Abstract
Huntington's disease (HD) is a fatal neurodegenerative disease, caused by expansion of polyglutamine repeats in the Huntingtin gene, with longer expansions leading to earlier ages of onset. The HD iPSC Consortium has recently reported a new in vitro model of HD based on the generation of induced pluripotent stem cells (iPSCs) from HD patients and controls. The current study has furthered the disease in a dish model of HD by generating new non-integrating HD and control iPSC lines. Both HD and control iPSC lines can be efficiently differentiated into neurons/glia; however, the HD-derived cells maintained a significantly greater number of nestin-expressing neural progenitor cells compared with control cells. This cell population showed enhanced vulnerability to brain-derived neurotrophic factor (BDNF) withdrawal in the juvenile-onset HD (JHD) lines, which appeared to be CAG repeat-dependent and mediated by the loss of signaling from the TrkB receptor. It was postulated that this increased death following BDNF withdrawal may be due to glutamate toxicity, as the N-methyl-d-aspartate (NMDA) receptor subunit NR2B was up-regulated in the cultures. Indeed, blocking glutamate signaling, not just through the NMDA but also mGlu and AMPA/Kainate receptors, completely reversed the cell death phenotype. This study suggests that the pathogenesis of JHD may involve in part a population of ‘persistent’ neural progenitors that are selectively vulnerable to BDNF withdrawal. Similar results were seen in adult hippocampal-derived neural progenitors isolated from the BACHD model mouse. Together, these results provide important insight into HD mechanisms at early developmental time points, which may suggest novel approaches to HD therapeutics.
Introduction
Huntington's disease (HD) is a fatal autosomal-dominant neurodegenerative disorder affecting ∼6 per 100 000 with the exception of Asia at 0.4 per 100 000 (1) and results in debilitating behavioral changes and uncontrolled movements that worsen over time. HD is caused by expanded polyglutamine (CAG) repeats in the Huntingtin (HTT) gene (2). Above 35 repeats cause the disease, with the number of repeats being inversely correlated to the onset age (2–4). Approximately 9% of patients have more than 60 CAG repeats, which results in an onset before age 20 years and is hence termed juvenile HD (JHD), also known as Westphal variant. JHD is generally associated with a more severe progression and phenotype. It is still unknown why the toxic-expanded HTT protein, which is a ubiquitously expressed, is initially toxic to striatal medium spiny neurons (MSNs) and later to cortical projection neurons. Therefore, finding ways to slow disease progression have been difficult. Developing new models of HD that could elucidate the mechanisms of this devastating disorder would facilitate the development of a successful treatment.
Reduced levels of brain-derived neurotropic factor (BDNF) and/or increased levels of glutamate can lead to the death of striatal MSNs in HD [for review, see (5)]. BDNF is a pro-survival factor that acts through the TrkB receptor (TrkBR) or p75NTR receptor (6–8). MSNs show enhanced susceptibility to BDNF loss in HD animal models (9). Conversely, BDNF has been shown to protect against excitotoxic-mediated HD-MSN death in vitro (10–12), in HD mouse corticostriatal slices (13) and in vivo (14). Glutamate, the main excitatory neurotransmitter, regulates cell survival, proliferation, migration, differentiation and forebrain neurogenesis (15–21). In addition, glutamate can be a neurotoxin, with striatal MSNs showing heightened susceptibility to glutamate-induced cell death (22–25). Glutamate acts through two receptor subtypes (26). The ionotropic receptor subtype is further subdivided into N-methyl-d-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainite receptors. The metabotropic receptor subtype (mGluR1-8) is further subdivided into group 1–3 receptors.
The association of BDNF and glutamate with HD is well studied, and yet has not provided a successful treatment to date. Therefore, a further understanding of HD is still required and may be achieved using HD-based tissue culture and animal models (27,28). Currently generated human tissue culture models often use fibroblasts and lymphoblasts as a readily available cell source from HD patients. While these cells show some HD-related defects (29,30), they do not harbor the characteristics of neurons undergoing neurodegenerative stresses. In contrast, rodent-derived striatal cultures and rodent models of HD that overexpress an expanded HTT both exhibit some characteristic HD phenotypes. One example is the transgenic BACHD mouse model using a bacterial artificial chromosome (BAC) to express the full-length human HTT gene with exon 1 containing an expanded polyglutamine stretch (31,32). However, these rodent-based models are limited as disease manifestation and response to treatments are often different from human patients (33–35). Therefore, while current models have contributed much to the field, human striatal-like neurons derived from an HD genetic background may be a more relevant cell type and source for disease modeling.
By expressing four genes found in embryonic stem cells (ESCs) (36), adult human fibroblasts can be reprogrammed to a primitive state with the regained capacity to differentiate into any cell in the body (37–39). These cells, termed induced pluripotent stem cells (iPSCs), are almost indistinguishable from ESCs but, importantly, come from an adult source. As such, fibroblasts from patients with genetic-based diseases, like HD, can now be reprogrammed to develop a novel disease in a dish model (40). We have previously used integrating viral vectors to generate iPSC lines derived from a range of HD patient and control fibroblasts (41). The cultured cells derived from these iPSC lines showed quantifiable and reproducible CAG repeat-expansion-associated phenotypes with various stressors, including BDNF withdrawal or repeated exposure to glutamate (41).
Here, we report on iPSCs generated from HD patient-derived fibroblasts using a newer non-integrating technology. While both HD and control iPSCs can be differentiated over time, at 42 days of differentiation, the HD-derived cells maintained a significantly greater number of nestin-expressing neural progenitor cells (NPCs) compared with control cells. Similar findings were seen in adult hippocampal NPCs from BACHD mice. Surprisingly, these persistent nestin-expressing NPCs, rather than emerging new neurons, showed increased cell death following an acute BDNF withdrawal, possibly due to the loss of signaling through the TrkB receptor. Furthermore, it was demonstrated that the cell death phenotype is due to the presence of mutant HTT (mtHTT) and may be mediated by enhanced susceptibility to glutamate toxicity in the absence of BDNF. This is the first report linking the loss of BDNF signaling to glutamate toxicity in neural progenitors from human HD patients.
Results
HD iPSC-derived cultures contain more nestin-expressing cells after differentiation
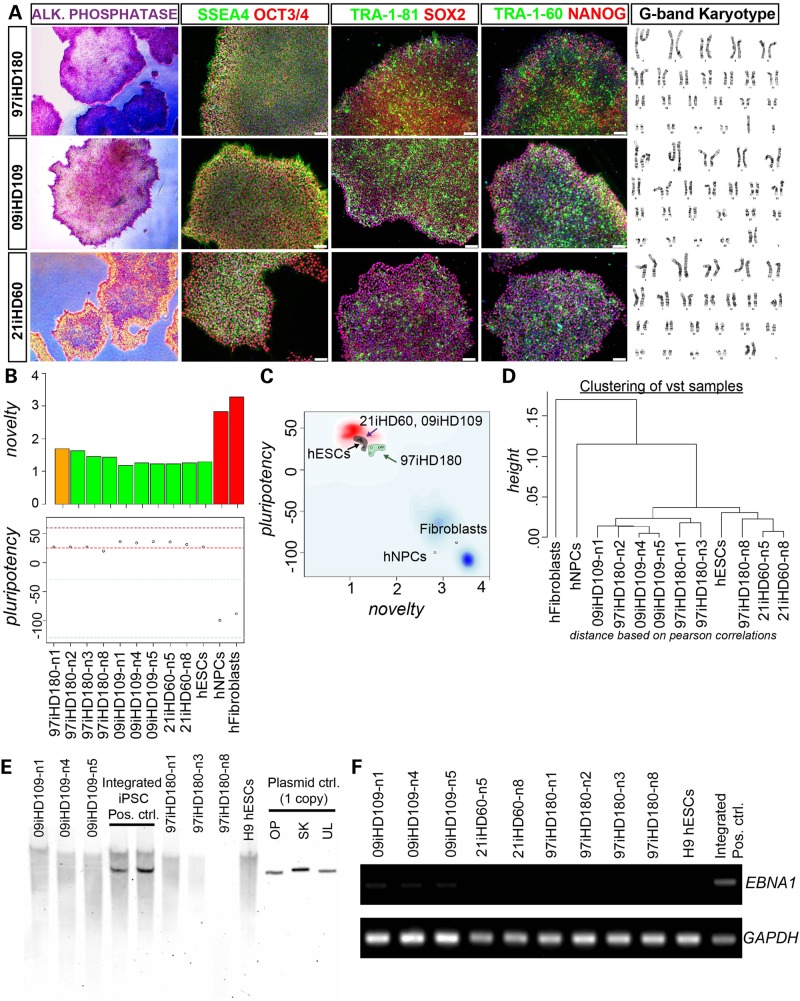
We previously generated HD and control iPSC lines using an integrating lentivirus to introduce pluripotency genes into fibroblasts (41). While these iPSC lines grew well in culture, remained karyotypically normal and could be differentiated into MSNs (41), there are drawbacks to using integrating viruses (42). Therefore, we have now implemented a non-integrating system to generate new iPSC lines from HD patients with 180, 109 and 60 CAGs and from control subjects with 33, 28 and 21 CAGs (Fig. 1 and Supplementary Material, Fig. S2). These iPSC lines were fully reprogrammed as demonstrated by staining for alkaline phosphatase and other pluripotency makers (Fig. 1A), passed the ‘PluriTest’ assessed by characterization of low ‘novelty’ and high ‘pluripotency’ gene expression (Fig. 1B and C) and grouped away from the original fibroblast source (Fig. 1D). Southern blotting (Fig. 1E) and genomic polymerase chain reaction (PCR) (Fig. 1F) analyses confirmed the absence of plasmid gene expression after several passages, confirming that there was no integration of the reprogramming plasmids. The iPSCs were also easy to grow, remained karyotypically normal over many passages (Fig. 1A and data not shown) and could be efficiently banked to provide resources for future experiments and collaborators.
Figure 1.
Generation and characterization of non-integrating HD iPSCs. (A) Bright-field images of the reprogrammed iPSC colonies from HD patient (containing 180, 109 and 60 CAG repeats) fibroblasts show a high nuclear-to-cytoplasmic ratio, are alkaline phosphatase positive and immunopositive for the surface antigens, SSEA4, TRA-1-60, TRA-1-81; and nuclear pluripotency markers OCT3/4, SOX2 and NANOG. Scale bar, 75 μm. All the HD-iPSC lines possessed a normal G-band karyotype as shown from representative karyotype spread images. (B) Novelty score graph: a score based in well-characterized pluripotent samples in the stem-cell model matrix. Samples are color-coded, green (pluripotent), red (not pluripotent or closer to somatic) and orange (need further evaluation in other pluripotency assays). All HD iPSCs (green and orange bars) were highly dissimilar to the non-pluripotent samples shown in red bars (fibroblasts and human NPCs) in the model matrix than a positive control human embryonic stem cell line (hESCs) pluripotent sample. The pluripotency score giving an indication if a sample contains a pluripotent signature above the red-dotted line shows that all HD-iPSCs and hESCs were pluripotent (43). (C) Chart combines pluripotency score on y-axis and novelty score on x-axis. The red and blue background hint to the empirical distribution of the pluripotent (red) and non-pluripotent (blue) samples. The HD-iPSCs were closer to the tested hESCs and other pluripotent stem cells (blue cloud) and diagonally opposite from differentiated (hNPCs) and somatic (fibroblast) cell types in the PluriTest test data set. (D) Hierarchical clustering of the samples based on PluriTest Gene Expression profile shows that HD-iPSCs clustered close to each other and away from hNPCs and fibroblasts. (E) Southern blot analysis of genomic DNA from clonal iPSC lines of 97iHD180 and 09iHD109 with a plasmid back-bone specific probe common to the reprogramming plasmids used shows lack of exogenous transgene-integration. Negative control: H9 hESC line. Positive controls: other integrating iPSC lines generated using the episomal plasmid technique showing plasmid-based gene integration. Technical control: a single copy of the plasmid DNA can be detected using this technique (OP = Oct4, shRNA-p53; SK = Sox2, Klf4; UL = L-Myc, Lin28). (F) Absence of episomal plasmid gene expression in HD iPSCs is also confirmed by genomic DNA PCR for reprogramming plasmids. OriP/EBNA1 reprogramming plasmids replicate extra-chromosomally once per cell cycle. Expression of EBNA1, a gene present on all reprogramming plasmids was monitored over multiple passages. Genomic DNA analyzed shown here eliminated the plasmids post-reprogramming, confirming that they are integration-free iPSC lines.
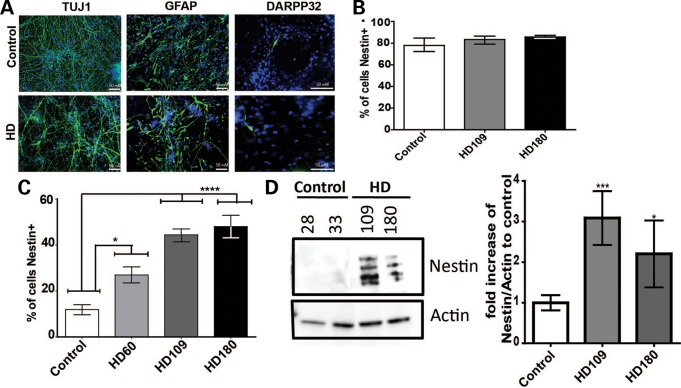
Upon differentiation, cultures consist of a mixed population of neural progenitors, astrocytes and neurons. We first used immunocytochemistry (ICC) to confirm that these non-integrating iPSCs could be differentiated in culture into nestin-expressing NPCs, glial fibrillary acidic protein (GFAP)-expressing astrocytes, TUJ1-expressing neurons and DARPP32-expressing cells (marker for MSN) (Fig. 2A). The quantification of cells expressing nestin at the early neural progenitor stage of differentiation showed that HD (180, 109 or 60 repeats) iPSC-derived cultures contained ∼80% nestin-expressing NPCs, which was comparable to control cultures (Fig. 2B). After 42 days of differentiation, both HD and control line cultures showed reduced levels of nestin-expressing cells as the neural progenitors were generating glia and neurons. However, at this time point, there were significantly more nestin+ cells in the HD cultures (∼40%) when compared with the control cultures (∼12%) (Fig. 2C). Western blotting at day 42 further confirmed an increased level of nestin protein in the HD lines (Fig. 2D). This was the only population observed to be significantly different between HD and control cultures, as quantitative reverse transcriptase–PCR (qRT–PCR) data showed that GFAP, MAP2 and DARPP-32 genes were not significantly different between HD and control cultures at this time point of differentiation (Supplementary Material, Fig. S2A–C). We next tested whether the HD iPSC differentiated cultures could have more nestin-expressing cells due to an overall increase in proliferation. Using Ki67 staining to examine proliferation levels revealed that there were similar numbers of Ki67-positive nuclei in the HD and control lines (Supplementary Material, Fig. S2D). Together, these results indicate that the HD-derived striatal-like cultures (180, 109 and 60 CAG repeats) accumulate NPCs that are not newly born cells, but rather that are a persistent cell population.
Figure 2.
There are significantly more HD iPSC-derived nestin expressing neural progenitors after differentiation into striatal-like cultures. (A) Striatal-like cultures derived from HD and control iPSCs express GFAP, DARPP32 and TUJ at day 42 of differentiation. (B) HD (180 or 109 repeats) iPSC-derived neural progenitors (at sphere stage in EGF/FGF containing media) had similar numbers of nestin-expressing cells as controls by ICC (one-way ANOVA). (C) HD iPSC-derived striatal cultures at day 42 of differentiation had significantly more nestin-expressing cells than controls by ICC (one-way ANOVA). (D) HD iPSC-derived striatal cultures at day 42 of differentiation had more nestin protein than controls by western blotting. Actin was used as an internal loading control. Graph represents average quantification of four separate western blots of independent differentiations, where ‘control’ is an average of the CTR33, CTR28 and CTR21 lines which were not significantly different from one another (*P < 0.05, *P < 0.001 Student's t-test).
JHD iPSC-derived cultures show increased susceptibility to BDNF withdrawal that is mediated through TrkBR signaling
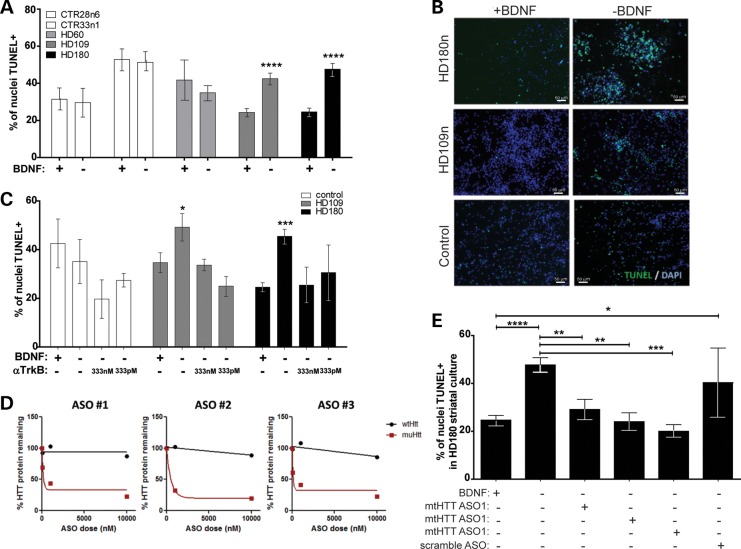
The previously generated integrating iPSC lines showed increased caspase 3/7 activity in the differentiated JHD lines upon BDNF withdrawal (41). To confirm this finding in our newly generated lines, a TUNEL assay was used to quantify the level of cell death following acute (24 h) BDNF withdrawal after 42 days of differentiation. Results showed that the percentage of TUNEL+ nuclei after BDNF withdrawal was significantly increased in the JHD cultures (180 and 109 repeats) but not in control cultures (33, 28 or 21 repeats) or in cultures with 60 CAG repeats (adult-onset HD) (Fig. 3A and B). To further understand the mechanism of action of BDNF withdrawal, we assessed whether the cell death in our JHD cultures was due to a loss of signaling through the TrkB receptor (TrkBR). HD cultures (180 or 109 repeats) were treated with a TrkBR agonist antibody during the 24 h BDNF withdrawal. We demonstrate in control iPSCs [CTR33i.8 line (41)] that this agonist activated the TrkB receptor, as phospho-TrkB was increased (Supplementary Material, Fig. S3A) along with the downstream signaling components phospho-AKT and phospho-ERK (Supplementary Material, Fig. S3B). Using the TrkBR antibody during the acute withdrawal successfully rescued the JHD cultures from increased cell death in the absence of BDNF (Fig. 3C). Taken together, these results confirm that JHD iPSC-derived striatal cultures are specifically susceptible to an acute BDNF withdrawal, which is likely mediated by the loss of signaling in the BDNF-TrkBR pathway.
Figure 3.
JHD iPSC-derived striatal-like cultures are specifically susceptible to the loss of BDNF-TrkBR signaling due to mtHTT expression. (A) There is a significant increase in TUNEL+ nuclei after an acute (24 h) BDNF withdrawal (+ versus −) in the JHD 180 CAG (black) and 109 CAG (grey) lines, while controls show no significant increase in cell death (white). (B) When a TrkBR agonist antibody was administered during an acute BDNF withdrawal, the cell death phenotype seen in the HD lines (grey and black) was reversed (*P < 0.05, **P < 0.01, unpaired Student's t-test when compared with -BDNF). (C) Representative images of TUNEL+ nuclei with or without a withdrawal of BDNF. (D) Three ASOs targeted toward an SNP in the mutant HTT are able to specifically knock-down HTT in a dose-dependent manner, as determined by western blotting and quantification. (E) After a 7-day pretreatment using ASOs to knock-down mtHTT, a significant decrease in cell death is seen in the HD 180 CAG line (black) exposed to an acute withdrawal of BDNF (one-way ANOVA).
Allele-specific antisense oligonucleotides can block the BDNF-withdrawal-induced cell death in JHD striatal cultures
We next used allele-specific antisense oligonucleotides (ASOs) in order to confirm that the cell death following BDNF withdrawal was truly an HD-dependent phenotype. These ASOs were designed to knock-down the expression from the mtHTT allele based on a unique single nucleotide polymorphism (SNP) found in the HD180 line. Three separate ASOs were tested by pre-treating the HD180 line for 7 days at 1 μm. mtHTT was shown to have an ∼60% knock-down, while the wild-type allele remained unaffected, showing that the ASOs were successfully optimized to be potent, highly allele selective and non-toxic to the cells (Fig. 3D). Next, the HD180 cultures were differentiated for 35 days and then treated for 7 days with the ASOs prior to the 24 h BDNF withdrawal. Treatment with mtHTT ASOs, but not a control ASO, reversed the increased percentage of cell death following acute BDNF withdrawal (Fig. 3E). These ASOs did not significantly affect the control lines (data not shown). These results confirm that the increased vulnerability to cell death in JHD striatal cultures is mtHTT-specific.
The ‘persistent’ HD nestin-expressing neural progenitor population is specifically susceptible to BDNF withdrawal
To assess which genes were changed upon BDNF withdrawal, qRT–PCR-based gene array that assessed 84 key genes thought to be involved in HD (Qiagen PAHS-123) was performed on differentiated striatal-like cultures before and after acute BDNF withdrawal. Results from the array showed that only the expression of Sox2, a transcription factor expressed in NPCs and previously implicated in HD (44), was altered with a trend toward increased expression in the HD180 line cultures followed by a trend toward decreased expression (P = 0.05558) after 24 h of BDNF withdrawal (data not shown). The fact that, from the panel of genes tested, only Sox2, gene expression changed following BDNF withdrawal suggested that neural progenitors may be the most affected cell type within the differentiated mixed cultures, which is very interesting given the enhanced population of neural progenitors in these cultures.
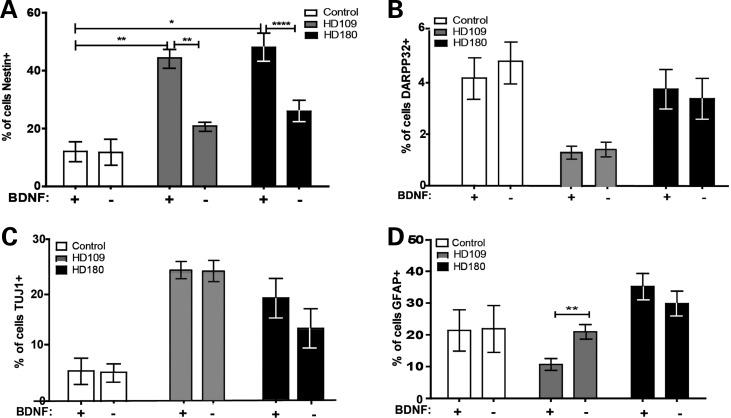
The selected array focused upon genes specifically thought to be altered in HD and did not examine neural markers other than Sox2. Therefore, we next performed ICC to pursue the initial array data showing that a neural progenitor marker was reduced after BDNF withdrawal. Interestingly, in the HD180 and 109 lines, the increase in nestin-expressing cells was almost completely lost following BDNF withdrawal (Fig. 4A and Supplementary Material, Fig. S4A), which corroborates the array result showing a loss of Sox2 expression. Together, this data set indicates that this originally increased population of neural progenitors may indeed be the population of cells susceptible to BDNF loss.
Figure 4.
An acute BDNF withdrawal significantly reduces the JHD neural progenitor population. (A) Upon acute BDNF withdrawal, this increase in nestin-expressing cells in the HD cultures was lost (Student's t-test). (B) A marker of MSNs DARPP32, (C) a pan-neural marker TUJ1 or (D) a glial marker GFAP were not decreased in HD cultures upon BDNF withdrawal.
To confirm that the BDNF withdrawal selectively reduced the accumulated HD neural progenitor population, ICC was performed to assess GFAP, TUJ1 and DARPP32-expressing cells. Cell quantification following BDNF withdrawal showed that DARPP32+ cells (Fig. 4B and Supplementary Material, Fig. S4B), neurons (Fig. 4C and Supplementary Material, Fig. S4C) and glia (Fig. 4D and Supplementary Material, Fig. S4D) were not reduced in either the HD180 line or control cultures. Unexpectedly, the HD109 line showed a significant increase in the percentage of GFAP+ cells after BDNF withdrawal, which may be an artifact of this particular line, as it was not replicated in the HD180 line. This may be explained partially because the HD109 line was derived from an earlier onset patient than the HD180 patient line (41), and therefore, this increase in astrocytes in culture after BDNF withdrawal may somehow be related to the increased gliosis seen in HD (45).
Nestin-expressing NPCs in the HD mouse brain are more abundant and specifically susceptible to BDNF withdrawal
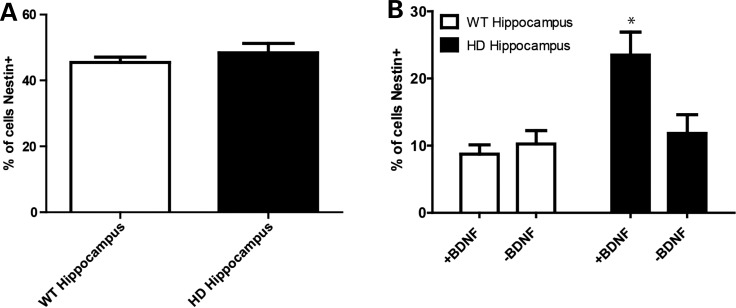
Demonstrating an increased number of nestin-expressing cells in the HD iPSC-derived striatal-like cultures is a novel and interesting in vitro developmental phenomenon. In order to expand this finding to adult HD, we next turned to HD mouse models, given that it is difficult to obtain adult human HD brains for culture derivation and given the reports that several transgenic HD mouse models have altered adult hippocampal neurogenesis (46–49). Neural progenitors were isolated from the hippocampus of BACHD (50) and control mice and maintained in an undifferentiated state in Epidermal Growth Factor (EGF)- and Fibroblast Growth Factor (FGF2)-containing media. Analyzing these undifferentiated cells by ICC showed that the undifferentiated control and HD hippocampal cultures had similar percentages of nestin-expressing cells (Fig. 5A and Supplementary Material, Fig. S4E). In contrast, when adult hippocampal progenitors were differentiated for 1 week in BDNF, the HD cultures had significantly more nestin-expressing cells (Fig. 5B and Supplementary Material, Fig. S4F). As such, differentiated cultures from both the human HD iPSCs and mouse BACHD hippocampus have an increased amount of persistent nestin-expressing neural progenitors in the presence of BDNF when compared with controls. Critically, these accumulated neural progenitors were then lost following BDNF withdrawal (Fig. 5B and Supplementary Material, Fig. S4F). Therefore, the ex vivo data mirror the in vitro cultures to replicate that the cell death induced by BDNF withdrawal appears to be specific to the nestin-expressing NPC population.
Figure 5.
HD mouse model hippocampal progenitors replicate the human HD iPSC-derived striatal-like culture findings. (A) Neural progenitors from the HD (BACHD) and control mouse hippocampus prior to the exposure of BDNF had similar percentages of nestin+ cells. (B) After 1 week in BDNF-containing media, the HD cultures had significantly more nestin+ cells and this population was lost after acute BDNF withdrawal.
The BDNF-withdrawal-induced death of JHD striatal cultures is due to increased response to glutamate
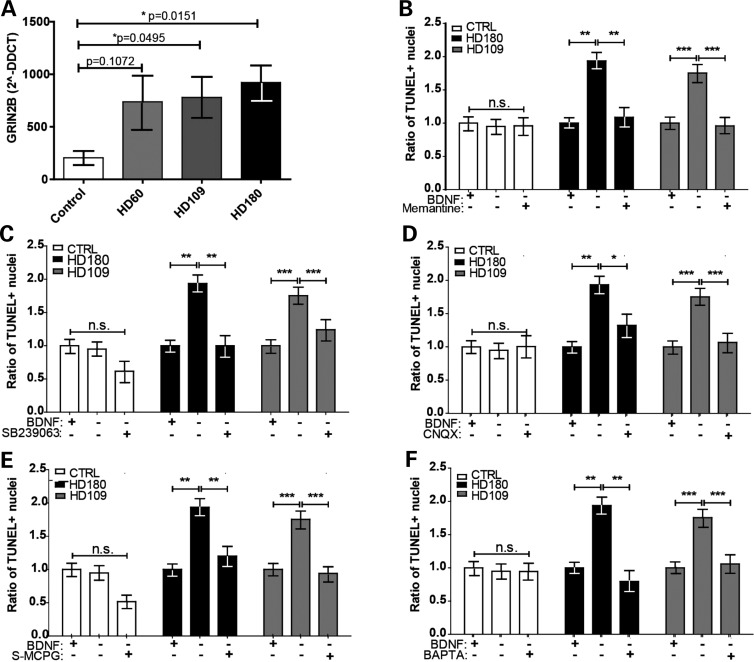
As integrating, and now non-integrating, JHD iPSC-derived striatal-like cultures have a pronounced cell death phenotype in response to BDNF withdrawal (41,51,52), we wanted to finally explore the mechanism of cell death. We have previously demonstrated that the integrating JHD iPSC-derived striatal cultures were susceptible to cell death upon repeated exposures to excess glutamate (41). A connection between reduced BDNF signaling and toxicity associated with the glutamate receptor NMDAR has been implicated in HD, although this was for neuronal rather than neural progenitor toxicity (10–12). In iPSC-derived striatal-like cultures maintained in basal media containing 0.05 mm glutamate (l-glutamic acid) and 0.25 mm glycine, the basal gene expression of the NMDAR subunits GRIN2A and GRIN2B was not significantly different between JHD and control lines (data not shown). However, upon acute BDNF withdrawal, the HD iPSC-derived cultures showed up-regulated levels of the GRIN2B subunit compared with control-derived cultures (Fig. 6A). This GRIN2B up-regulation and associated glutamate toxicity has also been reported in HD model mice (13). Therefore, we postulated that blocking the NMDAR during the acute BDNF withdrawal may prevent cell death.
Figure 6.
In the absence of BDNF-TrkBR signaling, the HD iPSC-derived striatal-like neural progenitors die due to glutamate toxicity. (A) Acute BDNF withdrawal in the HD iPSC-derived cultures (grey and black) up-regulated the GRIN2B subunit of the NMDA receptor (*P < 0.05, Student's t-test). (B) The cell death phenotype in the HD cultures (grey and black) is reversed using memantine, a small molecule blocker of NMDARs (*P < 0.05, **P < 0.001, ***P < 0.0001; one-way ANOVA). (C) The cell death phenotype in the HD cultures (grey and black) is reversed using SB23906, a small molecule blocker of cleaved p38 (*P < 0.05, **P < 0.001, ***P < 0.0001; one-way ANOVA). (D) The cell death phenotype in the HD cultures (grey and black) is reversed using CNQX, a small molecule blocker of the AMPA/Kainate receptors (*P < 0.05, **P < 0.001, ***P < 0.0001; one-way ANOVA). (E) The cell death phenotype in the HD cultures (grey and black) is reversed using s-MCPG, a small molecule blocker of group I/II mGluRs (*P < 0.05, **P < 0.001, ***P < 0.0001; one-way ANOVA). (F) The cell death phenotype in the HD cultures (grey and black) is reversed using BAPTA, a small molecule blocker of calcium influx (*P < 0.05, **P < 0.001, ***P < 0.0001; one-way ANOVA).
We assessed a small molecule antagonist of the NMDAR, memantine (53), which has been shown to increase striatal volume and ameliorate neurological and behavioral pathology in an HD model animal (54). Results showed that adding a low dose of memantine to the media during the 24 h BDNF withdrawal period was able to prevent the cell death in the HD cultures (Fig. 6B and Supplementary Material, Fig. S4G). Given this promising result, we assessed a small molecule inhibitor of the downstream signaling cascade within the NMDAR pathway. Indeed, adding SB239063, an inhibitor of p38 MAP kinase activation (55), prevented cell death following BDNF withdrawal in the JHD lines (Fig. 6C and Supplementary Material, Fig. S4G). Additional glutamate receptors were next examined in order to determine if this glutamate toxicity in the absence of BDNF was due solely to signaling through the NMDA receptor. Blocking the AMPA/Kainate glutamate receptors with CNQX (Fig. 6D and Supplementary Material, Fig. S4G) or the mGlu group I/II glutamate receptor with s-MCPG (Fig. 6E and Supplementary Material, Fig. S4G) also prevented cell death following BDNF withdrawal. Finally, as the ion channels of these glutamate receptors allow for an influx of calcium, and as we have previously demonstrated that the HD iPSC-derived cultures had increased percentage of cells with calcium dyshomeostasis after repeated pulsing with glutamate (41), we assessed the role of calcium influx in BDNF-withdrawal-induced cell death. Adding the calcium chelator, BAPTA, resulted in a reversal of the cell death phenotype seen in the JHD cultures (Fig. 6F and Supplementary Material, Fig. S4G). Collectively, these results implicate the NMDAR pathway along with calcium influx in the observed cell death.
Taken together, these data demonstrate that in the absence of BDNF-TrkBR signaling, JHD iPSC-derived NPCs in striatal-like cultures are left susceptible to all glutamate-mediated toxicity and that the ensuing cell death is potentially mediated through calcium influx.
Discussion
Using integrating iPSC lines from HD patients, the HD iPSC Consortium has previously shown a CAG-repeat-expansion-associated increase in cleaved caspases in response to BDNF withdrawal (41). However, the specific subpopulation of cells within the striatal-like cultures that was most vulnerable remained undefined. The current report has made non-integrating iPSC lines from HD patients with juvenile onset (180 and 109 CAG repeats) and with adult onset (60 CAG repeats) and from control subjects (33, 28, 21 CAG repeats). Using these new lines, this study has now defined a subpopulation of nestin-expressing neural progenitors that may be most vulnerable in juvenile-onset HD, and also has provided a mechanism for the enhanced cell death phenotype through glutamate toxicity. Collectively, this provides novel critical information to further understand and ultimately treat HD.
Persistent NPCs in HD striatal-like cultures from iPSCs
Most studies analyze cell death from onset to later time points in disease progression and show the specific death of striatal MSNs. However, the development of iPSC technology provides the opportunity to assess cells at early stages, as the diseased cells are taken back in time to a pluripotent state and then pushed forward again. These young pluripotent cells provided an ideal disease in a dish model of HD to address cells during the earliest stage of disease. As HD is a monogenic heterozygous dominant disorder that correlates with CAG repeat number, HD carriers can be identified even in utero. Novel studies of neurogenesis using HD iPSCs could allow us in the future to identify novel pathways as targets for therapeutic intervention at the earliest stages, decades before disease onset.
iPSC lines were differentiated into striatal-like cultures through the addition of defined developmental factors (40). Upon further characterization, however, it was discovered that while the percentages of neural progenitors between HD and control lines were similar upon starting neural differentiation, there was an increased proportion of nestin+ progenitors at 42 days of differentiation in the HD cells. Nestin is expressed in development throughout many tissues, including the central nervous system, and is then down-regulated in adulthood. Exceptions are the continued production of nestin-expressing NPCs in the hippocampus and subventricular zone that produce new neurons/glia throughout life (56) and the up-regulation of nestin in adult neurons/glia with traumatic injury (57) or neurodegenerative disease (58). It is believed that nestin reactivation may promote cell survival after injury or disease (57,58). The increased population of nestin cells that we observed in human cells from JHD patients has been previously reported in differentiated ESC cultures from an HD mouse model (54). This phenotype is most likely not selective to culture conditions, as there are several demonstrations of increased neurogenesis at early stages of HD in mouse models. Indeed, an HD mouse model (46) shows developmental abnormalities including delayed acquisition of early striatal cytoarchitecture, aberrant MSN neurogenesis (including overexpression of the pluripotency markers Sox2 and NANOG), delayed cell cycle exit between E13.5 and 15.5 and an expansion of intermediate progenitors. Therefore, the developmental abnormalities might compromise neuronal homeostasis and thereby leave MSNs more susceptible to stressors later in life (44). Altered levels of neural progenitors in HD are not only seen in embryonic development but also in neurogenic regions of the adult brain, with several HD rodent models showing changes in adult neurogenesis in the hippocampus (46,49,59). Our study using the BACHD mouse model of HD recapitulates these previous rodent studies by also demonstrating a defect of neural maturation. Importantly, for the first time, we show, using the iPSC technology, that these changes in neural cell development are not only seen in HD animal models but also transcend to human iPSC-derived neural precursors from HD patients.
The TrkBR mediates susceptibility to BDNF withdrawal in JHD iPSC-derived cultures and is CAG repeat-dependent
BDNF signaling through the TrkBR has been associated with neurogenesis and the survival of striatal MSNs in HD (60,61). Indeed, an in vitro model of HD using an immortalized rodent striatal progenitor cell line over-expressing an expanded CAG has shown that BDNF had a significant protective effect on glutamate toxicity (13). We, in collaboration with the HD iPSC Consortium, have previously reported a CAG-repeat-expansion-associated increase in caspase 3/7 activity in response to a withdrawal of BDNF in striatal-like cultures derived from HD iPSCs with 180 repeats (41). This has also been reported in a similar assay using iPSCs with 72 repeats (51,52). This phenotype was not significant in the 60 CAG repeat lines, indicating that this may be only seen in the lines derived from patients with the more severe JHD. Here, we present that these previously published findings are not just CAG-repeat-expansion-associated, but are dependent on specifically mtHTT expression, as the cell death phenotype was completely abolished when the JHD iPSC-derived striatal-like cultures were pre-treated with ASOs that specifically targeted an SNP in the mtHTT allele. We now confirmed that this cell death is mediated through the TrkB receptor as a TrkBR antibody successfully rescued the HD cultures from increased cell death. Therefore, it appears that the TrkB receptor signaling protects against both glutamate toxicity in rodent HD neural cells and BDNF withdrawal in human HD neural cells. The data from not only rodent cells but also human cells derived from HD patients help solidify the importance of BDNF signaling in HD. While effects of BDNF withdrawal were now clear, it was still unknown which particular cell type was vulnerable to BDNF withdrawal.
Persistent neural progenitors are selectively susceptible to BDNF withdrawal
In our initial publication (40), we did not directly assess which cellular subtype was undergoing cell death upon BDNF withdrawal. While striatal MSNs are the primary cell type affected upon HD onset, to our knowledge, no one has looked at what cell type is most affected at the earliest stages of HD. Using the iPSC technology to bring HD patient cells back in time allowed us to now address this. To our surprise, BDNF withdrawal from the differentiated cultures resulted in no significant differences in the numbers of DARPP32+ cells or even pan-neuronal cells in HD iPSC lines compared with control lines. Instead, the cell subpopulation found to be most vulnerable to BDNF withdrawal during early stages of the disease was the newly discovered persistent population of nestin-expressing NPCs.
Acute BDNF withdrawal studies done at an earlier time point (day 14) of differentiation did not display this same cell death phenotype (data not shown). This indicates that these same ‘persistent’ neural progenitors in the HD cultures at day 42 have an increased need for BDNF-TrkBR signaling for survival, demonstrated by the loss of the ‘persistent’ HD progenitor population upon an acute BDNF withdrawal. Even though the HD60 iPSC cultures have significantly more nestin-expressing NPCs, they do not demonstrate a significant cell death phenotype upon BDNF withdrawal. While this NPC death is most prominently seen in the JHD iPSC cultures, this requirement for BDNF by the NPCs also occurs in adult neurogenesis BACHD cultures as well. It is known that BDNF is a regulator of adult neurogenesis (62–64), which is decreased in multiple HD mouse models as well as human post-mortem brains (49,65,66). In HD model mice, viral delivery of BDNF was able to induce striatal neurogenesis (67), which were able to become yielded functional MSNs, delayed motor symptoms and increased survival. Taken together, this implicates the importance of BDNF in neurogenesis, especially in the presence of mtHTT. However, we are not discrediting that over a longer timeframe than 24 h, the lack of BDNF signaling would likely cause other adult cell types to die in the HD cultures, as has been reported in both HD animals and post-mortem patients (5). Additionally, in one of the lines (HD109), there was a significant increase in the number of GFAP+ cells post-BDNF withdrawal. We do not believe that this phenotype is CAG-repeat-associated, as the same increase was not seen in the HD180 lines. It is known in HD that while disease onset and progression correlates with CAG repeat number, there is also a significant effect of the genetic background of the patient. We hypothesize that, if we looked at more lines in the long repeat range, we may see some with increased GFAP and some without, but importantly all would have increased nestin. In addition, the populations of nestin-positive and other cell types and their sensitivity to BDNF may be different at later time points in culture, just as the developmental delay in HD striatum becomes normalized over time (44), potentially leaving neurons with increased susceptibility to cell death.
Until now, it has been possible to only assess human cells from patients after disease onset through to late stages of the disease. By using the new iPSC technology to model diseases in a dish at a very early stage, we were able to discover a novel subpopulation of cells that appeared most vulnerable to BDNF withdrawal.
BDNF-withdrawal-induced death is due to increased response to glutamate
BDNF has been shown to protect against glutamate-mediated toxicity in HD mouse striatal neurons (13). This mouse finding has been extended to human cells now, by showing that the JHD striatal-like cultures demonstrate cell death upon BDNF withdrawal and upon glutamate treatment (40). Collectively, these studies suggested that the JHD iPSC-derived progenitors might be susceptible to BDNF-withdrawal-induced death due to an increased response to glutamate. This was supported by the significant up-regulation of NMDA receptor subunits in the JHD iPSC-derived cultures that has been previously associated with toxicity in HD neurons (23–25). It has also previously been shown that mtHTT can activate NMDARs due to a decreased ability to bind the modular adaptor protein PSD95 (68). PSD95 is known to bind the NR2 subunit of NMDA, which recruits a tyrosine kinase that in turn phosphorylates NMDAR, thereby increasing its currents (69). Our data concur with this theory of increased activation of NMDARs in the presence of mtHTT.
While previous mouse data suggested that toxicity in the absence of BDNF signaling was mediated by the NMDA receptor, we found that toxicity was also occurring through other glutamate receptors, including the AMPA receptor and the mGluRs (group I/II). This makes sense, given that glutamate receptors are differentially expressed during development, with the AMPA/Kainate receptors being expressed first followed by the NMDA receptors (17,70). Therefore, it is logical that the HD iPSC cultures, including the aberrant Nestin+ population, would all have glutamate receptors. Additionally, it has been demonstrated that mtHTT interacts with group I mGluRs (71), but there are conflicting reports regarding mGluR involvement in HD (24,72–76). For example, mGluR5 antagonist treatment increases survival of an HD model mouse (76). Conversely, an mGluR group I agonist was shown to protect cortical neuronal cultures from NMDAR-mediated excitotoxicity (74,77–79).
Blocking one group of glutamate receptors at a time was sufficient to prevent toxicity, implying that the responsive signaling of all of the glutamate receptors in JHD progenitor cells play a role in the toxicity in the absence of BDNF-TrkB signaling. This could occur through calcium influx or release of intracellular calcium stores. There are a multitude of reasons for calcium influx into a cell, including both apoptosis and activation of glutamate receptors along with many other biological processes. Therefore BAPTA chelation of the calcium could simply be anti-apoptotic by preventing calcium influx, and is not necessarily correlated with the glutamate signaling. However, BAPTA also blocked the BDNF withdrawal phenotype. The BAPTA data support previous studies in both MSNs from HD model mice (80) and HEK293 cells (81), where mtHTT leads to potentiation of NR1/NR2B NMDA receptor activity, leading to an overloading influx of calcium. Together, these data point to the dual nature of BDNF in HD promoting neurogenesis as well as protecting against glutamate-mediated toxicity. As BDNF is down-regulated in HD (7,8), this finding is especially pertinent to further therapeutic investigations.
Relevance for JHD and possibly adult-onset HD
These studies have many implications for the biology of HD/JHD. First, this report indicates that certain populations of neural cells may be vulnerable to toxicity at ages much earlier than originally anticipated. This may predispose to later susceptibility to stressors in the CNS (44). This also may lead to later disturbances in adult neurogenesis. Histological and carbon-14 dating approaches have indicated that post-natally generated striatal neurons are preferentially depleted in patients with HD (82). Our data in mice raise the possibility that the decreased BDNF expression seen in HD (83) would leave these progenitors susceptible to glutamate toxicity. While these results in HD iPSCs implicate issues in neurodevelopment in HD, it has been demonstrated that administration of BDNF or TrkB agonists in adult HD model mice has beneficial effects, including less severe signs of neurological dysfunction, neuroprotection and extended lifespan (84–86). However, further investigations of HD neurogenesis and altered neurodevelopment may lead to novel therapeutic approaches for HD.
Materials and Methods
Generation of human non-integrating iPSCs using episomal plasmids
Unaffected and apparently healthy control (GM05400, 03814 and 02183) and HD (GM09197) human fibroblast cell lines was obtained from the Coriell Institute for Medical Research, under their consent and privacy guidelines. The parental fibroblast for 109 CAG repeat HD line were obtained from John's Hopkins University under Dr Russell Margolis' institutional review board (IRB) protocol # NA_00018358. All procedures were performed in accordance with the IRB guidelines at the Cedars-Sinai Medical Center under the auspice IRB-SCRO Protocols Pro00028429, Pro00021505 and Pro00032834. Upon iPSC generation at Cedars Sinai, lines were renamed CS00iCTR-21nXX, CS14iCTR-nXX, CS83iCTR-33nXX, CS97iHD-180nXX and CS109iHD-109nXX to reflect, (i) last two digits of parental lines identifier, (ii) control or HD line, (iii) CAG repeat number and (iv) XX is the clone number (87,88); see Supplementary Material, Table S1 for cell sources and naming convention. Fibroblasts were reprogrammed into non-integrating and virus-free iPSC lines using the Amaxa Human Dermal Fibroblast Nucleofector Kit to express episomal plasmids with six factors: OCT4, SOX2, KLF4, L-MYC, LIN28 and p53 shRNA (Addgene #27080, #27078, #27077, Cambridge, MA, USA) (89). This method has a significant advantage over viral transduction, because exogenously introduced genes do not integrate and are instead expressed episomally in a transient fashion. Briefly, fibroblasts (0.8 × 106 cells per nucleofection) were harvested, centrifuged at 200g for 5 min, re-suspended carefully in Nucleofector® Solution (VPD-1001, Lonza, Basel, Switzerland) and the U-023 program was applied. All cultures were maintained under norm-oxygen conditions (5% O2) during reprogramming, which further enhances the efficiency of iPSC generation. The media were kept on for 48 h and gradually changed to chemically defined mTeSR®1 medium (Stemcell Technologies, Cat# 05850, Vancouver, BC, Canada) containing small molecules to enhance reprogramming efficiency. The small molecules used were (i) sodium butyrate (0.5 mm) (Sigma-Aldrich, Saint Louis, MO, USA), (ii) glycogen synthase kinase 3β inhibitor of the Wnt/β-catenin signaling pathway (CHIR99021, 3 µm) (Selleck, Cat# CT99021, Houston, TX, USA), (iii) MEK pathway inhibitor (PD 0325901, 0.5 µm) (Tocris, Cat# 4192, Bristol, UK), (iv) selective inhibitor of TGF-β type I receptor ALK5 kinase, type I activin/nodal receptor ALK4 and type I nodal receptor ALK7 (A 83-01, 0.5 µm) (Tocris, Cat #2939). Individual iPSC colonies with ESC/iPSC-like morphology appeared between days 25 and 32 and those with best morphology were mechanically isolated, transferred onto 12-well plates fresh Matrigel™ Matrix (Corning, Cat # 354230, Corning, NY, USA) and maintained in mTeSR®1 medium (Stemcell Technologies, Cat# 05850). The iPSC clones were expanded for further analysis, then scaled up for cryopreservation.
Human iPSC characterization
Human iPSCs were rigorously characterized at the Cedars-Sinai iPSC Core using several assays. G-Band karyotyping (see below) ensured normal a karyotype, and genomic DNA PCR confirmed the absence of episomal plasmid genes, as previously described (88–90). Pluripotency was assessed by immunostaining with surface and nuclear pluripotency markers for subsequent flow cytometry quantification [>80% (R&D, Cat# FAB1435A, 1:10 dilution, Minneapolis, MN, USA) and Oct3/4 (BD Pharmingen, Cat# 560186, 1:5 dilution, San Jose, CA, USA) double positivity], by qRT–PCR of endogenous pluripotency genes, and by gene-chip and bioinformatics-based PluriTest assays (Muller; Illumina HT12v4, San Diego, CA, USA). Spontaneous embryoid body differentiation confirmed the capacity to form all germ layers.
Karyotype
Spheres were incubated in Colcemid (100 ng/ml; Life Technologies, Grand Island, NY, USA) for 30 min at 37°C and then dissociated using TrypLE (Life Technologies) for 10 min. They were then washed in phosphate-buffered saline (PBS) and incubated at 37°C in 5 ml hypotonic solution [33.5 mM KCl, 9.7 mM Na Citrate (Sigma-Aldrich)] for 30 min. The cells were centrifuged for 2.5 min at 300 RCF and resuspended in fixative (methanol:acetic acid, 3:1) at room temperature for 5 min. This was repeated twice before cells were resuspended in 500 µl of fixative solution and submitted to the Cedars-Sinai Clinical Cytogenetics Core for G-Band karyotyping.
iPSC striatal-like differentiations
iPSC colonies (Cedars-Sinai Medical Center IRB 21505) grown on Matrigel in TeSR media (feeder-free) were scraped into EGF/FGF2 (100 ng/ml each, Peprotech, Rocky Hill, NJ, USA) containing media (70:30 DMEM:F12 plus 2%B27; Life Technologies, Carlsbad, CA, USA) and grown as floating neural progenitor spheres for at least 9 passages (91). 8-10 spheres were then plated on poly-ornithine (PLO)/laminin-coated coverslips and differentiated in neural induction media (NIM; DMEM:F12 with 1% N2) for 5 days. BDNF (20 ng/ml; Peprotech 450-02, Rocky Hill, NJ, USA) was then added for 2 days. For the next 21 days, cells were differentiated in 20 ng/ml BDNF, rhShh (200 ng/ml; R&D 1845-SH) and Dkk1 (100 ng/ml; R&D 1096-DK-010) to promote a rostral forebrain fate. Cells were then matured in 20 ng/ml BDNF, dibutyryl cyclic AMP (dbcAMP, 0.5 mm; Sigma-Aldrich D0260) and valproic acid (VPA, 0.5 mm; Sigma-Aldrich P4546) for the rest of the remaining differentiation. Medium was half-changed twice per week or as needed.
BDNF withdrawal
Cells were differentiated toward a striatal fate for 42 days and then transferred into basic NIM or NIM plus 100 ng/ml BDNF for 24 h. dbcAMP and VPA were removed from the medium in the above experiments because they increase endogenous BDNF transcription but are not critical for cell survival (92). At the end of treatment, cells were fixed in 4% paraformaldehyde (PFA).
BDNF withdrawal rescue experiments
Cells were treated with mtHTT-targeted or control (targeted to a miRNA uninvolved with HD) ASO (1 μm) for 1 week prior to BDNF withdrawal (d42). ASOs were directly supplemented into the media and taken up passively by the cells (data not shown). ASOs were replaced during the tri-weekly half media changes and supplemented into the +/− BDNF media during the acute withdrawal. For all other rescue experiments [TrkB antibody 38B8 (333 nm or 333 pm); a kind gift from Pfizer, memantine (10 μm); Sigma-Aldrich, SB239063 (200 nm); Sigma-Aldrich, BAPTA (50 μm); Tocris, CNQX (10 μm); Tocris, or s-MCPG (250 μm); Abcam (Cambridge, MA, USA)], compounds were supplemented into the media without BDNF for the full 24 h withdrawal. Three separate lines were used for controls (CS83iCTR33n1, CS00iCTR21n1 and CS14iCTR28n6), two clones were used for the HD180 lines (CS97iHD180n1 and CS97iHD180n3) and three clones were used for HD109 lines (CS09iHD109n1, CS09iHD109n4, CS09HD109n5). As all lines/clones within a grouping (control, HD180 or HD109) were not significantly different from one another, they were compiled. Statistical analyses were examined using one-way ANOVA with Bonferroni's post hoc on the raw data after being compiled (the only exception being the scramble ASO, which was analyzed using an unpaired Student's t-test). For graphing purposes, the raw data are expressed as a ratio of the sample without withdrawal (+BDNF). All experiments were performed at least three separate times.
Allele-specific ASOs
HTT haplotypes of iPSC lines were determined as previously described (93). Briefly, DNA isolated from iPSCs or parental fibroblast lines was genotyped at 96 SNPs using a GoldenGate assay on the Illumina BeadArray platform and SNP calls phased to allele using PHASE 2.0. All lines were determined to be heterozygous at SNP rs7685686_A. Allele-selective ASOs used in this study were the most allele-selective ASOs developed to this target from previous studies (94,95). All ASOs employed the ‘gapmer’ design, i.e. 7–9 oligodeoxynucleotides in the central region flanked at both the 3′ and the 5′ ends with mixtures of 2′-O-methoxyethyl (MOE) and 2′-O-constrained ethyl (cEt)-modified nucleosides. All of the internucleosidic bonds were phosphorothioate to improve nuclease resistance and enhance cellular uptake (94). Oligonucleotides were synthesized as described previously (94). Oligo sequences are described in Supplementary Material, Table S2. All experiments were performed at least three separate times.
Western blot analyses
ASO-treated cell pellets were lysed by incubation in 20–40 µl SDP buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% Igepal, 0.1% SDS, 40 mm β-glycerophosphate, 10 mm NaF, 1× Roche complete protease inhibitor, 1 mm sodium orthovanadate and 800 mm PMSF) for 30 min on ice with vortexing every 5 min. Lysates were cleared by centrifugation (15 min, 20 000g, 4°C) and protein concentration was determined using the Biorad (Irvine, CA, USA) DC assay according to the manufacturer's specifications. 40 μg of total protein were then resolved on 10% low-bisacrylamide gels as previously described (96) and immunoblotted for HTT (1:1000, Millipore MAB2166, Darmstadt, Germany) and non-muscle Myosin IIA loading control (1:2000, Abcam ab24762). Proteins were detected with IR dye 800CW goat anti-mouse (Rockland 610-131-007, Limerick, PA, USA) and AlexaFluor 680 goat anti-rabbit (Molecular Probes A21076, Grand Island, NY, USA)-labeled secondary antibodies and the LiCor Odyssey Infrared Imaging system. Allelic HTT protein intensities were normalized to myosin loading control and then to the same allele for untreated cell pellets on the same membrane.
For the nestin blots, cells were differentiated for 42 days, washed in PBS, pelleted at 300 g, resuspended in JLB buffer (50 mm Tris–HCl, pH 9, 150 mm NaCl, 10% glycerol, 0.1% Roche PIC, 20 mm NaH2(PO)4, 25 mm NaF, 2 mm EDTA, 1% Triton X-100) and sonicated. Protein concentrations were determined using Bradford assay (Promega, Madison, WI, USA) according to manufacturer's directions. 50 µg of protein was loaded into Mini-PROTEAN® TGX Precast gels (Bio-Rad Laboratories, Inc.,), separated with 90 V for ∼1.5 h, electro-transferred onto PVDF membrane (BioRad Turbo transfer) in a semi-dry transfer system for 7 min at 1.3 A. The membrane was blocked in 6% dry non-fat milk in Tris-buffered saline plus 0.1% Tween 20 (Sigma-Aldrich) for 1 h at RT and then exposed to primary antibody against nestin (1:500, mouse, Millipore, Billerica, MA, USA) and β-actin (1:500, rabbit, Sigma-Aldrich) to ensure equal protein loading in block for 1.5 h at RT. Anti-mouse and anti-rabbit secondary antibodies conjugated to peroxidase (1:10 000, Jackson Labs, Bar Harbor, ME, USA) was applied in block for 1 h at RT, followed by exposure to chemiluminescence kit (Super Signal West Femto Maximum Sensitivity Substrate, Thermo Scientific, Waltham, MA, USA). Western blot was performed four separate times.
HD iPSCs differentiated in 6-well plates for 41–42 days were transferred to 1:1 DMEM:F12 plus 1% N2 for 48 h. Then, the cells were treated with either 300 nm 38B8 TrkB-activating antibodies or 20 ng/ml BDNF for 15 min and lysed with RIPA buffer-containing protease and phosphatase inhibitor cocktails (Thermo Scientific) on wet ice for western blotting. Protein amount was determined by BCA assay (Life Technologies) and equal protein amounts were loaded per well. Antibodies were used to phospho-TrkB (Abcamb), to total TrkB (Cell Signaling) and its regulated protein kinases Akt (Cell Signaling) and Erk-1,2 (Cell Signaling). Equal loading was verified by WB on β-actin (Sigma-Aldrich).
Immunocytochemistry
Cells were fixed in 4% PFA at room temperature, rinsed with PBS and permeabilized with 5% normal goat and/or donkey serum containing 0.2% Triton X-100 for 30 min at room temperature. Cells were then labeled with primary antibodies [Nestin 1:500 (Millipore mab5326 for HD iPSCs and Millipore mab353 for mouse NPCs), Tuj 1:1000 (Sigma-Aldrich T8660), DARPP32 1:400 (Cell Signaling 19A3, Beverly, MA, USA), GFAP 1:1000 (DAKO, Carpinteria, CA, USA)] for 60 min at room temperature or overnight at 4°C, and then the appropriate fluorescently tagged secondary antibodies for 60 min at room temperaturefollowed by hoescht nuclear dye. Cells were counted using stereological software (stereoinvestigator). Effects of BDNF withdrawal were assessed by quantifying TUNEL incorporation per total Hoechst-stained nuclei, according to the manufacturer's recommendations (Promega DeadEndFluorometric TUNEL System, G3250) using metamorph software. All experiments were performed at least three separate times.
RNA analyses
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and digested by DNase I (RQ1 DNase, Promega). Complementary DNA was generated from 0.5 mg total RNA using a Reverse Transcript System (Promega A3500). SYBR green primers were chosen from previously published manuscripts (41,97) or Primer Bank (http://pga.mgh.harvard.edu/primerbank/) and ordered from IDT. Primer sequences include: (GFAP: forward 5′-ATCGAGAAGGTTCGCTTCCTG-3′, forward 5′-TGTTGGCGGTGAGTTGATCG-3′; MAP2: forward 5′-AAAGCTGATGAGGGCAAGAA-3′, forward 5′-GGCCCCTGAATAAATTCCAT-3′; DARPP32: forward 5′-CCTGAAGGTCATCAGGCAGT-3′, forward 5′-GGTCTTCCACTTGGTCCTCA-3′; GRIN2B: forward 5′-TTCCGTAATGCTCAACATCATGG-3′, reverse 5′-TGCTGCGGATCTTGTTTACAAA-3′; GAPDH: forward 5′-GAGTCAACGGATTTGGTCGT-3′, reverse 5′-TTGATTTTGGAGGGATCTCG-3′) cDNA was generated from RNA samples using a reverse transcription kit (Promega A3500). qPCR was performed using a 384-well format (BioRad). GAPDH was included as an endogenous control. Data were analyzed using the 2−ΔΔCT method against H9 ESC colony RNA. Statistical analyses were performed using an unpaired Student's t-test. All experiments were performed at least three separate times.
For the Qiagen RT2 Profiler PCR Array for human HD (PAHS-123Z), RNA was isolated using the RNeasy Mini Kit, cDNA was generated using 0.5 μg of RNA and the RT2 First-strand kit (Qiagen), then cDNA was mixed with the RT2 SYBR green mastermix an aliquoted across the 384-well array plate. Data were analyzed using the 2−ΔΔCT method to the control with BDNF. Ratios were then determined by the log10 of −BDNF:+BDNF of the same line. Statistical analyses between HD and control were performed between the two groups using an unpaired one-tailed Student's t-test.
Mouse neural progenitors
BACHD model mice (Jackson labs, a kind gift from X. William Yang) were housed in accordance with the Guide of Care and Use of Experimental Animals of the American Council on Animal Care and the Institutional Animal Care and Use Committee of the Cedars-Sinai Medical Center (IRB/SCRO Pro00024899, IACUC 3648). The dentate gyrus of the hippocampus was isolated from euthanized adult BACHD model (n = 6, pooled), and non-HD control (n = 9, pooled), mice and dissociated to single cells with TrypLE for 5 min at 37°C. The cells were then placed into a T25 flask and allowed to aggregate into spheres in growth media (70:30 DMEM:F12, 1% anti-biotic/anti-mycotic (Life Technologies), 2% B27, 20 ng/ml EGF, 20 ng/ml FGF and 5 μg/ml Heparin). Cells were manually chopped weekly (98). For the experiments described, cells were dissociated using TrypLE, counted and plated on PLO/laminin-coated coverslips (30 000 cells/coverslip). Cells were either grown for 1 week in growth media or 1 week in 1:1 DMEM:F12, 1% N2, 1% PSA and 20 ng/ml BDNF. After 1 week, half of the cells grown in the presence of BDNF were given a complete media change with 1:1 DMEM:F12, 1% N2 and 1% PSA for 24 h (BDNF withdrawal) before fixation for ICC. All experiments were performed at least three separate times.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (NIH NS078370) and the Board of Governors Regenerative Medicine Institute at Cedars-Sinai Medical Center.
Supplementary Material
Acknowledgements
The authors would like to thank X. William Yang at UCLA for the generous gift of the BACHD model mice and Dr Soshana Svendsen for the critical review of the manuscript. The authors thank Pfizer for providing the monoclonal antibody for our studies.
Conflict of Interest statement. None declared.
References
- 1.Pringsheim T., Wiltshire K., Day L., Dykeman J., Steeves T., Jette N. (2012) The incidence and prevalence of Huntington's disease: a systematic review and meta-analysis. Mov. Disord., 27, 1083–1091. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 3.Duyao M., Ambrose C., Myers R., Novelletto A., Persichetti F., Frontali M., Folstein S., Ross C., Franz M., Abbott M., et al. (1993) Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat. Genet., 4, 387–392. [DOI] [PubMed] [Google Scholar]
- 4.Andrew S.E., Goldberg Y.P., Kremer B., Telenius H., Theilmann J., Adam S., Starr E., Squitieri F., Lin B., Kalchman M.A., et al. (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat. Genet., 4, 398–403. [DOI] [PubMed] [Google Scholar]
- 5.Estrada Sanchez A.M., Mejia-Toiber J., Massieu L. (2008) Excitotoxic neuronal death and the pathogenesis of Huntington's disease. Arch. Med. Res., 39, 265–276. [DOI] [PubMed] [Google Scholar]
- 6.Bergami M., Rimondini R., Santi S., Blum R., Gotz M., Canossa M. (2008) Deletion of TrkB in adult progenitors alters newborn neuron integration into hippocampal circuits and increases anxiety-like behavior. Proc. Natl Acad. Sci. USA, 105, 15570–15575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuccato C., Ciammola A., Rigamonti D., Leavitt B.R., Goffredo D., Conti L., MacDonald M.E., Friedlander R.M., Silani V., Hayden M.R., et al. (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science, 293, 493–498. [DOI] [PubMed] [Google Scholar]
- 8.Zuccato C., Cattaneo E. (2009) Brain-derived neurotrophic factor in neurodegenerative diseases. Nat. Rev. Neurol., 5, 311–322. [DOI] [PubMed] [Google Scholar]
- 9.Canals J.M., Pineda J.R., Torres-Peraza J.F., Bosch M., Martin-Ibanez R., Munoz M.T., Mengod G., Ernfors P., Alberch J. (2004) Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington's disease. J. Neurosci., 24, 7727–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidetti P., Luthi-Carter R.E., Augood S.J., Schwarcz R. (2004) Neostriatal and cortical quinolinate levels are increased in early grade Huntington's disease. Neurobiol. Dis., 17, 455–461. [DOI] [PubMed] [Google Scholar]
- 11.Schwarcz R., Coyle J.T. (1977) Neurochemical sequelae of kainate injections in corpus striatum and substantia nigra of the rat. Life Sci., 20, 431–436. [DOI] [PubMed] [Google Scholar]
- 12.McGeer E.G., McGeer P.L. (1976) Duplication of biochemical changes of Huntington's chorea by intrastriatal injections of glutamic and kainic acids. Nature, 263, 517–519. [DOI] [PubMed] [Google Scholar]
- 13.Martire A., Pepponi R., Domenici M.R., Ferrante A., Chiodi V., Popoli P. (2013) BDNF prevents NMDA-induced toxicity in models of Huntington's disease: the effects are genotype specific and adenosine A(2A) receptor is involved. J. Neurochem., 125, 225–235. [DOI] [PubMed] [Google Scholar]
- 14.Xie Y., Hayden M.R., Xu B. (2010) BDNF overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J. Neurosci., 30, 14708–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki M., Nelson A.D., Eickstaedt J.B., Wallace K., Wright L.S., Svendsen C.N. (2006) Glutamate enhances proliferation and neurogenesis in human neural progenitor cell cultures derived from the fetal cortex. Eur. J. Neurosci., 24, 645–653. [DOI] [PubMed] [Google Scholar]
- 16.Schlett K. (2006) Glutamate as a modulator of embryonic and adult neurogenesis. Curr. Top. Med. Chem., 6, 949–960. [DOI] [PubMed] [Google Scholar]
- 17.Sadikot A.F., Burhan A.M., Belanger M.C., Sasseville R. (1998) NMDA receptor antagonists influence early development of GABAergic interneurons in the mammalian striatum. Brain Res. Dev. Brain Res., 105, 35–42. [PubMed] [Google Scholar]
- 18.Luk K.C., Kennedy T.E., Sadikot A.F. (2003) Glutamate promotes proliferation of striatal neuronal progenitors by an NMDA receptor-mediated mechanism. J. Neurosci., 23, 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandhi R., Luk K.C., Rymar V.V., Sadikot A.F. (2008) Group I mGluR5 metabotropic glutamate receptors regulate proliferation of neuronal progenitors in specific forebrain developmental domains. J. Neurochem., 104, 155–172. [DOI] [PubMed] [Google Scholar]
- 20.Wagner W., Bork S., Horn P., Krunic D., Walenda T., Diehlmann A., Benes V., Blake J., Huber F.X., Eckstein V., et al. (2009) Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS One, 4, e5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pregno G., Zamburlin P., Gambarotta G., Farcito S., Licheri V., Fregnan F., Perroteau I., Lovisolo D., Bovolin P. (2011) Neuregulin1/ErbB4-induced migration in ST14A striatal progenitors: calcium-dependent mechanisms and modulation by NMDA receptor activation. BMC Neurosci., 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeiro F.M., Pires R.G., Ferguson S.S. (2011) Huntington's disease and Group I metabotropic glutamate receptors. Mol. Neurobiol., 43, 1–11. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H., Li Q., Graham R.K., Slow E., Hayden M.R., Bezprozvanny I. (2008) Full length mutant huntingtin is required for altered Ca2+ signaling and apoptosis of striatal neurons in the YAC mouse model of Huntington's disease. Neurobiol. Dis., 31, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeron M.M., Hansson O., Chen N., Wellington C.L., Leavitt B.R., Brundin P., Hayden M.R., Raymond L.A. (2002) Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron, 33, 849–860. [DOI] [PubMed] [Google Scholar]
- 25.Arzberger T., Krampfl K., Leimgruber S., Weindl A. (1997) Changes of NMDA receptor subunit (NR1, NR2B) and glutamate transporter (GLT1) mRNA expression in Huntington's disease—an in situ hybridization study. J. Neuropathol. Exp. Neurol., 56, 440–454. [DOI] [PubMed] [Google Scholar]
- 26.Palmada M., Centelles J.J. (1998) Excitatory amino acid neurotransmission. Pathways for metabolism, storage and reuptake of glutamate in brain. Front. Biosci., 3, d701–d718. [DOI] [PubMed] [Google Scholar]
- 27.Bard J., Wall M.D., Lazari O., Arjomand J., Munoz-Sanjuan I. (2014) Advances in huntington disease drug discovery: novel approaches to model disease phenotypes. J. Biomol. Screen, 19, 191–204. [DOI] [PubMed] [Google Scholar]
- 28.Yang X.W., Gray M. (2011) Neurobiology of Huntington‘s Disease. Mouse Models for Validating Preclinical Candidates for Huntington’s Disease. CRC Press, Boca Raton, FL. [PubMed] [Google Scholar]
- 29.Seo H., Sonntag K.C., Isacson O. (2004) Generalized brain and skin proteasome inhibition in Huntington's disease. Ann. Neurol., 56, 319–328. [DOI] [PubMed] [Google Scholar]
- 30.Varani K., Abbracchio M.P., Cannella M., Cislaghi G., Giallonardo P., Mariotti C., Cattabriga E., Cattabeni F., Borea P.A., Squitieri F., et al. (2003) Aberrant A2A receptor function in peripheral blood cells in Huntington's disease. FASEB J., 17, 2148–2150. [DOI] [PubMed] [Google Scholar]
- 31.Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. (1999) Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl Acad. Sci. USA, 96, 11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X.W., Model P., Heintz N. (1997) Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol., 15, 859–865. [DOI] [PubMed] [Google Scholar]
- 33.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. (1998) Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell, 95, 55–66. [DOI] [PubMed] [Google Scholar]
- 34.Sipione S., Rigamonti D., Valenza M., Zuccato C., Conti L., Pritchard J., Kooperberg C., Olson J.M., Cattaneo E. (2002) Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum. Mol. Genet., 11, 1953–1965. [DOI] [PubMed] [Google Scholar]
- 35.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. (2000) Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet., 9, 2799–2809. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663–676. [DOI] [PubMed] [Google Scholar]
- 37.Magnuson T., Epstein C.J., Silver L.M., Martin G.R. (1982) Pluripotent embryonic stem cell lines can be derived from tw5/tw5 blastocysts. Nature, 298, 750–753. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. [DOI] [PubMed] [Google Scholar]
- 39.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science, 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- 40.Mattis V.B., Svendsen C.N. (2011) Induced pluripotent stem cells: a new revolution for clinical neurology? Lancet Neurol., 10, 383–394. [DOI] [PubMed] [Google Scholar]
- 41.The HD iPSC Consortium. (2012) Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell, 11, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science, 324, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller F.-J., Brändl B., Loring J.F. (2012) Assessment of human pluripotent stem cells with PluriTest. StemBook, edn The Stem Cell Research Community, StemBook. [PubMed] [Google Scholar]
- 44.Molero A.E., Gokhan S., Gonzalez S., Feig J.L., Alexandre L.C., Mehler M.F. (2009) Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington's disease. Proc. Natl Acad. Sci. USA, 106, 21900–21905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selkoe D.J., Salazar F.J., Abraham C., Kosik K.S. (1982) Huntington's disease: changes in striatal proteins reflect astrocytic gliosis. Brain Res., 245, 117–125. [DOI] [PubMed] [Google Scholar]
- 46.Orvoen S., Pla P., Gardier A.M., Saudou F., David D.J. (2012) Huntington's disease knock-in male mice show specific anxiety-like behaviour and altered neuronal maturation. Neurosci. Lett., 507, 127–132. [DOI] [PubMed] [Google Scholar]
- 47.Simpson J.M., Gil-Mohapel J., Pouladi M.A., Ghilan M., Xie Y., Hayden M.R., Christie B.R. (2011) Altered adult hippocampal neurogenesis in the YAC128 transgenic mouse model of Huntington disease. Neurobiol. Dis., 41, 249–260. [DOI] [PubMed] [Google Scholar]
- 48.Grote H.E., Bull N.D., Howard M.L., van Dellen A., Blakemore C., Bartlett P.F., Hannan A.J. (2005) Cognitive disorders and neurogenesis deficits in Huntington's disease mice are rescued by fluoxetine. Eur. J. Neurosci., 22, 2081–2088. [DOI] [PubMed] [Google Scholar]
- 49.Phillips W., Morton A.J., Barker R.A. (2005) Abnormalities of neurogenesis in the R6/2 mouse model of Huntington's disease are attributable to the in vivo microenvironment. J. Neurosci., 25, 11564–11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray M., Shirasaki D.I., Cepeda C., Andre V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E., et al. (2008) Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci., 28, 6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An M.C., Zhang N., Scott G., Montoro D., Wittkop T., Mooney S., Melov S., Ellerby L.M. (2012) Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell, 11, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang N., An M.C., Montoro D., Ellerby L.M. (2010) Characterization of human Huntington's disease cell model from induced pluripotent stem cells. PLoS Curr., 2, RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kornhuber J., Mack-Burkhardt F., Kornhuber M.E., Riederer P. (1989) [3H]MK-801 binding sites in post-mortem human frontal cortex. Eur. J. Pharmacol., 162, 483–490. [DOI] [PubMed] [Google Scholar]
- 54.Okamoto S., Pouladi M.A., Talantova M., Yao D., Xia P., Ehrnhoefer D.E., Zaidi R., Clemente A., Kaul M., Graham R.K., et al. (2009) Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat. Med., 15, 1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan J., Gladding C.M., Wang L., Zhang L.Y., Kaufman A.M., Milnerwood A.J., Raymond L.A. (2012) P38 MAPK is involved in enhanced NMDA receptor-dependent excitotoxicity in YAC transgenic mouse model of Huntington disease. Neurobiol. Dis., 45, 999–1009. [DOI] [PubMed] [Google Scholar]
- 56.Rakic P. (1985) DNA synthesis and cell division in the adult primate brain. Ann. N. Y. Acad. Sci., 457, 193–211. [DOI] [PubMed] [Google Scholar]
- 57.Kaya S.S., Asim M., Li Y., Yavuz E., Chopp M. (1999) Expression of nestin after traumatic brain injury in rat brain. Brain Res., 840, 153–157. [DOI] [PubMed] [Google Scholar]
- 58.Mizuno Y., Ohama E., Hirato J., Nakazato Y., Takahashi H., Takatama M., Takeuchi T., Okamoto K. (2006) Nestin immunoreactivity of Purkinje cells in Creutzfeldt–Jakob disease. J. Neurol. Sci., 246, 131–137. [DOI] [PubMed] [Google Scholar]
- 59.Kandasamy M., Couillard-Despres S., Raber K.A., Stephan M., Lehner B., Winner B., Kohl Z., Rivera F.J., Nguyen H.P., Riess O., et al. (2010) Stem cell quiescence in the hippocampal neurogenic niche is associated with elevated transforming growth factor-beta signaling in an animal model of Huntington disease. J. Neuropathol. Exp. Neurol., 69, 717–728. [DOI] [PubMed] [Google Scholar]
- 60.Bath K.G., Mandairon N., Jing D., Rajagopal R., Kapoor R., Chen Z.Y., Khan T., Proenca C.C., Kraemer R., Cleland T.A., et al. (2008) Variant brain-derived neurotrophic factor (Val66Met) alters adult olfactory bulb neurogenesis and spontaneous olfactory discrimination. J. Neurosci., 28, 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sairanen M., Lucas G., Ernfors P., Castren M., Castren E. (2005) Brain-derived neurotrophic factor and antidepressant drugs have different but coordinated effects on neuronal turnover, proliferation, and survival in the adult dentate gyrus. J. Neurosci., 25, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scharfman H., Goodman J., Macleod A., Phani S., Antonelli C., Croll S. (2005) Increased neurogenesis and the ectopic granule cells after intrahippocampal BDNF infusion in adult rats. Exp. Neurol., 192, 348–356. [DOI] [PubMed] [Google Scholar]
- 63.Henry R.A., Hughes S.M., Connor B. (2007) AAV-mediated delivery of BDNF augments neurogenesis in the normal and quinolinic acid-lesioned adult rat brain. Eur. J. Neurosci., 25, 3513–3525. [DOI] [PubMed] [Google Scholar]
- 64.Bath K.G., Akins M.R., Lee F.S. (2012) BDNF control of adult SVZ neurogenesis. Dev. Psychobiol., 54, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Curtis M.A., Penney E.B., Pearson A.G., van Roon-Mom W.M., Butterworth N.J., Dragunow M., Connor B., Faull R.L. (2003) Increased cell proliferation and neurogenesis in the adult human Huntington's disease brain. Proc. Natl Acad. Sci. USA, 100, 9023–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gil J.M., Mohapel P., Araújo I.M., Popovic N., Li J.Y., Brundin P., Petersén A. (2005) Reduced hippocampal neurogenesis in R6/2 transgenic Huntington's disease mice. Neurobiol. Dis., 20, 744–751. [DOI] [PubMed] [Google Scholar]
- 67.Cho S.R., Benraiss A., Chmielnicki E., Samdani A., Economides A., Goldman S.A. (2007) Induction of neostriatal neurogenesis slows disease progression in a transgenic murine model of Huntington disease. J. Clin. Invest., 117, 2889–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Y., Savanenin A., Reddy P.H., Liu Y.F. (2001) Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D-aspartate receptors via post-synaptic density 95. J. Biol. Chem., 276, 24713–24718. [DOI] [PubMed] [Google Scholar]
- 69.Ali D.W., Salter M.W. (2001) NMDA receptor regulation by Src kinase signalling in excitatory synaptic transmission and plasticity. Curr. Opin. Neurobiol., 11, 336–342. [DOI] [PubMed] [Google Scholar]
- 70.LoTurco J.J., Owens D.F., Heath M.J., Davis M.B., Kriegstein A.R. (1995) GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron, 15, 1287–1298. [DOI] [PubMed] [Google Scholar]
- 71.Anborgh P.H., Godin C., Pampillo M., Dhami G.K., Dale L.B., Cregan S.P., Truant R., Ferguson S.S. (2005) Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. J. Biol. Chem., 280, 34840–34848. [DOI] [PubMed] [Google Scholar]
- 72.DiFiglia M. (1990) Excitotoxic injury of the neostriatum: a model for Huntington's disease. Trends Neurosci., 13, 286–289. [DOI] [PubMed] [Google Scholar]
- 73.Nicoletti F., Bruno V., Copani A., Casabona G., Knöpfel T. (1996) Metabotropic glutamate receptors: a new target for the therapy of neurodegenerative disorders? Trends Neurosci., 19, 267–271. [DOI] [PubMed] [Google Scholar]
- 74.Bruno V., Battaglia G., Copani A., D'Onofrio M., Di Iorio P., De Blasi A., Melchiorri D., Flor P.J., Nicoletti F. (2001) Metabotropic glutamate receptor subtypes as targets for neuroprotective drugs. J. Cereb. Blood Flow. Metab., 21, 1013–1033. [DOI] [PubMed] [Google Scholar]
- 75.Tang T.S., Tu H., Chan E.Y., Maximov A., Wang Z., Wellington C.L., Hayden M.R., Bezprozvanny I. (2003) Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron, 39, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schiefer J., Sprünken A., Puls C., Lüesse H.G., Milkereit A., Milkereit E., Johann V., Kosinski C.M. (2004) The metabotropic glutamate receptor 5 antagonist MPEP and the mGluR2 agonist LY379268 modify disease progression in a transgenic mouse model of Huntington's disease. Brain Res., 1019, 246–254. [DOI] [PubMed] [Google Scholar]
- 77.Baskys A., Fang L., Bayazitov I. (2005) Activation of neuroprotective pathways by metabotropic group I glutamate receptors: a potential target for drug discovery? Ann. N. Y. Acad. Sci., 1053, 55–73. [DOI] [PubMed] [Google Scholar]
- 78.Baskys A., Bayazitov I., Fang L., Blaabjerg M., Poulsen F.R., Zimmer J. (2005) Group I metabotropic glutamate receptors reduce excitotoxic injury and may facilitate neurogenesis. Neuropharmacology, 49 Suppl 1, 146–156. [DOI] [PubMed] [Google Scholar]
- 79.Baskys A., Blaabjerg M. (2005) Understanding regulation of nerve cell death by mGluRs as a method for development of successful neuroprotective strategies. J. Neurol. Sci., 229–230, 201–209. [DOI] [PubMed] [Google Scholar]
- 80.Zeron M.M., Fernandes H.B., Krebs C., Shehadeh J., Wellington C.L., Leavitt B.R., Baimbridge K.G., Hayden M.R., Raymond L.A. (2004) Potentiation of NMDA receptor-mediated excitotoxicity linked with intrinsic apoptotic pathway in YAC transgenic mouse model of Huntington's disease. Mol. Cell Neurosci., 25, 469–479. [DOI] [PubMed] [Google Scholar]
- 81.Chen N., Luo T., Wellington C., Metzler M., McCutcheon K., Hayden M.R., Raymond L.A. (1999) Subtype-specific enhancement of NMDA receptor currents by mutant huntingtin. J. Neurochem., 72, 1890–1898. [DOI] [PubMed] [Google Scholar]
- 82.Ernst A., Alkass K., Bernard S., Salehpour M., Perl S., Tisdale J., Possnert G., Druid H., Frisen J. (2014) Neurogenesis in the striatum of the adult human brain. Cell, 156, 1072–1083. [DOI] [PubMed] [Google Scholar]
- 83.Ferrer I., Goutan E., Marin C., Rey M.J., Ribalta T. (2000) Brain-derived neurotrophic factor in Huntington disease. Brain Res., 866, 257–261. [DOI] [PubMed] [Google Scholar]
- 84.Giampa C., Montagna E., Dato C., Melone M.A., Bernardi G., Fusco F.R. (2013) Systemic delivery of recombinant brain derived neurotrophic factor (BDNF) in the R6/2 mouse model of Huntington's disease. PLoS One, 8, e64037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang M., Peng Q., Liu X., Jin J., Hou Z., Zhang J., Mori S., Ross C.A., Ye K., Duan W. (2013) Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of Huntington's disease. Hum. Mol. Genet, 12, 2462–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simmons D.A., Belichenko N.P., Yang T., Condon C., Monbureau M., Shamloo M., Jing D., Massa S.M., Longo F.M. (2013) A small molecule TrkB ligand reduces motor impairment and neuropathology in R6/2 and BACHD mouse models of Huntington's disease. J. Neurosci., 48, 18712–18727 Erratum in: J Neurosci. (2014) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luong M.X., Auerbach J., Crook J.M., Daheron L., Hei D., Lomax G., Loring J.F., Ludwig T., Schlaeger T.M., Smith K.P., et al. (2011) A call for standardized naming and reporting of human ESC and iPSC lines. Cell Stem Cell, 8, 357–359. [DOI] [PubMed] [Google Scholar]
- 88.Sareen D., Ebert A.D., Heins B.M., McGivern J.V., Ornelas L., Svendsen C.N. (2012) Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS One, 7, e39113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K., et al. (2011) A more efficient method to generate integration-free human iPS cells. Nat. Methods, 8, 409–412. [DOI] [PubMed] [Google Scholar]
- 90.Muller F.J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O., et al. (2011) A bioinformatic assay for pluripotency in human cells. Nat. Methods, 8, 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ebert A.D., Shelley B.C., Hurley A.M., Onorati M., Castiglioni V., Patitucci T.N., Svendsen S.P., Mattis V.B., McGivern J.V., Schwab A.J., et al. (2013) EZ spheres: a stable and expandable culture system for the generation of pre-rosette multipotent stem cells from human ESCs and iPSCs. Stem Cell Res., 10, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pruunsild P., Sepp M., Orav E., Koppel I., Timmusk T. (2011) Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J. Neurosci., 31, 3295–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skotte N.H., Southwell A.L., Ostergaard M.E., Carroll J.B., Warby S.C., Doty C.N., Petoukhov E., Vaid K., Kordasiewicz H., Watt A.T., et al. (2014) Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all huntington disease patients. PLoS One, 9, e107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F., et al. (2013) Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res., 41, 9634–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Southwell A.L., Skotte N.H., Kordasiewicz H.B., Ostergaard M.E., Watt A.T., Carroll J.B., Doty C.N., Villanueva E.B., Petoukhov E., Vaid K., et al. (2014) In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther., 12, 2093–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. (2011) Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther., 19, 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aubry L., Bugi A., Lefort N., Rousseau F., Peschanski M., Perrier A.L. (2008) Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc. Natl Acad. Sci. USA, 105, 16707–16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ebert A.D., McMillan E.L., Svendsen C.N. (2008) Isolating, expanding, and infecting human and rodent fetal neural progenitor cells. Curr. Protoc. Stem Cell Biol., Chapter 2, Unit 2D 2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.