiagnosed Diseases Program, with two damaging variants in CAD (MIM-114010).
Results
Clinical presentation
UDP4003 is the younger of two male siblings born to non-consanguineous parents and was admitted to the NIH Clinical Center at 4 years of age; his parents gave written, informed consent. Gestation was normal and delivery occurred at 38 weeks via C-section due to a history of placenta accrete. Birth weight was 3402 g (25–50%); birth length 54.9 cm (75%); OFC 35.6 cm (25%); APGAR scores were 8 and 9. Diarrhea and poor weight gain led to a bowel biopsy at age 6 months that revealed pan-disaccharidase deficiency, treated with probiotics and digestive enzymes. Renal tubular acidosis was diagnosed at age 1 year and treated with citrate; it resolved at 21 months. Seizures occurred at 17 months of age.
When first seen at the NIH, his height was 100.2 cm (25%), weight 14.2 kg (10%), OFC 49 cm (10%). Delays in expressive language and fine motor development were recognized; mild hypotonia and a slightly wide-based gait had been noted at 17 months of age, at which time an MRI/MRS was normal.
Laboratory studies included a peripheral blood smear showing anisopoikilocytosis, acanthocytes and schistocytes; similar findings were present at 12 months of age. Fanconi and Shwachman-Diamond Syndromes (MIM-260400) were excluded. The bone marrow was dyserythropoietic, resembling Congenital Dyserthythropoietic Anemia Type II (MIM-224100) (Fig. 1B), but direct gene sequencing excluded mutations in SEC23B (MIM-610512). RBC enzymology was normal, but the RBC proteins RhAG and to a lesser extent Band-3 migrated abnormally on polyacrylamide gel electrophoresis, suggesting abnormal glycosylation (Fig. 1C).
Urine amino and organic acids, plasma amino acids (specifically no β-alanine abnormalities), uric acid, fasting lipid profile, lactate, pyruvate, acylcarnitine, free and total carnitine, quantitative analysis of immunoglobulins, vitamin D, vitamin E and cerebrospinal fluid examination were normal. Vitamin A was moderately decreased at 22 mcg/dl (normal, 36–120). Ammonia levels were 69 and 47 mg/dl (normal, 11–35).
Urine purines and pyrimidines (including uracil, uric acid, hypoxanthine or xanthine) were normal; previous urine purine testing for xanthine, inosine, guanosine, adenine, adenosine, deoxyadenosine and hypoxanthine showed only a mild elevation of guanosine. Urine orotic acid was normal at the NIH; a moderate decrease had been previously observed on one occasion.
Exome sequencing identifies novel mutations in CAD
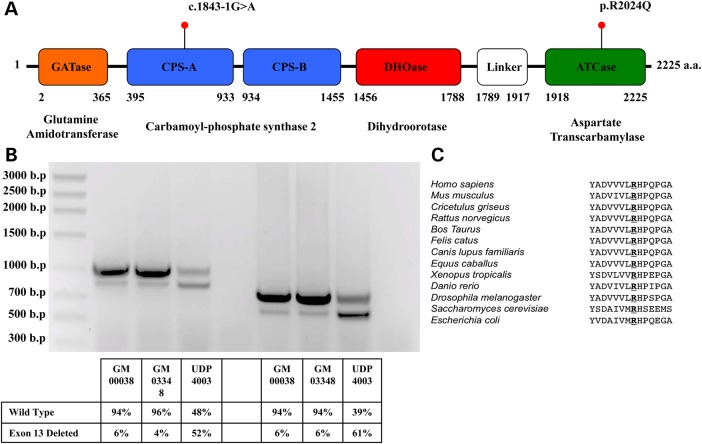
Exome sequencing (WES) identified two variants in CAD that were not present in the Single Nucleotide Polymorphism database, 1000 Genomes, the NHLBI Exome Variant Server (EVS) or any in-house derived exomes. One variant affects an essential splice site, c.1843-1G>A, upstream of exon-13 and the other causes a c.6071G>A change in exon-39 resulting in a missense variant, p.Arg2024Gln (Fig. 2A). A more expansive database, The Exome Aggregation Consortium (ExAC) browser, contained only a single heterozygous carrier for the p.Arg2024Gln out of 58 967 unrelated individuals (or 117 934 alleles). The c.1843-1G>A was not reported in those reads that passed filtering validation, however, a single ‘Non-Pass' heterozygous carrier was seen out of 60 569 unrelated individuals (or 121 138 alleles). Each parent was a carrier for one variant, while the healthy male sibling was a non-carrier. Both variants were confirmed in a Clinical Laboratory Improvement Amendments (CLIA) certified lab.
Figure 2.
Characterization of CAD Mutations identified in UDP4003. (A) Schematic for Human CAD (UniProt - P27708) with location of mutations identified by exome sequencing. The c.1843-1G>A results in an in-frame deletion of 63-amino acid in the CPSase A portion of Carbamoyl-phosphate synthase 2. (B) Abnormal splicing caused by the c.1843-1G>A was analyzed from two controls, GM00038 and GM03348 and UDP4003 using two different sets of primers. In both controls, a minor portion (4–6%) of CAD transcript lacks exon-13, whereas in UDP4003 there is substantially more (52–61%). (C) Conservation of human Arg2024 across other organisms.
The c.1843-1G>A is predicted to be deleterious according to American College of Medical Genetics (ACMG) recommendations for standards for interpretation of sequence variants (17). We confirmed that the c.1843-1G>A mutation affected splicing of CAD mRNA; cDNA analysis of UDP4003 fibroblasts revealed two PCR fragments (Fig. 2B). Sanger sequencing confirmed that the larger fragment was the reference transcript while the smaller, abnormally spliced fragment, was a transcript with an in-frame deletion of exon-13. This abnormal splicing results in a 63 amino acid deletion within the CPS2 domain of the CAD protein. Interestingly, extracts of two control fibroblast lines (GM00038 and GM03348) had small amounts of this in-frame deletion of exon 13 transcript, while UDP4003 had a significantly increased amount (Fig. 2B).
The p.Arg2024Gln missense mutation occurs within the ATCase domain of CAD at a position that is highly conserved (Fig. 2C). Structural and mutation analysis of E. coli ATCase enzyme shows that the orthologous p.Arg105 is essential for ATCase activity due to direct interactions with the substrate aspartate and the substrate analog phosphonoactamide (PAM) (18,19). The p.Arg2024Gln variant is predicted to be damaging by Polyphen2 with a HumDiv score of 0.998/1.00 and a HumVar score of 0.992/1.0. SIFT predicts it to be damaging with a score of 0.
Nucleotide sugar analysis in CHO-G9C and UDP4003
To determine pathogenicity of the CAD variants, we assayed two important processes of the de novo pyrimidine biosynthesis pathway: UDP nucleotide sugar levels and metabolic flux of aspartate into the de novo synthesis of DNA and RNA. One important product of the de novo pyrimidine biosynthesis, UTP, is used to form UDP-glucose (Glc), UDP-N-acetylglucosamine (GlcNAc), UDP-N-acetylgalactosamine (GalNAc), UDP-galactose (Gal) and UDP-glucoronic acid (GlcA); we predicted that loss of CAD would reduce these UDP nucleotide sugars. HPLC analysis of nucleotide sugars in the uridine-dependent Chinese hamster ovary mutant, CHO-G9C, confirmed that loss of cad reduced UDP-GlcNAc, UDP-GalNAc, UDP-Gal, UDP-Glc, as well as UTP and to a lesser extent CTP (Fig. 3A). Consistent with our model and previous reports of the pyrimidine pathway, addition of exogenous uridine restored the nucleotide sugar pools to normal levels (Fig. 3A).
Figure 3.
Analysis of purified nucleotide sugars by HPLC. (A) Nucleotide sugars were extracted and the purified material from CHO-K1 (wild-type) or CHO-G9C (CAD defective) cells with or without 30 µm uridine treatment were run on HPLC. (B) Nucleotide sugars were extracted in the same manner as above and the purified material from GM00038, GM03348 and UDP4003 with or without 30 µm uridine treatment was run on HPLC. The control sample represents an average of both GM00038 and GM03348. In both (A) and (B), the bar graph represents the peak area for each metabolite normalized to ATP levels, since ATP were consistently equal in multiple experiments. Error bars show standard deviation.
CHO-G9C is a haploid cell line selected to be auxotrophic for uridine and without exogenous uridine supplementation, will undergo growth arrest and eventual cell death (20). We optimized cell-culture conditions and treatment times so that the growth conditions alone did not adversely affect the outcome.
HPLC analysis of nucleotide sugars from two control fibroblast lines and UDP4003 fibroblasts showed that the patient's cells resembled CHO-G9C cells in having reduced levels of UDP-GlcNAc, UDP-GalNAc, UDP-Glc, UDP-Gal as well as the nucleotides UTP and CTP (Fig. 3B). Moreover, 30 µm uridine restored all the reduced metabolites in UDP4003 fibroblasts to control (n = 2) levels (Fig. 3B).
Aspartic acid flux in UDP4003
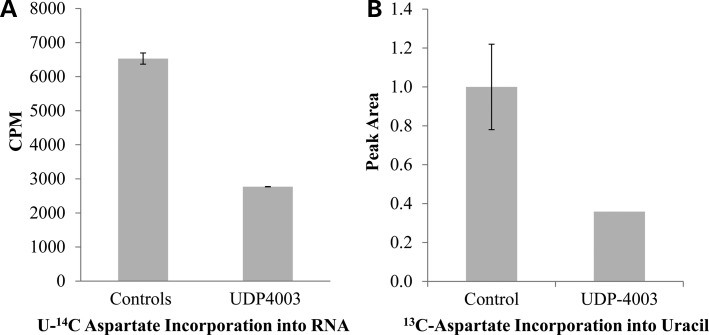
Loss of cad in CHO-G9C disrupts metabolic flux of aspartate through the de novo pyrimidine biosynthesis pathway (21,22). UDP4003 cells also showed reduced aspartate flux into RNA and DNA using two sets of isotopes: labeled 13C-15N-aspartate or l-[14C(U)-radiolabeled aspartate in combination with either 5,6-3H-uridine for RNA or 6-3H thymidine for DNA. In primary fibroblasts, incorporation of these labels was more efficient for RNA than DNA, but the trends were similar (DNA incorporation data not shown). Both methods revealed significantly less (>2-fold) labeled aspartate incorporation into the nucleic acids of UDP4003 fibroblasts compared with control cells (Fig. 4A and b); the level of reduction was similar to that for cad-deficient G9C CELls compared with the wild-type CHO-K1.
Figure 4.
Aspartate Flux. (A) Cells were serum starved for 24 h prior to stimulation with 100 nm recombinant Human Insulin for 7 h in the presence of 2uCi/ml U-14C aspartate and 1uCi/ml [5,6-3H] uridine. The radiolabeled U-14C aspartate incorporated into total RNA was normalized to the amount of [5,6-3H] uridine incorporated into RNA. Controls represent the averages of two separate fibroblast lines, GM00038 and GM03348. (B) Cells were serum starved for 24 h prior to stimulation with 100 nm recombinant Human Insulin for 7 h in the presence of 2 mm 13C,15N aspartate. Hydrolyzed samples were analyzed via GCMS. No difference was observed for incorporation of 13C aspartate into adenine. Controls represent the averages of two separate control fibroblast lines GM00038 and GM03348.
Expression of CAD mutations and growth complementation of CHO-G9C
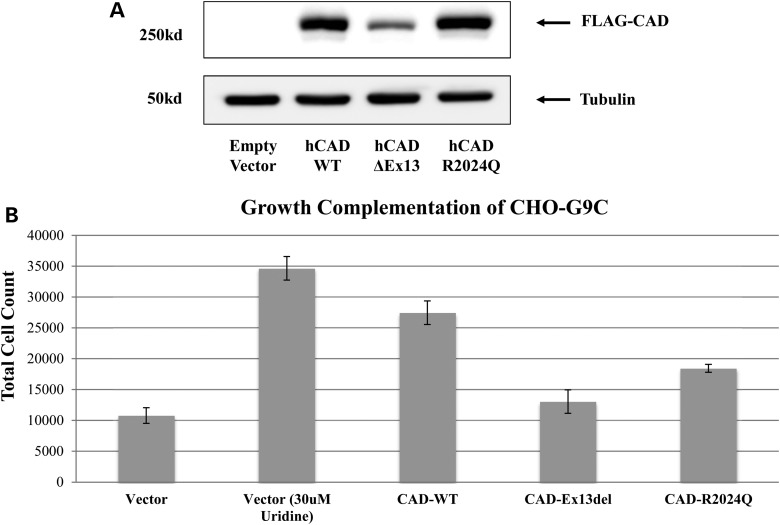
The c.1843-1G>A mutation in CAD causes an in-frame deletion of 63 amino acids, which, surprisingly, exists as an mRNA in normal tissue. This truncated CAD mRNA is present at low levels in normal primary fibroblasts, but we could not detect a truncated CAD protein, probably due to its relatively low abundance. We could transiently over express the truncated protein in HEK-293T cells, but its expression was less than that of wild-type CAD (Fig. 5A). This suggests that the truncated form of CAD is less stable than the wild-type. We were not able to resolve the ∼7 kDa difference between the exon-13 deleted CAD and the wild-type CAD (∼250 kDa). We speculate that the truncated CAD, which lacks the ATP grasp domain located within its CPS2 portion, may act as a metabolic sensor for regulating the de novo pyrimidine pathway. In this scenario, the truncated CAD would reduce cell growth because it binds ATP inefficiently, decreasing de novo pyrimidine synthesis.
Figure 5.
Expression and complementation assays. (A) Western blot analysis of HEK 293T cells transfected with FLAG tagged CAD expression constructs showing reduced expression of the exon-13 deleted CAD and normal expression of the pR2024Q CAD, when compared with wild-type CAD. (B) CHO-G9C cells transfected with FLAG tagged CAD constructs showing their ability to rescue the growth defect in the absence of uridine. In the non-rescued samples there is minimal growth, with any detectable growth is likely due to low levels of uridine with in the serum.
The p.Arg2024Gln mutation changes a highly conserved amino acid, considered essential for substrate binding based upon structural analyses of several organisms. This missense mutation does not affect protein expression under physiological conditions (Fig. 5A), but we cannot exclude the possibility that it may be thermolabile.
Transient over expression of the exon-13 deleted CAD was unable to rescue the defective growth phenotype in CHO-G9C, while the p.Arg2024Gln mutation partially rescued and, as expected, the wild-type fully rescued (Fig. 5B). One explanation is that since CAD is a tri-functional protein with three independent functions, G9C (which is haploid) likely has a hemizygous defect in only one of the three domains and complementation can occur only when the correct domain is expressed. For example, if G9C is mutated for CPS2, overexpressing wild-type ATCase or DHOase will have little or no effect.
Glycosylation changes in UDP4003
Finally, we investigated whether the changes in UDP nucleotide sugar pools affected overall protein glycosylation. Serum/plasma glycosylated transferrin was normal on several occasions. We did not see dramatic changes in either N-linked or O-linked glycosylation in fibroblast or serum from UDP4003 (data not shown). HPLC analysis of N-glycans from RBC ghosts detected only subtle differences in sialylation between UDP4003 and his non-carrier unaffected male sibling (data not shown).
Discussion
Nearly 2% of the human genome is estimated to encode glycosylation-related proteins, and mutations in over 100 genes produce congenital disorders of glycosylation that involve eight distinct pathways (23). Many of these include defects in precursor biosynthesis, but none of them impair de novo pyrimidine biosynthesis; CAD mutations do.
CAD expression must be ubiquitous since all cells require de novo synthesis of RNA and DNA, but there are likely other factors that contribute to the tissue-specific effects when CAD function or even UMPS or DHODH is lost. For example, why does loss of DHODH, but not UMPS or CAD, cause various malformations and limb defects? Why does loss of UMPS or CAD cause various forms of anemia, yet DHODH deficiency lacks anemia? These tissue-specific effects can best be seen in isolated mutants for several organisms including Drosophila melanogaster, Caenorhabditis elegans and Danio rerio.
The D. melanogaster CAD mutant rudimentary is lethal when grown on pyrimidine-deficient media, yet flies grown on pyrimidine containing media survive, but have smaller, abnormally shaped wings and females are sterile (24). The observation that uridine corrects this lethality, but not all the phenotypic defects, hints that certain tissues may have variable reliance on de novo pyrimidine synthesis versus salvage pathways.
Franks et al. (25) reported a C.elegans mutant with a p.His1602Gln missense mutation in the DHOase domain of CAD. Approximately 95% of late-stage embryos died, while the surviving 5% had pharyngeal morphogenesis abnormalities and appeared shorter when compared with wild-type worms (25). The lethality was avoided by providing exogenous uracil or uridine in the growth media. The pharyngeal morphogenesis abnormalities were likely due to disruption of the CAD-dependent synthesis of UDP sugars, which is critical for heparan sulfate proteoglycan production that controls morphogenesis (25).
Two independent studies describe zebrafish cad mutants. The initial mutant, perplexed, contains a missense mutation (p.Met1283Arg) within the CPS2 domain. Embryos homozygous for this mutation have abnormal retinal development along with growth and morphogenesis defects of the tectum, jaw and pectoral fins; they die at 8–12 days post fertilization (26). The second mutant, sl23, has a premature stop codon early within the CPS2 domain and a similar phenotype of small eyes and deformed jaw; they also die at 8–12 days post fertilization. However, the sl23 mutant also displays defects in the cranial sensory circuit formation (27). These papers show that either loss of UDP sugar synthesis/transport or blocking N-glycosylation via tunicamycin treatment recapitulates some of the phenotypic aspects of the cad-deficient fish.
These model organisms also suggest that homozygous loss of function or severely reduced activity in only one of the three CAD domains is not tolerated, since all are lethal. This may explain why more humans with CAD deficiency have not yet been identified. In addition, UDP4003 fibroblasts are not dependent upon uridine supplementation, as for the CHO-G9C cells or the cad-deficient model organisms mentioned earlier; this suggests that the mutations in UDP4003 may be hypomorphic. It should be noted that with only a single individual to characterize, it is impossible with absolute certainty, to assign all of UDP4003 clinical phenotypes solely to the loss of CAD activity. We acknowledge that some of these clinical phenotypes maybe due to other factors.
It is not surprising that there was not a substantial change in glycosylation, since the primary role of CAD in the de novo pyrimidine biosynthesis pathway is to provide precursors for RNA and DNA. It is likely that the contributions from the salvage and de novo pyrimidine biosynthesis pathways vary by cell type and that overall contributions to the glycosylation pathway are very small (28).
Biochemical screening typically identifies genetic disorders involving the purine and pyrimidine biosynthesis pathways. The accelerated use of next generations sequencing to solve rare genetic disorders has taught us that different mutations can influence clinical outcome as well as protein function. In this case, standard biochemical testing did not identify a pyrimidine disorder, likely because urine pyrimidine metabolites display a broad range of normal values, reflecting contributions from diet and cell breakdown. In contrast, WES identified causal mutations involving a critical component of de novo pyrimidine biosynthesis, and cellular studies confirmed the biochemical hallmarks of CAD deficiency. Uridine rescue of the biochemical defects suggest a potential therapy in CAD-deficient patients.
Methods and Materials
UDP clinical evaluation compliance
Biological parents provided written consent for both a clinical evaluation as well as whole exome sequencing under the protocol #76-HG-0238: diagnosis and Treatment of Patients with Inborn Errors of Metabolism and Other Genetic Disorders.
Exome sequencing
Performed as previously described (29).
cDNA analysis
Total RNA from control fibroblasts GM00038, GM03348 (Coriell Cell Repository) and UDP4003 was purified using TRIzol reagent (Life Technologies). cDNA was synthesized using 1 µg of total RNA and the SuperScript III First-Strand Synthesis System (Life Technologies). Two separate sets of PCR primers were used to amplify portions of the CAD mRNA surrounding exon-13.
HPLC analysis of nucleotide sugars
Nucleotide sugars were extracted from control, patient fibroblast, CHO-K1 and CHO-G9C pellets that had been stored at −20°C. Cell pellets were thawed on ice then resuspended in 900 μl ice cold 85% ethanol and 120 μl PBS. After brief sonication, cellular debris was removed by centrifugation at 13 000 rpm for 10 min. The supernatant was evaporated in a SpeedVac concentrator and resuspended in 1 ml water. Insoluble material was removed by an additional centrifugation step and the resulting cleared supernatant mixed with 50 μl 0.2 M NH4HCO3. The sample was further purified using an Envi-Carb carbon column according to the method of Rabina et al. (30). Nucleotide sugar pools were measured by reverse phase HPLC on an Inertsil ODS-4 column according to Nakajima et al. (31).
Aspartic acid flux
Expression and western blot analysis of CAD mutations in HEK-293T cells
HEK 293T cells were transfected with either 2 µg of the expression vectors pcmv6 - empty vector, FLAG hCAD wild-type, FLAG hCAD del Exon-13 or FLAG hCAD p.R2024Q in six-well plates and collected after 48 h post transfection. Cells were lyzed in 62.5 mm Tris pH 6.8, 10% glycerol and 2% SDS and boiled at 100°C for 15 min. Normalized extracts were separated on a 6% SDS–PAGE gel and FLAG-proteins were detected using a monoclonal M2 FLAG antibody (SIGMA). Western blot analysis with a monoclonal antibody against alpha tubulin (12G10) (Developmental Studies Hybridoma Bank University of Iowa) was used to confirm equal loading.
Growth defect complementation of CHO-G9C cells
CHO-G9C cells plated at 5000 cells per well and allowed to attach overnight before being transfected in 12-well plates with 1.5 µg of appropriate expression plasmid for 48 h in the presence of 5 µm uridine. Each data point was done in biological duplicates. After the initial 48 h period, cells were washed and grown with or without uridine for 72 h. Cell growth was measured by counting total cell number for each well.
Web Resources
1000 Genomes Project, http://www.1000genomes.org/
Exome Aggregation Consortium (ExAC), Cambridge, MA, http://exac.broadinstitute.org (November, 2014)
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
The Single Nucleotide Polymorphism database (dbSNP), http://www.ncbi.nlm.nih.gov/SNP/.
Funding
Supported in part by The Rocket Fund, National Institutes of Health (NIH) grants (R01DK099551 and R01DK55615 to H.H.F), the Common Fund of the Office of the Director, NIH and the Intramural Research Program of the National Human Genome Research Institute.
Acknowledgements
We thank the family for their participation and support. We also thank Dr David Patterson for the Chinese hamster ovary mutant G9C and Dr Mohandas Narla for analysis of blood samples.
Conflict of Interest statement. None declared.
References
- 1.Bulter T., Elling L. (1999) Enzymatic synthesis of nucleotide sugars. Glycoconj. J., 16, 147–159. [DOI] [PubMed] [Google Scholar]
- 2.Huang M., Graves L.M. (2003) De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci., 60, 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K.C., Vannais D.B., Jones C., Patterson D., Davidson J.N. (1989) Mapping of the gene encoding the multifunctional protein carrying out the first three steps of pyrimidine biosynthesis to human chromosome 2. Hum. Genet., 82, 40–44. [DOI] [PubMed] [Google Scholar]
- 4.Simmer J.P., Kelly R.E., Rinker A.G., Jr., Scully J.L., Evans D.R. (1990) Mammalian carbamyl phosphate synthetase (CPS). DNA sequence and evolution of the CPS domain of the Syrian hamster multifunctional protein CAD. J. Biol. Chem., 265, 10395–10402. [PubMed] [Google Scholar]
- 5.Grayson D.R., Evans D.R. (1983) The isolation and characterization of the aspartate transcarbamylase domain of the multifunctional protein, CAD. J. Biol. Chem., 258, 4123–4129. [PubMed] [Google Scholar]
- 6.Kelly R.E., Mally M.I., Evans D.R. (1986) The dihydroorotase domain of the multifunctional protein CAD. Subunit structure, zinc content, and kinetics. J. Biol. Chem., 261, 6073–6083. [PubMed] [Google Scholar]
- 7.Hines V., Keys L.D., III, Johnston M. (1986) Purification and properties of the bovine liver mitochondrial dihydroorotate dehydrogenase. J. Biol. Chem., 261, 11386–11392. [PubMed] [Google Scholar]
- 8.Floyd E.E., Jones M.E. (1985) Isolation and characterization of the orotidine 5′-monophosphate decarboxylase domain of the multifunctional protein uridine 5′-monophosphate synthase. J. Biol. Chem., 260, 9443–9451. [PubMed] [Google Scholar]
- 9.Suttle D.P., Bugg B.Y., Winkler J.K., Kanalas J.J. (1988) Molecular cloning and nucleotide sequence for the complete coding region of human UMP synthase. Proc. Natl Acad. Sci. USA, 85, 1754–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyhan W.L. (2005) Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab., 86, 25–33. [DOI] [PubMed] [Google Scholar]
- 11.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet., 42, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogilvy-Stuart A.L., Parsons A.C. (1991) Miller syndrome (postaxial acrofacial dysostosis): further evidence for autosomal recessive inheritance and expansion of the phenotype. J. Med. Genet., 28, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rainger J., Bengani H., Campbell L., Anderson E., Sokhi K., Lam W., Riess A., Ansari M., Smithson S., Lees M., et al. (2012) Miller (Genee-Wiedemann) syndrome represents a clinically and biochemically distinct subgroup of postaxial acrofacial dysostosis associated with partial deficiency of DHODH. Hum. Mol. Genet., 21, 3969–3983. [DOI] [PubMed] [Google Scholar]
- 14.Suchi M., Mizuno H., Kawai Y., Tsuboi T., Sumi S., Okajima K., Hodgson M.E., Ogawa H., Wada Y. (1997) Molecular cloning of the human UMP synthase gene and characterization of point mutations in two hereditary orotic aciduria families. Am. J. Hum. Genet., 60, 525–539. [PMC free article] [PubMed] [Google Scholar]
- 15.Girot R., Hamet M., Perignon J.L., Guesnu M., Fox R.M., Cartier P., Durandy A., Griscelli C. (1983) Cellular immune deficiency in two siblings with hereditary orotic aciduria. N. Engl. J. Med., 308, 700–704. [DOI] [PubMed] [Google Scholar]
- 16.Schwenger B., Schober S., Simon D. (1993) DUMPS cattle carry a point mutation in the uridine monophosphate synthase gene. Genomics, 16, 241–244. [DOI] [PubMed] [Google Scholar]
- 17.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E. and Molecular Subcommittee of the, A.L.Q.A.C. (2008) ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med., 10, 294–300. [DOI] [PubMed] [Google Scholar]
- 18.Stieglitz K.A., Pastra-Landis S.C., Xia J., Tsuruta H., Kantrowitz E.R. (2005) A single amino acid substitution in the active site of Escherichia coli aspartate transcarbamoylase prevents the allosteric transition. J. Mol. Biol., 349, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Eldo J., Kantrowitz E.R. (2007) Structural model of the R state of Escherichia coli aspartate transcarbamoylase with substrates bound. J. Mol. Biol., 371, 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musmanno L.A., Jamison R.S., Barnett R.S., Buford E., Davidson J.N. (1992) Complete hamster CAD protein and the carbamylphosphate synthetase domain of CAD complement mammalian cell mutants defective in de novo pyrimidine biosynthesis. Somat. Cell Mol. Genet., 18, 309–318. [DOI] [PubMed] [Google Scholar]
- 21.Robitaille A.M., Christen S., Shimobayashi M., Cornu M., Fava L.L., Moes S., Prescianotto-Baschong C., Sauer U., Jenoe P., Hall M.N. (2013) Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science, 339, 1320–1323. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Sahra I., Howell J.J., Asara J.M., Manning B.D. (2013) Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science, 339, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeze H.H., Chong J.X., Bamshad M.J., Ng B.G. (2014) Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet., 94, 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norby S. (1970) A specific nutritional requirement for pyrimidines in rudimentary mutants of Drosophila melanogaster. Hereditas, 66, 205–214. [DOI] [PubMed] [Google Scholar]
- 25.Franks D.M., Izumikawa T., Kitagawa H., Sugahara K., Okkema P.G. (2006) C. elegans pharyngeal morphogenesis requires both de novo synthesis of pyrimidines and synthesis of heparan sulfate proteoglycans. Dev. Biol., 296, 409–420. [DOI] [PubMed] [Google Scholar]
- 26.Willer G.B., Lee V.M., Gregg R.G., Link B.A. (2005) Analysis of the Zebrafish perplexed mutation reveals tissue-specific roles for de novo pyrimidine synthesis during development. Genetics, 170, 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox J.A., LaMora A., Johnson S.L., Voigt M.M. (2014) Novel role for carbamoyl phosphate synthetase 2 in cranial sensory circuit formation. Int. J. Dev. Neurosci., 33, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichikawa M., Scott D.A., Losfeld M.E., Freeze H.H. (2014) The metabolic origins of mannose in glycoproteins. J. Biol. Chem., 289, 6751–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markello T.C., Adams D.R. (2013) Genome-scale sequencing to identify genes involved in Mendelian disorders. Curr. Protoc. Hum. Genet., 79, Unit 6.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabina J., Maki M., Savilahti E.M., Jarvinen N., Penttila L., Renkonen R. (2001) Analysis of nucleotide sugars from cell lysates by ion-pair solid-phase extraction and reversed-phase high-performance liquid chromatography. Glycoconj. J., 18, 799–805. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima K., Kitazume S., Angata T., Fujinawa R., Ohtsubo K., Miyoshi E., Taniguchi N. (2010) Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology, 20, 865–871. [DOI] [PubMed] [Google Scholar]