Abstract
Lysosomes are crucial cellular organelles for human health that function in digestion and recycling of extracellular and intracellular macromolecules. We describe a signaling role for lysosomes that affects aging. In the worm, Caenorhabditis elegans, the lysosomal acid lipase LIPL-4 triggered nuclear translocalization of a lysosomal lipid chaperone LBP-8, consequently promoting longevity by activating the nuclear hormone receptors NHR-49 and NHR-80. We used high-throughput metabolomic analysis to identify several lipids whose abundance was increased in worms constitutively over-expressing LIPL-4. Among them, oleoylethanolamide directly bound to LBP-8 and NHR-80 proteins, activated transcription of target genes of NHR-49 and NHR-80, and promoted longevity in C. elegans. These findings reveal a lysosome-to-nucleus signaling pathway that promotes longevity and suggest a function of lysosomes as signaling organelles in metazoans.
Lysosomes contain acid hydrolytic enzymes, digesting macromolecules taken up by endocytosis and recycling dysfunctional cellular components during autophagy (1). Lysosomal deficiency is associated with human diseases. For example, loss of human lysosomal acid lipase, LIPA, results in severe systemic metabolic malfunction known as infantile Wolman disease (2). Here, we explored how lysosomes might generate signaling molecules that regulate aging by influencing nuclear transcription.
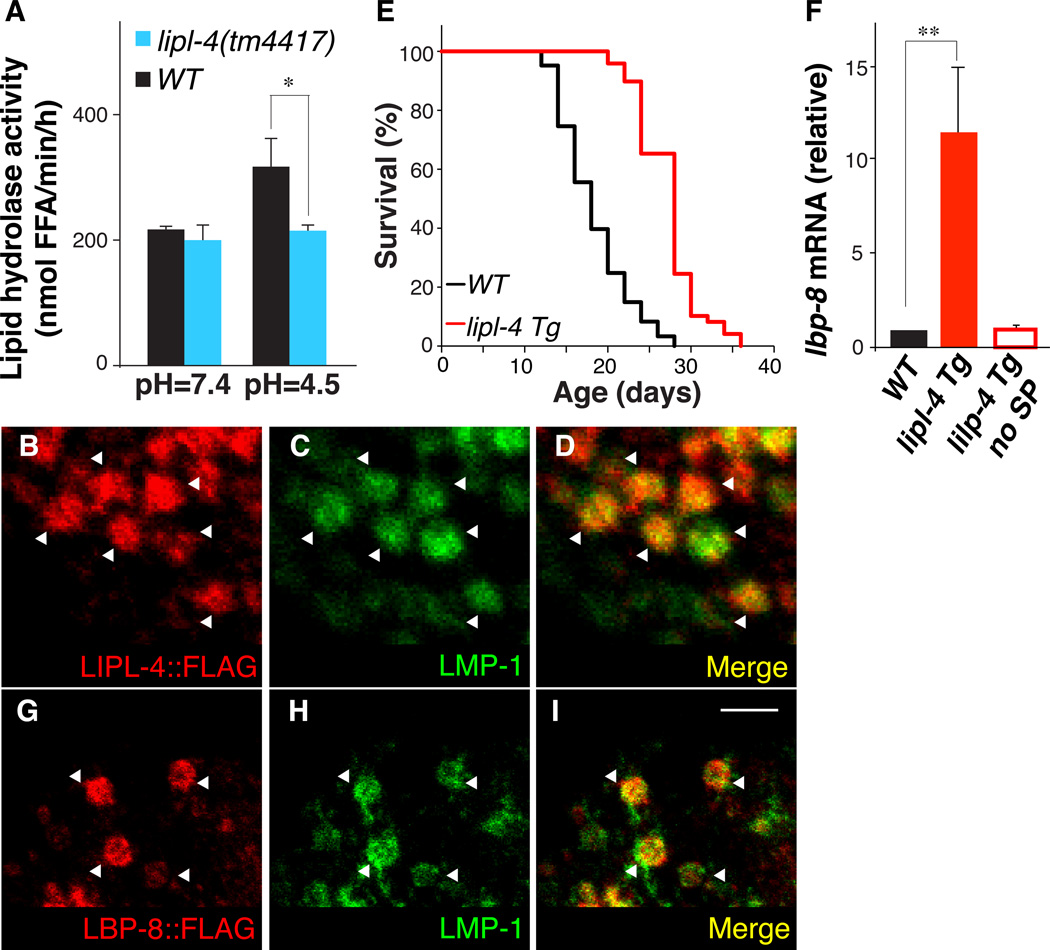
We analyzed a C. elegans longevity-promoting lipase, LIPL-4, which has sequence and functional similarities to human LIPA (fig. S1). Lipid hydrolysis activity was decreased in lipl-4(tm4417) loss-of-function mutants at pH 4.5 but not at pH 7.4 (Fig. 1A). FLAG-tagged LIPL-4 protein was localized to intestinal lysosomes (Fig. 1, B–D and fig. S2). Increased lipl-4 expression is associated with longevity (3). A transgenic strain (lipl-4 Tg) that constitutively expressed lipl-4 in the intestine had 55% mean lifespan increase compared to wild-type (WT) animals (Fig. 1E and table S1) and delayed age-related decline of physical activity (fig. S3A). Constitutive expression of LIPL-4 without the signal peptide (lipl-4 Tg no SP), which was not targeted to the lysosome, caused little extension of lifespan (fig. S4 and table S1), suggesting that the lysosomal activity of LIPL-4 is essential for its longevity effect.
Figure 1. Lysosomal lipid chaperone is increased in long-lived worms.
(A) The amount of free fatty acids (FFA) liberated from 3H-triolein is significantly decreased in the lipl-4(tm4417) loss-of-function mutant compared to wild-type (WT) at pH = 4.5 but not at pH = 7.4. *p<0.05, Student’s t-test. (B–D) Adult worms (raxEx20[ges-1p::lipl-4::3xFLAG]) were stained with anti-FLAG and anti-LMP-1 antibodies. LIPL-4 co-localizes with LMP-1, an established protein marker of lysosomes (18). Scale bar = 10 µm. (E) Mean lifespan is increased 55% in lipl-4 Tg worms (raxIs3[ges-1p::lipl-4::sl2gfp]) compared to WT, p<0.0001, Log-rank-test. (F) lbp-8 mRNA amounts were increased in lipl4 Tg compared to WT, but not in the transgenic strain over-expressing lipl-4 that lacks the signal peptide for lysosomal expression (lipl-4 Tg no SP). Error bars represent standard deviation (SD). **p<0.001, Student’s t-test. (G–I) Adult worms (raxEx31[lbp-8p::lbp-8::3xFLAG]) were stained with anti-FLAG and anti-LMP-1 antibodies. LBP-8 co-localizes with LMP-1 in intestinal lysosomes. Scale bar = 10 µm.
To elucidate whether lipid signals are affected by the LIPL-4 lipase, we examined fatty acid-binding proteins (FABPs) that are intracellular lipid chaperones shuttling lipid molecules between cellular compartments for different functions (4, 5). Of the nine C. elegans FABP homologues, only amounts of lbp-8 were increased in lipl-4 Tg animals, but not in the lipl-4 Tg no SP strain (Fig. 1F). A green fluorescent protein (GFP) reporter strain showed that lbp-8 was exclusively expressed in the intestine (fig. S5A). Both FLAG- and mCherry-tagged LBP-8 proteins were predominantly localized to intestinal lysosomes (Fig. 1, G–I and fig. S5, B–J).
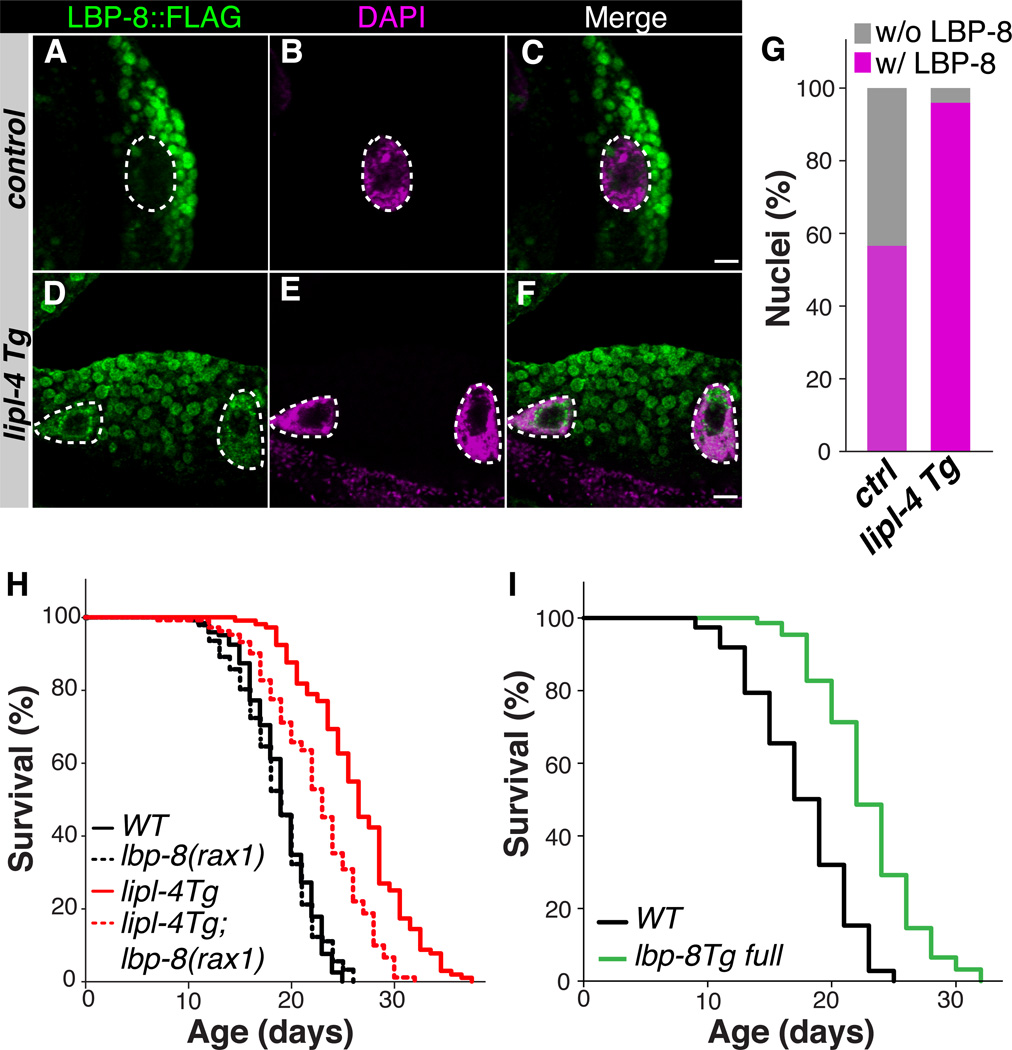
We also detected partial nuclear localization of LBP-8 in the intestine, which was enhanced in lipl-4 Tg animals (Fig. 2, A–G and fig. S6, A–F). LBP-8 contains an N-terminal nuclear localization signal (NLS) (fig. S6G), and was present in both cytoplasmic and nuclear fractions of total worm lysate (fig. S6H). Both RNA interference (RNAi)-mediated depletion of LBP-8 and a newly isolated deletion mutant, lbp-8(rax1), suppressed the lifespan extension in lipl-4 Tg animals without affecting WT lifespan (Fig. 2H, fig. S7 and table S1). Thus, LBP-8 appears to be required for LIPL-4 lysosomal activity to confer longevity.
Figure 2. Lysosomal lipid chaperone promotes longevity.
(A–F) Adult worms expressing LBP-8::3xFLAG were stained with anti-FLAG and 4',6-diamidino-2-phenylindole (DAPI). More LBP-8 nuclear staining in lipl-4 Tg versus control worms. Scale bar = 10 µm. (G) Quantification of the percentage of nuclei positive for LBP-8::3xFLAG staining. n=100. (H) The lbp-8(rax1) loss-of-function mutation reduces mean lifespan extension in lipl-4 Tg by 46% (p<0.0001), but has no effect on the lifespan of WT (p>0.05). Log-rank-test. (I) Mean lifespan is increased 30% in lbp-8 Tg (raxIs4[lbp-8p::lbp-8::sl2gfp]) compared to WT, p<0.0001, Log-rank-test.
We found that a transgenic strain (lbp-8 Tg) constitutively expressing lbp-8 had a 30% increase in mean lifespan compared to WT animals (Fig. 2I and table S1) and improved maintenance of physical activity in old age (fig. S3B). However, a transgenic strain that constitutively expresses LBP-8 lacking NLS (lbp-8 Tg no NLS) was excluded from nuclei and showed little or no lifespan extension (fig. S8 and table S1). Thus, LBP-8 may function as a lysosomal lipid chaperone transducing lipid signals to the nucleus.
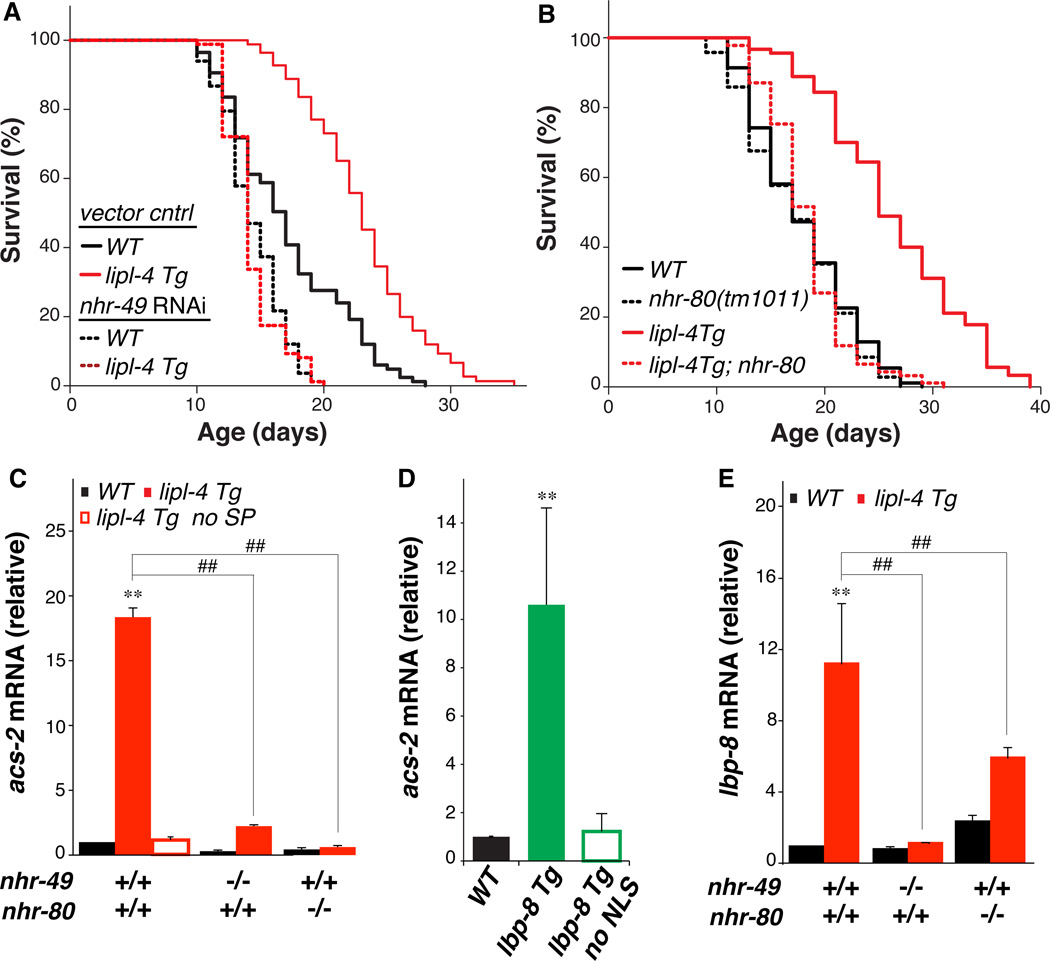
To test whether lysosomal signals might influence nuclear transcription, we screened several transcription factors implicated in longevity regulation (6–11). Nuclear hormone receptors nhr-49 and nhr-80, previously demonstrated to physically interact (10), were both required for lipl-4–and lbp-8–mediated longevity. RNAi-mediated inactivation of nhr-49 in adult worms shortened the lifespan of WT worms but also completely suppressed longevity extension in lipl-4 Tg and lbp-8 Tg worms (Fig. 3A, fig. S9A and table S1). The loss-of-function mutation nhr-80(tm1011) abrogated longevity extension without affecting the lifespan of WT worms (Fig. 3B, fig. S9B and table S1). Neither nhr-49 nor nhr-80 is required for dietary restriction-induced longevity (6, 12), suggesting that the LIPL-4-mediated longevity mechanism may act independently of dietary restriction. Concordantly, the longevity extensions by lipl-4 Tg and eat-2(ad1116), a genetic model of dietary restriction in C. elegans (13), were additive (fig. S10).
Figure 3. Nuclear receptors act in lysosomal longevity signaling.
(A and B) With adult-only RNAi inactivation of nhr-49 or with the nhr-80(tm1011) loss-of-function mutation, the mean lifespan of lipl-4 Tg and WT are not significantly different, p>0.5, Log-rank-test. (C) Increased mRNA amount of NHR-49 target gene acs-2 is suppressed by the nhr-49(nr2041) or nhr-80(tm1011) loss-of-function mutation in lipl-4 Tg, and is absent in lipl-4 Tg no SP. (D) acs-2 mRNA amount is increased in lbp-8 Tg, but not in the transgenic strain expressing lbp-8 without the N-terminal NLS (lbp-8 Tg no NLS). (E) Increased mRNA amount of lbp-8 in lipl-4 Tg is suppressed by the nhr-49(nr2041) or nhr-80(tm1011) mutant.
Error bars represent SD. **p<0.01 by Student’s t-test; ##p<0.01 by two-way ANOVA.
acs-2 encodes an acyl-CoA synthetase required for mitochondrial β-oxidation and is a target gene of NHR-49 (11). acs-2 transcription was increased more than 15-fold in lipl-4 Tg animals; this effect was dependent on nhr-49 and nhr-80, and absent in the lipl-4 Tg no SP strain (Fig. 3C). Transcription of acs-2 was also increased over 10-fold in lbp-8 Tg but not in lbp-8 Tg no NLS animals (Fig. 3D). Thus, LIPL-4–induced activation of NHR-49 and NHR-80 can be reproduced by nuclear action of LBP-8. Transcriptional increase of lbp-8 by lipl-4 Tg was in turn mediated by NHR-49 and NHR-80 (Fig. 3E).
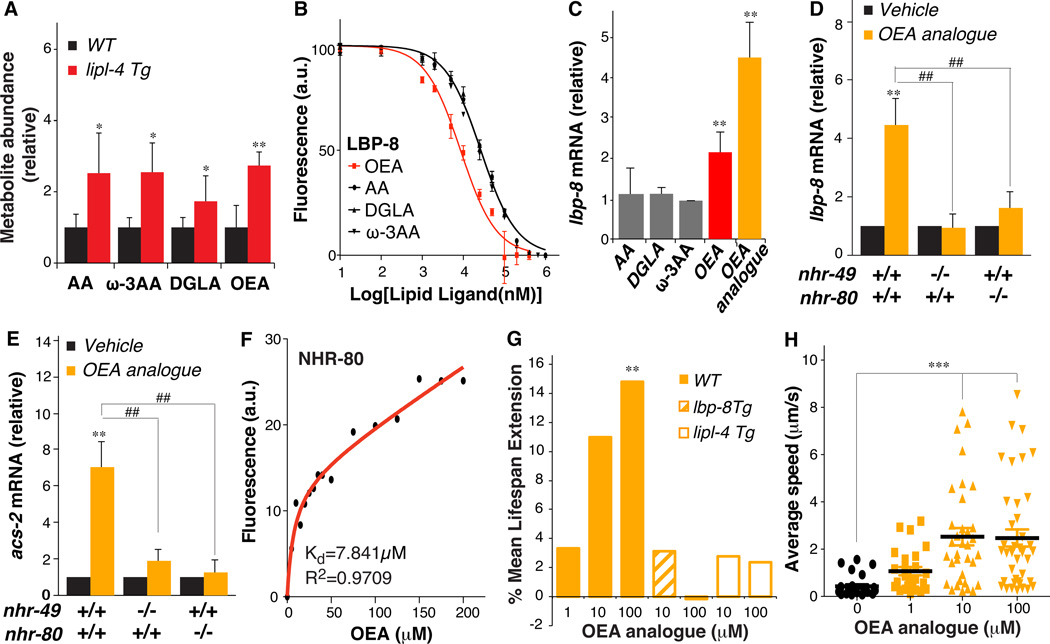
To identify lipid molecules that might function in this lysosome-to-nucleus lipid signaling, we performed high-throughput metabolomic profiling analyses on WT and lipl-4 Tg worms. Among 352 metabolites detected, 71 had significantly altered abundance in lipl-4 Tg animals (table S2). Long-chain fatty acids and their derivatives are likely binding partners of FABPs (4). Thus, we focused our analysis on three C20 fatty acids—arachidonic acid (AA), ω-3 arachidonic acid (ω-3 AA), and dihomo-γ-linolenic acid (DGLA)—and oleoylethanolamide (OEA), an N-acylethanolamine fatty acid derivative (Fig. 4A and table S2). In fluorescence-based binding assays, all four lipids bound to LBP-8, and the binding affinity of OEA for LBP-8 was 3 times higher than that of the fatty acids (Fig. 4B).
Figure 4. OEA activates nuclear receptors and promotes longevity.
(A) Increased levels of arachidonic acid (AA), ω-3 arachidonic acid (ω-3 AA), dihomo-γ-linoleic acid (DGLA), and oleoylethanolamide (OEA) in lipl-4 Tg compared to WT, *p<0.05; **p<0.001, Welch’s t-test. (B) Decreased fluorescence derived from binding of 1-anilinonaphthalene-8-sulfonic acid (1,8-ANS) to LBP-8 by increasing OEA, AA, DGLA, and ω-3 AA competition. OEA has 3-fold higher binding affinity than the other lipids. (C) lbp-8 mRNA amounts are increased in WT supplemented with OEA or OEA analogue, but not AA, DGLA or ω-3 AA, **p<0.01, Student’s t-test. (D and E) mRNA amounts of lbp-8 and acs-2 are increased by OEA analogue supplementation in WT, but not in the nhr-49(nr2041) or nhr-80(tm1011) mutant. **p<0.01, Student’s t-test. ##p<0.01, two-way ANOVA. (F) The intrinsic fluorescence intensity of GST-NHR-80 fusion proteins is decreased with increasing concentration of OEA (dissociation constant (Kd) of the binding reaction, 7.841 ± 4.065µM). (G) Supplementation of OEA analogue increases mean lifespan in WT, but not in lipl-4 Tg or lbp-8 Tg. **p<0.01, Log-rank-test. (H) Mean locomotion velocity is increased in WT treated with OEA analogue at age day 18. ***p<0.0001, Student’s t-test.
Error bars represent SD.
Next, we tested the effects of the four lipids on transcription when directly applied to WT adult worms. We also used an OEA analogue, KDS-5104 that is more resistant to hydrolysis than OEA (14). Only OEA and its analogue were sufficient to increase the transcription of lbp-8 in WT worms, and the analogue exerted a stronger effect (Fig. 4C). After 3 hours treatment with OEA analogue, transcription of lbp-8 and acs-2 was increased more than 4- and 7-fold above the control levels, respectively (Fig. 4, D and E). This effect was abrogated in the nhr-49(nr2041) or nhr-80(tm1011) mutant (Fig. 4, D and E). Thus, accumulation of OEA in response to LIPL-4 may act to promote transcription via NHR-49/NHR-80.
To test whether OEA directly bind to NHR-49 or NHR-80 or both, we measured intrinsic fluorescence changes of GST-NHR fusion proteins in the presence of OEA. OEA binding significantly decreased the fluorescence intensity of the NHR-80 fusion protein in a dose-dependent manner [equilibrium dissociation constant (Kd), 7.841 µM] (Fig. 4F). In a differential protease-sensitivity assay, chymotrypsin digestion of [35S]NHR-80 in the presence of OEA analogue resulted in protease-resistant fragments of approximately 45 kD and 35 kD (fig. S11), indicating direct binding between NHR-80 and OEA analogue. However, no binding was detected between NHR-49 and OEA or OEA analogue (fig. S12). Thus, NHR-80 appears to act as a direct nuclear receptor of OEA and NHR-49 may function as a co-factor of NHR-80.
N-acylphosphatidylethanolamine–specific phospholipase D (NAPE-PLD) mediates OEA synthesis (15). In C. elegans, nape-1 and nape-2 encode NAPE-PLD (16). The nape-1(tm3860) loss-of-function mutation suppressed the lifespan extension in lipl-4 Tg and lbp-8 Tg by half (fig. S13 and table S1). Additionally, a loss-of-function mutant lipl-4(tm4417) reduced the longevity of lbp-8 Tg by 68% (fig. S14), supporting the possibility that LIPL-4 activity promotes the generation of longevity-promoting OEA carried by LBP-8.
Direct treatment of WT worms with OEA analogue prolonged lifespan (Fig. 4G and table S1), and improved physical activity maintenance in aged animals (Fig. 4H). In contrast, neither lipl-4 Tg nor lbp-8 Tg lifespan was affected by OEA analogue supplementation (Fig. 4G and table S1), suggesting that OEA may promote longevity by the same mechanism as occurs in lipl-4 Tg and lbp-8 Tg animals. Interestingly, OEA supplementation decreased lifespan in the nhr-80(tm1011) mutant (fig. S15 and table S1), indicating that OEA requires NHR-80 to promote longevity, and it can have detrimental effects in the absence of NHR-80. Thus, OEA may act as a lipid messenger to transduce lysosome-to-nucleus signaling in promoting longevity.
Overall, our studies suggest that bioactive lipid messengers and lipid chaperones link lysosomal activity and nuclear transcription to promote longevity. All the components of this lysosome-to-nucleus signaling pathway are well conserved in mammals. Interestingly, mammalian PPARα is activated by OEA (17), whereas NHR-80 is homologous to mammalian HNF4, suggesting that different nuclear receptors bind same ligands despite divergent ligand binding domains. Considering that FABPs are quite promiscuous in ligand binding (4), there may be other lipid molecules binding to LBP-8 and functioning in this longevity pathway.
Supplementary Material
Acknowledgments
We thank H.Y. Mak, A. Antebi for providing strains; A. Dervisefendic, H. Jen for experimental support; N. Timchenko, J. Wang, Z. Yu, D. Chow for instrumental support; H. Dierick, C. Herman, H. Zheng, F. Xia, S. Rosenberg, H. Zoghbi for critical reading of the manuscript; P.P. Metoyer for scientific editing. Supported by NIH grants T32GM008602 (E.H.A), RO1DK095750 (E.A.O), T32HD055200 (A.F.), F30AG046043 (A.F.), RO0AG034988 (M.C.W), RO1AG045183(M.C.W.); Ellison New Scholar Award (M.C.W.); ERC-Advanced grant (R.Z.); Fondation Leducq (R.Z.)
References
- 1.Settembre C, Fraldi A, Medina DL, Ballabio A. Nature Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patrick AD, Lake BD. Nature. 1969;222:1067–1068. doi: 10.1038/2221067a0. [DOI] [PubMed] [Google Scholar]
- 3.Wang MC, O'Rourke EJ, Ruvkun G. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furuhashi M, Hotamisligil GS. Nature Rev. Drug Discovery. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storch J, Corsico B. Annu. Rev. Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 6.Goudeau J, et al. PLOS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 8.Ogg S, et al. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 9.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 10.Pathare PP, Lin A, Bornfeldt KE, Taubert S, Van Gilst MR. PLOS Genet. 2012;8:e1002645. doi: 10.1371/journal.pgen.1002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. PLOS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heestand BN, et al. PLOS Genet. 2013;9:e1003651. doi: 10.1371/journal.pgen.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakowski B, Hekimi S. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astarita G, et al. J. Pharmacol. Exp. Ther. 2006;318:563–570. doi: 10.1124/jpet.106.105221. [DOI] [PubMed] [Google Scholar]
- 15.Fu J, et al. J. Biol. Chem. 2007;282:1518–1528. doi: 10.1074/jbc.M607809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucanic M, et al. Nature. 2011;473:226–229. doi: 10.1038/nature10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, et al. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 18.Hadwiger G, Dour S, Arur S, Fox P, Nonet ML. PLOS ONE. 2010;5:e10161. doi: 10.1371/journal.pone.0010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.