Abstract
Chromosomal translocation is a common cause of leukaemia1 and the most common chromosome translocations found in leukaemia patients involve the mixed lineage leukaemia (MLL) gene2,3. AF10 is one of more than 30 MLL fusion partners in leukaemia4. We have recently demonstrated that the H3K79 methyltransferase hDOT1L contributes to MLL–AF10-mediated leukaemogenesis through its interaction with AF10 (ref. 5). In addition to MLL, AF10 has also been reported to fuse to CALM (clathrin-assembly protein-like lymphoid–myeloid) in patients with T-cell acute lymphoblastic leukaemia (T-ALL) and acute myeloid leukaemia (AML)6,7. Here, we analysed the molecular mechanism of leukaemogenesis by CALM–AF10. We demonstrate that CALM–AF10 fusion is both necessary and sufficient for leukaemic transformation. Additionally, we provide evidence that hDOT1L has an important role in the transformation process. hDOT1L contributes to CALM–AF10-mediated leukaemic transformation by preventing nuclear export of CALM–AF10 and by upregulating the Hoxa5 gene through H3K79 methylation. Thus, our study establishes CALM–AF10 fusion as a cause of leukaemia and reveals that mistargeting of hDOT1L and upregulation of Hoxa5 through H3K79 methylation is the underlying mechanism behind leukaemia caused by CALM–AF10 fusion.
Our interest in CALM–AF10 fusion protein stems from our recent demonstration that hDOT1L, a histone H3K79 methyltransferase8, has an important role in MLL–AF10-mediated leukaemogenesis5. We demonstrated that mistargeting of hDOT1L to the Hoxa9 gene by MLL–AF10 results in H3K79 methylation and Hoxa9 upregulation, which contributes to leukaemic transformation5. We further demonstrated that the hDOT1L and MLL–AF10 interaction involves the octapeptide motif-leucine zipper (OM–LZ) region of AF10, this is also required for MLL–AF10-mediated leukaemic transformation9. The observation that the OM–LZ region is retained in the CALM–AF10 fusion protein raises the possibility that hDOT1L may also function in CALM–AF10-mediated leukaemia. To examine the potential role for hDOT1L in CALM–AF10-mediated leukaemia, we first attempted to establish a causal relationship between the CALM–AF10 fusion protein and leukaemia.
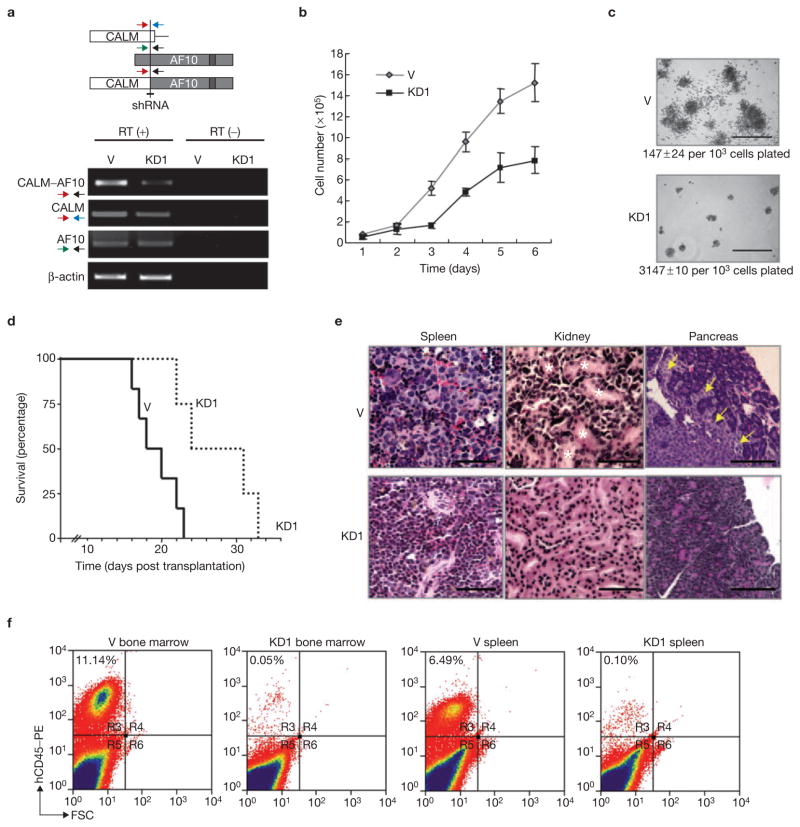
Previous studies have established that the human monocytic leukemia cell line U937 expresses the CALM–AF10 fusion protein10,11. To determine whether CALM–AF10 is relevant for cell proliferation and transformation, CALM–AF10 was knocked down in U937 cells using a vector-based RNA interference (RNAi) approach5. Results shown in Fig. 1a indicate that we were able to generate a stable U937 derivative cell line with significant CALM–AF10 knockdown, whereas neither the CALM nor AF10 mRNA is affect by the RNAi. Compared with the vector-transduced control, the knockdown (KD1) cells not only proliferated more slowly in liquid RPMI media (Fig. 1b), but also resulted in fewer and smaller colonies when cultured on methylcellulose (Fig. 1c). Similar results were obtained using a second knockdown clone KD2 (data not shown). These results suggest that knockdown of CALM–AF10 affects cell proliferation in vitro.
Figure 1.
Knockdown of CALM–AF10 in U937 cells impairs their proliferation and leukaemogenesis in vitro and in vivo. (a) Schematic representation of the fusion protein and the target region for knockdown. The arrows indicate the position of primers used for RT–PCR. The RT–PCR results evaluating the knockdown efficiency are also shown. β-actin served as a control. V, vector control; KD1, knockdown clone 1. (b, c) Effects of CALM–AF10 knockdown on cell proliferation in RPMI medium (b) and methylcellulose (c). Only colonies having over 50 cells were counted. The numbers below the pictures in c represent colony numbers ± s.d. The experiment was repeated twice. n = 4 for V and KD1 in both experiments. The scale bar represents 1 mm. (d–f) Effects of CALM–AF10 knockdown in vivo. Knockdown CALM–AF10 extends survival time of transplanted mice when compared with those receiving transplantation of parental U937 cells (d). Haematoxylin and eosin staining demonstrating that mice transplanted with CALM–AF10 knockdown cells less infiltration of leukaemic cells in spleen, kidney and pancreas when compared with mice transplanted with control U937 cells (e). Asterisks indicate renal tubules and arrows indicate infiltration of transplanted U937 cells in the pancreatic tissue. Original magnification: 40× for spleen, 20× for others. The scale bar represents 50 μm. FACS analysis of proliferation in bone marrow and spleen of transplanted U937 hCD45-positive cells (f). CALM–AF10 knockdown affects U937 cell proliferation in both bone marrow and spleen.
To evaluate the role of CALM–AF10 in vivo, 1 × 107 knockdown and control cells were transplanted into NOD–SCID mice. CALM–AF10 knockdown prolonged the survival time of the transplanted mice (Fig. 1d). Histological analysis of the mice transplanted with the vector-transduced control cells revealed infiltration of leukaemic cells in multiple organs at the terminal stage (Fig. 1e). In contrast, leukaemic cells were barely detectable in the organs of the mice transplanted with CALM–AF10 knockdown cells when analysed on the same day of post-transplantation (Fig. 1e). FACS analysis of cells isolated from bone marrow and spleen revealed that transplantation with CALM–AF10 knockdown cells resulted in a significantly lower percentage of human cells when compared with control (0.05% versus 11.14% in bone marrow, 0.10% versus 6.49% in spleen), even though equal numbers of cells were transplanted (Fig. 1f). Collectively, these data indicate that the CALM–AF10 fusion protein contributes to cellular proliferation and maintenance of the transformed state of leukaemic cells in vitro and in vivo.
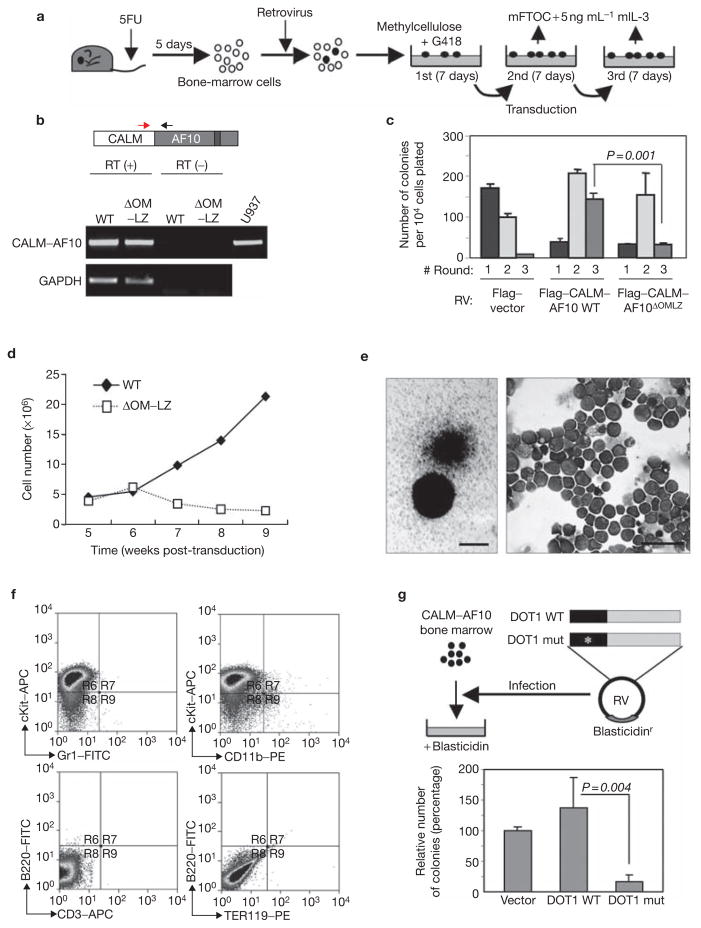
We next analysed whether expression of CALM–AF10 is sufficient to cause bone-marrow cell transformation. For this purpose, a methylcellulose-colony serial-plating assay was used (Fig. 2a). To examine the role of hDOT1L in CALM–AF10-mediated bone marrow transformation, we first established that hDOT1L can interact with CALM–AF10 and that the interaction is dependent on the OM–LZ region of AF10 (see Supplementary Information, Fig. S1). A colony serial-plating assay was then performed by transduction of mouse bone-marrow cells with a wild-type or a CALM–AF10 mutant that lacked the hDOT1L interaction region, OM–LZ. RT–PCR analysis confirmed expression of both fusion proteins in cells derived from the second round of colonies (Fig. 2b). The colony serial-plating assay indicated that the capacity to generate a significant number of third round colonies is dependent on the ability of CALM–AF10 to interact with hDOT1L (Fig. 2c). Although the colony numbers did not increase in the third round of plating when compared with the second round (Fig. 2c), the cells were able to proliferate continuously in mouse fetal thymus organ culture (mFTOC) media supplemented with mouse interleukin-3 (mIL-3) for more than 6 months (Fig. 2d and data not shown). In contrast, cells that expressed the OM–LZ deletion mutant of CALM–AF10 did not generate a significant number of third round colonies in the methylcellulose assay and did not continue to proliferate in mFTOC media after 6 weeks of culture (Fig. 2c, d). The colonies exhibit typical granulocyte–macrophage-like morphology (Fig. 2e), which mainly consisted of immature myeloid cells and a few macrophages (Fig. 2e). Consistently, FACS analysis indicated that most of the cells express high levels of c-Kit (Fig. 2f), although a small population of c-Kitlow CD11b+ cells were also observed. Cells expressing other lineage-specific markers, such as Gr-1, B-220, CD3 and TER-119, were not observed (Fig. 2f), indicating that these c-Kithigh cells are likely to be early progenitor bone-marrow cells that have not committed to any haematopoietic cell lineages. Based on these results, we conclude that expression of CALM–AF10 fusion protein is sufficient to transform progenitor bone-marrow cells and that the hDOT1L interaction region is required for the transformation capacity of CALM–AF10. The observation that the hDOT1L interaction region of AF10 is required for the transformation capacity of CALM–AF10 strongly suggests that hDOT1L has an important role in CALM–AF10-mediated leukaemogenesis. To further demonstrate the role of hDOT1L mediated H3K79 methylation in proliferation and leukaemic transformation by CALM–AF10, wild-type and H3K79 methyltransferase-defective hDOT1L was expressed in CALM–AF10-transformed bone-marrow cells and their effects on colony formation in methylcellulose were analysed. Expression of the wild-type hDOT1L enhanced the colony formation, whereas expression of the mutant hDOT1L strongly suppressed the colony-forming capacity of the CALM–AF10-expressing cells (Fig. 2g). Taken together, these results allow us to conclude that hDOT1L, and its associated H3K79 methyltransferase activity, have an important role in CALM–AF10-mediated leukaemic transformation.
Figure 2.
Expression of CALM–AF10 is sufficient to cause mouse bone-marrow cell transformation and hDOT1L has an important role in this process. (a) Schematic representation of the retroviral transduction procedures described in the Methods. (b) Confirmation of expression of the transduced genes by RT–PCR. U937 cells serve as a positive control. GAPDH serves as a control for equal input of RNA in RT–PCR. (c) Serial colony-plating assay of bone-marrow cells transduced by wild-type and mutant CALM–AF10 with vector control. The average colony numbers ± s.d. of three independent experiments are shown. The P value of the third round colony numbers between the wild-type and the OM–LZ deletion mutant is also indicated. (d) Growth curve of the transduced cells in mFTOC culture. (e) Morphology of colonies formed in the third round of methylcellulose assays transduced by CALM–AF10. The scale bar represents 0.5 mm. Wright-Giemsa-stained cytospin preparation of cells from the third round of colonies is also shown. The scale bar represents 50 μm. (f) Immunophenotyping by FACS of CALM–AF10 transformed cells. The cell surface markers used in the analysis are indicated. (g) Schematic representation of the second transduction procedure and the effect of wild-type and catalytic-deficient hDOT1L on colony formation of CALM–AF10 transformed cells. RV, retrovirus.
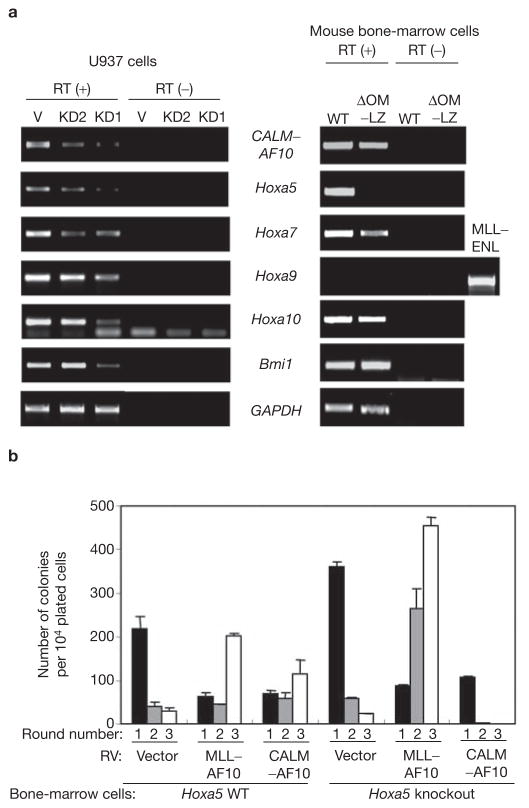
cDNA microarray studies have revealed that overexpression of late Hoxa genes and Bmi-1 are a characteristic of leukaemias involving CALM–AF10 and some MLL-fusion proteins12–14. In particular, over-expression of Hoxa9 has been demonstrated to be crucial for leukaemias involving MLL–ENL and MLL–AF10 fusion proteins5,15,16. To understand the molecular mechanism by which hDOT1L and H3K79 methylation contribute to leukaemogenesis by CALM–AF10, late Hoxa genes and Bmi-1 expression was examined by RT–PCR. To avoid potential non-specific effects of small-interfering RNA (siRNA), two independent clones expressing siRNAs that target the CALM–AF10 junction (Fig. 1a) were analysed. Similar results were obtained in the two independent knockdown clones, KD1 and KD2, and both resulted in decreased expression of late Hoxa and Bmi-1 genes when compared with the parental U937 cells (Fig. 3a). Consistent with this result, overexpression of CALM–AF10 in mouse bone-marrow cells resulted in activation of these genes (Fig. 3a). The hDOT1L interaction region did not seem to have a role in Bmi-1 upregulation, although it did affect expression of late Hoxa genes (Fig. 3a). Interestingly, the expression level of Hoxa5 was most closely correlated with that of CALM–AF10 in U937 cells, and Hoxa5 was significantly upregulated in mouse bone-marrow cells transduced by CALM–AF10 when compared with cells transduced by the OM–LZ deletion mutant (Fig. 3a), suggesting that hDOT1L functions in Hoxa5 upregulation. Given that the expression levels of Hoxa9 were undetectable in the CALM–AF10 transformed cells, Hoxa5 upregulation may have a key role in the transformation process. To determine whether Hoxa5 is necessary for CALM–AF10-mediated leukaemic transformation, we performed colony-replating assays using bone-marrow cells isolated from Hoxa5 knockout mice. In parallel, we also analysed the effect of Hoxa5 knockout on the transformation capability of MLL–AF10. Although CALM–AF10 can transform bone-marrow cells derived from the wild-type mice, as evident from increased colony numbers in the third round of plating (Fig. 3b), it fails to transform bone-marrow cells deficient for Hoxa5. In contrast, Hoxa5 knockout does not affect the transforming capability of MLL–AF10. Based on these results, we conclude that Hoxa5 upregulation is critical for CALM–AF10 mediated leukaemic transformation.
Figure 3.
Hoxa5 is critical for leukaemic transformation by CALM–AF10. (a) RT–PCR analysis of the expression of late Hoxa genes and Bmi-1 in vector control (V) and two independent CALM–AF10 knockdown clones (KD1 and KD2) derived from U937 cells, and in mouse bone-marrow cells derived from second round colonies and transduced by wild-type and OM–LZ deletion CALM–AF10 mutants. Lack of detectable signal for Hoxa9 is not due to primer failure as the same primers detected Hoxa9 expression in MLL–ENL transduced cells. (b) Hoxa5 knockout attenuates the transformation capability of CALM–AF10, but has no effect on that of MLL–AF10. Serial colony-plating assays were performed as in Fig. 2a. The average colony numbers ± s.d. of three independent experiments is shown. Due to embryonic lethality of Hoxa5 knockout in C57B6 strain, the bone-marrow cells used in the assays were derived from Hoxa5 knockout and wild-type littermates of 129Sv/Ev-MF1 strain24.
CALM is a cytoplasmic protein known to function in clathrin assembly17,18. It contains a putative CRM1-dependent nuclear-exporting signal (NES) sequence at its carboxy-terminus, which is retained in the CALM–AF10 fusion protein19 (see Supplementary Information, Fig. S2). On the other hand, AF10 is a nuclear protein and its nuclear-localization signal (NLS) is also retained in the CALM–AF10 fusion protein (see Supplementary Information, Fig. S2). Depending on the subcellular localization, CALM–AF10 may directly or indirectly contribute to Hoxa5 upregulation. To determine the subcellular localization of CALM–AF10, plasmid DNA that encodes Flag-tagged CALM–AF10 was transfected into U2OS cells. In parallel, two constructs encoding CALM–AF10 with deletion on the OM–LZ region and Flag-tagged CALM were also transfected. Immunofluorescence microscopy revealed that most of the overexpressed CALM–AF10 protein exhibited cytoplasmic distribution (Fig. 4a). However, nuclear distribution in wild-type, but not in mutant CALM–AF10 transfected cells was also noticeable (Fig. 4a, b). In contrast, CALM exhibited a mesh-like cytoplasmic distribution (Fig. 4a), which is characteristic of clathrin-related proteins18. Interestingly, when CALM–AF10 is coexpressed with hDOT1L, CALM–AF10 becomes localized to the nucleus (Fig. 4c, d). High resolution analysis of the proteins indicates that they colocalize in the nucleus (Fig. 4e). Importantly, this change in CALM–AF10 localization depends on its ability to interact with hDOT1L, as coexpression of hDOT1L with a CALM–AF10 mutant that lacks the OM–LZ region has no effect on the localization of the fusion protein (Fig. 4c, d). In addition, hDOT1L did not affect CALM localization (Figs. 4c, d). These results strongly suggest that hDOT1L interacts with and retains CALM–AF10 in the nucleus and that the nuclear CALM–AF10 observed in Fig. 4a and b is likely to be due to the interaction of CALM–AF10 with endogenous hDOT1L.
Figure 4.
hDOT1L retains CALM–AF10 in the nucleus. (a) Subcellular distribution of wild-type and OM–LZ deletion mutants, Flag–CALM–AF10 and CALM in U2OS cells. Transfected cells (red) are indicated with arrows. Nuclei are stained with DAPI (blue). (b) Analysis of nuclear staining by Flag antibody with confocal microscopy. (c) Immnofluorescent staining of U2OS cells cotransfected with hDOT1L (green) and Flag–CALM–AF10 (red). (d) Quantification of the cells presented in c. N, cells expressed in the nucleus; C, cells expressed in the cytoplasm. (e) Confocal microscopy analysis of the nuclear localization of hDOT1L and CALM–AF10 when coexpressed. The two staining patterns observed are shown. The pattern shown in the lower panels with more dots was predominant (~80%). The scale bar represents 10 μm in a, b and e, and 20 μm in c.
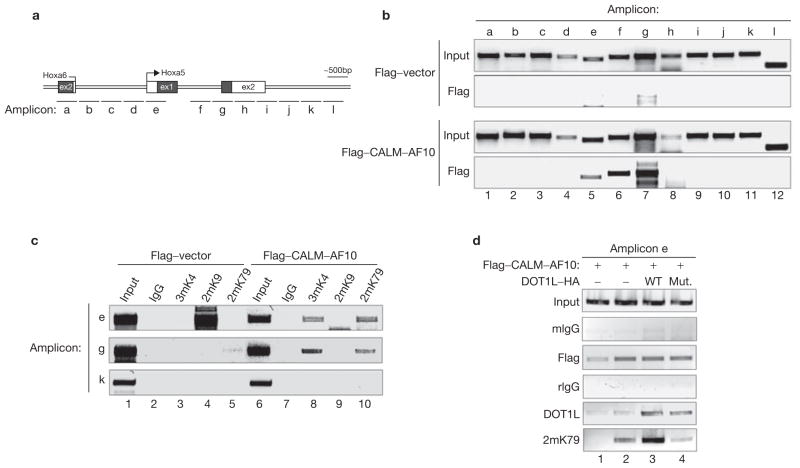
The observation that CALM–AF10 can localize to the nucleus and interact with hDOT1L raises the possibility that Hoxa5 might be a direct target of CALM–AF10. To address this possibility, binding of CALM–AF10 across the entire Hoxa5 gene was analysed by chromatin immunprecipitation (ChIP) assay using the CALM–AF10 transformed cells described in Fig. 2. The fusion proteins bound to a region covered by amplicons e–g (Fig. 5b, lanes 5–7). The observed binding is specific, as no signal is observed in a parallel ChIP experiment using cells transduced by an empty vector. Given that CALM–AF10 interacts with the H3K79 methyltransferase hDOT1L, we expect the CALM–AF10 binding sites to be enriched for H3K79 methylation in the CALM–AF10 transformed cells. ChIP results presented in Fig. 5c demonstrate that H3K79 methylation correlates with the presence of CALM–AF10 in the transformed cells but not in the control cells. Consistent with the hypothesis that H3K4 methylation correlates with gene activation and H3K9 methylation correlates with gene repression, H3K9 methylation was observed at the Hoxa5 gene promoter when it is silenced in non-transformed bone-marrow cells (Fig. 5c), whereas H3K4 methylation was observed when the gene is activated in CALM–AF10 transformed cells (Fig. 5c). All the ChIP signals are specific as they are not present in a control region covered by amplicon k.
Figure 5.
CALM–AF10 in association with hDOT1L binds to Hoxa5 in both the promoter and downstream regions to mediate local H3K79 methylation. (a) A schematic representation of the Hoxa5 gene locus and amplicons used for ChIP assays. Ex, exon. (b) ChIP analysis of the location of CALM–AF10 across the Hoxa5 locus in CALM–AF10 transduced and control bone-marrow cells. Input = 0.5%. (c) ChIP analysis of histone modifications in selected regions of the Hoxa5 gene. Cells and antibodies used in the assay are indicated. Input = 2%. (d) ChIP analysis of the DOT1L-dependent H3K79 methylation in amplicon e of the Hoxa5 gene in control and transduced U937 cells. Input = 0.5%.
To examine whether H3K79 methylation of the Hoxa5 gene in CALM–AF10 transformed cells depends on hDOT1L recruitment and its enzymatic activity, we focused on the transcription start site encompassed by amplicon e. As antibodies that can specifically ChIP CALM–AF10 are not available, U937 cells were transduced with retroviruses expressing Flag-tagged CALM–AF10 in the presence or absence of cotransduction of a HA-tagged wild-type or mutant hDOT1L. After selection, the cells were subjected to ChIP analysis. Overexpression of CALM-AF10 lead to recruitment of hDOT1L, concomitant with increased H3K79 methylation and overexpression of hDOT1L significantly increased the hDOT1L recruitment concomitant with increased levels of H3K79 methylation (Fig. 5d). In contrast, overexpression of the enzymatically defective hDOT1L mutant, although increased its recruitment, resulted in lower level of H3K79 methylation when compared with that of the cells transduced by CALM–AF10 alone, supporting the notion that the mutant hDOT1L functions in a dominant–negative fashion (Fig. 2g). These results are consistent with a model in which CALM–AF10, in association with hDOT1L, is targeted to the Hoxa5 gene. This targeting results in upregulation of the Hoxa5 gene, which in turn contributes to leukaemic transformation.
Using bone-marrow transformation and transplantation assays, we demonstrate that CALM–AF10 fusion is both necessary and sufficient for leukaemogenesis (Figs 1 and 2). In addition, we provide evidence that hDOT1L has an important function in CALM–AF10 mediated leukaemic transformation (Fig. 2c, d, g). Furthermore, we demonstrated that hDOT1L contributes to CALM–AF10-mediated leukaemic transformation by preventing nuclear export of CALM–AF10 (Fig. 4), and by methylation of H3K79 at the Hoxa5 gene (Fig. 5), which contributes to its activation. Finally, we showed that Hoxa5 is specifically required for CALM–AF10 mediated leukaemic transformation (Fig. 3b). Thus, our studies not only establish CALM–AF10 as a cause of leukaemic transformation, but also reveal that Hoxa5 and hDOT1L as important players in leukaemias that involve CALM–AF10 fusion.
We have previously demonstrated that hDOT1L also contributes to leukaemogenesis by MLL–AF10 (ref. 5). Although leukaemogenesis mediated by MLL–AF10 and CALM–AF10 both involve hDOT1L and Hox gene activation, the underlying mechanisms are not the same. Unlike Hoxa9, Hoxa5 does not seem to have a key role in MLL–AF10-mediated leukaemic transformation (Fig. 3b). It worth noting that although Hoxa9 overexpression has been observed in a number of leukemias, its overexpression is not always required for leukaemic transformation20. This highlights the importance of distinguishing over-expressed Hoxa genes that are necessary for leukaemic transformation from those not essential for transformation. A second difference lies in their distinct immunophenotypes. Whereas CALM–AF10 transformed bone-marrow cells are mainly lineage-uncommitted progenitors (positive only for c-Kit), the cells transformed by MLL–AF10 tend to differentiate into myeloid lineage with expression of lineage markers (Fig. 2f and see Supplementary Information, Fig. S3)5. This observation is consistent with the fact that leukaemia patients with CALM–AF10 often develop haematologic malignancy of multi-lineages — an indication that the leukaemic cells have originated from a very early stage of haematopoietic development21. Upregulation of different Hoxa genes may be linked to these phenotypical differences. For example, constitutive expression of Hoxa5 in human CD34+ cells prevents differentiation toward erythrocytes and increases myelopoiesis22. Although there is currently no report that overexpression of Hoxa5 in haematopoitic stem cells can directly cause leukaemic transformation, many leukaemia cell lines without MLL-fusion have a higher incidence of overexpressing Hoxa5 than overexpressing Hoxa9 (ref. 23), indicating that upregulation of Hoxa5 may not be limited to leukaemia mediated by CALM–AF10. The third difference is that DOT1L also contributes to the retention of CALM–AF10 in nucleus, which is necessary for targeting CALM–AF10 to the Hoxa5 gene. Despite the difference in the particular Hox genes involved, the catalytic activity of hDOT1L and the interaction between hDOT1L and AF10 are both important in leukaemogenesis involving CALM–AF10 and MLL–AF10. Therefore, inhibition of the catalytic activity of hDOT1L or interaction between hDOT1L and AF10 may constitue a useful therapeutic treatment for leukaemia patients with CALM–AF10 or MLL–AF10 fusion.
METHODS
CALM–AF10 knockdown in U937 cells and transplantation
DNA encoding a 21 bp siRNA (5′-ATCAGGAGCACAGAGATGTGA-3′) that targets the junction of CALM and AF10 was subcloned into pMSCV–puro with an H1 gene promoter. Transduced cells were selected by puromycin (0.5 μg ml−1) and cloned by limiting dilution. Knockdown efficiency was evaluated by RT–PCR. Five to ten week old NOD–SCID mice were irradiated (300 rad) and after 6 h, CALM–AF10 knockdown and vector-transduced U937 cells were intraorbitally injected (1 × 107 cells per mouse, n = 6 for each group). Mice were monitored and killed at the terminal stage for histology and FACS analysis. Some knockdown mice were killed before the terminal stage for control experiments.
Mouse bone-marrow cell isolation and transduction
Four to ten week old C57BL/6 mice were used to harvest bone-marrow cells. Hoxa5-deficient bone-marrow cells were obtained from 8–10 week old 129Sv/Ev–MF1 mice24. Mice were treated with 150 mg kg−1 5-fluorouracil (5FU) for 5 days before bone-marrow cells were harvested. Lin− cells were enriched using EasySep mouse haematopoietic progenitor cell enrichment kit (Stem Cell Technology, Vancouver, Canada) and used for retroviral transduction. cDNA of CALM–AF10 was cloned from U937 cells by RT–PCR. Wild-type and OM–LZ deletion mutant CALM–AF10 were subcloned into pMSCV–neo downstream of a Flag-tag. Retrovirus preparation, transduction and colony assays were performed as previously described5. In the experiments presented in Fig. 3b, G418 (1 mg ml−1) was added to the methylcellulose until the 3rd round, to prevent the growth of untransduced cells. Colonies on methylcellulose were picked and further cultured in mFTOC (10% FBS in RPMI1640, 1 mM MEM sodium pyruvate, 1% MEM non-essential amino acid, 10 mM HEPES at pH 7.3, 50 μm 2-mercaptoethanol with 5 ng ml−1 mIL-3 (Peprotech, Rocky Hill, NJ). Transduction of DOT1L with blasticidin-resistant retrovirus was previously described5. For transduction of U937 cells, retroviruses expressing hDOT1L–HA with a neomycin-resistant gene were used.
Cell culture, transfection, immunostaining and immunoprecipitation
U937 cells were maintained in RPMI 1640 supplemented with 10% fetal calf serum. 293T or U2OS cell were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Transfection was performed by using Fugene6 (Roche, Indianapolis, IN). Twenty-four hours after transfection, U2OS cells were fixed with 3% paraformaldehyde and permiabilized with 0.1% Triton X-100 for 5 min. Flag M2 (Sigma, St Louis, MO) and hDOT1L5 antibodies were used as primary antibodies for immunostaining. DAPI was used for nuclear staining.
For immunoprecipitation, 293T cells were harvested 36 h after transfection and washed with ice-cold PBS before being lysed with lysis buffer (50 mM HEPES–KOH at pH 7.9, 420 mM KCl, 0.1 mM EDTA at pH 8.0, 5 mM MgCl2, 20% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 μg aprotinin per ml, 0.5 μg leupeptin per ml and 0.7 μg pepstatin per ml). After incubation for 1 h at 4 °C, cells were gently sonicated and the cell debris was removed by centrifugation at 14,000g for 30 min. Anti-Flag M2 agarose beads (Sigma) were added to 0.5 mg protein extracts and incubated for 3 h at 4 °C. After five washes with lysis buffer containing 500 mM KCl, the immunoprecipitates were analysed by western blotting.
FACS analysis
Cells harvested from NOD–SCID mice and the mouse bone-marrow cells transduced by CALM–AF10 were incubated for 20 min on ice with CD16–CD32 Fc block (BD Pharmingen, Franklin Lakes, NJ) and subsequently stained for 30 min on ice with phycoerythrin-, fluorescein isothiocyanate- and allophycocyanin-conjugated isotype controls and monoclonal antibodies against human CD45, mouse CD11b, Gr-1, c-kit, B220, CD3 and Ter119 (BD Pharmingen). Cells were then washed with 2% FBS-containing PBS and analysed using a FACSCalibur (BD Pharmingen). Data collected were analysed with Summit V3.1 (Cytomation Inc., Fort Collins, CO).
RT–PCR analysis of Hox genes, Bmi-1 and CALM–AF10
Total RNA was isolated from vector-transduced or CALM–AF10-knockdown U937 cells or colonies of mouse bone-marrow cells derived from second and third round of wild-type or mutant CALM–AF10 transduced cells using RNeasy (Qiagen, Valencia, CA). Up to 1 μg of total RNA was treated with RNase-free DNase I and applied for reverse transcription using ImProm-II (Promega, Madison, WI) according to manufacturer’s protocol. Half a microliter of cDNA from the 20 μl RT reaction was used as template for PCR amplification using Platinum Taq Polymerase (Invitrogen, Carlsbad, CA) with 33–35 cycles. One tenth of the cDNA was used for β-actin and GAPDH amplification. Primer sequences for Hoxa and Bmi-1 are shown in the Supplementary Information, Table S1. Primers for detection of CALM and AF10 used in Fig. 1a do not amplify the CALM–AF10 gene and the primers for CALM–AF10 used in Figs 1a, 2b and 3a do not amplify CALM or AF10. The primer sequences are shown in the Supplementary Information, Table S1.
ChIP assay
For modification-specific anti-histone and anti-Flag antibody Chip, 1 × 106 cells were used. For immunoprecipitation, 1.5 μg of each antibody was used. The procedure used is similar to a previous publication5. For ChIP of U937 cells using anti-DOT1L and anti-H3K79me2 antibodies 5 × 106 cells were used. Lysis buffer containing 0.5% NP-40 and 0.5% Triton X-100, instead of 1% SDS, was used. After RNase A and proteinase K treatment, DNA was purified by PCR purification kit (Qiagen) and amplified by PCR using either Platinum Taq Polymerase (Invitrogen) or Ex-Taq (TaKaRa, Otsu, Japan), with 35 cycles for modification-specific histone antibodies and 37–38 cycles for Flag and DOT1L antibodies. Modification-specific histone antibodies used are as follows: anti-H3K4me3 (Abcam, Cambridge, UK); anti-H3K9me2 (Upstate, Lake Placid, NY); and anti-H3K79me2 (ref. 8). Primer sequences are listed in the Supplementary Information, Table S1.
Supplementary Material
Figure S1. CALM-AF10 and hDOT1L interaction is mediated by the OM-LZ region of AF10. (a) Diagram of the constructs used in transfection. (b) Co-immunoprecipitation of CALM-AF10 and hDOT1L in 293T cells. Transfected cells were harvested 36 hours post-transfection. Total cell extracts were prepared with lysis buffer containing 420 mM KCl. M2 agarose beads were used for immunoprecipitation, and the beads were washed with lysis buffer containing 500 mM KCl. The result was analyzed by Western blotting using hDOT1L (top panel) and Flag (bottom panel) antibodies. In: Input, S: Supernatant, IP: Immunoprecipitate.
Figure S2. a, Schematic representation of CALM and AF10 proteins. Potential functional motifs, including NES and NLS are indicated. Red arrows indicate the major breakpoint for the CALM-AF10 fusion present in U937 cells. The green arrow indicates breakpoint of AF10 in MLL-AF10 fusion. Numbers represent amino acid position. b, Alignment of a putative NES sequence in CALM. This sequence is conserved in AP180, an endocytotic accessory protein similar to CALM.
Figure S3. FACS analysis demonstrating the immunophenotypic differences between CALM-AF10 and MLL-AF10-tranduced bone marrow cells. Cells derived from second round colonies of wild-type Hoxa5 bone marrow cells were stained with c-Kit, Gr1, and CD11b antibodies before FACS analysis.
Acknowledgments
We thank J.A. Frelinger for providing NOD–SCID mice and E. Kallin for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (NIH) to Y.Z. (GM68804) and L.S. (AI48407, HL72240). Y.O. is a research fellow of Japan Society for the Promotion of Science. L.J. holds a Chercheur National Award from the Fonds de la recherche en Sante du Quebec. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nature Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 2.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 3.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 4.Chaplin T, et al. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood. 1995;86:2073–2076. [PubMed] [Google Scholar]
- 5.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Carlson KM, et al. Identification and molecular characterization of CALM/AF10fusion products in T cell acute lymphoblastic leukemia and acute myeloid leukemia. Leukemia. 2000;14:100–104. doi: 10.1038/sj.leu.2401629. [DOI] [PubMed] [Google Scholar]
- 7.Dreyling MH, et al. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood. 1998;91:4662–4667. [PubMed] [Google Scholar]
- 8.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 9.DiMartino JF, et al. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL–AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 10.Dreyling MH, et al. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi H, Thirman MJ, Rowley JD. U937 cell line has a t(10;11)(p13–14;q14–21) rather than a deletion of 11q. Genes Chromosomes Cancer. 1995;13:217–218. doi: 10.1002/gcc.2870130312. [DOI] [PubMed] [Google Scholar]
- 12.Dik WA, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19:1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 13.Drabkin HA, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 14.Soulier J, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106:274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 15.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisig BB, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyerholz A, et al. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 18.Tebar F, Bohlander SK, Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell. 1999;10:2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vecchi M, et al. Nucleocytoplasmic shuttling of endocytic proteins. J Cell Biol. 2001;153:1511–1517. doi: 10.1083/jcb.153.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi H, et al. Hematologic malignancies with the t(10;11) (p13;q21) have the same molecular event and a variety of morphologic or immunologic phenotypes. Genes Chromosomes Cancer. 1997;20:253–259. doi: 10.1002/(sici)1098-2264(199711)20:3<253::aid-gcc5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Crooks GM, et al. Constitutive HOXA5 expression inhibits erythropoiesis and increases myelopoiesis from human hematopoietic progenitors. Blood. 1999;94:519–528. [PubMed] [Google Scholar]
- 23.Quentmeier H, et al. Expression of HOX genes in acute leukemia cell lines with and without MLL translocations. Leuk Lymphoma. 2004;45:567–574. doi: 10.1080/10428190310001609942. [DOI] [PubMed] [Google Scholar]
- 24.Jeannotte L, Lemieux M, Charron J, Poirier F, Robertson EJ. Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Dev. 1993;7:2085–2096. doi: 10.1101/gad.7.11.2085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. CALM-AF10 and hDOT1L interaction is mediated by the OM-LZ region of AF10. (a) Diagram of the constructs used in transfection. (b) Co-immunoprecipitation of CALM-AF10 and hDOT1L in 293T cells. Transfected cells were harvested 36 hours post-transfection. Total cell extracts were prepared with lysis buffer containing 420 mM KCl. M2 agarose beads were used for immunoprecipitation, and the beads were washed with lysis buffer containing 500 mM KCl. The result was analyzed by Western blotting using hDOT1L (top panel) and Flag (bottom panel) antibodies. In: Input, S: Supernatant, IP: Immunoprecipitate.
Figure S2. a, Schematic representation of CALM and AF10 proteins. Potential functional motifs, including NES and NLS are indicated. Red arrows indicate the major breakpoint for the CALM-AF10 fusion present in U937 cells. The green arrow indicates breakpoint of AF10 in MLL-AF10 fusion. Numbers represent amino acid position. b, Alignment of a putative NES sequence in CALM. This sequence is conserved in AP180, an endocytotic accessory protein similar to CALM.
Figure S3. FACS analysis demonstrating the immunophenotypic differences between CALM-AF10 and MLL-AF10-tranduced bone marrow cells. Cells derived from second round colonies of wild-type Hoxa5 bone marrow cells were stained with c-Kit, Gr1, and CD11b antibodies before FACS analysis.