Abstract
mRNA decay is an essential and active process that allows cells to continuously adapt gene expression to internal and environmental cues. There are two mRNA degradation pathways: 3′ to 5′ and 5′ to 3′. The DCPS protein is the scavenger mRNA decapping enzyme which functions in the last step of the 3′ end mRNA decay pathway. We have identified a DCPS pathogenic mutation in a large family with three affected individuals presenting with a novel recessive syndrome consisting of craniofacial anomalies, intellectual disability and neuromuscular defects. Using patient's primary cells, we show that this homozygous splice mutation results in a DCPS loss-of-function allele. Diagnostic biochemical analyses using various m7G cap derivatives as substrates reveal no DCPS enzymatic activity in patient's cells. Our results implicate DCPS and more generally RNA catabolism, as a critical cellular process for neurological development, normal cognition and organismal homeostasis in humans.
Introduction
Human mRNAs contain a modified 5′ cap that is generated co-transcriptionally. This structure is made of a guanosine methylated at position 7 linked to the first transcribed nucleotide by a 5′-5′ triphosphate (1). The cap structure, marking the bona fide 5′ end of cellular and some viral mRNAs, plays an important role by facilitating pre-mRNA splicing and mRNA exportation to the cytoplasm, before promoting translation initiation (2). This landmark also protects mRNA from degradation by nucleases (3). mRNA decay is, however, an essential process that allows cells to continuously adapt gene expression to internal and environmental changes. The cap structure needs to be eliminated as part of RNA turnover giving rise to free m7G cap (di-) nucleotides as end-products. The two major mRNA degradation mechanisms described in human cells result in the production of different cap derivatives (4). The first process involves the severing of the triphosphate linkage of the cap by the DCP2 (MIM609844) decapping enzyme yielding m7GDP (5–7). The second process entails the 3′-5′ exonucleolytic degradation of the mRNA by the multi-subunit exosome (8), releasing free cap (m7GpppN) which is degraded by the scavenger decapping enzyme DCPS (MIM610534) (9). This hydrolase, belonging to the HIT protein family, cleaves m7GpppN into m7GMP and NDP (10). DCPS was also recently shown to be involved in the elimination of m7GDP formed by DCP2 (11). DCPS is thus a major factor involved in the elimination of cap residues generated by mRNA degradation with an estimated number of 250 molecules being produced per minute in mammalian cells (12,13). This enzyme is evolutionarily conserved in eukaryotes, underlying its importance for basic cellular RNA homeostasis. No diseases or mutations have yet been reported in the DCP2 or DCPS enzymes in humans.
Results
Clinical presentations of patients diagnosed with a novel autosomal recessive disorder
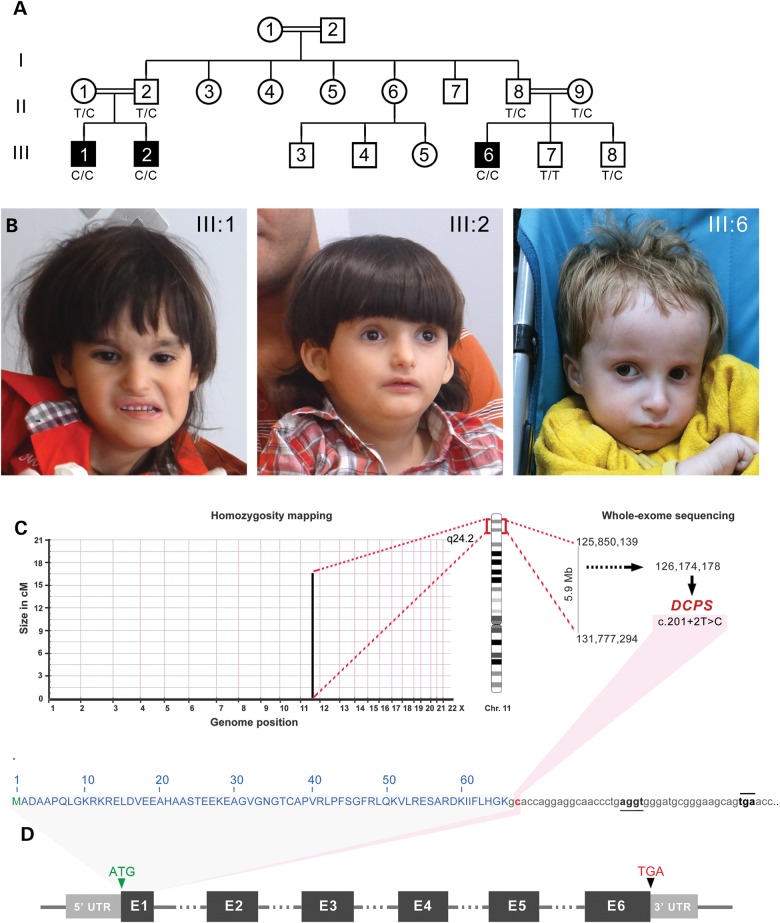
We studied a Jordanian inbred family consisting of three affected boys III-1, III-2 and III-6 with an unknown syndrome born to unaffected consanguineous parents (Fig. 1A). The three propositi had birth weights below the 5th percentile ranging from 2.3 and 2.9 kg. All presented with similar congenital muscle hypotonia with severe growth delay as measured by height, weight and occipital head circumference at birth and during infancy. Brain structures were normal by MRI scans although all affected cases had mild microcephaly (Table 1 and Supplementary Material, Fig. S1). Developmental milestones including motor skills such as sitting, walking and linguistic acquisition were also significantly delayed. At age 7, proband (III:1) only spoke a few words, walked with ataxic gait and could not stand for long periods of time (Supplementary Material, Video S1). Mild facial dysmorphisms including flat face with deep-set eyes, low-set ears, simple helices, small nose and mouth were observed (Fig. 1B and Supplementary Material, Fig. S1). Other symptoms documented in all three patients include joint laxity, partial syndactyly, brachydactyly and sandal gap pointing to skeletal anomalies (Supplementary Material, Fig. S1). Skin hypopigmentation and atrial septal defects were also recorded for all three propositi (Table 1). None of these clinical phenotypes were observed in the non-affected family members. The overall clinical presentation consisting of severe growth delay, craniofacial anomalies, cognitive impairment and mild skeletal defects did not match with previously described syndromes suggesting that this may be an unrecognized congenital recessive disorder.
Figure 1.
Phenotypic characteristics of three male propositi with an autosomal recessive Al-Raqad syndrome caused by a splice site mutation in DCPS. (A) Pedigree of Jordanian inbred family with three affected children (III:1, III:2 and III:6) and two unaffected sibs (III:7 and III:8) born to first-cousin parents. (B) Head shot of the three propositi with syndromic craniofacial anomalies, including deep set eyes, thin upper lip, small nose and mild microcephaly. (C) Homozygousity mapping delineates a single candidate region of 5.9 Mb on chromosome 11q. Whole-exome sequencing revealed a private mutation in DCPS in the first splice donor of intron 1: c.201 + 2T > C in the homozygous state. (D) Schematic representation of DCPS exons and location of the mapped mutation in its intron 1 (highlighted in red). Alternative cryptic splice site (underlined) and an in-frame premature termination codon (overlined) are detected 19 and 40 bp downstream of exon 1, respectively.
Table 1.
Clinical characteristics of the three affected children
|
Propositi with Al-Raqad Syndrome (ARS) |
|||
|---|---|---|---|
| III:1 | III:2 | III:6 | |
| Gender, year of birth | Male, 2006 | Male, 2008 | Male, 2011 |
| Birth weight (kg)/length (cm), (%a) | 2.4 (<5th)/51 cm (75th) | 2.3 (<5th)/48 cm (10th–25th) | 2.9 (5th)/NA |
| Birth OFC, cm (%a) | 31 (<5th) | 33 (10th) | NA |
| Follow-up weight, kg | 5 (<5th) at 8 months | 6.5 (50th–75th) at 2 months | 5.1 (<3rd) at 6 months |
| 7.65 (<5th) at 13 months | 6.48 (<5th) at 4 months | 5.8 (<3rd) at 10 months | |
| 17.5 (25th) at 5 years | 11.5 (5th) at 3 years | 7 (<3rd) at 2 months | |
| 20.9 (10th) at 8 years | 17.6 (5th) at 6.5 years | 8 (<3rd) at 3.5 years | |
| Follow-up height, cm | 107 (25th) at 5 years | 80 (<5th) at 2 years | 64.5 (<3rd) at 6 months |
| 125.5 (30th) at 8 years | 109.5 (<5th) at 6.5 years | 84 (<3rd) at 3.5 years | |
| Follow-up OFC, cm | 40.5 (<3rd) at 8 months | 48 (10th) at 2 years | 41 (5th) at 6 months |
| 51 (25th) at 8 years | 51 (10th) at 6.5 years | 46 (<3rd) at 3.5 years | |
| Dysmorphic facial features | |||
| Deep set eyes | + | + | + |
| Low set ears | + | + | + |
| Simple helices | + | + | + |
| Small nose | +/− | + | + |
| Small mouth | +/− | + | + |
| Flat face | + | + | + |
| Microcephaly | + | + | + |
| Developmental delay and intellectual disabilities | |||
| Sitting at age | 4 years | 2.5 years | 2.5 years |
| Walking at age | 6.5 years with ataxic gait | 3.5 years | − |
| Speaking at age | Few separate words at 7 years | Few separate words at 4.5 years | − |
| Others | Unable to stand for long periods | Unable to stand for long periods | Crawling only |
| Others | |||
| Age at onset of infections | At birth and resolved after 2 years | At birth and resolved after 2 years | − |
| Neurological findings | Hypotonia, convulsion, epilepsy | Hypotonia | Hypotonia |
| Early constipation | + | + | + |
| Joint laxity | + | + | + |
| Partial syndactyly | + | + | + |
| Brachydactyly | + | + | + |
| Sandal gap | + | + | + |
| Hypopigmentation of skin | + | + | +++ |
| Blond hair | +/− | +/− | +++ |
| Involuntary papillary movement | + | − | + |
| Atrial septal defects | + | + | + |
NA, not available; OFC, occipitofrontal circumference.
aAge percentiles according to WHO Child Growth Standards.
Mapping and deep sequencing identifies a recessive splice site mutation in DCPS gene
To find the causative allele for this apparently novel syndrome, we performed identical by descent homozygosity mapping following single-nucleotide polymorphism (SNP) genotyping of nine individuals consisting of the four unaffected parents and their five children. Only one identical-by-descent (IBD) locus consisting of 5.9 Mb on chromosome 11q was found to be homozygous and shared in the three affected cases but not in parents or unaffected sibs (Fig. 1C). Whole-exome sequencing of proband (III:1), followed by bioinformatics filtering, identified 33 protein-changing variants with population minor allele frequencies <1%. Only one variant was located in the candidate Chr. 11 q24.2 locus (Supplementary Material, Table S1). This private SNP resulted from the transition of a thymine into cytosine in the first splice donor site (c.201+2T>C) of intron 1 of the DCPS gene. This mutation disrupts a restriction site for endonuclease Kpn1. Sanger sequencing and Kpn1 restriction digest confirmed complete segregation with the disease. The unaffected sib (III:7) is homozygous non-carrier, unaffected sib (III:8) and parents are heterozygous carriers, and the three propositi are homozygous mutant. This nucleotide is evolutionarily conserved in all DCPS homologues and is part of the canonical GU sequence required for the recruitment of the major spliceosome. Our mapping and sequencing results suggest that this potentially novel congenital syndrome may be caused by a pathogenic recessive splice site mutation in DCPS, encoding the scavenger decapping enzyme.
The DCPS mutation abrogates transcription of canonical, but not all, DCPS isoforms
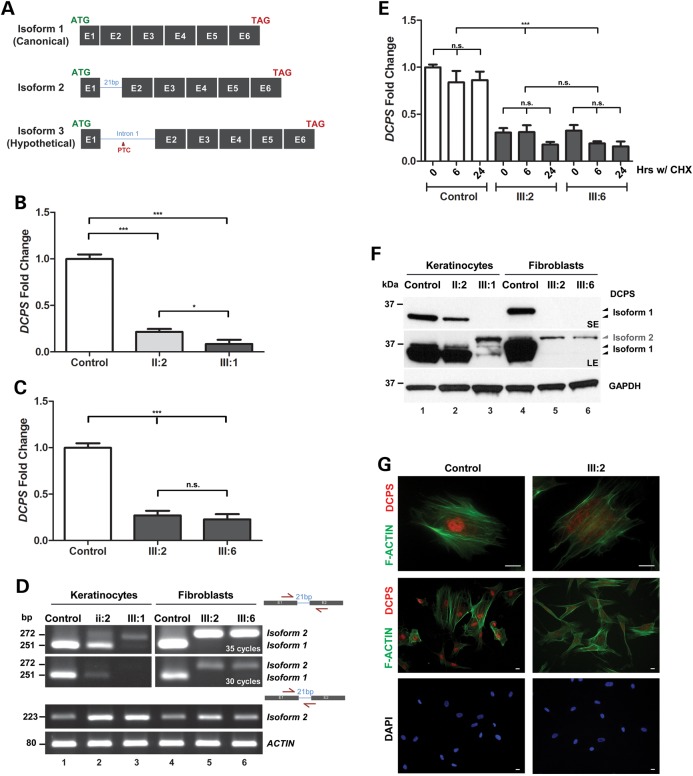
A single spliced DCPS transcript is reported in public databases and comprises all six annotated exons (Fig. 1D). The presence of this homozygous donor splice site mutation is likely to give rise to different transcript isoforms. Relative to the canonical transcript referred to as isoform 1, a putative second isoform 2 may arise due to the presence of a second cryptic splice donor sequence 19 nucleotides downstream of exon 1 (Figs 1D and 2A) which would result in the in-frame insertion of 7 amino acids (p.K67_V68insAPGGNPE). In the event that this second splice site was not recognized by the spliceosome, a hypothetical isoform 3 may be transcribed until the in-frame stop codon located 40 nucleotides downstream of exon1 (Figs 1D and 2A).
Figure 2.
Patient cells are devoid of major DCPS transcript/protein but minor isoform remains. (A) Schematic diagram showing the three different possible DCPS transcript isoforms. Isoform 1 represents the canonical transcript. Isoform 2 is a newly identified splice variant transcript predicted to encode a DCPS protein with additional 7 amino acids in-frame. Isoform 3 remains a hypothetical transcript which could not be documented. (B) QPCR detects reduced DPCS transcript levels, normalized to HPRT1, in mutant or carrier keratinocytes relative to control cells. Student t-tests were performed ***P < 0.0001 and *P < 0.01. (C) QPCR detects reduced DPCS transcript levels, normalized to HPRT1, in mutant primary fibroblasts relative to control cells. Student t-tests were performed ***P < 0.0001 and *P < 0.01. (D) RTPCR using primer sets designed to distinguish isoforms 1, 2 and 3 were performed on control and patients' cells. Sanger sequencing confirmed the identity of transcript isoform 2 as a normal cryptic splice form present in both control and patients' cells. (E) DCPS transcript isoform 3 cannot be detected in patient fibroblasts after blocking the NMD pathway with cycloheximide (CHX) treatment. (F) Western blot against DCPS detects different DCPS protein isoforms. Top panel shows a short exposure with loss of major DCPS isoform 1 in mutant cells relative to control or carrier cells. Under long exposure, minor isoform 2 becomes apparent (middle panel). Bottom panel shows equal loading across samples. (G) Immunofluorescent staining of patient-derived dermal primary fibroblasts showing loss of DCPS staining (red) in nucleus relative to control cells. F-ACTIN (green) is stained with phalloidin, scale bar: 40 μm.
Using primary cells derived from skin biopsies taken from each affected patients and one unaffected carrier (II-2), we performed a series of cellular assays to determine the pathogenicity of this splice site DCPS mutation. Quantitative PCR performed on dermal keratinocytes detected reduced levels of DCPS transcripts in heterozygous and homozygous mutant cells relative to control cells. The differences observed were consistent with the DCPS genotype (Fig. 2B). Likewise, QPCR performed on dermal fibroblasts confirmed the significant reduction of endogenous DCPS levels in both patients III:2 and III:6 relative to control cells which were taken from an age-and gender-matched healthy Jordanian (Fig. 2C). To identify what are the DCPS transcripts present in the patients' cells, primer sets were designed to distinguish isoforms 1, 2 and 3 (Fig. 2A and D and Supplementary Material, Table S2). Sanger sequencing of all amplicons derived from patients' and control cDNA revealed the presence of the major canonical isoform 1 only in control cells but not in mutant cells. Unexpectedly, isoform 2 was seen in all genotypes (Fig. 2D). This indicates that the second cryptic splice site can be recognized in both cell types even when the canonical donor splice site of exon1 is not mutated. While it represents a minor form in control cells, isoform 2 is the sole transcript seen in patients' cells (Fig. 2D). Isoform 3 (Fig. 2D and Data not shown) could not be amplified, but we reasoned that this may be due to the action of nonsense-mediated decay (NMD) resulting from the inclusion of a premature termination codon (PTC) located in intron 1. To test this hypothesis, patients' and control fibroblasts were treated with cycloheximide (CHX), a translation inhibitor that blocks the NMD pathway (14). Levels of DCPS transcripts monitored by QPCR did not change, suggesting that isoform 3 may not exist (Fig. 2E).
We next assessed the consequences of having two DCPS isoforms on the synthesis of the DCPS enzyme in patients' cells. By western blotting using a rabbit polyclonal anti-DCPS antibody, DCPS corresponding to the canonical isoform 1 was readily detected in control keratinocytes and fibroblasts but was entirely absent in mutant cells (Fig. 2F). A slight change in its apparent size was seen between the two control cell types which may be explained by DCPS-specific post-translational modifications such as acetylation (15–17). The father's cells (II:2) exhibited nearly half the amount of DCPS isoform 1 relative to control cells (Fig. 2F). A longer exposure of the same blots revealed trace amounts of a slightly larger DCPS isoform likely representing the 7 amino-acid in-frame insertion translated from isoform 2 (Fig. 2F). Mass spectrometry performed on cell lysates of patient III:6 confirmed that it was indeed DCPS (data not shown). These results suggest that the discovered splice site mutation completely abolished the translation of the canonical and most abundant DCPS isoform 1. However, we find that the minor isoform 2 is still produced resulting in the synthesis of a larger DCPS enzyme which differs only by one residue (p.V68A) between control and mutant cells. We conclude that the splicing machinery can occasionally skip the canonical donor site and splice at the newly identified cryptic site, albeit at a much lower frequency. By immunofluorescence, we found that in primary fibroblasts, staining for DCPS in the nucleus was lost in mutant cells relative to control cells (Fig. 2G). Taken together, these results suggest that this rare splice site mutation in DCPS causes aberrant splicing resulting in the complete absence of the major DCPS isoform 1, leaving trace amounts of a novel DCPS isoform 2 carrying a homozygous (p.V68A) missense mutation.
Patients' cells lack detectable DCPS enzymatic activities
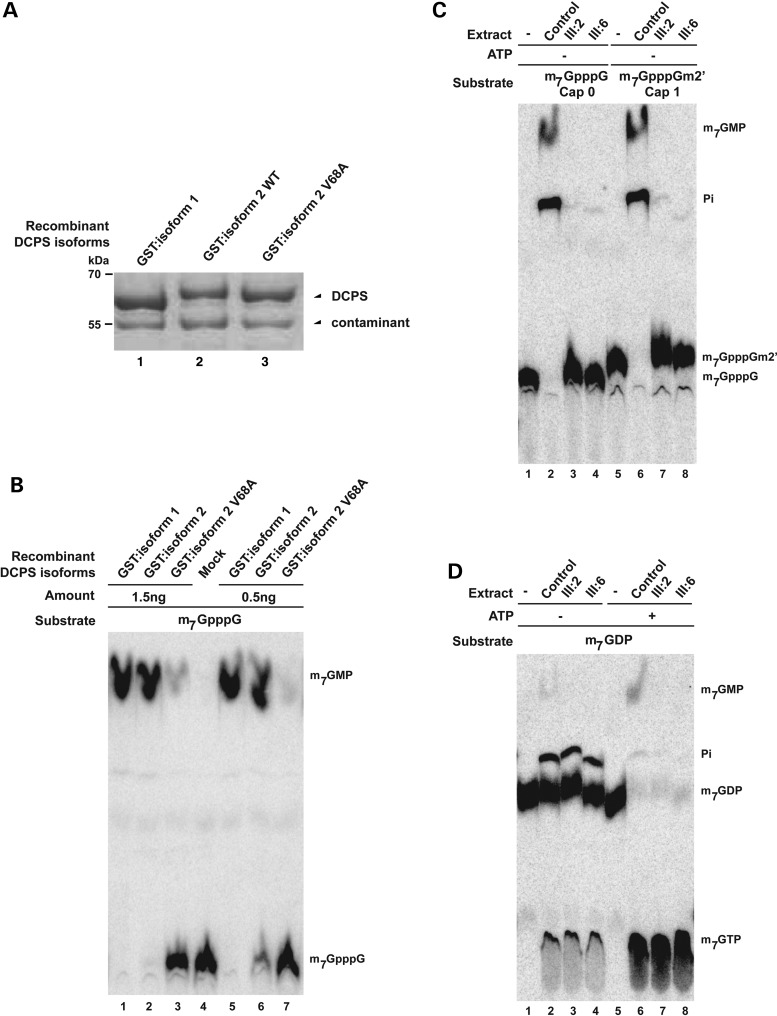
To determine if the minor DCPS isoform 2, either wild-type and p.V68A mutant form, have any biological function, we compared their enzymatic activities relative to that of the major DCPS isoform 1. To this end, we overexpressed and purified the recombinant N-terminally GST tagged isoforms from Escherichia coli (Fig. 3A). The mutant p.V68A isoform 2 of DCPS displayed a strongly reduced in vitro activity towards its substrate compared with that of canonical DCPS isoform 1 (Fig. 3B). The wild-type isoform 2 was only slightly more active than the mutant isoform 2 and clearly less active than isoform 1 (Fig. 3B).
Figure 3.
Patients' cells have no detectable enzymatic DCPS activity. (A) Protein profiles of purified recombinant GST:DCPS isoform 1 and GST:DCPS isoform 2 and isoform 2 V68A on SDS–PAGE. The contaminant migrating below GST-DCPS serves as a loading control. Positions of migration of molecular weight markers are indicated on the left side. (B) Enzymatic assay demonstrates the reduced activity of the DCPS when 7 amino acids is inserted between exon 1 and exon 2 of DCPS protein, especially in the V68A mutant. (C) Enzymatic assay demonstrates the loss of DCPS activity in patient's cells using m7GpppG and m7GpppGm2′ substrates. Purified substrates are presented in lanes 1 and 5. (D) In vitro assays demonstrate that the absence of DCPS-dependent conversion of m7GTP in extracts of cells from two affected individuals. m7GTP is efficiently formed from m7GDP in the presence of ATP (lanes 5–8), demonstrating that all extracts are similarly active. Trace of ATP in extract explains the residual formation of m7GTP in the absence of added ATP (lanes 1–4). The purified substrate (+/− ATP) is presented in lanes 1 and 5.
To obtain definitive evidence that this allele abrogates endogenous DCPS enzymatic activity, we next analyzed the fate of exogenous cap molecules incubated in extracts from patients or control cells. Extract from control cells hydrolyzed radioactively labeled m7GpppG, producing m7GMP (Fig. 3C, lane 2) which was further processed into inorganic phosphate as previously reported (9). In contrast, m7GpppG incubated in lysates obtained from two affected individuals remained stable (Fig. 3C, lanes 3 and 4) indicative of an absence of detectable DCPS enzymatic activity. Similar results were obtained using radiolabeled m7GpppGm2′ (Fig. 3C, lanes 7–8) which is the natural cap of human mRNAs (1). These results demonstrate that natural cap derivatives generated by the 3′–5′ mRNA decay mechanism is a substrate of DCPS and that it cannot be catabolized in the extract of fibroblasts from affected patients.
In the presence of ATP, incubation of m7GDP with cell extracts containing functional DCP2 results in efficient conversion into m7GTP (Fig. 3D, lanes 6–8). Secondary to its known function of degrading the free cap m7GpppN, DCPS is also known to cleave m7GTP into m7GMP (11,18,19). While the control extract were capable of cleaving m7GTP into m7GMP, such observations were absent in extracts taken from affected individuals (Fig. 3D, compare lane 6 with 7 and 8). This result demonstrates that all extracts are functional being able to phosphorylate m7GDP in m7GTP. However, patient extracts are specifically defective in DCPS activity that cleaves m7GTP into m7GMP. Altogether, these biochemical results demonstrate that cells from affected patients lack detectable DCPS enzymatic activity resulting in defective processing of cap derivatives generated by the 3′-5′ and 5′-3′ RNA decay mechanisms.
Discussion
In this report, we delineate a novel clinical entity which we suggest to refer to as Al-Raqad syndrome (ARS). ARS is a recessive congenital disorder with anomalies in multiple organs. Its most salient phenotypes comprise severe growth delay, neurological defects, cognitive impairment, skeletal and cardiac anomalies. Its genetic etiology can be attributed to homozygous loss-of-function alleles in the DCPS gene (MIM608183). The accompanying report by I. Ahmed et al. (2015) independently confirmed that a distinct homozygous splice mutation in intron 4 (c.636+1G>A) or a compound heterozygous mutation in DCPS also cause neurological deficits in three affected individuals of Pakistani origin. We note that the patient's phenotypes differ in severity between the two families, possibly reflecting on the hypomorphic nature of the Pakistani allele. A similar situation exists in mice where a Dcps mouse knockout is reported to be embryonic lethal (20) while a mutant Dcps mouse line, referenced at the Mouse Genome Informatics (MGI:1916555) is viable but phenotypic information is unavailable. No DCPS mutants have been reported for Drosophila or zebrafish but in Caenorhabditis elegans, knockdown of its homologue dcs-1 increased spontaneous mutation rate favoring the accumulation of genetic lesions (21). Recent findings indicate that a second enzyme FHIT (MIM601153) is also working in the same mRNA scavenging pathway (11). Partial redundancy between DCPS and FHIT might limit the phenotypic consequences of a DCPS loss-of-function mutation. Given the implication of FHIT in cancer (22) and the fact that dcs-1 increases genomic instability in C. elegans (21), it will be of interest to test whether inactivation of both DCPS and FHIT results in higher mutation rates and/or of cancer incidence.
At the molecular level, the absence of DCPS will result in accumulation of cap derivatives generated by mRNA decay. These by-products may impair splicing, mRNA nuclear export and/or translation (2), thus impacting on cell function. It is possible that neurons have less capacity to eliminate these by-products and/or are more sensitive to the long-term consequences of the cellular damage that they produce. Moreover, m7GTP, generated by conversion of m7GDP produced by the 5′-3′ mRNA decay mechanism, could affect GTP-dependent processes, in particular some affecting neurons, such as tubulin polymerization and/or signal transduction by GTPases. The presence of m7GTP may also lead to the synthesis of mRNA containing modified bases. Interestingly, accumulation of m7G in mRNA was noticed in cells from patients suffering from Huntington's disease (23) connecting the presence of this abnormal base in nucleic acid with neuronal degeneration leading to motor and cognitive troubles. DCPS activity has also been associated with the regulation of mRNA degradation by XRN1 (24,25) and the function of the SMN protein (26). In particular, inhibition of DCPS activity was reported to improve the motor function of mouse models of SMA (27,28). It is possible that alteration of DCPS in a context were SMN is fully functional in all tissues negatively impact on the balance of DCPS/SMN activities with detrimental consequences on neuronal functions.
Materials and Methods
Patients and clinical assessments
The affected children were initially diagnosed at Queen Rania Paediatric Hospital in Amman (Jordan) by Dr Mohammad Al-Raqad. Saliva samples were collected from nine members of these kindred and genomic DNAs were extracted using Oragene OG-500 kit. Skin biopsies were obtained from the three affected patients (III:1, III:2 and III:6) and the unaffected father (II-2). Parents gave their informed consent and the study was approved by the local ethic commissions in Jordan and Singapore.
Genotyping and homozygosity mapping
The four unaffected parents and their five offspring were genotyped using Illumina HumanCoreExome-12v1 BeadChips following manufacturer's instructions. Call rates were above 99%, gender and relationship were verified using Illumina GenomeStudio software. Identical-By-Descent (IBD) mapping was performed by searching for shared regions that are homozygous in the three affected individuals using custom programs written in Mathematica (Wofram Research, Inc.). Allowing 1% error rate, all homozygous regions that were >1 Mb were examined. Candidate regions were further refined by exclusion of common homozygous segments with any unaffected family members. A single shared homozygous region totaling 5.9 Mb was found on chromosome 11: 125 850 139–131 777 294 (hg19).
Whole-exome sequencing
Whole exome sequencing of patient III:1 was performed on the Illumina HiSeq2000 platform according to the manufacturer's condition to get 100 bp paired-end reads. The sequence reads were aligned to the human genome reference UCSC version hg19 (build 37). After the duplicated reads were removed, Genome Analysis Toolkit (GATK, v1.1) (29) was used to re-align indels and improve alignment accuracy, followed by variant calling across the RefSeq protein coding exons and flanking exon-intron junctions. An average coverage of ×64 was achieved across the exome with 86% of these bases covered at >×9. A total of 19 100 small nucleotide variants and 870 indels were identified. Of these, only 33 homozygous variants were protein-changing variants with population minor allele frequencies <1%.
Mutation analysis
Direct Sanger sequencing of genomic DNA flanking the DCPS mutation was performed according to its sequence found on GenBank and Ensembl databases. The primer sequences for DCPS (NM_014026) mutation screening are given in Supplementary Material, Table S2. Sequence analysis was done with the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA), and products were run on a 3730 DNA Analyzer (Applied Biosystems).
Cell culture
Primary patient skin fibroblasts were cultivated in high glucose DMEM (Lonza) supplemented with 10% fetal bovin serum (FBS) (Lonza), ×1 penicillin/streptomycin and 2 mm l-glutamine. Primary dermal keratinocytes were grown in DermaLife®K (Life Line Cell Tech). Media was replaced every other day. Fibroblasts were treated with media supplemented with 100 μg/ml cyclohexidine (CHX) and samples were collected at 0, 6 and 24 h.
Quantitative PCR
Total RNA was isolated from patient's fibroblasts and keratinocytes using the Trizol (Invitrogen) followed by RNeasy Mini Kit (Qiagen). One microgram of RNA was reverse transcribed using Iscript™ cDNA Synthesis Kit (Bio-Rad) and transcript levels were determined using the ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems). Gene expression was normalized to the housekeeping gene HPRT1 or ACTB. Primer sequences can be found in Supplementary Material, Table S2.
Statistical analysis
Each experiment was repeated at least three times and data were expressed as mean ± SD. Differences among treatments were analyzed by Student's t-test. Significant differences were considered as those with a P-value of ***<0.0001, **<0.001, *<0.01.
Western blot
Cells were lysed in RIPA extraction buffer supplemented with 1 mm dithiothreitol (DTT) and 1X Protease Inhibitor Cocktail (Roche). Protein concentrations of cleared lysates were measured using the BCA assay before equal amount of protein were loaded on precast 10% SDS–polyacrylamide gels (Bio-Rad). Transferred PVDF membranes were blotted using antibodies against: 1:4000 mouse anti-GAPDH (Santa-Cruz; sc-47724) and polyclonal rabbit anti-DCPS (Sigma; HPA039632). Secondary anti-mouse-HRP and anti-rabbit-HRP were used at 1:4000 dilution before visualization on X-ray film with SuperSignal West Dura Chemiluminescent Substrate (ThermoScientific).
Immunofluoresence analysis
Primary cells were grown on a Nunc™ Lab-Tek™ 8-well glass chamber slide and fixed for 15 min in ice cold 4% (w/v) paraformaldehyde. Permeabilization using 0.6% (v/v) Triton-X in 1× PBS was performed for 15 min then incubated with 1:200 rabbit anti-DCPS (Sigma; HPA039632) overnight at 4°C in 4% (v/v) FBS diluted with 1× PBS. For visualization, secondary antibody conjugated to Alexa Fluor 594 (Molecular probes) was applied. Counter staining for nuclei and F-ACTIN are performed using 1× Hoeschst stain and 1:50 phalloidin-488, respectively. Images were captured using the MetaMorph software and a stereomicroscope M205 FA equipped with an ICD camera from Leica.
Protein expression and purification
Plasmid pBS2498 was used for expression of GST-DCPS-His6 (18). The DCPS 7 amino acids insertion was introduced using the QuickChange Mutagenesis kit (Agilent). Oligonucleotides OBS6783–OBS6784 were used for the DCPS isoform 2 generating pBS5281 and OBS6789-OBS6790 for the DCPS isoform 2 V68A mutant yielding pBS5282. Primer sequences can be found in Supplementary Material, Table S2. Recombinant proteins were expressed in E. coli BL21 (codon +) strain, grown in 200 ml of auto induction media terrific broth (Formedium™), and purified on Glutathione Sepharose 4B beads essentially following the supplier recommendation (GE Healthcare). Briefly, cell pellets were dissolved in 10 ml PBS 1× and, after cell break, clarified lysates were applied to 100 μl (bed volume) of beads. After 1 h incubation at 4°C with mild shaking, beads were washed three times with 10 ml PBS 1× and proteins eluted with 200 μl of reduced glutathione 10 mm.
Preparation of cell extracts for DCPS assays
To prepare extracts, primary fibroblast cells from two affected patients and an unaffected sibling were grown to confluence in two 10-cm diameter plates. Cells were washed twice with cold PBS and resuspended in 500 µl buffer A (10 mm HEPES–KOH pH 7.6, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, 0.5 mm PMSF, 2 mm Benzamidine, 1 µm Leupeptin, 2 µm Pepstatin A, 4 µm Chymostatin, 2.6 µm Aprotinin). Cell were broken in a Dounce homogenizer and lysates were centrifuged at 3000g (Beckman JA-25.50 rotor) for 5 min at 4°C, 450 µl of the supernatant were collected, mixed with 50 µl of 2 m KCl and centrifuged at 20 000g in a table-top Eppendorf centrifuge for 2 min at 4°C. The supernatant was further centrifuged at 110 000g for 20 min at 4°C in TLA 120.2 rotor in Beckman TL-100 Ultracentrifuge. Supernatants were transferred in dialysis tubing (MWCO 12–14 000 Daltons) and dialyzed two times, 60 min each; against 2 l of cold buffer D (20 mm HEPES–KOH, pH 7.6, 50 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, 20% glycerol, 0.5 mm PMSF, 2 mm benzamidine). The resulting extracts were aliquoted and stored at −80°C. Protein concentrations of cleared lysates were measured using the Bradford assay.
In vitro DCPS enzymatic activity assays
Synthesis of RNA carrying a radioactively labeled cap and in vitro decapping reactions were performed as described (6). To obtain a cap1 structure (m7GpppGm2′), capped RNA was incubated with mRNA Cap 2′-O-Methyltransferase (NEB BioLabs) essentially according to the manufacturer's suggestions. To obtain radiolabeled m7GDP, m7GpppG or m7GpppGm2′, the appropriate cap-labeled RNAs were incubated with recombinant hDcp2 or nuclease P1 (Sigma-Aldrich), respectively. Reaction products were separated by Thin Layer Chromatography (TLC) in a developing solution made of 0.3 M LiCl, 1 M formic acid. m7GDP, m7GpppG or m7GpppGm2′ spots were scraped off from the TLC plates and eluted in decapping buffer (45 mm Tris–HCl, pH 8, 27 mm (NH4)2SO4, 49 mm MgAc) for 40 min at room temperature with constant shaking. Insoluble TLC material was removed by centrifugation. For activity assays, 1000 cpm of m7GDP, m7GpppG or m7GpppGm2′ were incubated with 1.5 µg of cellular extract for 60 min, or with the specified amount of recombinant protein for 10 min at 37°C in a 10 µl final volume. When indicated, ATP (final concentration 2 mm) was added to the reactions. Reactions were stopped by phenol–chloroform extraction. Five microliters of supernatants were analyzed by TLC using the conditions described earlier. Chromatograms were visualized with a PhosphorImager.
Supplementary Material
Funding
Work in BS laboratory was supported by the Ligue Contre le Cancer (Equipe Labellisée 2014), the Centre National pour la Recherche Scientifique, the CERBM-IGBMC, the project Decapping from Agence Nationale pour la Recherche (grant number ANR-11-BSV8-009-02 and ANR-10-LABX-0030-INRT managed under the program Investissements d'Avenir ANR-10-IDEX- 0002-02). Partial support of this study was provided by NIH/National Institute of Arthritis Musculoskeletal and Skin (NIAMS) NIH-P30-5P30AR057230 through the Center for Duchenne Muscular Dystrophy. This work was supported by grants from the Society in Science Branco Weiss Foundation and the EMBO Young Investigator to B.R. A Strategic Positioning Fund on Genetic Orphan Diseases (grant number SPF2012/005) from A*STAR, Singapore partly funded this work. Funding to pay the Open Access publication charges for this article was provided by A*STAR, Singapore.
Supplementary Material
Acknowledgements
We are indebted to the family for kindly partaking in this study. We are grateful to our group members for discussion and advice. V.T. and B.S. thank the IGBMC cell-culture service for support.
Conflict of Interest statement. None declared.
References
- 1.Ghosh A., Lima C.D. (2010) Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA, 1, 152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topisirovic I., Svitkin Y.V., Sonenberg N., Shatkin A.J. (2011) Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA, 2, 277–298. [DOI] [PubMed] [Google Scholar]
- 3.Cougot N., van Dijk E., Babajko S., Seraphin B. (2004) ‘Cap-tabolism’. Trends Biochem. Sci., 29, 436–444. [DOI] [PubMed] [Google Scholar]
- 4.Meyer S., Temme C., Wahle E. (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol., 39, 197–216. [DOI] [PubMed] [Google Scholar]
- 5.Lykke-Andersen J. (2002) Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol, 22, 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B. (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J., 21, 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Jiao X., Carr-Schmid A., Kiledjian M. (2002) The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl Acad. Sci. USA, 99, 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chlebowski A., Tomecki R., Lopez M.E., Seraphin B., Dziembowski A. (2010) Catalytic properties of the eukaryotic exosome. Adv. Exp. Med. Biol., 702, 63–78. [PubMed] [Google Scholar]
- 9.Wang Z., Kiledjian M. (2001) Functional link between the mammalian exosome and mRNA decapping. Cell, 107, 751–762. [DOI] [PubMed] [Google Scholar]
- 10.Liu H., Rodgers N.D., Jiao X., Kiledjian M. (2002) The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J., 21, 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taverniti V., Séraphin B. (2015) Elimination of cap structures generated by mRNA decay involves the new scavenger mRNA decapping enzyme Aph1/FHIT together with DcpS. Nucl. Acids Res., 43, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro E., Biezuner T., Linnarsson S. (2013) Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet., 14, 618–630. [DOI] [PubMed] [Google Scholar]
- 13.Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E., Jr (2003) Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res., 13, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- 15.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science, 325, 834–840. [DOI] [PubMed] [Google Scholar]
- 16.Gauci S., Helbig A.O., Slijper M., Krijgsveld J., Heck A.J., Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem, 81, 4493–4501. [DOI] [PubMed] [Google Scholar]
- 17.Van Damme P., Lasa M., Polevoda B., Gazquez C., Elosegui-Artola A., Kim D.S., De Juan-Pardo E., Demeyer K., Hole K., Larrea E., et al. (2012) N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl Acad. Sci. USA, 109, 12449–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk E., Le Hir H., Seraphin B. (2003) DcpS can act in the 5′-3′ mRNA decay pathway in addition to the 3′-5′ pathway. Proc. Natl Acad. Sci. USA, 100, 12081–12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wypijewska A., Bojarska E., Lukaszewicz M., Stepinski J., Jemielity J., Davis R.E., Darzynkiewicz E. (2012) 7-methylguanosine diphosphate (m(7)GDP) is not hydrolyzed but strongly bound by decapping scavenger (DcpS) enzymes and potently inhibits their activity. Biochemistry, 51, 8003–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bail S., Kiledjian M. (2008) DcpS, a general modulator of cap-binding protein-dependent processes? RNA Biol., 5, 216–219. [DOI] [PubMed] [Google Scholar]
- 21.Kwasnicka D.A., Krakowiak A., Thacker C., Brenner C., Vincent S.R. (2003) Coordinate expression of NADPH-dependent flavin reductase, Fre-1, and Hint-related 7meGMP-directed hydrolase, DCS-1. J. Biol. Chem., 278, 39051–39058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin J., St-Pierre M.V., Dufour J.F. (2011) Hit proteins, mitochondria and cancer. Biochim. Biophys. Acta, 1807, 626–632. [DOI] [PubMed] [Google Scholar]
- 23.Thomas B., Matson S., Chopra V., Sun L., Sharma S., Hersch S., Rosas H.D., Scherzer C., Ferrante R., Matson W. (2013) A novel method for detecting 7-methyl guanine reveals aberrant methylation levels in Huntington disease. Anal. Biochem., 436, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosse G.D., Ruegger S., Ow M.C., Vasquez-Rifo A., Rondeau E.L., Ambros V.R., Grosshans H., Simard M.J. (2013) The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell, 50, 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H., Kiledjian M. (2005) Scavenger decapping activity facilitates 5′ to 3′ mRNA decay. Mol. Cell Biol., 25, 9764–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh J., Salcius M., Liu S.W., Staker B.L., Mishra R., Thurmond J., Michaud G., Mattoon D.R., Printen J., Christensen J., et al. (2008) DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem. Biol., 3, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gogliotti R.G., Cardona H., Singh J., Bail S., Emery C., Kuntz N., Jorgensen M., Durens M., Xia B., Barlow C., et al. (2013) The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum. Mol. Genet., 22, 4084–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Meerbeke J.P., Gibbs R.M., Plasterer H.L., Miao W., Feng Z., Lin M.Y., Rucki A.A., Wee C.D., Xia B., Sharma S., et al. (2013) The DcpS inhibitor RG3039 improves motor function in SMA mice. Hum. Mol. Genet., 22, 4074–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. (2010) Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. The Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.