Abstract
Equine infectious anemia virus (EIAV) exhibits a high rate of genetic variation in vivo, and results in a clinically variable disease in infected horses. In vivo populations of EIAV have been characterized by the presence of distinct, genetic subpopulations of Rev that differ in phenotype and fluctuate in dominance in a manner coincident with each clinical stage of disease. This study examined the specific mutations that arose in vivo and altered the phenotype. The Rev protein was found to be highly conserved, and only 10 aa mutations were observed at a frequency greater than 10 % in the sample population. Nine of these mutations were capable of significantly altering Rev activity, either as single mutations in the context of the founder variant, or in the context of cumulatively fixed mutations. The results indicated that limited genetic variation outside the essential functional domains of Rev can alter the phenotype and may confer a selective advantage in vivo.
Lentiviruses are characterized by high rates of mutation, recombination and replication, resulting in multiple, diverse populations of viral variants that rapidly adapt to changes in the host environment (Coffin, 1995). Understanding virus and host factors that shape the evolution and selection of viral variants in vivo is an essential component of preventive and therapeutic strategies to control lentivirus infections in humans and animals. Infection of horses with equine infectious anemia virus (EIAV) results in a dynamic disease course characterized by recurrent cycles of fever, viraemia and thrombocytopenia. Most animals eventually gain control of viral replication, progressing to a clinically inapparent stage of disease, yet remain carriers of the virus for life. The dynamics of clinical disease and immune control make EIAV a good model to study the role of both host and viral mechanisms contributing to lentiviral persistence and pathogenesis.
High genetic variation has been observed in the EIAV rev/ tm overlapping reading frames, which encode the regulatory protein Rev and the cytoplasmic tail of the transmembrane (TM) protein (Alexandersen & Carpenter, 1991; Leroux et al., 1997; Belshan et al., 1998). Rev is an essential regulatory protein required for nucleocytoplasmic transport of incompletely spliced viral mRNAs encoding structural proteins. Variation in human immunodeficiency virus type 1 (HIV-1) Rev has been shown to downregulate the expression of viral late genes and alter sensitivity to Gag-specific cytotoxic-T-lymphocytes (CTL) (Bobbitt et al., 2003). In addition, CTL epitopes have been identified within HIV-1 Rev (Addo et al., 2001), as well as within EIAV Rev (Mealey et al., 2003). In EIAV-infected horses, non-progressors exhibited a strong-avidity CTL response to epitopes within Rev, while progressors did not (Mealey et al., 2003). Genetic changes within rev may facilitate immune evasion directly by altering CTL epitopes in Rev, and/or indirectly through altering Rev nuclear-export activity and decreasing expression of structural proteins.
Previously, we undertook longitudinal analyses of EIAV Rev variation throughout a clinically dynamic disease course in one pony experimentally infected with the virulent EIAVWYO2078 (Belshan et al., 2001; Baccam et al., 2003). This pony exhibited a classical disease course, with an acute stage of disease followed by a chronic stage of recurrent febrile episodes, a prolonged inapparent stage of disease and two late febrile episodes. Analysis of EIAV sequences sampled over time identified multiple sub-populations of EIAV Rev that differed in phenotype and fluctuated in dominance coincident with each clinical stage of disease (Baccam et al., 2003). A subpopulation with high Rev phenotype was dominant during the chronic and late chronic stages of disease, whereas a subpopulation with lower Rev phenotype was dominant during the inapparent stage of disease. These studies indicated that in vivo selection on EIAV may drive genetic and phenotypic variation in Rev. In the present study, we used genetic and biological analyses to identify specific changes in Rev genotype that altered phenotype and may have contributed to Rev selection in vivo.
The experimental infection and identification of Rev variants was described previously (Belshan et al., 2001). Briefly, the virulent Wyoming strain of EIAV was used to infect pony 524, and sequential sera samples were collected from different stages of clinical disease. This inoculum has been maintained by serial in vivo passage and contains a heterogeneous population of EIAV, similar to a natural infection. Virion RNA was isolated from the inoculum and from sera samples collected at sequential times post-infection (p.i.), and the rev exon 2/tm overlapping region of EIAV was amplified, cloned and sequenced. All sequences were translated in the Rev open reading frame, and amino acid variants were named in the order they were identified, with identical variants given the same name, e.g. R1. Nucleotide variants were named based on the corresponding amino acid variant name, e.g. R1A, R1B. The consensus rev sequence from the inoculum was the nucleotide variant R1A and Rev amino acid variant R1. This variant was used as the reference strain in all analyses.
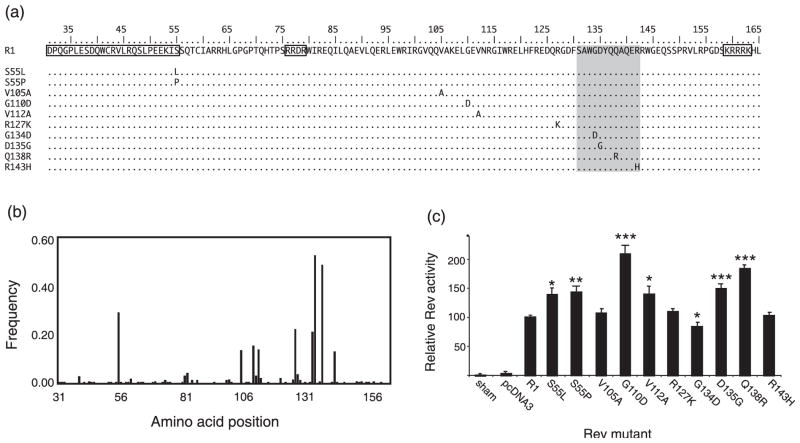
Analyses of Rev evolution and selection are complicated by the fact that the second exon of Rev, which contains the functional domains required for nuclear-export activity (Fridell et al., 1993; Mancuso et al., 1994; Harris et al., 1998; Lee et al., 2006) (Fig. 1a), overlaps with the cytoplasmic tail of TM. To characterize the genetic variation in the population of Rev sequences, we examined the frequency of non-consensus amino acids at each position (Fig. 1b). Genetic variation was observed in 121 of 135 Rev codons; however, the mutations at these positions occurred at a low frequency within the entire population of 320 clones (less than 2 %). Most mutations were observed only in a single clone, and many of these were synonymous changes; 70 aa positions were 100 % conserved. Despite this strong conservation of most amino acid positions, we identified nine highly variable amino acid positions in Rev, which (together) experienced a total of 10 distinct high frequency amino acid mutations from the consensus R1 (Fig. 1a and b). Nine amino acid positions varied in more than 10 % of the sequences, and at eight of these nine positions the nucleotide substitution resulting in amino acid change in Rev was also non-synonymous in TM. One Rev codon, at position 55, experienced a second high frequency mutation that was non-synonymous in both reading frames. Except for this case, the other sites displayed only one high frequency non-consensus amino acid. All but one of the highly variable amino acid positions observed in vivo were found outside the known functional domains of EIAV Rev (Fridell et al., 1993; Mancuso et al., 1994; Harris et al., 1998; Lee et al., 2006) (Fig. 1b). Nonetheless, changes at these positions were almost certainly responsible for the Rev phenotype variation we observed previously (Belshan et al., 2001; Baccam et al., 2003).
Fig. 1.
Identification and functional characterization of high frequency mutations of EIAV rev in vivo. (a) Amino acid sequence of Rev exon 2. The functional domains required for Rev activity are boxed and include the nuclear-export signal (aa 31–55) and the RNA binding/nuclear localization signals (RRDR and KRRRK). The shaded area indicates a region not essential for Rev nuclear-export activity (Lee et al., 2006). The location and identity of the high frequency amino acid changes introduced into the backbone of R1 cDNA are indicated. (b) The frequency and location of non-consensus amino acids in Rev exon 2, relative to the consensus variant, R1. (c) Rev nuclear-export activity of single amino acid mutants depicted in (a). The results are expressed relative to R1, and represent the mean activity of at least six independent transfections ± standard error. Variants that differed significantly from the activity of R1, according to a Student’s t-test with unequal variances, are indicated by asterisks: *P<0.05, ** P<0.005, ***P<0.0005.
To determine the molecular basis of Rev phenotypic variation, each of the 10 single amino acid mutations was introduced into the backbone of an R1 expression vector using PCR-based mutagenesis (Fig. 1a). Rev nuclear-export activity was assayed in transient transfection assays as described previously (Belshan et al., 1998; Harris et al., 1998; Baccam et al., 2003). This reporter assay reflects net Rev activity, which includes the cumulative activity of individual functional domains required for nuclear import, RNA binding and nuclear export. Seven of the 10 aa mutations significantly altered Rev phenotype: six increased Rev activity and one decreased Rev activity (Fig. 1c). The two mutations at position 55 were the only high frequency mutations located within a known functional domain of Rev, at the C-terminal end of the nuclear-export signal. Both S55L and S55P significantly increased Rev activity compared with R1. Surprisingly, three of the four mutations located within a non-essential region of Rev, aa 131–143 (Lee et al., 2006), resulted in significant changes in activity; D135G and Q138R increased activity, whereas G134D decreased activity. The phenotype analyses demonstrated that the majority of Rev mutations observed at a high frequency in vivo were alone sufficient to cause significant changes in nuclear-export activity. Moreover, EIAV Rev activity was found to be highly sensitive to point mutations outside essential functional domains. The presence of multiple mutational pathways to high Rev activity could confer flexibility on a protein whose evolution is constrained by an overlapping reading frame and/or immune epitopes.
It was interesting to note that the majority of genetic changes in a non-essential region of Rev led to marked differences in Rev phenotype, either enhancing or attenuating Rev activity. The non-essential region is also the site of a CTL epitope previously associated with high avidity response in a non-progressor pony (Mealey et al., 2003). The CTL epitope contained the D135G and Q138R mutations, which conferred high Rev activity in our assays. In the non-progressor pony, genetic variation in the CTL epitope included the simultaneous appearance of 135D and 138Q (Mealey et al., 2003). These changes would potentially facilitate immune escape through variation in a high-avidity CTL epitope and/or decreased Rev activity and downregulation of viral gene expression. The non-essential region in Rev may function as a variable, regulatory domain that accommodates a high rate of genetic and phenotypic variation to facilitate immune escape.
To gain insight into how the genetic mutations were related to the evolution and selection of EIAV Rev in vivo, we determined the temporal order of the 10 mutations. Four of the 10 mutations pre-existed in the inoculum, and six mutations arose throughout the course of disease in vivo (Table 1), including the second mutation at position 55. R1 was detected at all stages of disease, but was the dominant variant in the inoculum, as well as during the acute and inapparent stages of disease. Four of the high frequency mutations (S55L, G134D, D135G and Q138R) pre-existed in the inoculum and persisted throughout the course of disease. The S55L single mutation observed in the inoculum persisted during the acute and inapparent stages of disease and did not accumulate any more high frequency mutations. Five of the high frequency mutations arose sequentially during the course of infection in the background of the D135G/Q138R mutations: R127K became dominant (frequency >50 %) at 118 days p.i.; G110D at 201 days p.i.; followed by the simultaneous dominance of S55P, V105A and R143H at 754 days p.i. At 800 days p.i., 91 % of the variants sampled contained these 7 aa changes, which resulted in a significant increase in Rev activity (Belshan et al., 2001). The V112A mutation was observed in the background of the G134D mutation at 35 days p.i., near the end of the acute stage of disease. Although no further high frequency mutations were observed in this background, G134D/V112A variants persisted through the last time point of the inapparent stage of disease.
Table 1.
Temporal appearance of high frequency mutations during disease progression in vivo
| Mutation
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | S55L | D135G | Q138R | R127K* | G110D* | V105A* | S55P* | R143H* | G134D | V112A† | |
| Group‡ | B | B | A | A | A | A | A | A | A | A | A |
| Days p.i.§ | Inoc | Inoc | Inoc | Inoc | 118 | 201 | 754 | 754 | 754 | Inoc | 35 |
Mutations most frequently observed in the background of D135G/Q138R.
Mutation most frequently observed in the background of G134D.
Group assignment of high frequency mutations is based on previous phylogenetic and cluster analyses of Rev populations observed in vivo (Baccam et al., 2003). Variants in Group A have overall higher Rev activity than variants in Group B.
The first day post-inoculation each high frequency Rev mutation was observed at greater than 50 % frequency in the sampled population.
It was of interest to determine if the cumulative appearance of mutations in the D135G/Q138R background conferred progressively higher Rev activity, possibly indicative of increasing fitness during disease. A series of constructs was created that reflected the temporal appearance of the high frequency mutations through 800 days p.i. (Fig. 2). Rev phenotype was quantified in transient expression assays and results were expressed as activity relative to R1. All sequential mutants had Rev activity significantly greater than the variant R1; however, there did not appear to be selection for ever-increasing relative Rev activity. Pre-existing mutations, Q138R and D135G, both alone and together, conferred a dramatic increase in Rev activity relative to R1 (Figs 1 and 2). Many of the five novel mutations that arose during infection substantially altered Rev phenotype, but none significantly decreased Rev activity below that of Q138R. The maintenance of high Rev activity, despite continued mutation, suggests that high Rev activity is important for the virus, especially during febrile stages of disease.
Fig. 2.
Genetic and phenotypic variation in Rev over time. The order of appearance of high frequency Rev mutations was determined by the day each mutant first dominated (frequency >50 %) the sampled population. Rev nuclear-export activity is reported relative to R1. Variants that differed significantly from R1 are indicated by asterisks: *P<0.05, **P<0.005, ***P<0.0005. #, Indicates significant difference from the prior ancestral sequence.
In several instances, the effect of specific mutations on Rev phenotype was dependent on the sequence context of the mutation. R127K and V105A showed no effect on Rev activity when introduced singly in the backbone of R1 (Fig. 1). However, R127K significantly decreased activity in the context of Q138R/D135G (Fig. 2), and V105A significantly increased activity on the background of G134D (data not shown). Overall, nine of the 10 aa changes that occurred at a high frequency in vivo were found to significantly alter Rev activity, either as single mutations or in the context of cumulative mutations. Therefore, specific sites outside the functional domains of Rev can not only accommodate genetic variation, but can also have a positive, negative or neutral effect on Rev phenotype, depending on the sequence context of that change.
Rev overlaps the intracytoplasmic tail (ICT) of TM, and eight of the nine positions non-synonymous in Rev were also non-synonymous in TM. Therefore, selection in vivo may act on non-synonymous changes in TM. The ICT of lentiviruses is unusually long and analyses of primate lentiviruses indicate that the ICT affects multiple steps in virus replication, including infectivity, cytopathicity and assembly (Lee et al., 1989; Gabuzda et al., 1992; Dubay et al., 1992; Kalia et al., 2003; Freed & Martin, 1996; Cosson, 1996). In addition, the ICT has been shown to be a locus for simian immunodeficiency virus attenuation in vivo (Shacklett et al., 2000; Fultz et al., 2001). Limited analyses of EIAV rev/tm variants in the context of infectious molecular clones correlated Rev activity in transient expression assays with replication phenotype in vitro (Baccam et al., 2003).
The success of virus in vivo is a function of its ability to evade immune recognition and elimination as well as its ability to replicate (replicative capacity). Populations of virus in vivo reflect a balance between positive selection for genetic change to escape virus-specific immune responses, and purifying selection to maintain the optimal structure and function of viral proteins needed for replication. Accumulating data suggest that selective factors driving virus genetic diversity include adaptations in replicative capacity that enable the virus to survive in an immunocompetent host (Allen et al., 2005; Liu et al., 2007). An immune evasion strategy that involves Rev-mediated downregulation of protein expression may allow the virus to evade CTL and neutralizing antibody responses that target-specific epitopes from which immune escape exacts a fitness cost. In our studies, the dominance of Rev-attenuated variants during the inapparent stage of infection coincided with the appearance of broadly neutralizing antibody and selection of neutralization-resistant surface glycoprotein variants (Belshan et al., 2001; Sponseller et al., 2007). A vaccine strategy that also targets Rev may abrogate this evasion mechanism and prolong the effectiveness of responses directed at more traditional targets in Gag and Env.
Acknowledgments
The authors thank Yvonne Wannemuehler and Susan Vleck for excellent technical assistance. This work was supported in part by funding from the NIH grant CA97936 and the USDA CREES NRI grant 2002-35204-12699. W. O. S. was partially supported by USDA HEP National Needs Fellowship 2000-3842-8824.
References
- Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Phillips MN, Habeeb K, Khatri A, Brander C, Robbins GK other authors & HIV Controller Study Collaboration. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci U S A. 2001;98:1781–1786. doi: 10.1073/pnas.98.4.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersen S, Carpenter S. Characterization of variable regions in the envelope and S3 open reading frame of equine infectious anemia virus. J Virol. 1991;65:4255–4262. doi: 10.1128/jvi.65.8.4255-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O’Sullivan KM, Desouza I, Feeney ME, Eldridge RL, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam P, Thompson RJ, Li Y, Sparks WO, Belshan M, Dorman KS, Wannemuehler Y, Oaks JL, Cornette JL, Carpenter S. Subpopulations of equine infectious anemia virus Rev coexist in vivo and differ in phenotype. J Virol. 2003;77:12122–12131. doi: 10.1128/JVI.77.22.12122-12131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshan M, Harris ME, Shoemaker AE, Hope TJ, Carpenter S. Biological characterization of Rev variation in equine infectious anemia virus. J Virol. 1998;72:4421–4426. doi: 10.1128/jvi.72.5.4421-4426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshan M, Baccam P, Oaks JL, Sponseller BA, Murphy SC, Cornette J, Carpenter S. Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology. 2001;279:185–200. doi: 10.1006/viro.2000.0696. [DOI] [PubMed] [Google Scholar]
- Bobbitt KR, Addo MM, Altfeld M, Filzen T, Onafuwa AA, Walker BD, Collins KL. Rev activity determines sensitivity of HIV-1-infected primary T cells to CTL killing. Immunity. 2003;18:289–299. doi: 10.1016/s1074-7613(03)00031-1. [DOI] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- Dubay JW, Roberts SJ, Hahn BH, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell RA, Partin KM, Carpenter S, Cullen BR. Identification of the activation domain of equine infectious anemia virus rev. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz PN, Vance PJ, Endres MJ, Tao B, Dvorin JD, Davis IC, Lifson JD, Montefiori DC, Marsh M, et al. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J Virol. 2001;75:278–291. doi: 10.1128/JVI.75.1.278-291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda DH, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Gontarek RR, Derse D, Hope TJ. Differential requirements for alternative splicing and nuclear export functions of equine infectious anemia virus Rev protein. Mol Cell Biol. 1998;18:3889–3899. doi: 10.1128/mcb.18.7.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Gupta P, Montelaro RC. Rational site-directed mutations of LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J Virol. 2003;77:3634–3646. doi: 10.1128/JVI.77.6.3634-3646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hu W, Fisher AG, Looney DJ, Kao VF, Mitsuya H, Ratner L, Wong-Staal F. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res Hum Retroviruses. 1989;5:441–449. doi: 10.1089/aid.1989.5.441. [DOI] [PubMed] [Google Scholar]
- Lee JH, Murphy SC, Belshan M, Sparks WO, Wannemuehler Y, Liu S, Hope TJ, Dobbs D, Carpenter S. Characterization of functional domains of equine infectious anemia virus Rev suggests a bipartite RNA-binding domain. J Virol. 2006;80:3844–3852. doi: 10.1128/JVI.80.8.3844-3852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux C, Issel CJ, Montelaro RC. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, McNevin J, Zhao H, Tebit DM, Troyer RM, McSweyn M, Ghosh AK, Shriner D, Arts EJ, et al. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J Virol. 2007;81:12179–12188. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso VA, Hope TJ, Zhu L, Derse D, Phillips T, Parslow TG. Posttranscriptional effector domains in the rev proteins of feline immunodeficiency virus and equine infectious anemia virus. J Virol. 1994;68:1998–2001. doi: 10.1128/jvi.68.3.1998-2001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey RH, Zhang B, Leib SR, Littke MH, McGuire TC. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology. 2003;313:537–552. doi: 10.1016/s0042-6822(03)00344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacklett BL, Weber CJ, Shaw KES, Keddie EM, Gardner MB, Sonigo P, Luciw PA. The intracytoplasmic domain of the Env transmembrane protein is a locus for attenuation of simian immunodeficiency virus SIVmac in rhesus macaques. J Virol. 2000;74:5836–5844. doi: 10.1128/jvi.74.13.5836-5844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponseller BA, Sparks WO, Wannemuehler Y, Li Y, Antons AK, Oaks JL, Carpenter S. Immune selection of equine infectious anemia virus env variants during the long-term inapparent stage of disease. Virology. 2007;363:156–165. doi: 10.1016/j.virol.2007.01.037. [DOI] [PubMed] [Google Scholar]