Abstract
Recent studies of Escherichia coli thymidylate synthase (ecTSase) showed that a highly conserved residue, Y209, that is located 8 Å away from the reaction site, plays a key role in the protein’s dynamics. Those crystallographic studies indicated that Y209W mutant is a structurally identical but dynamically altered relative to the wild type (WT) enzyme, and that its turnover catalytic rate governed by a slow hydride-transfer has been affected. The most challenging test of an examination of a fast chemical conversion that precedes the rate-limiting step has been achieved here. The physical nature of both fast and slow C-H bond activations have been compared between the WT and mutant by means of observed and intrinsic kinetic isotope effects (KIEs) and their temperature dependence. The findings indicate that the proton abstraction step has not been altered as much as the hydride transfer step. Additionally, the comparison indicated that other kinetic steps in the TSase catalyzed reaction were substantially affected, including the order of the substrate binding. Enigmatically, although Y209 is H-bonded to 3'-OH of 2'-deoxyuridine-5'-monophosphate (dUMP), its altered dynamics is more pronounced on the binding of the remote cofactor, (6R)-N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4folate), revealing the importance of long-range dynamics of the enzymatic complex and its catalytic function.
Keywords: TSase, KIEs, kinetic isotope effects, WT, wild-type, TRS, tunneling ready state
1. Introduction
The role of protein dynamics in enzyme catalysis is one of the open questions in enzymology today. In particular, the participation of residues distal to the active site in catalyzing bond-activation at the active site is a topic of interest as it combines several enigmas: the long range communication in the enzyme, its “holistic” nature, and the different catalytic strategies applied by the same active site to catalyze different chemical conversions. Recent experimental evidence suggests that mutations distant from the active site affect chemical bond activations in numerous enzymatic studies including dihydrofolate reductase [1,2,3,4], human purine nucleoside phosphorylase [5,6], and soybean lipoxygenase-1 [7].
Thymidylate synthase (TSase, EC 2.1.1.45) catalyzes the synthesis of 2'-deoxythymidine-5'-monophosphate (dTMP), using (6R)-N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4folate) as a cofactor and 2'-deoxyuridine-5'-mono-phosphate (dUMP) as the substrate [8]. The essential role of this enzyme in de novo synthesis of a precursor of DNA, thymidine, has made this enzyme an outstanding target for the development of antiproliferative therapeutics for several decades [9,10]. In fact, one of the most commonly used pyrimidine analog drugs in the treatment of cancer is 5-fluorouracil, which is a covalent inhibitor of TSase [10]. Due to its biological and pharmacological importance, TSase has been studied kinetically and structurally over many years in many theoretical and experimental studies [8,11,12,13].
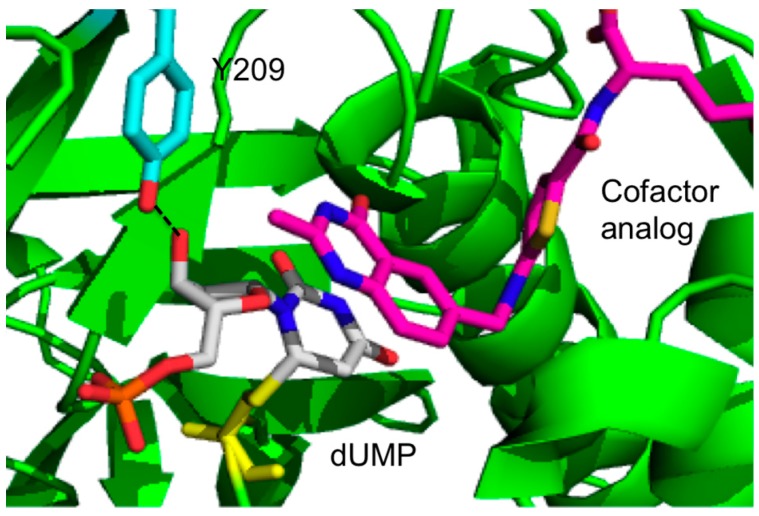
Newby et al. have used kinetic and X-ray crystallography experiments to study the role of a highly conserved residue Y209 of Escherichia coli TSase (ecTSase), 8 Å away from where the chemistry takes place, which contributes one of only two hydrogen bonds to the ribosyl 3'-hydroxyl group of dUMP (Figure 1) [14].
Figure 1.
Active site crystal structure of the wild type (WT) ecTSase (green; PDB ID 2KCE) with 2'-deoxyuridine-5'-mono-phosphate (dUMP) (gray) and CH2H4folate analog, Raltitrexed (magenta). Tyr209 is shown in cyan. H-bond between 3'-OH of the dUMP and Tyr209 is shown in dashed lines. TSase chain is shown in cartoon representation (green). Residue Y209, the ligands dUMP and the cofactor analog, raltitrexed and the thio-ether bond between C-6 of dUMP and the C146 are in sticks. Color code for Y209: cyan (carbons), red (oxygen); Color code for dUMP: blue (nitrogen), grey (carbon), red (oxygen), orange (phosphorous); Color code for raltitrexed: blue (nitrogen), magenta (carbon), yellow (sulfur), red (oxygen), and the atoms in the thio-ether bond are shown in yellow.
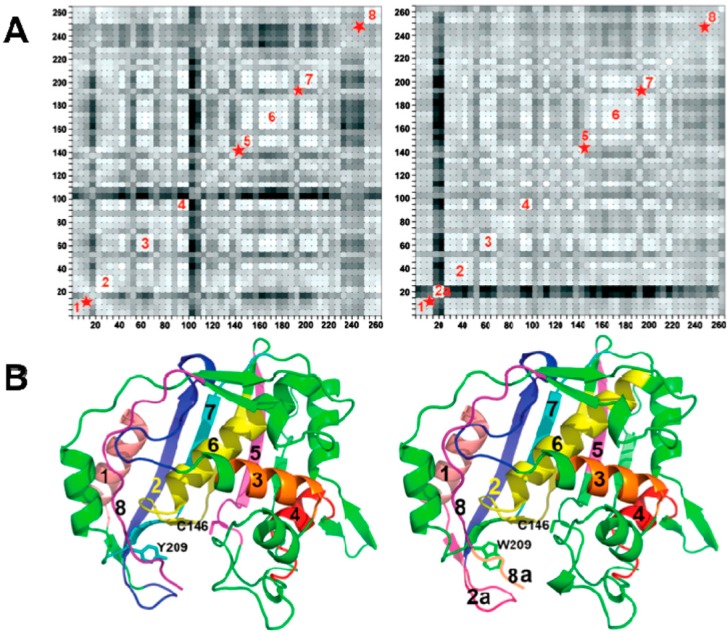
Succinctly, these investigators used the crystal structures of the wild type (WT) and Y209W ternary complexes with dUMP and CB3717, an analogue of the cofactor (PDB IDs 2G80 and 2G8M, respectively). Those studies showed that the crystal structure of Y209W complex is strikingly similar to WT-dUMP-CB3717 at a resolution of 1.3 Å [14,15]. The most conspicuous difference between these two crystal structures was in the anisotropic B-factors, the refined anisotropic B factors of the crystal structures revealed that some protein segments of Y209W have disrupted rigid-body vibrations compared to the WT (Figure 2). Anisotropic B-factors provide the information of the directionality of the atomic mean square displacements in a given crystal. The anisotropic B factors of atoms in several loops across the WT protein are all oriented in the same direction. This suggests a concerted movement of these loops in the WT TSase. However, in Y209W, the anisotropic B factors of those segments (marked by red stars in Figure 2A) are randomly oriented, indicating a disruption of the correlated atomic vibrations of protein residues in the mutant.
Figure 2.
(A) The residue-based matrix plots showing correlations of anisotropic B-factor displacements for Escherichia coli TSase(ecTSase) atom pairs identified by the (x,y) grid coordinates of the plot. The left plot is for the WT ecTSase and the right plot is for Y209W ecTSase (Reproduced from [14] with permission from the American Chemical Society (ACS)). The degree of correlation is represented by color-coding, where the lighter shades of grey indicate greater correlation. The blocks of light-colored squares along the diagonals of the plots indicate the protein residues that vibrate as rigid bodies. The red stars indicate segments with disrupted rigid body vibrations of the Y209W mutation. Segment 2A in the right plot is the phosphate-binding loop with relatively higher B factors; (B) Ribbon diagrams of WT (left) and Y209W (right). The segments labeled in (A) are colored. The cofactor analog (CB3717), dUMP and the mutated residue and catalytic cysteine are shown as sticks. Segment 8A represents the C-terminal. (adapted from [15] with permission from the ACS).
The higher KM of CH2H4folate is in accordance with the conformation of Y209W mutant prior to hydride transfer being more disordered and further away from the dUMP than in the WT. This is in agreement with the significant changes in the average B factors of several loops in Y209W crystal structure compared to the WT. Greater mobility of these loops could impair the binding of the substrate and cofactor. Subsequently, the effect of Y209W mutation on the mobility of this loop (segment 2a, the phosphate-binding loop) can propagate to the other regions in the active site cavity.
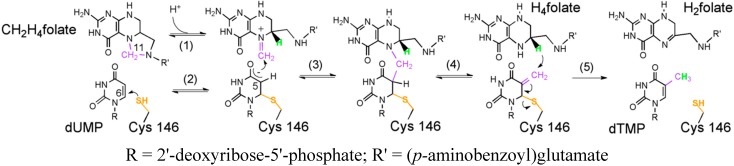
The substitution of tyrosine to tryptophan dramatically affected the catalytic rate and the KM values for the substrate and cofactor. Interestingly, the trend was more pronounced for the cofactor of the enzyme, CH2H4folate, although its binding site is even more remote from the mutation’s site. The lower affinity for CH2H4folate indicates a need of modulating contacts between different segments of the protein. Furthermore, those findings suggest that Y209W mutation change protein dynamics that play a direct role in certain catalytic steps of TSase. However, the analysis performed in [14] did not identify the specific steps affected by the mutation. The mechanism of TSase (Scheme 1) involves several chemical conversions including two C–H bond activations: a rate limiting hydride transfer (step 5) and a much faster proton transfer (step 4). Both these H-transfers have been studied in the WT enzyme via examination of the temperature dependence of intrinsic kinetic isotope effects (KIEintS) [16].
Scheme 1.
The proposed mechanism for thymidylate synthase (adapted from [17] with copyright permission from the ACS).
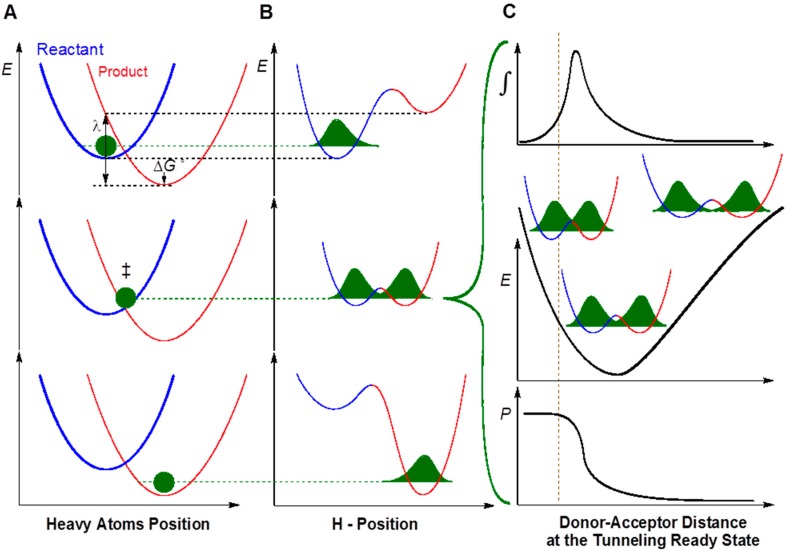
In recent years, the KIEs and their temperature dependence have been used as a gold standard to probe the physical nature of enzyme- catalyzed H-transfer reactions in various studies [18,19,20,21,22]. Models that are used to rationalize temperature dependence of KIEs are sometimes called Marcus-like models [23,24,25,26]. In such a model (Figure 3), hydrogen transfers solely by nuclear quantum mechanical (QM) tunneling that is much faster than the motions of other atoms in the system (surrounding solvents and the protein environment). In the non-adiabatic limit, the reaction coordinate is comprised of two designate orthogonal coordinates, the heavy atom coordinate (Panel A) and the H-atom position (Panel B). Heavy atom reorganization brings the system to the tunneling ready state (TRS), which is the QM delocalized transition state and is the crossing point in middle panels A and B. When the system is at the TRS, hydrogen can tunnel between the donor and the acceptor.
Figure 3.
An example of a model used to analyze kinetic isotope effects (KIE) for H-transfer reactions (sometimes called Marcus-like models). Three slices of the potential energy surface (PES) along components of the collective reaction coordinate showing the effect of heavy-atom motions on the zero point energy in the reactant (blue) and product (red) potential well. (A) Presents the heavy atom coordinate (also known as the Marcus parabolas); and (B) shows the H-atom position, which is orthogonal to the heavy-atom coordinate. In the top panels, the hydrogen is localized in the reactant well, and the zero point energy of the product state is higher than that of the reactant state. Heavy atom reorganization brings the system to the tunneling ready state (TRS, middle panels A and B), where the zero point energy in the reactant and product wells are degenerate and the hydrogen can tunnel between the wells. Further heavy atom reorganization breaks the transient degeneracy and traps the hydrogen in the product state (bottom panels). The rate of reaching the TRS depends on the reorganization energy (λ) and driving force (ΔG°), which are indicated in the top panel (see [24] for more details and equations); (C) shows the effect of Donor–Acceptor Distance (DAD) sampling on the wave function overlap at the TRS (middle panel). Transmission probability (P) is a function of the overlap integral of the hydrogen wave functions in the reactant (blue) and product (red) wells (bottom panel C). The top panel C presents the contribution to H-transfer at each DAD as a function of the P and the population at each DAD. The vertical dashed line represents the DAD under which the Zero Point Energy (ZPE) is greater than the barrier height. At such distances, the process of a wave function spreading from reactant well to product well is no longer “tunneling”, but one can still use the particle’s transmission probability analogously to the tunneling probability at longer DADs.
In the non-adiabatic approximation (Figure 3), the heavy atom motion toward the TRS can be calculated as two parabolas (Marcus-parabolas). In such a case, the rate of reaching TRS is governed by the driving force (ΔG°) and the reorganization energy (λ) which are indicated in the top panel A. Please note that this term has little isotopic sensitivity to the mass of the particle being transferred, and thus it does not affect the model whether the system is electronically adiabatic or not. Panel C shows the effect of donor-acceptor distance (DAD) fluctuations on the wave function overlap at the TRS. This part is isotopically and temperature sensitive, but the difference between nuclear non-adiabatic H-transfer (tunneling) or the adiabatic one (over the barrier), is not substantial, and simply depends on the technical procedure used to calculate the mixing term between the reactant and product states. The top panel C presents a fraction of H-transfer events at each DAD as an integral of the transmission probability (P, bottom panel) and the population as function of DAD (middle panel). This model suggests temperature independent KIEs (as found for most natural, well-evolved enzymes) that result from TRS that is well defined. This means a narrow distribution of DADs, where the DAD’s sampling function (middle panel C) is of such high frequency that the integral is not affected by temperature (within the 35 °C range tested). For mutants [27], non-physiological substrates or conditions [28], or for primitive enzymes [29], a larger temperature dependence of the KIEintS is interpreted as resulting from a broader DAD-distribution. Therefore, the temperature dependence of KIEintS provides unique information on the chemical step of reactions.
In a recent study [15], we used both KIEs and X-ray crystallography experiments to study the correlation between the protein motions at various time scales that influence the hydride transfer reaction of the same TSase mutant Y209W under study here. It was found that with Y209W mutant, the reaction’s intermediate that precedes the hydride transfer forms a thiol-trapped by-product. This finding indicated that the mutation no longer forced the H-donor and acceptor close together, allowing small thiol to compete for that intermediate. Furthermore, the altered dynamics of the H-donor and acceptor at the TRS were probed by temperature dependence of KIEint, indicating disrupted DAD distribution.
Toward the broad goal of understanding in greater detail the impact of protein motions on different catalytic steps, we sought to study another C–H activation in the catalytic cycle of TSase, i.e., the catalyzed proton transfer (step 4 in Scheme 1). In particular, we explore the kinetic properties of Y209W mutant further by comparing the temperature dependence of KIEints on WT vs. Y209W-catalyzed proton transfer reactions. Along with the X-ray crystallographic data, we could correlate the kinetic findings and distinguish the effect of distal mutations on protein motions at different timescales that impact two different H-transfer reactions as well as other catalytic steps.
2. Results and Discussion
2.1. Binding Mechanism of Y209W Mutant
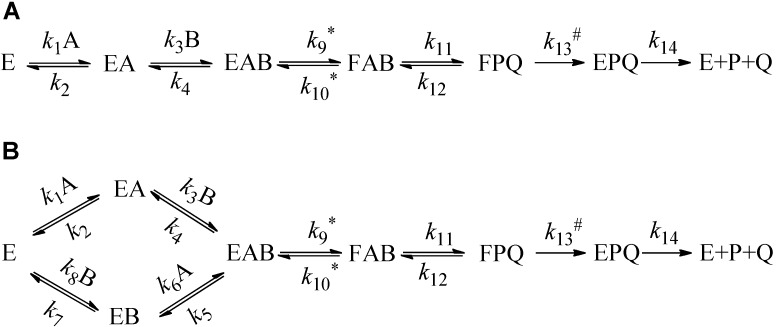
The wealth of information available for the WT ecTSase in terms of steady state kinetics, product inhibition studies, and structure suggests a bi–bi ordered mechanism of reactants binding and product release (Figure 4A) [13,30].
Figure 4.
Binding Scheme for a Sequential Ordered Mechanism (A); and random binding mechanism (B). In the case of TSase, substrate A would be dUMP and B would be CH2H4folate. The rate constant with an asterisk represents isotopically sensitive steps in the proton abstraction experiments, using labeled dUMP and rate constants with hash signs is isotopically sensitive in the hydride transfer experiments using labeled CH2H4folate.
The observed KIEs on the second order rate constant (V/KA or V/KB) for the individual substrates (A: dUMP and B: CH2H4folate in TSase) depend on the mechanism of the substrates’ binding with differences indicated in the expression for the forward commitment factor (Cf, Equations (2)–(4)). The forward commitment factor is the ratio between the rate constants for the forward isotopically sensitive step and the isotopically insensitive steps proceeding backward. Therefore, the KIEs on V/K could discriminate the ordered and random sequential binding mechanisms [31,32]. However, the value of V/KA is always dependent on the concentration of B, while the value V/KB only depends on the concentration of A in the random mechanism (Figure 4B) but not in the ordered mechanism (Figure 4A). Therefore, we could use the substrate dUMP (A), labeled with tritium at C5 with varied concentrations of CH2H4folate, to distinguish between these two mechanisms.
To elucidate the binding mechanism of Y209W, we employed the following equation, which describes the relationship between the observed KIE (KIEobs) for H/T KIEobs on kcat/KM (TV/K) and the KIE after the formation of the ternary EAB complex in Figure 4 (H/T KIE on k9, or Tk9) [31,33].
| (1) |
where EIE (Tkeq.) is the equilibrium isotope effect on the proton abstraction step, Cf and Cr are the forward and reverse commitments to catalysis, respectively.
The Cr for the proton abstraction step represents the competition between the trace tritium and the water protons. Since the tritium released is diluted into ~100 M proton in the reaction medium, Cr is assumed to be zero. This is because the re-association of tritium is negligible given that its concentration is much less than that of protons in water. Thus, Equation (1) can be reduced to Equation (2):
| (2) |
In an ordered mechanism, Figure 4A, the commitment Cf is described by Equation (3):
| (3) |
The commitment Cf for the Figure 4B is described by Equation (4):
| (4) |
where [B] is the concentration of substrate B (CH2H4folate). For ordered mechanisms (Figure 4A), it is apparent from Equations (3) and (4) that Cf varies from infinity at infinite [B] to k9/k4 when [B] goes to zero. In the random mechanism (Figure 4B), Cf goes to k9/k5 at infinite [B] and to k9/(k4 + k5) at zero [B]. Consequently, the KIEobs in the ordered mechanism vary between Tk9 at zero [B] to unity at high [B], while in the random mechanisms, KIEobs vary between two finite values.
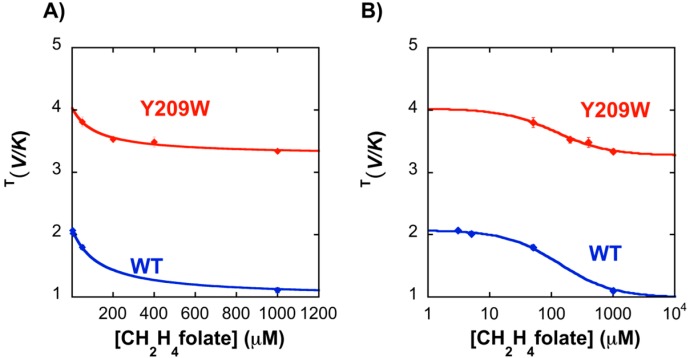
In the current study, KIEobs on the proton abstraction step of the Y209W mutant was studied as a function of the concentration of CH2H4folate. As apparent from Figure 5, the KIEobs for the WT approaches unity at higher concentrations (1 mM) of CH2H4folate, suggesting a strictly ordered binding mechanism in agreement with previous product inhibition and steady-state studies [34]. However, the binding mechanism for the Y209W mutant is more random, as suggested by the non-unity KIE at high CH2H4folate concentrations.
Figure 5.
Observed H/T KIE for proton abstraction as a function of CH2H4folate concentration ((A) on linear scale and (B) on logarithmic scale). Data with WT (Blue) and Y209W (Red) are compared. The lines are fitted to Equation (2).
The observation that Y209W has a less ordered binding mechanism compared to WT ecTSase is further supported by the KM values measured for the substrate and the cofactor for Y209W [15]. In Y209W, KM for dUMP is increased by ~5-fold, and the KM for CH2H4folate is increased by ~16-fold compared to WT, which is in accordance with the observed less ordered binding mechanism for the mutant.
2.2. Temperature Dependence of Intrinsic KIEs in the Y209W Mutant
To investigate the effects of Y209W mutation on the TRS of hydride and the proton transfer steps, we determined the temperature dependence of their intrinsic KIEs. We measured the observed KIEs on the second order rate constant (V/K) competitively and used the Northrop method to extract the KIEintS as described before [32,35]. The KIEintS are presented in Figure 6, where the lines represent the non-linear fit to the Arrhenius equation for KIE:
| (5) |
where k is the microscopic rate constant of the isotopic sensitive step for the light (L) and heavy (T) isotopes, respectively; ∆Ea is the difference in the energy of activation between the light and heavy isotopes (∆Ea = EaL − EaT); R is the gas constant; T is the absolute temperature; and AL/AT is the isotope effect on the pre-exponential factors.
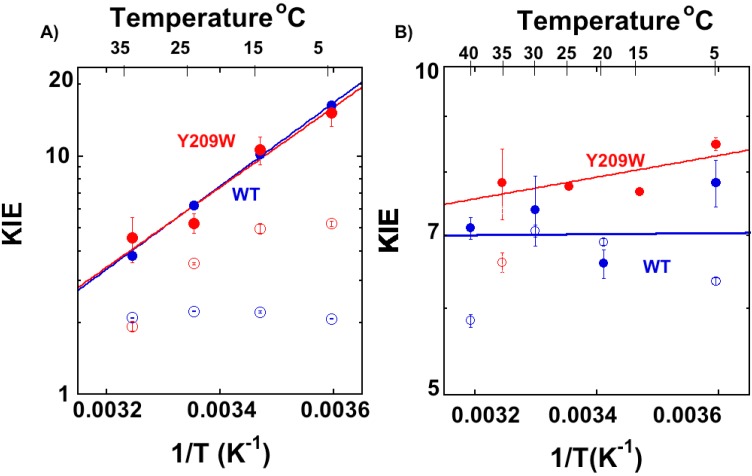
Figure 6.
Arrhenius plots of the primary H/T KIEs on the proton abstraction (A) and the hydride transfer (B) catalyzed by the WT [16,25] (blue) and Y209W [15] (red). The empty circles represent the observed KIEs, and filled circles represent the intrinsic KIEs. The lines represent the least-squares nonlinear regression of KIEintS to Equation (5). Figure S1 presents Arrhenius plots of both H/T and D/T KIEs on proton abstraction and hydride transfer steps.
Table 1 summarizes the H/T and D/T isotope effects on the Arrhenius parameters of both the WT- and Y209W-catalyzed proton abstraction and hydride transfer steps. Importantly, the ∆Ea of WT and Y209W on hydride transfer are different, while upon proton transfer, they are similar within the experimental errors. Figure 6A shows the H/T KIEs on proton transfer and 6B the hydride transfer for both the WT (blue) and Y209W (red). A similar trend was observed for D/T KIEs and are presented in Figure S1 and the H/T and D/T KIE values on proton transfer is presented in Table S2.
Table 1.
Isotope effects on Arrhenius parameters of wild type (WT) and Y209W Escherichia coli thymidylate synthase(ecTSase) on the proton abstraction and the hydride transfer.
| Proton Abstraction | Hydride Transfer | |||
|---|---|---|---|---|
| WT a | Y209W | WT b | Y209W c | |
| AH/AT | 8.3 (±1.0) ×10−6 | 1.4 (±0.1) ×10−6 | 6.8 (±2.8) | 3.6 (±0.9) |
| ∆Ea H–T (kcal/mol) | −8.0 (±0.1) | −7.70 (±0.40) | −0.02 (±0.25) | −0.5 (±0.1) |
Apparently, the intrinsic KIEs on the hydride transfer of WT are temperature-independent while that of Y209W is more temperature-dependent [15]. The lack of temperature dependence of intrinsic KIEs is rationalized as a rearrangement of the heavy atoms (protein and solvent) toward narrow DAD distribution at the TRS. However, in the Y209W mutant, less perfect reorganization seems to occur, as reflected by KIEs with larger temperature dependence. The KIEintS on the proton transfer, on the other hand, are similar and temperature dependent in both the WT and mutant. The fact that proton transfer has temperature dependent KIEs is in accordance with the faster proton transfer which lacks the enzyme induced “fine-tuning” that is critical for the hydride transfer. This has been rationalized as the differences between the difficulty to catalyze the hydride transfer and the ease of catalyzing the proton abstraction [16].
2.3. Effect of Y209W Mutation on Catalytic Steps Other than the Two C–H Activations
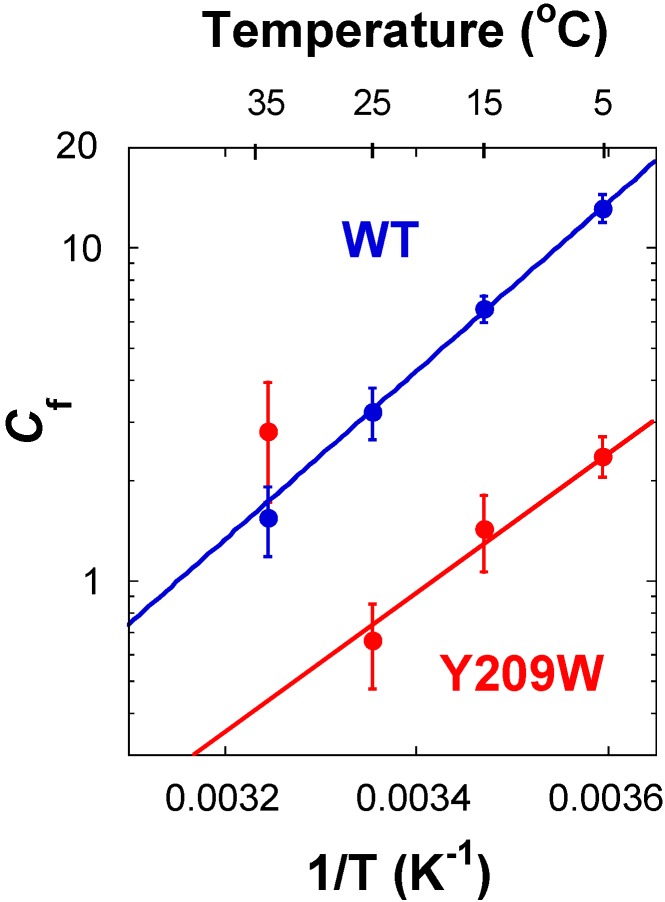
For the proton transfer studied here and the hydride transfer at high and low temperatures, the observed KIEs are smaller than their corresponding intrinsic KIEs (Figure 7) due to kinetic complexity (i.e., Cf > 0). However, the magnitudes of the observed KIEs on proton transfer are higher compared to WT, at the temperature range of 5–25 °C, indicating a reduced kinetic complexity in Y209W compared to WT, as evident from their prospective CfS.
Figure 7.
Comparison of commitment to catalysis (Cf) on the V/K of proton abstraction for WT [16] (blue) and Y209W (red) ecTSase. The lines represent the exponential fit to all data points excluding Y209W at 35 °C.
Figure 6B reveals that there is no commitment on hydride transfer at 25 and 15 °C, indicating it is rate limiting for V/K. It is also true for kcat at 25 °C as reported in [34]. The proton transfer, on the other hand, has non-zero commitments throughout the temperature range. Figure 7 presents the Cf values on the proton transfer of the WT and Y209W mutant as Arrhenius plots (logarithmic scale of Cf vs. the reciprocal of the absolute temperature). The exponential plots for the proton transfers suggest that essentially a single kinetic step is responsible for the Cf, with the exception of 35 °C for the mutant.
At low concentrations of CH2H4folate, Cf for the proton transfer’s T(V/K) approaches k9/(k4 + k5), i.e., the ratio between the rates of the isotopically sensitive proton abstraction step and the rates of dissociation of either dUMP (A) or CH2H4folate (B) from the ternary enzyme–dUMP–CH2H4folate complex (the EAB complex in Figure 4). In addition, Cf on the hydride transfer’s T(V/K) in the mutant case indicates that a step preceding the hydride transfer (most likely the proton abstraction) became more rate limiting. In contrast to the Cf of proton transfer with the WT, Cf with the mutant was significantly lower, which is in accordance with that step becoming more rate limiting overall.
3. Experimental Section
3.1. Materials
[2-14C] dUMP (specific radioactivity 53 Ci/mol) and [5-3H] dUMP (specific radioactivity 13.6 Ci/mmol) were from Moravek Biochemicals. Unlabeled CH2H4folate was a generous gift from Merck and Cie (Weisshausmatte, Switzerland). The WT and Y209W ecTSase enzymes were expressed and purified following a published procedure [36]. Ultima Gold liquid scintillation cocktails were from Perkin Elmer (Waltham, MA, USA). Liquid scintillation vials were from Research Products International Corp. (Mount Prospect, IL, USA). All other materials were purchased from Sigma (St. Louis, MO, USA). All the purifications and analytical separations were performed using an Agilent Technologies model 1100 HPLC system (Santa Clara, CA, USA). The column was from Supelco (C18, 250 × 4.6 mm, 5 μm, Discovery®; by Sigma, St. Louis, MO, USA). The radioactive samples were analyzed using a flow scintillation analyzer (Model RT505 from Packard, now Perkin Elmer Biosciences, Waltham, MA, USA) or a Liquid Scintillation Counter (LSC, from Perkin Elmer Biosciences, Waltham, MA, USA).
3.2. Synthesis of [2-14C,5-2H] dUMP(>99.5% D)
The synthesis of [2-14C, 5-2H] dUMP was carried out by the published procedure of Wataya and Hayatsu [37,38,39]. Briefly, the reaction mixture contained 1 M l-cysteine (pD = 8.8) and 1 mM [2-14C] dUMP in D2O solution (>99.96% D). The reaction mixture was incubated at 37 °C for 7 days until complete deuteration (>99.5% D) was achieved and verified by 1H NMR measurements.
3.3. Competitive KIEs on the Proton Abstraction (Step 4 in Scheme 1)
The competitive method for the primary H/T KIE on the proton transfer step was used according to the procedure published for the WT enzyme with modified experimental conditions [16,40]. The reaction mixture contained 100 mM tris(hydroxymethyl)aminomethane (Tris)/HCl buffer (pH = 7.5), 2 mM TCEP, 1 mM EDTA, 5 mM HCHO, 50 mM MgCl2, 0.5 Mdpm [2-14C] dUMP, and 1.5 Mdpm [5-3H] dUMP. Varied concentrations of CH2H4folate (50–1000 μM) were added to the reaction mixture and preincubated at 25 °C. All the experiments were performed following the previously published procedure with a modified HPLC method as given in Table S1. Three infinity time points (t∞) were obtained by adding concentrated WT TSase. Two independent time points with no enzymes (t0) were obtained as controls for the experiment. The competitive observed KIEs on the second order rate constant were calculated from the following equation [41].
| (6) |
where Rt and R∞ are the 3H/14C ratio of products (3H2O and dTMP) at each time point and at the infinity time points respectively, and f is the fraction conversion typically ranging from 20% to 80%. The fraction conversion f was calculated by [25,40]:
| (7) |
To determine the intrinsic KIEs for the proton abstraction step, 200 μM CH2H4folate was used for both H/T and D/T KIE experiments at the desired temperatures at 5, 15, 25 and 35 °C. The observed D/T KIE was measured the exact same way except using [2-14C, 5-2H] dUMP instead of [2-14C] dUMP. The intrinsic KIEs for the proton abstraction step were calculated using the Northrop method as in Equation (8) [31,35,42]:
| (8) |
where ki is the rate constant for the reaction involving isotope I and T(V/K)Hobs and T(V/K)Dobs are the observed competitive KIE values on the second order rate constant. Although kT/kH, the reciprocal of kH/kT (intrinsic KIE) is the only unknown in Equation (8), it cannot be solved analytically. This equation was solved numerically using the program developed in our group. This program is available on our web site, http://cricket.chem.uiowa.edu/~kohen under “Tools”.
4. Conclusions
The role of long-range amino acid communications and enzyme dynamics across proteins and its catalytic function is of significant contemporary interest and controversy. In the current study, we examined a mutation Y209W ecTSase that is 8 Å from the site where chemistry took place in the enzyme active site. This mutant has been defined as a dynamically altered mutant, based on overlapping crystal structures (down to 1.3 Å resolution) with the WT enzyme and altered anisotropic B-factors. We compared the temperature dependence of intrinsic KIEs on two consecutive steps in the enzyme’s catalytic cycle: the fast and non-rate limiting proton abstraction (step 4 in Scheme 1), and the slow and mostly rate limiting hydride transfer step (step 5 in Scheme 1) of both the WT and Y209W mutant. The findings suggest that this dynamically altered mutant partly distorts the well-defined reactive state of the WT toward the hydride transfer but has no measurable effect on the proton abstraction. This finding can be rationalized in light of the fact that the hydride transfer is the most challenging chemical conversion in this system and has no relevant uncatalyzed equivalent model. Thus, the enzyme had to evolve for accurate rearrangement of the donor and acceptor toward the TRS and those dynamics involved Y209, as evident from the distorted TRS in the mutant.
The proton abstraction step, on the other hand, is not nearly as challenging following the Michael addition of Cys to C6 of the substrate (step 2 in Scheme 1). Indeed, we can easily mimic that step by incubating dUMP with a high concentration of cysteine in solution [37,38,39]. As the enzyme does not need to carefully orient the donor and acceptor in this case, the mutant’s altered dynamics have little to no effect on the nature of the H-transfer of that step. The comparison of intrinsic and observed KIEs, and the analysis of their kinetic complexity (via Cf calculations) indicated that other steps apart from the two C–H bond activations in question have also been altered. The commitments (Cf on V/K) of both H-transfers show a break above 25 °C, indicating an effect of Y209W that could be common to both steps. This observation would be in accordance with a step that precedes the proton transfer that is affected by the mutation. Such a step could be the release of CH2H4folate from the ternary complex, which would also be in accordance with higher KM for that substrate, and the thiols attack on the dUMP intermediately prior to hydride transfer, indicating poor folate orientation in the active site.
In summary, despite its remote location relative to the site of chemical conversion catalyzed by this enzyme, the mutation of Y209 extends in the overall architecture of the enzyme, altering its dynamics (as indicated by anisotropic B-factors analysis), and affects several kinetic steps along the complex catalytic cascade of this enzyme. These results support a dynamic coupling of a remote residue to different kinetic events across the enzyme. The results further emphasize the importance of amino acid residues controlling long-range protein dynamics critical for the enzyme function.
Acknowledgments
This work was supported by NIH GM065368 and NSF CHE-1149023.
Supplementary Files
Author Contributions
Thelma Abeysinghe and Amnon Kohen designed the experiments; Thelma Abeysinghe conducted the experiments under Amnon Kohen supervision; both authors wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wong K.F., Selzer T., Benkovic S.J., Hammes-Schiffer S. Impact of distal mutations on the network of coupled motions correlated to hydride transfer in dihydrofolate reductase. Proc. Natl. Acad. Sci. USA. 2005;102:6807–6812. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Goodey N.M., Benkovic S.J., Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc. Natl. Acad. Sci. USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francis K., Stojković V., Kohen A. Preservation of protein dynamics in dihydrofolate reductase evolution. J. Biol. Chem. 2013;288:35961–35968. doi: 10.1074/jbc.M113.507632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh P., Sen A., Francis K., Kohen A. Extension and limits of the network of coupled motions correlated to hydride transfer in dihydrofolate reductase. J. Am. Chem. Soc. 2014;136:2575–2582. doi: 10.1021/ja411998h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghanem M., Li L., Wing C., Schramm V.L. Altered thermodynamics from remote mutations altering human toward bovine purine nucleoside phosphorylase. Biochemistry. 2008;47:2559–2564. doi: 10.1021/bi702132e. [DOI] [PubMed] [Google Scholar]
- 6.Saen-Oon S., Ghanem M., Schramm V.L., Schwartz S.D. Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase. Biophys. J. 2008;94:4078–4088. doi: 10.1529/biophysj.107.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer M.P., Tomchick D.R., Klinman J.P. Enzyme structure and dynamics affect hydrogen tunneling: The impact of a remote side chain (I553) in soybean lipoxygenase-1. Proc. Natl. Acad. Sci. USA. 2008;105:1146–1151. doi: 10.1073/pnas.0710643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carreras C.W., Santi D.V. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 9.Kanaan N., Marti S., Moliner V., Kohen A. A quantum mechanics/molecular mechanics study of the catalytic mechanism of the thymidylate synthase. Biochemistry. 2007;46:3704–3713. doi: 10.1021/bi061953y. [DOI] [PubMed] [Google Scholar]
- 10.Phan J., Steadman D.J., Koli S., Ding W.C., Minor W., Dunlap R.B., Berger S.H., Lebioda L. Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J. Biol. Chem. 2001;276:14170–14177. doi: 10.1074/jbc.M009493200. [DOI] [PubMed] [Google Scholar]
- 11.Kanaan N., Marti S., Moliner V., Kohen A. QM/MM study of thymidylate synthase: Enzymatic Motions and the temperature dependence of the rate limiting step. J. Phys. Chem. A. 2009;113:2176–2182. doi: 10.1021/jp810548d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Sapienza P.J., Abeysinghe T., Luzum C., Lee A.L., Finer-Moore J.S., Stroud R.M., Kohen A. Mg2+ binds to the surface of thymidylate synthase and affects hydride transfer at the interior active site. J. Am. Chem. Soc. 2013;135:7583–7592. doi: 10.1021/ja400761x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finer-Moore J.S., Santi D.V., Stroud R.M. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: Thymidylate synthase mutagenesis, function, and structure. Biochemistry. 2002;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 14.Newby Z., Lee T.T., Morse R.J., Liu Y., Liu L., Venkatraman P., Santi D.V., Finer-Moore J.S., Stroud R.M. The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261. Biochemistry. 2006;45:7415–7428. doi: 10.1021/bi060152s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Abeysinghe T., Finer-Moore J.S., Stroud R.M., Kohen A. A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase. J. Am. Chem. Soc. 2012;134:17722–17730. doi: 10.1021/ja307859m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Kohen A. Thymidylate synthase catalyzed H-transfers: Two chapters in one tale. J. Am. Chem. Soc. 2010;132:9820–9825. doi: 10.1021/ja103010b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishanina T.V., Koehn E.M., Conrad J.A., Palfey B.A., Lesley S.A., Kohen A. Trapping of an intermediate in the reaction catalyzed by flavin-dependent thymidylate synthase. J. Am. Chem. Soc. 2012;134:4442–4448. doi: 10.1021/ja2120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel Z.D., Klinman J.P. Update 1 of: Tunneling and dynamics in enzymatic hydride transfer. Chem. Rev. 2010;110:PR41–PR67. doi: 10.1021/cr1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Roston D., Kohen A. Experimental and theoretical studies of enzyme-catalyzed hydrogen-transfer reactions. Adv. Protein Chem. Struct. Biol. 2012;87:155–180. doi: 10.1016/B978-0-12-398312-1.00006-8. [DOI] [PubMed] [Google Scholar]
- 20.Klinman J.P., Kohen A. Hydrogen tunneling links protein dynamics to enzyme catalysis. Annu. Rev. Biochem. 2013;82:471–496. doi: 10.1146/annurev-biochem-051710-133623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loveridge E.J., Allemann R.K. The temperature dependence of the kinetic isotope effects of dihydrofolate reductase from thermotoga maritima is influenced by intersubunit interactions. Biochemistry. 2010;49:5390–5396. doi: 10.1021/bi100761x. [DOI] [PubMed] [Google Scholar]
- 22.Sutcliffe M.J., Masgrau L., Roujeinikova A., Johannissen L.O., Hothi P., Basran J., Ranaghan K.E., Mulholland A.J., Leys D., Scrutton N.S. Hydrogen tunnelling in enzyme-catalyzed H-transfer reactions: Flavoprotein and quinoprotein systems. Philos. Trans. R. Soc. B. 2006;361:1375–1386. doi: 10.1098/rstb.2006.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen A., Kohen A. Enzymatic tunneling and kinetic isotope effects: Chemistry at the crossroads. J. Phys. Org. Chem. 2010;23:613–619. doi: 10.1002/poc.1633. [DOI] [Google Scholar]
- 24.Roston D., Islam Z., Kohen A. Isotope effects as probes for enzyme catalyzed hydrogen-transfer reactions. Molecules. 2013;18:5543–5567. doi: 10.3390/molecules18055543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal N., Hong B., Mihai C., Kohen A. Vibrationally enhanced hydrogen tunneling in the Escherichia coli thymidylate synthase catalyzed reaction. Biochemistry. 2004;43:1998–2006. doi: 10.1021/bi036124g. [DOI] [PubMed] [Google Scholar]
- 26.Roston D., Cheatum C.M., Kohen A. Hydrogen donor–acceptor fluctuations from kinetic isotope effects: A phenomenological model. Biochemistry. 2012;51:6860–6870. doi: 10.1021/bi300613e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojkovic V., Perissinotti L.L., Willmer D., Benkovic S.J., Kohen A. Effects of the donor–acceptor distance and dynamics on hydride tunneling in the dihydrofolate reductase catalyzed reaction. J. Am. Chem. Soc. 2012;134:1738–1745. doi: 10.1021/ja209425w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohen A., Cannio R., Bartolucci S., Klinman J.P., Klinman J.P. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature. 1999;399:496–499. doi: 10.1038/20981. [DOI] [PubMed] [Google Scholar]
- 29.Yahashiri A., Howell E.E., Kohen A. Tuning of the H-transfer coordinate in primitive versus well-evolved enzymes. Chem. Phys. Chem. 2008;9:980–982. doi: 10.1002/cphc.200800067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stroud R.M., Finer-Moore J.S. Conformational dynamics along an enzymatic reaction pathway: Thymidylate synthase, “the movie”. Biochemistry. 2003;42:239–247. doi: 10.1021/bi020598i. [DOI] [PubMed] [Google Scholar]
- 31.Northrop D.B. In: Enzyme Mechanisms from Isotope Effects. Cook P.F., editor. CRC Press; Boca Raton, FL, USA: 1991. pp. 181–202. [Google Scholar]
- 32.Cook P.F., Cleland W.W. In Enzyme Kinetics and Mechanism. Garland Science; London, UK; New York, NY, USA: 2007. pp. 253–324. [Google Scholar]
- 33.Cook P.F. In: Enzyme Mechanisms from Isotope Effects. Cook P.F., editor. CRC Press; Boca Raton, FL, USA: 1991. pp. 203–320. [Google Scholar]
- 34.Spencer H.T., Villafranca J.E., Appleman J.R. Kinetic scheme for thymidylate synthase from Escherichia coli: Determination from measurements of ligand binding, primary and secondary isotope effects, and pre-steady-state catalysis. Biochemistry. 1997;36:4212–4222. doi: 10.1021/bi961794q. [DOI] [PubMed] [Google Scholar]
- 35.Northrop D.B. Determining the Absolute Magnitude of Hydrogen Isotope Effects. University Park Press; Baltimore, MD, USA: 1977. pp. 122–152. [Google Scholar]
- 36.Changchien L.-M., Garibian A., Frasca V., Lobo A., Maley G.F., Maley F. High-level expression of Escherichia coli and Bacillus subtilis thymidylate synthases. Protein Expr. Purif. 2000;19:265–270. doi: 10.1006/prep.2000.1245. [DOI] [PubMed] [Google Scholar]
- 37.Hayatsu H., Wataya Y., Kai K., Iida S. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry. 1970;9:2858–2865. doi: 10.1021/bi00816a016. [DOI] [PubMed] [Google Scholar]
- 38.Wataya Y., Hayatsu H. Effect of amines on the bisulfite-catalyzed hydrogen isotope exchange at the 5 position of uridine. Biochemistry. 1972;11:3583–3588. doi: 10.1021/bi00769a014. [DOI] [PubMed] [Google Scholar]
- 39.Wataya Y., Hayatsu H., Kawazoe Y. Cysteine-catalyzed hydrogen isotope exchange at the 5 position of uridylic acid. J. Am. Chem. Soc. 1972;94:8927–8928. doi: 10.1021/ja00780a059. [DOI] [PubMed] [Google Scholar]
- 40.Hong B., Maley F., Kohen A. Role of Y94 in proton and hydride transfers catalyzed by thymidylate synthase. Biochemistry. 2007;46:14188–14197. doi: 10.1021/bi701363s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melander L., Saunders W.H. Reaction Rates of Isotopic Molecules. R.E. Krieger Publication Co.; Malabar, FL, USA: 1987. [Google Scholar]
- 42.Northrop D.B. Steady-state analysis of kinetic isotope effects in enzymic reactions. Biochemistry. 1975;14:2644–2651. doi: 10.1021/bi00683a013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.