Introduction
One key obstacle to the translation of advances in cancer research into the clinic is a deficiency of adequate preclinical models that recapitulate human disease. The development and application of validated preclinical models that reflect patient histological, cellular, and molecular characteristics is needed. Current preclinical models rely heavily on conventional cell line xenograft models which are established by engrafting human tumor cell lines cultured in the laboratory into mice. This model is widely acknowledged to provide useful, but unreliable predictive capacity for anti-tumor activity in humans (Sharpless and Depinho 2006). One possible explanation for the unreliability of cell line xenograft results translating to the clinic, is that these cells represent clonal tumor populations that have selectively grown on plastic and have adapted to growth outside of the natural tumor microenvironment (Frese and Tuveson 2007; Tentler et al. 2012). Because cell line xenograft models lack stromal cells, which are increasingly recognized as a critical element for tumorigenesis, these models fail to accurately recapitulate tumor biology and tumor response to therapy (Bhowmick et al. 2004; Sharpless and Depinho 2006; Frese and Tuveson 2007).
To overcome these disadvantages patient-derived xenografts (PDX), which are established by engrafting fresh patient tumor tissue into immunocompromised mice, have been developed (Figure 1). PDX models are advantageous because they capture tumor heterogeneity and architecture (Sausville and Burger 2006; Siolas and Hannon 2013). PDX models have been shown to be better predictive models for the evaluation of novel therapeutics than cell line xenografts across multiple tumor types (Tentler et al. 2012). A large retrospective review comparing preclinical PDX response rates with Phase II clinical trial response rates found that the PDX models were reliable in predicting response for non-small cell lung cancer and ovarian cancer (Voskoglou-Nomikos et al. 2003). In another study, a panel of 80 PDX (breast, lung, ovarian, testicular, and colon cancer) was shown to have a high clinical predictive value for treatment sensitivity and resistance (Fiebig et al. 2004). Furthermore, data obtained using PDX models have already been successfully translated into the design of clinical trials (Furman et al. 1999; Hidalgo et al. 2011). Given this strong correlation there is much excitement to use PDX models for the study of novel therapies and biomarkers (Bang et al. 2013; Neel et al. 2014). These studies reinforce the vital role that PDX play in the understanding of the biology of human disease and their potential utility to translating results into clinical practice.
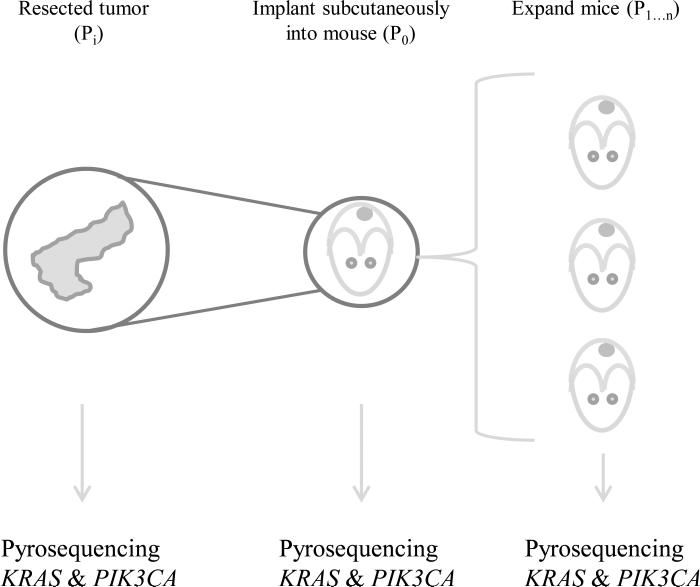
Figure 1. Establishment of patient derived xenograft mouse models.
Tumor pieces (Pi) are implanted subcutaneously into immunocompromised mice (P0). After tumors are established they are harvested, split, and passaged into additional mice (P1...n). Tumor sections are flash frozen and DNA isolated for pyrosequencing initially and at each passage to evaluate KRAS and PIK3CA mutational status.
One key advantage of PDX models is their availability as a renewable resource. Thus multiple therapies may be simultaneously evaluated on the same PDX tumor line. Examination of PDX across multiple passages has found that histologic and gene expression profiles are retained (Siolas and Hannon 2013). Studies of early passage (fewer than three passages) PDX models of multiple solid tumors show that mutations of the source patient tumor are retained (Rubio-Viqueira et al. 2006; Fichtner et al. 2008; Sivanand et al. 2012; Zhang et al. 2013). Although many studies show overall genomic stability across passages whether specific mutations are retained in later passages has not been well studied (Julien et al. 2012; Laurent et al. 2013; Zhang et al. 2013). There is concern that selective pressure and genetic instability could lead to mutational drift over multiple passages, and thus late passage PDX could be an inaccurate reflection of patient tumors (Tentler et al. 2012). Therefore in this study we evaluated if KRAS and PIK3CA mutations were retained at late passages in primary colorectal cancer (1°C CRC), metastatic colorectal cancer (mCRC), and primary pancreatic ductal adenocarcinoma (PDAC) PDX and whether mutational frequency is reflective of patient populations.
Materials and Methods
PDX Expansion
PDAC, 1°C CRC, and mCRC tumor tissue from de-identified patients were engrafted subcutaneously into the flanks of immunocompromised mice, expanded, and passaged over time. All animal experiments were carried out under protocols approved by the University of North Carolina Institutional Animal Care and Use Committee.
DNA Isolation
Tumors were harvested and flash frozen. DNA was isolated using the AllPrep Kit (Qiagen).
Mutational analysis of KRAS by pyrosequencing
Polymerase chain reaction (PCR) of exon 2 to detect KRAS codon 12 and 13 mutations was performed using the following primers: 5’ – CGATGGAGGAGTTTGTAAATGAA – 3’ and 5’ - /BioTEG/TTCGTCCACAAAATGATTCTGA – 3’. PCR amplification was done for 55 cycles with an annealing temperature of 58 C. PCR products were analyzed using pyrosequencing with the Pyromark MD (Qiagen) using the internal primer 5’ – AAACTTGTGGTAGTTGGA – 3’.
Mutational analysis of PIK3CA by pyrosequencing
PCR of exon 9 to detect PIK3CA codon 542 and 545 mutations was performed using the following primers: 5’ – CCATTTTAGCACTTACCTGTGAC – 3’ and 5’ - /BioTEG/ATTTCTACACGAGATCCTCTCTCT – 3’. PCR amplification was done for 55 cycles with an annealing temperature of 62 C. PCR products were analyzed with pyrosequencing using the internal primer 5’ – TTCTCCTGCTCAGTGAT – 3’ for codon 542 and the internal primer 5’ – TAGAAAATCTTTCTCCTG – 3’ for codon 545. PCR of exon 20 to detect PIK3CA codon 1047 mutations was performed using the following primers: 5’ – TGAGCAAGAGGCTTTGGAGTAT – 3’ and 5’ - /BioTEG/TGCTGTTTAATTGTGTGGAAGATC – 3’. PCR amplification was done for 55 cycles with an annealing temperature of 62 C. PCR products were analyzed with pyrosequencing using the internal primer 5’ – GAAACAAATGAATGATGC – 3’.
Results KRAS mutations in PDAC and CRC PDXs
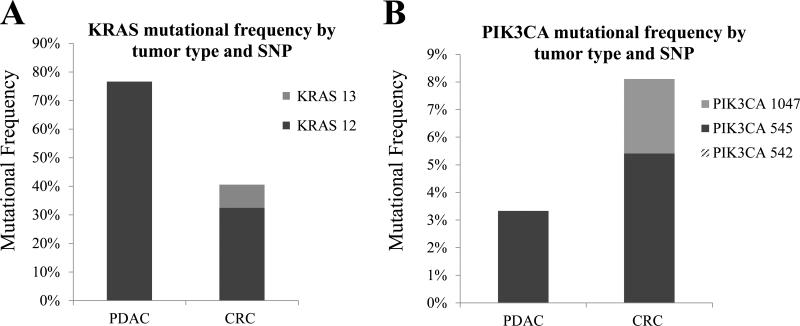
We examined mutations in KRAS codons 12 and 13 in 30 PDAC, 32 mCRC and five 1°C CRC PDX. We found that the frequency of KRAS mutations in PDAC PDX was 77% (23 of 30) (Figure 2a). All mutations were located at KRAS codon 12 and were either G12V (13 of 23 (57%)) or G12D (10 of 23 (43%)). KRAS mutations were found in 41% (15 of 37) of 1°C CRC and mCRC PDX (Figure 2a). Mutations identified in mCRC were KRAS G12V (3 of 14 (21%)), G12D (4 of 14 (29%)), G12S (3 of 14 (21%)), G13D (3 of 14 (21%)), and G12A (1 of 14 (7%)). One of five (20%) 1°C CRC PDX showed a KRAS G12V mutation.
Figure 2. Frequency of KRAS and PIK3CA mutations in PDAC and CRC PDX.
(A) KRAS codon 12 and 13 mutations in PDAC and CRC PDX (B) PIK3CA codon 542, 545, and 1047 mutations in PDAC and CRC PDX. (PDAC = pancreatic ductal adenocarcinoma, CRC = primary and metastatic colorectal cancer, SNP = single nucleotide polymorphism)
PIK3CA mutations in PDAC and CRC PDXs
PDX tumors were examined for mutations in PIK3CA codons 542, 545 and 1047. A single PIK3CA E545K mutation was found in 1 of 30 PDAC PDX (3%) (Figure 2b). PIK3CA mutations were found in 8% (3 of 37) of 1°C CRC and mCRC PDX (Figure 2b). Two of 32 (6%) mCRC PDX had E545K mutations. Both were associated with a G13D KRAS mutation. One mutation was identified in 1°C CRC PIK3CA H1047R (1 of 5 (20%)) and was not associated with a KRAS mutation.
KRAS and PIK3CA mutational status retained across passages
KRAS and PIK3CA mutations were evaluated at later passages (Figures 3 and 4). PDAC PDX passages 2 - 8 were analyzed. KRAS and PIK3CA mutational status was retained in all passages examined in PDAC PDX. 1°C CRC and mCRC PDX passages 2 - 4 were analyzed. KRAS and PIK3CA mutational status was retained across all passages evaluated in 1°C CRC and mCRC PDX.
Figure 3. KRAS mutations are stable across passages.
KRAS mutation status across passages for metastatic CRC (mCRC), primary CRC (1°C CRC), and pancreatic ductal adenocarcinoma (PDAC) PDXs. (ND = not done)
Figure 4. PIK3CA mutations are stable across passages.
PIK3CA mutation status across passages for metastatic CRC (mCRC), primary CRC (1°C CRC), and pancreatic ductal adenocarcinoma (PDAC) PDXs. (ND = not done)
Discussion
The aim of this study was to evaluate if the mutational frequency of key mutations, KRAS and PIK3CA, in 1°C CRC, mCRC, and PDAC PDX, remained stable across late passages and was reflective of patient populations. Our analysis demonstrated that the frequency of KRAS mutations in PDAC was 77%. This correlated with previous studies that have documented that the KRAS mutational frequency in PDAC is 71-100% (Almoguera et al. 1988; Hruban et al. 1993; Pellegata et al. 1994; Hidalgo 2010; Schultz et al. 2012). It is well known that KRAS is one of the key early driver mutations in PDAC (Hingorani et al. 2003; Hezel et al. 2006). Activating mutations in KRAS impair its intrinsic GTPase activity, thus resulting in a protein that is constitutively active, stimulating multiple key kinase pathways integral to cellular survival and proliferation (Hidalgo 2010). This has been confirmed using genetically engineered mouse models in which an activating KRAS mutation was sufficient for the development of precursor pancreatic cancer lesions known as pancreatic intraepithelial neoplasia (Hingorani et al. 2003). KRAS mutational status in CRC has been shown to be an important predictive biomarker of resistance to anti-EGFR therapy and an early mutation in the genetics of CRC (Vogelstein et al. 1988; Amado et al. 2008; Karapetis et al. 2008; Allegra et al. 2009; Walther et al. 2009). In this study we also evaluated KRAS mutational frequency in CRC PDX. We demonstrated that the frequency of KRAS mutations in CRC was 41% which correlated with previous studies that have documented a mutational frequency of 35 - 51% (De Roock et al. 2010; Janku et al. 2011; Tan and Du 2012).
We found a frequency of PIK3CA mutations in PDAC PDX of 3%. This correlates with previous studies that have documented a mutational frequency of 0-11% (Janku et al. 2011). Our analysis demonstrated that the frequency of PIK3CA mutations in CRC PDX was 8%. This result was slightly lower than the 12-21% frequency reported previously. Samuels et al. analyzed 234 CRC tumors for PIK3CA mutational status (Samuels et al. 2004). The overall frequency of PIK3CA mutations identified was 32%. This may be explained by the fact that they evaluated all known PIK3CA single nucleotide polymorphisms (SNPs), whereas our analysis only evaluated the three most common sites (codons 542, 545, and 1047). Janku et al. evaluated 54 CRC tumors for PIK3CA mutational status and identified a 14% PIK3CA mutational frequency for codons 542, 545, and 1047 (Janku et al. 2011). Similarly, De Roock et al. analyzed 773 CRC tumors and identified a 12% PIK3CA mutational frequency for codons 542, 545, and 1047 (De Roock et al. 2010). Our results were not significantly different than those reported in Janku et al. and De Roock et al. (p = NS). While the mutational frequency of 8% that we identified is slightly lower than previously published studies our sample size is small. This result may also be reflective of the population seen in this single institution study.
Of the three PIK3CA mutations identified in CRC the two mutations in exon 9 were associated with KRAS mutations, whereas the sole exon 20 mutation identified was not. This finding is in agreement with previous reports that show associations between exon 9 of PIK3CA and KRAS mutations and not exon 20 (De Roock et al. 2010).
There is concern that genetic drift can occur over late passages in PDX (Julien et al. 2012; Tentler et al. 2012). Because of this, late passage PDX are not routinely used for preclinical drug evaluation (Tentler et al. 2012; Mattie et al. 2013). To evaluate this possibility we characterized PDX genetic stability over late passages. We found that the mutational status of KRAS and PIK3CA was 100% preserved across both early and late passages analyzed in PDAC and CRC PDX. This finding suggests that genetic profiles remain stable over late passages despite potential selection pressures and reinforces the utility of late passage PDX in preclinical experiments.
Conclusion
In conclusion, mutational frequencies in 1°C CRC, mCRC and PDAC PDX closely parallel that of patient populations and crucial mutations remain stable across late passages. The accurate mirroring and stability of genetic changes in PDX models compared to patient tumors suggest that these models are good preclinical surrogates for patient tumors.
Acknowledgements
The authors thank Charlene Ross, the Animal Studies Core Facility, Anne Dvorak and the Harold Mcleod lab for their assistance.
Funding Sources
American College of Surgeons: Resident Research Scholarship (CJT) T32 CA009688 (CJT)
Womack Society Research and Outreach Grant (CJT)
R01 CA14024 (JJY)
References
- 1.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 2.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 3.Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66:3351–3354. doi: 10.1158/0008-5472.CAN-05-3627. discussion 3354. [DOI] [PubMed] [Google Scholar]
- 6.Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–4239. [PubMed] [Google Scholar]
- 8.Fiebig HH, Maier A, Burger AM. Clonogenic assay with established human tumour xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur J Cancer. 2004;40:802–820. doi: 10.1016/j.ejca.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Furman WL, Stewart CF, Poquette CA, et al. Direct translation of a protracted irinotecan schedule from a xenograft model to a phase I trial in children. J Clin Oncol. 1999;17:1815–1824. doi: 10.1200/JCO.1999.17.6.1815. [DOI] [PubMed] [Google Scholar]
- 10.Hidalgo M, Bruckheimer E, Rajeshkumar NV, et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther. 2011;10:1311–1316. doi: 10.1158/1535-7163.MCT-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang D, Wilson W, Ryan M, et al. GSK-3alpha promotes oncogenic KRAS function in pancreatic cancer via TAK1-TAB stabilization and regulation of noncanonical NF-kappaB. Cancer Discov. 2013;3:690–703. doi: 10.1158/2159-8290.CD-12-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neel NF, Stratford JK, Shinde V, et al. Response to MLN8237 in Pancreatic Cancer Is Not Dependent on RalA Phosphorylation. Mol Cancer Ther. 2014;13:122–133. doi: 10.1158/1535-7163.MCT-12-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubio-Viqueira B, Jimeno A, Cusatis G, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 14.Fichtner I, Rolff J, Soong R, et al. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 15.Sivanand S, Pena-Llopis S, Zhao H, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4:137ra175. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Claerhout S, Prat A, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien S, Merino-Trigo A, Lacroix L, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18:5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- 18.Laurent C, Gentien D, Piperno-Neumann S, et al. Patient-derived xenografts recapitulate molecular features of human uveal melanomas. Molecular Oncology. 2013;7:625–636. doi: 10.1016/j.molonc.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 20.Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 21.Pellegata NS, Sessa F, Renault B, et al. K-ras and p53 gene mutations in pancreatic cancer: ductal and nonductal tumors progress through different genetic lesions. Cancer Res. 1994;54:1556–1560. [PubMed] [Google Scholar]
- 22.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 23.Schultz NA, Roslind A, Christensen IJ, et al. Frequencies and prognostic role of KRAS and BRAF mutations in patients with localized pancreatic and ampullary adenocarcinomas. Pancreas. 2012;41:759–766. doi: 10.1097/MPA.0b013e31823cd9df. [DOI] [PubMed] [Google Scholar]
- 24.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 25.Hezel AF, Kimmelman AC, Stanger BZ, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 27.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 28.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 29.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 30.Walther A, Johnstone E, Swanton C, et al. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 31.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 32.Janku F, Lee JJ, Tsimberidou AM, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol. 2012;18:5171–5180. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 35.Mattie M, Christensen A, Chang MS, et al. Molecular characterization of patient-derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia. 2013;15:1138–1150. doi: 10.1593/neo.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]