Abstract
Introduction
Early identification of microbial organisms from respiratory secretions of patients with cystic fibrosis (CF) is important to guide therapeutic decisions. The objective was to compare the accuracy of matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) relative to the conventional phenotypic method in identifying common bacterial isolates, including nonfermenting Gram-negative bacteria, in a cohort of patients with CF.
Methods
A total of 123 isolates from 50 patients with CF representing 14 bacterial species from respiratory specimens were identified using MALDI-TOF MS in parallel with conventional phenotypic methods. Discrepancies were confirmed by 16S ribosomal RNA (rRNA) gene sequencing in five Gram-negative isolates.
Results
The MALDI-TOF MS managed to identify 122/123 (99.2%) bacterial isolates to the genus level and 118/123 (95.9%) were identified to the species level. The MALDI-TOF MS results were 100% consistent to the species level with conventional phenotypic identification for isolates of Staphylococcus aureus, Pseudomonas aeruginosa, Haemophilus influenzae, Streptococcus pyogenes, Achromobacter xylosoxidans, Stenotrophomonas maltophilia, and other uncommon organisms such as Chryseobacterium gleum and Enterobacter cloacae. The 5/123 (4.6%) isolates misidentified were all Gram-negative bacteria. The isolation of E. cloacae and Haemophilus paraphrohaemolyticus may extend the potentially pathogenic list of organisms isolated from patients with CF.
Conclusion
Although the technique provides an early identification and antimicrobial therapy approach in patients with CF, limitation in the diagnosis of uncommon Gram-negative bacteria may exist.
Keywords: cystic fibrosis, MALDI-TOF MS, bacteria, respiratory secretions, molecular identification
Introduction
Cystic fibrosis (CF) is the most common life-threatening recessive genetic disease among Caucasian races, with a frequency of about 1 in 2,500 live births.1 The underlying basic defect in CF, the restriction of infection to the lung, and the chronic nature of infection characterize the vicious cycle of inflammation and respiratory infections. Patients with CF acquire a unique set of bacterial pathogens that are frequently isolated from the respiratory tract in an age-dependent sequence. Staphylococcus aureus is a frequent isolate and may be cultured early in infancy, Haemophilus influenzae is associated with childhood and has been reported to be the most common CF pathogen at the age of 1 year, and Pseudomonas aeruginosa is the most common pathogen in CF and the prevalence increases with age.2 With the improved survival, new emergent pathogens in the CF lung, such as other nonfermenting Gram-negative bacilli of undetermined clinical significance that can be isolated include Achromobacter xylosoxidans, Stenotroph-omonas maltophilia, or species belonging to the Burkhold-eria cepacia complex.3 Other uncommon bacterial species, such as Ralstonia mannitolilytica,4 Pandoraea apista,5 and Inquilinus limosus,6 have also been isolated from patients with CF. Bacteria isolated from patients with CF do not have the same virulence, and different bacterial species display distinct degrees of pathogenicity, thus requiring different clinical management approaches.7 Correct identification of these bacteria by conventional microbiology methods is often limited due to phenotypic misidentification.8
The development of 16S ribosomal RNA (rRNA) gene sequencing has succeeded in providing reliable results. However, it is still more expensive than most traditional identification methods and, therefore, its application is limited to large research and reference laboratories.9 Moreover, the emergence of new bacterial species in respiratory secretions of patients with CF requires rapid identification tools to start early antibacterial therapy. Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) has been recently introduced in clinical microbiology laboratories as a newly emerging technology applied to the problem of bacterial species identification.10 Several studies have reported the use of MALDI-TOF MS for identification of bacteria, including nonfermenting Gram-negative bacteria (NFGNB).11,12 In addition, MALDI-TOF MS is a useful tool for the characterization of microorganisms that are difficult to identify using routine methods.
MALDI-TOF MS has proven to be cost effective and accurate for the identification of microorganisms, obviating the need for routine biochemical methods.13 Thus, in the present work, we used MALDI-TOF MS technology to identify common bacterial pathogens, including NFGNB, from patients with CF, to compare the outcome with phenotypic identification and confirm its discriminatory power for discrepant organisms with that of 16S rRNA gene sequence analysis.
Materials and methods
Patients and specimen collection
Fifty patients with CF were enrolled between July 1, 2013 and December 31, 2013. Sputum samples or deep-oropharyngeal swabs were prospectively collected from both pediatric and adult patients with CF at regular clinical examinations (patients were reviewed at 3-month intervals) as well as during admission for acute exacerbation. The research and ethics committee of Hamad Medical Corporation (reference number 12177/12) granted approval to conduct this study.
Routine identification
The organisms were cultivated on a variety of different media (Remel, Lenexa, KS, USA), including trypticase soy agar with 5% sheep blood, chocolate agar, MacConkey agar, mannitol salt agar, and B. cepacia-selective agar. Plates were incubated in ambient air or 5% CO2 at 35°C for 48 hours. After Gram staining, bacteria were further identified using catalase and oxidase tests. A small inoculum from a single colony of each isolate was used to prepare the inocula for biochemical identification by Vitek II Compact® (BioMérieux, Marcy l’Etoile, France), and Phoenix™ (BD Diagnostics, Sparks Glencoe, MD, USA) as recommended by the manufacturer. In parallel, one single colony was directly deposited on a MALDI-TOF MS target plate (Bruker Daltonik GmbH, Bremen, Germany).
MALDI-TOF MS measurement
MALDI-TOF MS measurements were performed with a 60-Hz nitrogen laser and the spectra were analyzed over a mass range of 2–20 kDa. The strains from both reference and clinical sets were measured and identified by MALDI Biotyper RTC software 3.0 (Bruker Daltonik GmbH). Each series of measurements was preceded by calibration with a bacterial test standard (Bruker Daltonik GmbH). All specimens were processed as per manufacturer’s guidelines. Each sample was tested in duplicate to ensure reproducibility of spectra. Identification scores were interpreted according to the manufacturer’s recommended criteria: a score of >2.0 indicated species-level identification, a score of 1.70–1.999 indicated identification at the genus level, and a score of <1.70 was interpreted as “not reliable identification.” The identification of the tested strain corresponds to the species of the reference strain with the best match in the database. When MALDI-TOF MS and phenotypic identifications agreed, no further analyses were performed. Discrepancies at genus or species level were resolved with a molecular technique based on sequence analysis of 16S rRNA, which is considered the “gold standard” identification method.9
16S rRNA gene sequencing
Further genomic studies were conducted to identify the five discrepant isolates using sequencing studies. Briefly, DNA from pure cultures was obtained using Prepman DNA extraction kits with the MicroSEQ® ID software (Applied Biosystems, Foster City, CA, USA), and DNA estimation was done using the nanodrop method (NanoDrop ND 1000, Thermo Scientific, Waltham, MA, USA). For molecular identification of bacterial isolates, universal primers B27F and 16R1492 and B27F and 1522 R based on the 16S rRNA gene were used in a polymerase chain reaction (PCR) to amplify the nearly complete 1,500-bp sequence. The PCR product was purified using Exo SAP-IT (Affymetrix, Inc., Santa Clara, CA, USA) and sequenced in both forward and reverse directions in duplicate using an ABI PRISM model 3130 automatic DNA sequencer and Big Dye Terminator cycle sequencing kit (Applied Biosystems). The 16S rRNA gene sequence was subjected to BLAST sequence similarity search.
Results
The 50 patients enrolled in this study comprised 29 males and 21 females; their mean age was 14.2±9.9 years. Thirty-one patients with CF were <18 years of age and 19 patients with CF were of ≥18 years of age. A total of 123 isolates from 83 samples representing 14 bacterial species were identified.
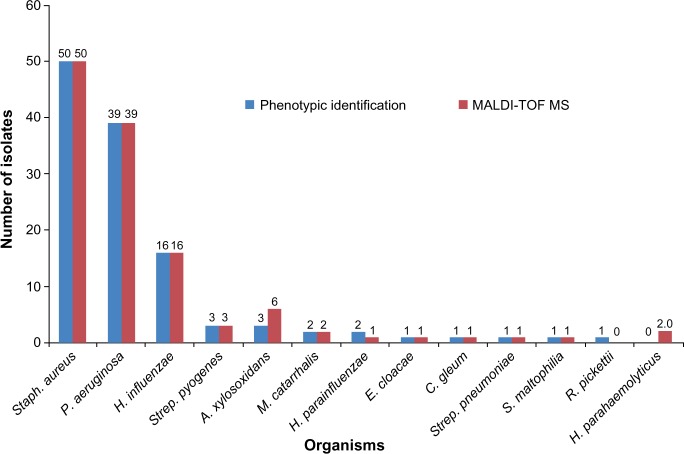
Conventional phenotypic and MALDI-TOF MS identification results are shown in Figure 1. The performance of the MALDI-TOF MS system was, compared to routine conventional methodology for commonly isolated bacteria, faster requiring shorter time for identification. The MALDI-TOF MS correctly identified 122/123 (99.2%) [log (score): <2 and ≥1.7] of the studied isolates at the genus level when compared to routine biochemical phenotypic tests, but the accuracy decreased to 95.9% (118/123) [log (score): ≥2] at the species level. Both methods showed full concordance for 50 isolates of S. aureus, 39 P. aeruginosa, 16 H. influenzae, three Streptococcus pyogenes, three A. xylosoxidans, two Moraxella catarrhalis, and one isolate each of Haemophilus parain-fluenzae, Haemophilus parahaemolyticus, S. maltophilia, Chryseobacterium gleum, and Enterobacter cloacae. A low level of discrepancy was obtained between MALDI-TOF MS and routine biochemical phenotypic tests for Haemophilus, Achromobacter, and Ralstonia species, as no reliable distinction at the species level was observed. Two isolates that were misidentified by routine biochemical phenotypic tests as H. parainfluenzae and Haemophilus species were identified by MALDI-TOF MS as H. parahaemolyticus and by 16S rRNA sequencing analysis as Haemophilus paraphrohaemolyticus (Table 1). Two Achromobacter species were identified as A. xylosoxidans by MALDI-TOF MS, which was later confirmed by 16S rRNA sequencing. One NFGN organism was identified by phenotypic method as Ralstonia pickettii but was identified by MALDI-TOF MS as Achromobacter xylosoxidans and by 16S rRNA sequencing as A. insolitus.
Figure 1.
Identification of bacterial species by conventional phenotypic and MALDI-TOF MS methods.
Notes: The organisms listed are Staphylococcus aureus, Pseudomonas aeruginosa, Haemophilus influenzae, Streptococcus pyogenes, Achromobacter xylosoxidans, Moraxella catarrhalis, Haemophilus parainfluenzae, Enterobacter cloacae, Chryseobacterium gleum, Streptococcus pneumoniae, Stenotrophomonas maltophilia, Ralstonia pickettii, and Haemophilus parahaemolyticus.
Abbreviation: MALDI-TOF MS, matrix-assisted laser desorption/ionization-time of flight mass spectrometry.
Table 1.
Discrepancy in the identification of bacterial isolates by different methods
| Phenotypic identification (strain number) | MALDI-TOF MS identification | 16S rRNA gene sequence | GenBank accession number |
|---|---|---|---|
| Haemophilus parainfluenzae (1612506366) | Haemophilus parahaemolyticus | Haemophilus paraphrohaemolyticus | KJ128299 |
| Haemophilus species (1612505787) | Haemophilus parahaemolyticus | Haemophilus paraphrohaemolyticus | KJ128300 |
| Achromobacter species I (1612508591) | Achromobacter xylosoxidans | Achromobacter xylosoxidans | KJ128296 |
| Achromobacter species II (1612509043) | Achromobacter xylosoxidans | Achromobacter xylosoxidans | KJ128298 |
| Ralstonia pickettii (1612507054) | Achromobacter xylosoxidans | Achromobacter insolitus | KJ128297 |
Abbreviations: MALDI-TOF MS, matrix-assisted laser desorption/ionization-time of flight mass spectrometry; rRNA, ribosomal RNA.
Discussion
Many clinical laboratories have recently and enthusiastically adopted MALDI-TOF MS as a diagnostic method because of its high throughput, relatively low cost, and adaptability to the laboratory workflow; in addition, it was approved by the US Food and Drug Administration in the USA in November 2013 for the identification of Gram-negative bacterial colonies cultured from human specimens. The use of MALDI-TOF MS technique has the advantage of rapid microbial identification and has contributed to decreases in health care expenditures.14
Compared to conventional phenotypic identification methods, by using the database for MALDI-TOF MS-based identification of common pathogens from patients with CF, we were able to show accurate results. Identification to the species level typically requires numerous consecutive steps based on defined phenotypic assays and conventional approaches, which require 24–36 hours after isolation for obtaining definitive results. One hundred percent of our tested common CF isolates, which included S. aureus, P. aeruginosa, H. influenzae, S. pyogenes, Streptococcus pneumoniae, H. parainfluenzae, A. xylosoxidans, S. maltophilia, C. gleum, and E. cloacae, matched to the species level. This is consistent with other reports showing improvement in cost and time when using MALDI-TOF MS-based methods for common routine bacterial identification.15
In the present study, S. aureus was the most common Gram-positive organism isolated from patients with CF, which is in agreement with previous studies that demonstrated the utility of MALDI-TOF MS for the identification of specific groups of Gram-positive bacteria, including S. aureus, coagulase-negative group streptococci.15,16
Correct identification of Haemophilus species based on phenotypic characterization can be challenging and standard phenotypic methods do not reliably distinguish among Haemophilus species.17,18 With the MALDI-TOF MS technology, the rarely encountered species of Haemophilus can be identified.18 However, identification of some strains will be problematic, necessitating DNA sequencing of multiple housekeeping gene fragments or full-length 16S rRNA genes. It has been reported that the discrimination of closely related species H. influenzae, H. parainfluenzae, and Haemophilus haemolyticus is of diagnostic importance because of their striking differences in pathogenicity.17
Although highly sophisticated molecular procedures have been described for a reliable discrimination between H. haemolyticus and H. influenzae,19,20 they are hardly suitable for use under routine diagnostic conditions. In the present study, we demonstrated 100% correct identifications of H. influenzae and H. parainfluenzae by MALDI-TOF MS. Two isolates were identified as H. parahaemolyticus by MALDI-TOF MS and as H. paraphrohaemolyticus by 16S rRNA gene sequencing. Although H. paraphrohaemolyticus has been isolated from a variety of clinical specimens,21 it has not been previously reported in patients with CF. In a previous study, MALDI-TOF MS analysis led to 100% correct identifications of H. influenzae and H. parahaemolyticus, but the technique failed to separate H. parainfluenzae and H. haemolyticus.22
Correct identification of these bacteria by conventional microbiology methods is often limited due to low biochemical reactivity.8
In our study, P. aeruginosa and S. maltophilia were correctly identified (100%) by both routine conventional methods and MALDI-TOF MS. However, performance of the MALDI-TOF MS system was more reliable for identification than conventional routine biochemical phenotypic tests in the case of other Gram-negative bacteria. Overall, five isolates of Achromobacter were correctly identified by MALDI-TOF MS to species level as A. xylosoxidans. Two discrepancies (Achromobacter species) of routine phenotypic and MALDI-TOF MS identifications were resolved by 16S rRNA gene sequencing in favor of the last one. Discrepancy in Achromobacter has also been reported in a recent study for identification of uncommon glucose NFGNB associated with CF.23
Our analyses showed that isolates of species such as C. gleum, which is rarely reported in patients with CF, can be identified by both conventional and MALDI-TOF MS methods.24 This organism has been reported to be a source of nosocomial infections, capable of producing Ambler class B carbapenem-hydrolyzing β-lactamase,25 which might cause treatment failure when β-lactam antibiotics are used as a first-line treatment. It had been thought that a limited spectrum of respiratory pathogens was seen in CF,26 but increasing numbers of other species are being recognized as potential pathogens. The isolation of E. cloacae from a patient with CF in this study may extend the potentially pathogenic list of organisms isolated from this group of patients.
The advantage of the MALDI-TOF MS technique is in providing rapid and early identification of both common and NFGNB strains recovered from patients with CF, apart from being a potential cost-saving method with high reliability.14,27 In addition, it is useful for detecting microorganisms that are difficult to identify using routine biochemical methods. However, the performance of MALDI-TOF MS was not perfect for rare microorganisms due to the low number of isolates obtained. The system cannot determine the antimicrobial resistance patterns. Our results revealed important information on the performance of MALDI-TOF MS in the rapid diagnosis of both common and uncommon bacteria isolated from the lower respiratory secretions of patients with CF.
Acknowledgments
This publication was made possible by the Undergraduate Research Experience Program (grant number UREP12-057-3-011) from the Qatar National Research Fund (a member of Qatar Foundation). The statements made herein are solely the responsibility of the authors.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ratjen F, Doring G. Cystic fibrosis. Lancet. 2003;361(9358):681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 2.Cystic Fibrosis Foundation . 2012 Annual Data Report. Bethesda, MD: Cystic Fibrosis Foundation Patient Registry; 2013. [Google Scholar]
- 3.Lambiase A, Raia V, Del Pezzo M, Sepe A, Carnovale V, Rossano F. Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis. 2006;6:4. doi: 10.1186/1471-2334-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye T, Vandamme P, LiPuma JJ. Infection by Ralstonia species in cystic fibrosis patients: identification of Ralstonia pickettii and R. mannitolilytica by polymerase chain reaction. Emerg Infect Dis. 2002;8(1):692–696. doi: 10.3201/eid0807.010472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segonds C, Paute S, Chabanon G. Use of amplified ribosomal DNA restriction analysis for identification of Ralstonia and Pandoraea species: interest in determination of the respiratory bacterial flora in patients with cystic fibrosis. J Clin Microbiol. 2003;41(7):3415–3418. doi: 10.1128/JCM.41.7.3415-3418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellinghausen N, Essig A, Sommerburg O. Inquilinus limosus in patients with cystic fibrosis, Germany. Emerg Infect Dis. 2005;11(3):457–459. doi: 10.3201/eid1103.041078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LiPuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23(2):299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosshard PP, Zbinden R, Abels S, Böddinghaus B, Altwegg M, Böttger EC. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J Clin Microbiol. 2006;44(4):1359–1366. doi: 10.1128/JCM.44.4.1359-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferroni A, Sermet-Gaudelus I, Abachin E, et al. Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center. J Clin Microbiol. 2002;40(10):3793–3797. doi: 10.1128/JCM.40.10.3793-3797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49(4):543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 11.Sandrin TR, Goldstein JE, Schumaker S. MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom Rev. 2013;32(3):188–217. doi: 10.1002/mas.21359. [DOI] [PubMed] [Google Scholar]
- 12.Fernández-Olmos A, García-Castillo M, Morosini MI, Lamas A, Máiz L, Cantón R. MALDI-TOF MS improves routine identification of non-fermenting Gram negative isolates from cystic fibrosis patients. J Cyst Fibros. 2012;11(1):59–62. doi: 10.1016/j.jcf.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Gaillot O, Blondiaux N, Loiez C, et al. Cost-effectiveness of switching to matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine bacterial identification. J Clin Microbiol. 2011;49(12):4412. doi: 10.1128/JCM.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai AP, Stanley T, Atuan M, et al. Use of matrix assisted laser desorption ionisation-time of flight mass spectrometry in a paediatric clinical laboratory for identification of bacteria commonly isolated from cystic fibrosis patients. J Clin Pathol. 2012;65(9):835–838. doi: 10.1136/jclinpath-2012-200772. [DOI] [PubMed] [Google Scholar]
- 15.Bizzini A, Durussel C, Bille J, Greub G, Prod’hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48(5):1549–1554. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McElvania Tekippe E, Shuey S, Winkler DW, Butler MA, Burnham CA. Optimizing identification of clinically relevant Gram-positive organisms by use of the Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system. J Clinc Microbiol. 2013;51(5):1421–1427. doi: 10.1128/JCM.02680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy TF, Brauer AL, Sethi S, Kilian M, Cai X, Lesse AJ. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenza. J Infect Dis. 2007;195(1):81–89. doi: 10.1086/509824. [DOI] [PubMed] [Google Scholar]
- 18.Nørskov-Lauritsen N. Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans. Clin Microbiol Rev. 2014;27(2):214–240. doi: 10.1128/CMR.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukundan D, Ecevit Z, Patel M, Marrs CF, Gilsdorf JR. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J Clinc Microbiol. 2007;45(10):3207–3217. doi: 10.1128/JCM.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson R, Wang X, Briere EC, et al. Haemophilus haemolyticus isolates causing clinical disease. J Clin Microbiol. 2012;50(7):2462–2465. doi: 10.1128/JCM.06575-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedegaard J, Okkels H, Bruun B, Kilian M, Mortensen KK, Nørskov-Lauritsen N. Phylogeny of the genus Haemophilus as determined by comparison of partial infB sequences. Microbiology. 2001;147(pt 9):2599–2609. doi: 10.1099/00221287-147-9-2599. [DOI] [PubMed] [Google Scholar]
- 22.Frickmann H, Christner M, Donat M, et al. Rapid discrimination of Haemophilus influenzae, H. parainfluenzae, and H. haemolyticus by fluorescence in situ hybridization (FISH) and two matrix-assisted laser-desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) platforms. PLoS One. 2013;8(4):e63222. doi: 10.1371/journal.pone.0063222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homem de Mello de Souza HA, Dalla-Costa LM, Vicenzi FJ, et al. MALDI-TOF: a useful tool for laboratory identification of uncommon glucose non-fermenting Gram-negative bacteria associated with cystic fibrosis. J Med Microbiol. 2014;63(pt 9):1148–1153. doi: 10.1099/jmm.0.076869-0. [DOI] [PubMed] [Google Scholar]
- 24.Coenye T, Goris J, Spilker T, Vandamme P, LiPuma JJ. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol. 2002;40(6):2062–2069. doi: 10.1128/JCM.40.6.2062-2069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellais S, Naas T, Nordmann P. Genetic and biochemical characterization of CGB-1, an Ambler class B carbapenem-hydrolyzing β-lactamase from Chryseobacterium gleum. Antimicrob Agents Chemother. 2002;46:2791–2796. doi: 10.1128/AAC.46.9.2791-2796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cystic Fibrosis Trust Laboratory Standards for Processing Microbiological Samples from People with Cystic Fibrosis: Report of the UK Cystic Fibrosis Trust Microbiology Laboratory Standards Working Group. 2010. Available from https://www.cysticfibrosis.org.uk/media/82034/CD_Laboratory_Standards_Sep_10.pdf.
- 27.Alby K, Gilligan PH, Miller MB. Comparison of matrix-assisted laser desorption-time of flight (MALDI-TOF) mass spectrometry platforms for the identification of Gram-negative rods from patients with cystic fibrosis. J Clin Microbiol. 2013;51(11):3852–3854. doi: 10.1128/JCM.01618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]