Abstract
Bone mesenchymal stem cell (BMSC) age-related changes include decreased osteogenesis and increased adipogenesis. Rev-erbα and the Wnt/β-catenin signaling pathway were known to play important roles in BMSC aging. In this study, we have aimed to elucidate whether Rev-erbα and Wnt/β-catenin signaling interact during BMSC proliferation and osteogenesis. Our results showed that Rev-erbα expression gradually dropped during BMSC osteogenesis, and overexpression of Rev-erbα in BMSCs inhibited cell proliferation and osteogenesis. The inhibition of cell proliferation induced by Rev-erbα overexpression was partially reversed when Wnt/β-catenin signaling was activated. These results suggested that Rev-erbα could promote BMSC aging and may be the negative regulator during the late stage of osteogenesis. The clock gene Rev-erbα and Wnt/β-catenin signaling interact in the regulation of cell proliferation.
Introduction
Age-related changes to cell proliferation and differentiation capacity are properties of the basic cellular processes and the main cause of organic aging. Mesenchymal stem cells (MSCs) are multipotent stem cells in the bone marrow, and MSC aging-related changes are closely related to bone aging. The quality and quantity of MSCs can change with age, contributing to their reduced capacity for proliferation and osteodifferentiation. All of these factors reduce the number of osteoblasts and result in the degeneration of bone formation and aging [1].
There is still disagreement regarding the changing mechanism of MSC differentiation with age (eg, increased adipogenesis and decreased osteogenesis or the inhibition of both processes); however, most scholars have supported the idea that MSCs from aged donors have decreased proliferation and osteogenesis [2,3]. Zhou found that compared with young human bone marrow stromal cells (hBMSCs), hBMSCs from the elderly are four-fold more senescence-associated β-galactosidase (SA-β-gal) positive (an index of cell aging) and the doubling period is 1.7 times that of the young hBMSCs; furthermore, alkaline phosphatase (ALP) activation and the declining expression of osteo-related genes showed that osteogenesis decreased with age [3]. The colony-forming unit-fibroblast (CFU-F) refers to a population of adherent cells from the bone marrow grown in culture. A related study found that the number of CFU-Fs in MSCs from old mice was decreased compared with MSCs from young mice [4], while there were fewer colony-forming unit-osteoblasts and more colony-forming unit-lipoblasts for MSCs from old mice [2], indicating the varying trend in differentiation with age.
The mechanism underlying MSC aging changes has not been elucidated, and the genes and signaling molecules involved in the process may be key elements. Clock genes, which could lead to the aging variations by affecting the metabolism of basic life, may be an important part of this process. Clock genes regulate the biological rhythm through feedback signaling formed by an oscillator system and are expressed in peripheral tissue, such as long bone, calvarial bone, hematopoietic stem cell, and even the whole marrow [5,6]. Researchers found that mice lacking the core clock genes, Bmal1 (Bmal1−/−), Clock (Clock−/−), or Per2 (Per2−/−), showed early aging symptoms compared with their wild-type littermates [7–9], especially the Bmal1−/− mice, in which early aging symptoms were more obvious [10]. Studies also found that endochondral ossification was under control by clock genes in chondrocytes [11].
Our previous studies demonstrated that in aged mice the decreased proliferation and osteogenesis capacity is positively related to the decreased Bmal1 expression. Overexpression of Bmal1 could promote bone mesenchymal stem cell (BMSC) proliferation, and this process was relatively independent of Bmal1 regulation of circadian rhythm, testifying that Bmal1 could inhibit cell aging [12]. Furthermore, the expression of β-catenin, the key factor of Wnt/β-catenin signaling, increased when Bmal1 was overexpressed [13], and another study showed that β-catenin expression decreased with aging [14]. As the main signaling pathway in development, Wnt/β-catenin signaling may play vital roles in the aging process [15,16]. Based on these studies, clock genes and Wnt/β-catenin signaling both play important roles in the aging process of BMSCs and we deduced that there is a cross talk between them, which was explored in our later study.
The expression of Bmal1 is regulated by the Retinoic acid-related orphan receptors (Rorα) and Rev-erbα: Rorα activates and Rev-erbα restrains the expression of Bmal1. Bmal1, Rorα, and Rev-erbα compose a closed loop that controls the stability of the clock system [17]. Compared with Rorα, Rev-erbα has a pivotal role in metabolism and was speculated to be the molecular junction for communicating clock gene signals to the metabolism [18,19]. Moreover, Yu et al. found that the expression of Rev-erbα in Rhesus macaques declines with age [20].
However, previous studies of Rev-erbα focused more on its effects on adipogenesis. These studies demonstrated that Rev-erbα had a positive effect on adipogenesis, as progenitor cells were inclined to undergo adipogenesis when Rev-erbα was overexpressed, and Rev-erbα expression increased during adipogenesis [21,22]. During BMSC aging, decreased osteogenesis is always accompanied with increased adipogenesis [2], and maybe relates to the increased expression of Rev-erbα. This prompts the idea that age-related BMSC changes may be associated with the expression of Rev-erbα.
As a clock gene, Rev-erbα decreases Bmal1 expression through glycogen synthase kinase (GSK)-3β-mediated phosphorylation [23]. At the same time, GSK-3β is an important component of Wnt/β-catenin signaling as GSK-3β and the scaffolding proteins, Axin and adenomatous polyposis coil, compose the degradation complex that induces β-catenin phosphorylation. This process, which degrades β-catenin, leads to the suppression of Wnt/β-catenin signaling [24], implying that GSK-3β may be the connection between the clock genes and Wnt/β-catenin signaling during BMSC aging.
The influence of Rev-erbα on Wnt signaling is more direct than that of Bmal1. As the main negative regulator of Bmal1 and having a close relationship with adipogenesis, Rev-erbα may be a key player in Bmal1-induced cellular variation. We designed the present study to explore the interaction between Rev-erbα and Wnt/β-catenin in BMSC proliferation and osteogenesis. The proliferation rate, Bmal1 and GSK-3β expression, and Wnt/β-catenin signaling activity were studied in BMSCs with overexpressing Rev-erbα, and the results were compared with BMSCs that had been treated with exogenous Wnt3a protein to activate Wnt signaling to show the interaction and mechanism between Rev-erbα and Wnt signaling with regard to BMSC proliferation. The expression of Rev-erbα was detected during osteogenesis to explore the influence of osteogenesis on Rev-erbα expression. Following Rev-erbα overexpression, the mRNA levels of osteo-related factors ALP, Osterix (OSX), and bone sialoprotein (BSP) were measured along with the activity of Wnt/β-catenin signaling.
Materials and Methods
Cell culture
BMSCs (passage 5) isolated from 4-week-old C57/BL6 male mice were purchased from Cyagen Biosciences, Inc. The identification results from the vendor showed that Passage 12 BMSCs had the good ability of osteogenesis and adipogenesis, and cell surface molecules of BMSCs (CD29 and CD44 were positive over 96%, while CD117 and CD31 were negative below 1%) were explicit as measured by flow cytometry. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2, and alpha-minimum essential medium (α-MEM) (Gibco) media containing 10% fetal bovine serum (FBS) (Hyclone) were changed every other day. At 80%–90% confluence, BMSCs were detached using 0.25% trypsin in 0.01% ethylenediamine-tetraacetic acid (EDTA) and then passaged.
Osteoinduction media (OS media) were α-MEM containing 10% FBS and 10 mM β-glycerophosphate, 10 nM dexamethasone, and 50 mg/L l-ascorbic acid (Sigma-Aldrich). Wnt3a-conditioned media were prepared by the addition of Wnt3a protein powder isolated from Wnt3a recombinant mice (R&D Systems, Inc.) to α-MEM media at a final concentration of 10 ng/mL [25].
Rev-erbα overexpression lentiviral vector transfection
Passage 8 BMSCs were cultured in 12-well plastic plates in basic media at an initial density of 1×104 cells/cm2. Approximately 48 h after plating, cells were transfected with the Rev-erbα overexpression lentiviral vector (synthesized by Hanheng bio.) (Rev-erbα group) or enhanced green fluorescent protein (EGFP)-expressing lentiviral vector (EGFP group) at a multiplicity of infection (MOI) of 30. The fluorescence level was observed using an inverted fluorescent microscope after 48 h, and the transfection efficiencies were calculated. Reverse transcription–polymerase chain reaction (RT-PCR) and western blotting assays were used to test the effects of transfection with Rev-erbα-expressing vector. As a circadian clock gene, Rev-erbα expressed in an oscillatory manner in vivo. However, without serum shock in vitro, the rhythmic expression of Rev-erbα should not be induced [26]; no rigorous time point was used to obtain the cells for test.
Flow cytometry assay of BMSCs following Rev-erbα transfection
BMSCs were divided into six groups. Cells cultured in basic media with Rev-erbα transfection (Rev-erbα group), Wnt3a-conditioned media with Rev-erbα transfection (Rev-erbα +Wnt3a group), basic media with EGFP transfection (EGFP group), Wnt3a-conditioned media with EGFP transfection (EGFP+Wnt3a group), basic media with no transfection (control group), or Wnt3a-conditioned media with no transfection (Wnt3a group). BMSCs were seeded onto six-well plates at a density of 1×104 cells/cm2. Following lentiviral infection, basic and Wnt3a-conditioned media were added to certain groups. When cells reached about 70% confluency, they were detached with trypsin and EDTA after washing twice with phosphate-buffered saline (PBS), centrifuged at 1,000g for 5 min, and then fixed with cool 70% alcohol and frozen below −20°C. Two hours before detection, the cells were centrifuged to remove the alcohol and were washed with cool PBS. After incubating with RNase at 37°C for 30 min, propidium iodide staining detection dye was infused into the cells. The cell density was ∼1×106 cells/mL upon detection. One tube was incubated only with the steam buffer as the control to eliminate the interference. The content of DNA cells was detected by flow cytometry (Beckman Coulter), and the S-phase fraction (SPF) and DNA Proliferation Index (PI) of the total cells in each sample were calculated according to the following formulae:
SPF (%)=S(G0/G1+S+G2/M)×100%
PI (%)=(S+G2/M)/(G0/G1+S+G2/M)×100%
Assessment of SA-β-gal staining
Passage 8 BMSCs grouped as described in the flow cytometry assay were seeded into a six-well plate at a density of 1×104 cells/cm2. After 48 h of culture under a humidified atmosphere containing 5% CO2, the media were discarded and the cells were washed once with PBS. Subsequently, 1 mL of fixative was added to each well for 15 min, and the wells were then rinsed thrice. The cells were incubated in 1 mL working solution of β-galactosidase with X-Gal per well overnight at 37°C. A SA-β-gal staining kit was obtained from Beyotime. Senescent cells were observed using an optical microscope and counted in three random fields of vision of 200 cells per field for calculation of the positive rate.
Real-time RT-PCR analysis
Passage 8 BMSCs were seeded into a six-well plate at a density of 1×104 cells/cm2, grouped as described in the flow cytometry assay, and the RNA was tested following 2 days of culture. BMSCs in the OS medium were grouped as Rev-erbα (Rev-erbα transfection), EGFP (EGFP transfection), and control. The RNA was tested following 0, 7, and 14 days of culture. The cells were obtained and maintained in an RNA preservation solution (RNA safeguard). A simple P total RNA extraction kit (Bioer) was used to extract total RNA. Total RNA was quantified using a spectrophotometer to measure the absorbance (A) at 260 nm, and the RNA samples used had an A260:A280 ratio of 2.0 to ensure high purity. Real-time PCR was performed in 20 μL reactions in triplicate using an ABI PRISM 7300 Real-time PCR System according to the manufacturer's instructions. The initial copy numbers of the unknown samples were calculated using a 7300 System SDS Software (Applied Biosystems) and a standard curve. Table 1 contains the primer sequences used for RT-PCR analysis.
Table 1.
The Primers Used for the RT-PCR Analysis
| Target gene | Primers | Sequence | Fragment size (bp) |
|---|---|---|---|
| β-Actin | Forward | 5′-GGGCTGTATTCCCCTCCATCG-3′ | 201 |
| Reverse | 5′-GCAGCTCATTGTAGAAGGTGTGGTG-3′ | ||
| Rev-erbα | Forward | 5′-ATTCGGGAGGTGGTAGAGTTTGC-3′ | 148 |
| Reverse | 5′-ATCACTGTCTGGTCCTTCACGTTG-3′ | ||
| Bmal1 | Forward | 5′-AACCTTCCCGCAGCTAACAG-3′ | 79 |
| Reverse | 5′-AGTCCTCTTTGGGCCACCTT-3′ | ||
| Wnt3a | Forward | 5′-CTTAGTGCTCTGCAGCCTGA-3′ | 92 |
| Reverse | 5′-GAGTGCTCAGAGAGGAGTACTGG-3′ | ||
| β-Catenin | Forward | 5′-GGGTCCTCTGTGAACTTGCTC-3′ | 165 |
| Reverse | 5′-TGTAATCCTGTGGCTTGTCCTC-3′ | ||
| GSK-3β | Forward | 5′-CCTTATCCCTCCACATGCTCG-3′ | 103 |
| Reverse | 5′-GTTATTGGTCTGTCCACGTCTC-3′ | ||
| OSX | Forward | 5′-TGCCTACTTACCCATCTGACTT-3′ | 134 |
| Reverse | 5′-TTGCCCACTATTGCCAACC-3′ | ||
| ALP | Forward | 5′-CCCCCCGTGGCAACTCTATCTT-3′ | 272 |
| Reverse | 5′-GTAGTTCTGCTCGTGGACGCCG-3′ | ||
| BSP | Forward | 5′-CACAAGCAGACACTTTCACTCC-3′ | 77 |
| Reverse | 5′-TCCATAAGCCAAGCTATCACC-3′ | ||
| c-Myc | Forward | 5′-CTGTATGTGGAGCGGTTTCTC-3′ | 90 |
| Reverse | 5′-AGGCTGGTGCTGTCTTTGC-3′ | ||
| TCF1 | Forward | 5′-CTACAGCGACGAGCACTTTTCTC-3′ | 115 |
| Reverse | 5′-GTAGAAGGTGGGGATTTCAGGAG-3′ |
ALP, alkaline phosphatase; BSP, bone sialoprotein; GSK, glycogen synthase kinase; OSX, Osterix; RT-PCR, reverse transcription–polymerase chain reaction; TCF, T cell factor.
Western blotting
Passage 8 BMSCs were seeded into a six-well plate at a density of 1×104 cells/cm2 and grouped as described in the flow cytometry assay, and whole-cell protein was obtained after 2 days of culture. BMSCs in the OS medium were grouped as described in the RT-PCR analysis, and whole-cell protein was obtained following 0, 7, and 14 days of culture. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference. After being cultured for a certain time, the cells were washed twice with ice-cold PBS and then lysed in lysis buffer from the Keygen total protein extraction kit (Keygen Biotech). Following centrifugation at 14,000g at 4°C for 15 min, the supernatant was collected and quantitatively assayed using the bicinchonininc acid (BCA) method. The total protein extracts were separated using standard sodium dodecylsulfonate-polyacrylate gel electrophoresis and subsequently transferred to a polyvinylidene fluoride membrane. After blocking, the membranes were probed with anti-Bmal1 primary antibodies (Abcam) or anti-β-catenin primary antibodies (Cell Signaling), followed by the addition of horseradish peroxidase-conjugated secondary antibodies (1:5,000). Immunoreactive proteins were visualized with a chemiluminescence kit (Millipore) and the band intensities were determined using a ChemiDoc XRS Gel documentation system and Quantity One software (Bio-Rad).
Statistical analysis
Measurements are expressed as the mean±SD. Statistical comparisons were performed using factorial analysis of variance, followed by multiple comparisons using the Student–Newman–Keuls test. A P value of<0.05 was considered to be statistically significant.
Results
BMSC proliferation in Wnt3a-conditioned medium after transfection with Rev-erbα
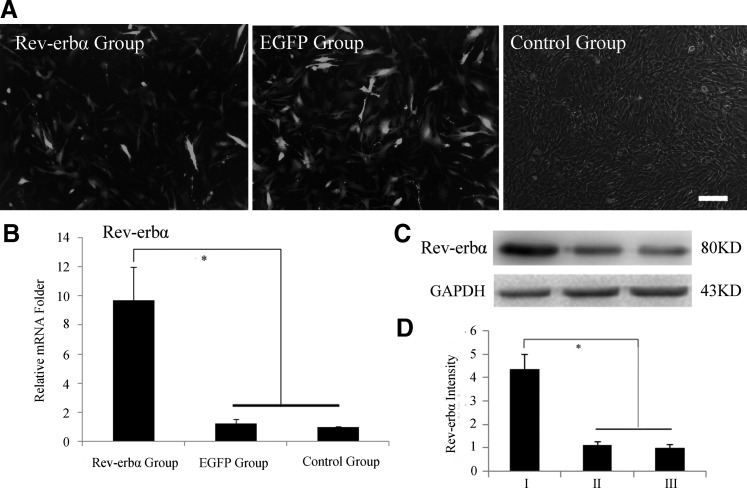
After 48 h of lentivirus infection, the expression of EGFP was observed under an inverted fluorescent microscope. Since the target gene has been established and packaged to lentivirus, the fluorescence level, about 50%, in the Rev-erbα group was slightly below the level, about 60%, in the EGFP group. RT-PCR and western blotting assays showed overexpressing of Rev-erbα in mRNA and protein levels (Fig. 1).
FIG. 1.
Overexpression of Rev-erbα in bone mesenchymal stem cells (BMSCs). (A) The expression of enhanced green fluorescent protein (EGFP) in BMSCs 48 h after infection with recombinant lentivirus [multiplicity of infection (MOI)=30]. The expression of EGFP in the EGFP group was slightly higher than that of the Rev-erbα group, 100×, scale bar=100 μm. (B) Reverse transcription–polymerase chain reaction (RT-PCR) analysis showed that the Rev-erbα mRNA levels were ∼10-fold higher than the other two groups following transfection. (C) Western blotting analysis showed the enhanced Rev-erbα protein levels 48 h after transfection. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an endogenous loading reference. Rev-erbα protein levels of the (I) Rev-erbα, (II) EGFP, and (III) control groups. (D) Quantified using densitometry by Quantity One software, the ratio of Rev-erbα relative to GAPDH is expressed as the relative content of Rev-erbα protein. Data represent the mean±SD (n=3), and the * symbol denotes a significant difference (P<0.05).
Passage 8 BMSCs were detected by flow cytometry to determine the effect of Rev-erbα and Wnt3a on cell proliferation. The number of S-phase cells in the Rev-erbα and Rev-erbα +Wnt3a groups was obviously lower than that of other groups, but no exact cycle arrest appeared. Furthermore, SPF and PI indices were detected to tell the specific difference of proliferation. The value of SPF and PI when Rev-erbα overexpressed was smaller with a statistically significant difference (P<0.01), and the value of the Rev-erbα group was the smallest one (Fig. 2).
FIG. 2.
BMSC growth rates as determined by flow cytometry assays. (A) Cell cycle of the Rev-erbα group by flow cytometry assay. (B) Cell cycle of the Rev-erbα +Wnt3a group. (C) Cell cycle of the EGFP group. (D) Cell cycle of the EGFP+Wnt3a group. (E) Cell cycle of the control group. (F) Cell cycle of the Wnt3a group. (G) The S-phase fraction (SPF) value of the groups. (H) Proliferation index (PI) value of the groups. Data represent mean±SD (n=3), and the * symbol denotes a significant difference (P<0.05). The # symbol indicates P<0.05 compared with the other groups.
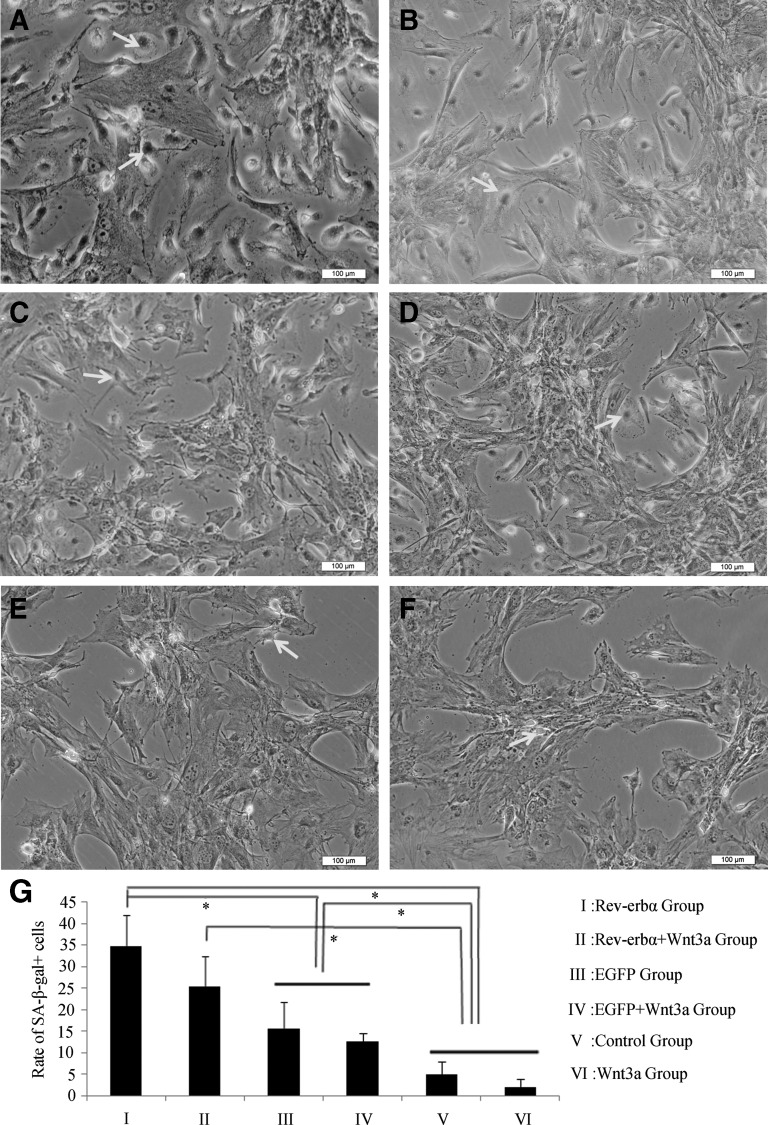
SA-β-gal staining indicated that more aged cells were present in the Rev-erbα and Rev-erbα +Wnt3a groups (Fig. 3A, B) compared with the EGFP and EGFP+Wnt3a groups (Fig. 3C, D), while only a few aged cells could be found in the control and Wnt3a groups (Fig. 3E, F). These results show that the Rev-erbα group had the most SA-β-gal+ cells, followed by the Rev-erbα +Wnt3a group, and was significantly different (P<0.05) from other groups (Fig. 3G).
FIG. 3.
Senescence-associated β-galactosidase (SA-β-gal) staining of BMSCs following Rev-erbα overexpression and Wnt3a protein addition. (A) There are relatively more SA-β-gal+ cells (arrow) in the Rev-erbα group. (B) The rate of SA-β-gal+ cells for the Rev-erbα +Wnt3a group was a little bit lower than for the Rev-erbα group. (C) There were fewer SA-β-gal+ cells in the EGFP group. (D) The number of SA-β-gal+ cells in the EGFP+Wnt3a group was nearly the same as in the EGFP group. (E) There were only a few SA-β-gal+ cells in the control group. (F) No obvious SA-β-gal+ cells were seen in the Wnt3a group. 100×, scale bar=100 μm. (G) Statistical analysis of SA-β-gal+ cells in all groups. SA-β-gal+ cells were counted in three different fields using light microscopy and averaged. The * symbol denotes a significant difference (P<0.05).
From this part of the experiment, we found that the SPF and PI values of the Rev-erbα and Rev-erbα +Wnt3a groups were lower than values of other groups. The number of SA-β-gal+ cells increased when Rev-erbα was overexpressed, demonstrating Rev-erbα-mediated inhibition of cell proliferation. Moreover, the situation could be partially reversed to a certain extent by the addition of Wnt3a protein, while Wnt3a-mediated promotion of cell proliferation was not obvious for groups lacking Rev-erbα overexpression.
Bmal1 and Wnt-related gene expression in Wnt3a-conditioned media after Rev-erbα transfection
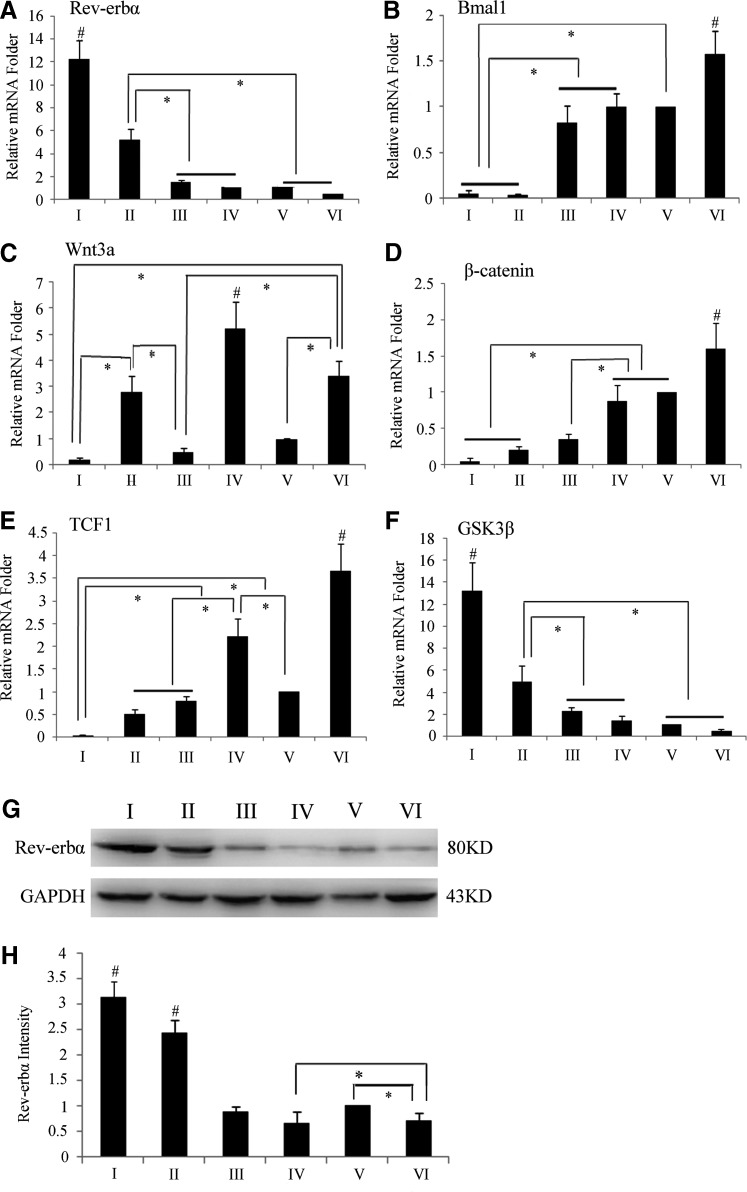
As the RT-PCR results show, the mRNA levels of Rev-erbα in the Rev-erbα and Rev-erbα +Wnt3a groups increased, but decreased when Wnt3a protein was added; however, this variation was not evident in the EGFP or EGFP+Wnt3a groups (Fig. 4A). The expression of Bmal1 was quite different as it was greatest in the Wnt3a group and lowest for the Rev-erbα overexpression group (Fig. 4B). The transcription of Wnt3a increased in groups with exogenous Wnt3a protein and decreased when Rev-erbα was overexpressed (Fig. 4C). The levels of β-catenin mRNA decreased after transfection, especially for the Rev-erbα and Rev-erbα +Wnt3a groups (Fig. 4D). The highest T cell factor 1 (TCF1) expression was observed after Wnt3a protein was added (Fig. 4E). The expression trend of GSK-3β was similar to that of Rev-erbα, with highest expression in the Rev-erbα group (Fig. 4F).
FIG. 4.
Transcriptional changes in Wnt signaling and Clock genes after Rev-erbα overexpression and addition of Wnt3a protein. (A) Differences in Rev-erbα mRNA levels among the groups. The transcriptional levels of Rev-erbα decreased when Wnt3a was added (Rev-erbα +Wt3a group). (B) Differences in Bmal1 mRNA levels among the groups. The trend was nearly opposite that observed for Rev-erbα. (C) The transcriptional level of Wnt3a among the groups. Less Wnt3a mRNA was expressed in the Rev-erbα group. (D) The transcriptional level of β-catenin among the groups. The least expression was observed in the Rev-erbα group, and the most expression was seen in the Wnt3a group. (E) The expression of T cell factor 1 (TCF1) mRNA among groups. The expression was decreased following Rev-erbα overexpression and was increased when Wnt3a protein was added. (F) The expression of glycogen synthase kinase (GSK)-3β mRNA among groups. The Rev-erbα mRNA expression trend was nearly unchanged. (G) Western blot results of Rev-erbα protein changes among the groups. The trend was almost the same as the Rev-erbα mRNA expression trend. Rev-erbα protein levels for the (I) Rev-erbα, (II) Rev-erbα +Wnt3a, (III) EGFP, (IV) EGFP+Wnt3a, (V) control, and (VI) Wnt3a groups. (H) Quantified using densitometry by Quantity One software, the ratio of Rev-erbα relative to GAPDH is expressed as the relative content of Rev-erbα protein. Data represent the mean±SD (n=3), and the * symbol denotes a significant difference (P<0.05). The # symbol indicates P<0.05 compared with the other groups.
Rev-erbα protein levels are shown in Figure 4G and H, and its expression levels in the Rev-erbα and Rev-erbα +Wnt3a groups were distinctly higher than for other groups. While Rev-erbα protein levels were decreased when Wnt3a protein added, the trend was similar to the Rev-erbα mRNA levels.
Rev-erbα expression during BMSC osteogenesis
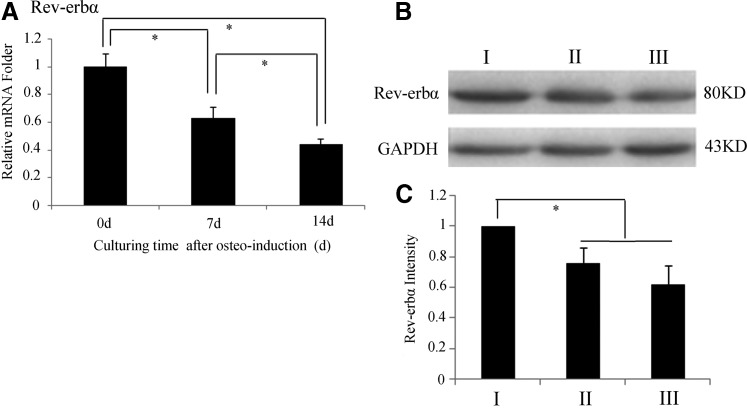
The Rev-erbα mRNA and protein levels were measured following 0, 7, and 14 days of osteoinduction. The results showed that Rev-erbα transcription decreased with culture time, as the mRNA levels declined by half after 14 days of osteogenesis, and there was a statistically significant difference (P<0.05) between the culture times (Fig. 5A). The variation in Rev-erbα protein levels showed a similar trend, but no statistically significant difference (P>0.05) between 7 and 14 days of osteoinduction (Fig. 5B, C). These data demonstrate that the expression of Rev-erbα decreased in osteogenesis with culture time.
FIG. 5.
The expression changes of Rev-erbα during osteogenesis. (A) The expression of Rev-erbα mRNA declined gradually during osteogenesis. (B) Western blot analysis indicates that Rev-erbα protein levels changed during osteoinduction and displayed the same trend as Rev-erbα mRNA levels. (I) 0 days of osteogenesis, (II) 7 days of osteogenesis, and (III) 14 days of osteogenesis. (C) Quantified using densitometry by Quantity One software, the ratio of Rev-erbα relative to GAPDH is expressed as the relative content of Rev-erbα protein. Data represent mean±SD (n=3), and the * symbol denotes a significant difference (P<0.05).
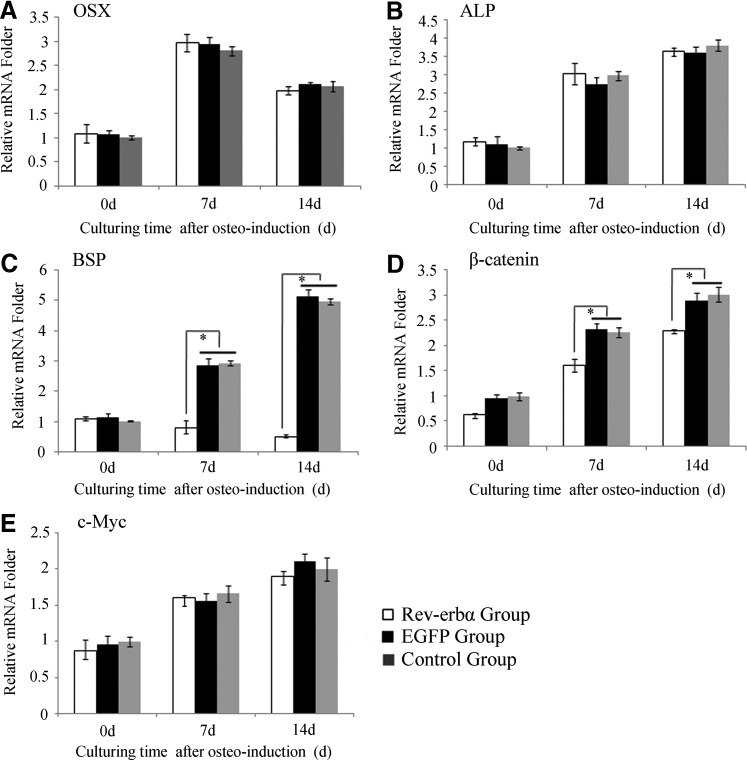
Transcriptional changes in osteofactors and Wnt signaling after the overexpression of Rev-erbα in Wnt3a-conditioned media during osteoinduction
RT-PCR was used to quantify the mRNA levels of osteogenic markers. OSX served as the anosteoblast progenitor marker, ALP activity level served as an early osteoblast marker, and BSP served as a late osteogenic marker [27,28]. The highest OSX expression levels occurred after 7 days of osteoinduction, and then decreased after 14 days of induction, with no statistically significant differences between the groups (Fig. 6A). The transcription levels of ALP were increased with osteoinduction, but no statistically significant differences (P>0.05) were observed between the groups. ALP mRNA levels in the EGFP and control groups increased with osteoinduction (Fig. 6B). In the Rev-erbα group, BSP transcription levels decreased with osteoinduction (P<0.05) (Fig. 6C). These data show that the expression of BSP is inhibited when Rev-erbα is overexpressed. To measure Wnt/β-catenin signaling activity, the mRNA levels of intracellular signal transducer β-catenin and the downstream gene c-Myc were quantified. The β-catenin mRNA level was increased with osteoinduction in the Rev-erbα group, but was relatively lower than those for the EGFP and control groups (Fig. 6D), and β-catenin transcription was reduced when Rev-erbα was overexpressed. The levels of c-Myc mRNA were increased with osteoinduction, but the differences among the groups were not statistically significant (Fig. 6E). These results show that Wnt/β-catenin signaling is active in osteogenesis. Moreover, overexpressed Rev-erbα could inhibit the expression of β-catenin, but not c-Myc.
FIG. 6.
Transcriptional changes to Wnt signaling and osteo-related genes during osteoinduction. (A) Differences in Osterix (OSX) mRNA levels among the groups during osteogenesis. The changing trend was similar among groups, and the highest OSX mRNA expression occurred on the seventh day of osteogenesis. (B) There were no significant differences in alkaline phosphatase (ALP) mRNA expression among the groups, but it did gradually increase during osteogenesis. (C) The bone sialoprotein (BSP) transcription levels among the groups. The BSP mRNA expression levels were much lower in the Rev-erbα group and the low level was maintained, while BSP mRNA expression in the other two groups increased markedly during osteogenesis. (D) The β-catenin mRNA expression among groups showed the same trend as that observed for BSP. (E) Differences in c-Myc mRNA levels among the groups. No expression differences could be found among the groups, and the expression gradually increased during osteogenesis. Data represent the mean±SD (n=3), and the * symbol denotes a significant difference (P<0.05).
Discussion
Changes in metabolic activity promote the changes associated with aging, and aging-related genes affect cell proliferation and differentiation. Recently, studies showed that Rev-erbα expression was increased, while Bmal1 expression was decreased in older BMSCs in vitro without the induction of circadian rhythm by dexamethasone exposure or serum shock [20,29–31]. As a unit, the clock genes may regulate the aging process of cells in some way. Originally, it was thought that the control of cell proliferation by circadian clock genes arose from their roles in circadian rhythm [32,33]. Recently, a study has suggested that a more complex mechanism, including cell metabolism and related signaling pathways, might be involved in this process [34]. As the core component of clock genes, Bmal1 has been reported to play important roles in the regulation of adipogenic differentiation in mature adipocytes [35]. NOC, another circadian-regulated protein, can stimulate adipogenesis while suppressing osteogenesis [36]. Previous studies have focused on the relationship between Rev-erbα and adipogenesis, while little is known about Rev-erbα and osteogenesis. Researchers have shown that Rev-erbα is a target gene of peroxisome proliferator-activated receptor gamma (PPAR-γ), which is key in the regulation of adipogenesis and osteogenesis [22]. Another study showed that the expression of PPAR-γ increased in senescent cells [2] and that BMSC aging led to enhanced adipogenesis and decreased osteogenesis. Therefore, the changes to cell metabolism caused by Rev-erbα may play vital roles in promoting BMSC aging.
During the BMSC aging process, alterations to proliferation capacity appear more direct and prominent than changes to the differentiation ability. A recent study found that the overexpression of Rev-erbα dramatically inhibited cell growth and caused cycle arrest in the G2/M phase; furthermore, this reduced rate of growth was reversed upon the addition of doxycycline to block Rev-erbα expression [37]. These results agree with our experiments as two indicators of cell senescence, decreased proliferative activity and an increased β-galactosidase positive rate, were correlated with Rev-erbα overexpression.
We found that Rev-erbα mRNA levels declined during BMSC osteoinduction, and western blotting analysis showed that Rev-erbα protein levels also declined. Following overexpression of Rev-erbα, the expression of ALP and BSP mRNAs increased gradually during osteoblastic induction of BMSCs. The OSX mRNA levels peaked at day 7 and declined at day 14. The ALP and OSX mRNA levels showed no statistically significant differences among the three groups, suggesting that Rev-erbα cannot regulate the expression of osteogenic markers during early osteoinduction. Overexpression of Rev-erbα obviously restrained the expression of BSP in the Rev-erbα group, indicating that Rev-erbα may repress osteogenesis by regulating the expression of BSP, an osteogenic marker seen during later osteodifferentiation. Considering the decline of Rev-erbα expression during osteoinduction, Rev-erbα may repress osteogenesis during late osteoinduction. Together with the result that BMSC proliferation declined when Rev-erbα was overexpressed, this result shows that Rev-erbα may accelerate the aging process by promoting cell senescence with decreased proliferation and osteogenesis.
In our experiments, the overexpression of Rev-erbα reduced Wnt signaling activity as shown by lower Wnt3a, β-catenin, and TCF1 mRNA levels and higher GSK-3β mRNA levels in BMSC proliferation. After adding exogenous Wnt3a protein to BMSCs that were overexpressing Rev-erbα, Wnt signaling activity recovered to a certain level as shown by higher β-catenin and TCF1 mRNA levels. Activation of Wnt signaling partially reversed the effects of Rev-erbα overexpression, suggesting an interaction between Rev-erbα and Wnt signaling in cell proliferation. This interaction was confirmed by western blotting as Rev-erbα protein levels were reduced after the addition of exogenousWnt3a protein.
From the flow cytometry assay, we can deduce that the activation of Wnt signaling promoted proliferation to some degree as it reversed the inhibition of BMSC proliferation induced by Rev-erbα overexpression. Compared with the Rev-erbα group, addition of exogenous Wnt3a protein alone had little effect on Rev-erbα and Bmal1 expression, and the cell proliferation rate was unaffected. These outcomes may result from the fact that in this study a low concentration of Wnt3a protein had little effect on the proliferation rate of cells with good cellular activity.
Among the Wnt signaling factors, the expression pattern of GSK-3β was similar to that of Rev-erbα. Researchers showed that the GSK-3β−/− genotype promoted the expression of Bmal1 in mouse embryonic fibroblast cells, and Bmal1 protein levels declined after rescuing GSK-3β expression in GSK-3β−/− cells. GSK-3β and Bmal1 transcription cycles displayed nearly opposite phases [38]. As an inhibitor of Wnt signaling and an important clock gene component, GSK-3β can phosphorylate Bmal1 to maintain the robustness of the circadian clock. These roles led us to speculate that GSK-3β may be a key factor coordinating the circadian clock and Wnt signaling.
We detected increased β-catenin and c-Myc mRNA levels in each group during osteoinduction, indicating that Wnt/β-catenin signaling was activated during BMSC osteogenesis. At the same time, ALP, OSX, and BSP mRNA levels increased gradually in agreement with Bennett's study showing that the activation of Wnt/β-catenin signaling promoted the expression of osteo-related genes, Runx2 and OSX [27]. Although there have been many studies focusing on the relationship between osteogenesis and Wnt signaling [28,39,40], whether Wnt signaling would repress or promote osteogenesis is still debated. Gong's research showed that adding Wnt3a enhanced the activity of ALP [41]; however, Boland's research showed decreased ALP activity following the addition of Wnt3a [42]. These different results most likely arise from differences in the Wnt signaling pathway activation status. Other studies have shown that canonical Wnt signaling promotes MSC differentiation to osteoblasts while repressing the collagen matrix secretion of osteoblasts [43,44], indicating that the different effects of Wnt/β-catenin signaling on osteogenesis may also be due to the varied MSC differentiation stages.
However, the expression of β-catenin, but not c-Myc, was restrained when Rev-erbα was overexpressed, suggesting that Wnt/β-catenin signaling may not be suppressed by Rev-erbα during osteoinduction. The transcription of downstream Wnt/β-catenin signaling target genes is activated by the binding of β-catenin to T cell factor/lymphoid enhancer factor (TCF/LEF). Therefore, it is most likely that the expression of c-Myc was not restrained as a result of relatively high TCF/LEF expression. Wnt/β-catenin signaling regulates BMSC osteogenesis by controlling the transcription of early-stage osteo-related markers, such as Runx2, OSX, and ALP [45]. Our experiments showed that Rev-erbα might influence late stages of BMSC osteogenesis by repressing the expression of BSP. Wnt/β-catenin signaling and Rev-erbα may regulate the different stages of the osteogenesis process. With regard to the mechanism of cell aging, the interaction between clock genes and developmental signaling requires further studies. Moreover, Rev-erbα, as the important physiological regulator of cell metabolism [46], is likely to be the drug target to treat bone-related diseases in the near future [47].
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (no. 81371113).
Author Disclosure Statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- 1.Sethe S, Scutt A. and Stolzing A. (2006). Aging of mesenchymal stem cells. Ageing Res Rev 5:91–116 [DOI] [PubMed] [Google Scholar]
- 2.Moerman EJ, Teng K, Lipschitz DA. and Lecka-Czernik B. (2004). Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell 3:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS. and Glowacki J. (2008). Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell 7:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, DiGirolamo CM, Navarro PA, Blasco MA. and Keefe DL. (2004). Telomerase deficiency impairs differentiation of mesenchymal stem cells. Exp Cell Res 294:1–8 [DOI] [PubMed] [Google Scholar]
- 5.Kondratov RV. (2007). A role of the circadian system and circadian proteins in aging. Ageing Res Rev 6:12–27 [DOI] [PubMed] [Google Scholar]
- 6.Zvonic S, Ptitsyn AA, Kilroy G, Wu X, Conrad SA, Scott LK, Guilak F, Pelled G, Gazit D. and Gimble JM. (2007). Circadian oscillation of gene expression in murine calvarial bone. J Bone Miner Res 22:357–365 [DOI] [PubMed] [Google Scholar]
- 7.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV. and Antoch MP. (2006). Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 20:1868–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrovsky YV, Samsa WE. and Kondratov RV. (2010). Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging (Albany NY) 2:936–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maronde E, Schilling AF, Seitz S, Schinke T, Schmutz I, van der HG, Amling M. and Albrecht U. (2010). The clock genes period 2 and cryptochrome 2 differentially balance bone formation. PLoS One 5:e11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu EA. and Weaver DR. (2011). Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 3:479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takarada T, Kodama A, Hotta S, Mieda M, Shimba S, Hinoi E. and Yoneda Y. (2012). Clock genes influence gene expression in growth plate and endochondral ossification in mice. J Biol Chem 287:36081–36095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin F, Chen Y, Li X, Zhao Q. and Tan Z. (2013). Over-expression of circadian clock gene Bmal1 affects proliferation and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem Funct 31:166–172 [DOI] [PubMed] [Google Scholar]
- 13.He Y, Chen Y, Zhao Q. and Tan Z. (2013). The roles of Brain and muscle ARNT-like 1 and Wnt antagonist Dkk1 during osteogenesis of bone marrow stromal cells. Cell Prolif 46:644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Lu W, Zhao Y, Rong P, Cao R, Gu W, Xiao J, Miao D, Lappe J, Recker R. and Xiao GG. (2011). Adipocytes derived from human bone marrow mesenchymal stem cells exert inhibitory effects on osteoblastogenesis. Curr Mol Med 11:489–502 [DOI] [PubMed] [Google Scholar]
- 15.DeCarolis NA, Wharton KA., Jr and Eisch AJ. (2008). Which way does the Wnt blow?. Exploring the duality of canonical Wnt signalling on cellular aging. Bioessays 30:102–106 [DOI] [PubMed] [Google Scholar]
- 16.Hoffman J, Kuhnert F, Davis CR. and Kuo CJ. (2004). Wnts as essential growth factors for the adult small intestine and colon. Cell Cycle 3:554–557 [PubMed] [Google Scholar]
- 17.Guillaumond F, Dardente H, Giguère V. and Cermakian N. (2005). Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 20:391–403 [DOI] [PubMed] [Google Scholar]
- 18.Duez H. and Staels B. (2008). Rev-erb alpha gives a time cue to metabolism. FEBS Lett 582:19–25 [DOI] [PubMed] [Google Scholar]
- 19.Duez H. and Staels B. (2009). Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol 107:1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu JM, Wu X, Gimble JM, Guan X, Freitas MA. and Bunnell BA. (2011). Age-related changes in mesenchymal stem cells derived from rhesus macaque bone marrow. Aging Cell 10:66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J. and Lazar MA. (2008). Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol 28:2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart NJ. and Staels B. (2003). The orphan nuclear receptor Rev-erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipo-cyte differentiation. J Biol Chem 278:37672–37680 [DOI] [PubMed] [Google Scholar]
- 23.Iitaka C, Miyazaki K, Akaike T. and Ishida N. (2005). A role for glycogen synthase kinase-3 beta in the mammalian circadian clock. J Biol Chem 280:29397–29402 [DOI] [PubMed] [Google Scholar]
- 24.Jennifer JW, Rachel AK. and Tania MS. (2004). Wnt signaling in osteoblasts and bone diseases (review). Gene 314:19–39 [DOI] [PubMed] [Google Scholar]
- 25.Spencer GJ, Utting JC, Etheridge SL, Arnett TR. and Genever PG. (2006). Wnt signalling in osteobalsts regulates expression of the receptor activator of NFκB ligand and inhibits osteoclastogenesis in vitro. J Cell Sci 119:1283–1296 [DOI] [PubMed] [Google Scholar]
- 26.Janich P, Pascual G, Merlos-Suárez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, DiCroce L. and Benitah SA. (2011). The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480:209–214 [DOI] [PubMed] [Google Scholar]
- 27.Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD. and MacDougald OA. (2005). Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A 102:3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosoi T. (2010). Cytokines in bone diseases. Wnt signal and excessive bone formation. Clin Calcium 20:1526–1531 [PubMed] [Google Scholar]
- 29.Chen Y, Xu X, Tan Z, Ye C, Zhao Q. and Chen Y. (2012). Age-related BMAL1 change affects mouse bone marrow stromal cell proliferation and osteo-differetiation potential. Arch Med Sci 8:30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu XL, Yu G, Parks H, Hebert T, Goh BC, Dietrich MA, Pelled G, Izadpanah R, Gazit D, Bunnell BA. and Gimble JM. (2008). Circadian mechanisms in murine and human bone marrow mesenchymal stem cells following dexamethasone exposure. Bone 42:861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang TS, Grodeland G, Sleire L, Wang MY, Kvalheim G. and Laerum OD. (2009). Induction of circadian rhythm in cultured human mesenchymal stem cells by serum shock and cAMP analogs in vitro. Chronobiol Int 26:242–257 [DOI] [PubMed] [Google Scholar]
- 32.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F. and Schibler U. (2004). Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass to daughter cells. Cell 119:693–705 [DOI] [PubMed] [Google Scholar]
- 33.Welsh DK, Yoo SH, Liu AC, Takahashi JS. and Kay SA. (2004). Bioluminesence imaging of individual reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14:2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheer FA, Hilton MF, Mantzoros CS. and Shea SA. (2009). Adverse metabolic and cardiovascular consequences of circadian misalignment. Prco Natl Acad Sci U S A 106:4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T. and Tezuka M. (2005). Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A 102:12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, et al. (2010). A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A 107:10508–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu N, Yin L, Hanniman EA, Joshi S. and Lazar MA. (2009). Negative feedback maintenance if heme homeostasis by its receptor, Eev-reb-α. Gene Dev 23:2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahar S, Zocchi L, Kinoshita C, Borrelli E. and Sassone CP. (2010). Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One 5:e8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galli C, Passeri G. and Macaluso GM. (2010). Osteocytes and WNT: the mechanical control of bone formation. J Dent Res 89:331–343 [DOI] [PubMed] [Google Scholar]
- 40.Caronia G, Wilcoxon J, Feldman P. and Grove EA. (2010). Bone morphogenetic protein signaling in the developing telencephalon controls formation of the hippocampal dentate gyrus and modifies fear-related behavior. J Neurosci 30:6291–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, et al. ; Osteoporosis-Pseudoglioma Syndrome Collaborative Group. (2001). LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107:513–523 [DOI] [PubMed] [Google Scholar]
- 42.Boland GM, Perkins G, Hall DJ. and Tuan RS. (2004). Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem 93:1210–1230 [DOI] [PubMed] [Google Scholar]
- 43.Kahler RA. and Westendorf JJ. (2003). Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J Biol Chem 278:11937–11944 [DOI] [PubMed] [Google Scholar]
- 44.Eijken M, Meijer IM, Westbroek I, Koedam M, Chiba H, Uitterlinden AG, Pols HA. and van Leeuwen JP. (2008). Wnt signaling acts and is regulated in a human osteoblast differentiation dependent manner. J Cell Biochem 104:568–579 [DOI] [PubMed] [Google Scholar]
- 45.Hill TP, Spater D, Taketo MM, Birchmeier W. and Hartmann C. (2005). Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiation into chondrocytes. Dev Cell 8:727–738 [DOI] [PubMed] [Google Scholar]
- 46.Estelle W, Yasmine S, Laura AS, Christian D, Steve L, Jerome E, Matthijs KH, Charlotte P, Stephane D, et al. (2013). Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 19:1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douglas JK. and Thomas PB. (2014). REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov 13:197–216 [DOI] [PMC free article] [PubMed] [Google Scholar]