Abstract
Enzymes underpin physiological function and exhibit dysregulation in many disease-associated microenvironments and aberrant cell processes. Exploiting altered enzyme activity and expression for diagnostics, drug targeting, and drug release is tremendously promising. When combined with booming research in nanobiotechnology, enzyme-responsive nanomaterials for controlled drug release have achieved significant development and been studied as an important class of drug delivery devices in nanomedicine. In this review, we describe enzymes such as proteases, phospholipase and oxidoreductases that serve as delivery triggers. Subsequently, we explore recently developed enzyme-responsive nanomaterials with versatile applications for extracellular and intracellular drug delivery. We conclude by discussing future opportunities and challenges in this area.
1. Introduction
The advent of nanomaterials-based drug delivery systems has made a seminal impact to the field of drug delivery.1, 2 Therapeutics and diagnostic agents incorporated in versatile nanoscale particles composed of dynamic nanomaterials have been developed for the diagnosis and treatment of cancer,3 diabetes,4, 5 bacterial infections,6 etc. Well recognized advantages of nanoscale particles include sustained drug delivery,7 hydrophilic and hydrophobic multi-drug co-incorporation and programmed-delivery,8, 9 prolonged circulation time in vivo, the ability to be engineered with functionalized ligands10, 11 and stimuli-responsive controlled drug release.12, 13 Among myriads of successful applications, stimuli-responsive “smart” nanomaterials have emerged as especially promising materials in comparison to conventional nanoscale materials due to their bio-responsive physicochemical nature. For example, pH-responsive nanomaterials have been used to design sensitive nano-systems for drug delivery in cancer therapy as they can stabilize the drug at physiological pH and release the drug when the pH trigger point is reached.14 Due to the lower pH values in the tumor microenviroment compared to normal tissue, pH-responsive nanomaterials are triggered and release their cargoes (e.g. drugs and diagnostic agents) specifically at the tumor site, therefore reducing the unwanted side effects.15, 16 Other stimuli-triggered drug delivery systems have been exploited extensively in biomedical research which exhibit reversible or irreversible changes in chemical structures and/or physical properties in response to a specific stimulus such as temperature,17 redox,18 ionic strength,19 electric,20 magnetic fields21 and enzyme activity22.
A developing arena in stimuli-responsive “smart” nanomaterials is the design of nanomaterials whose chemical structures and/or physical properties are responsive to the biocatalytic action of an enzyme.23 Enzymes play critical roles in all biological and metabolic processes and dysregulation of enzyme expression and activity underpins the pathology of many diseases. Dysregulated enzymes are promising biological triggers in therapeutics.24 Exploiting enzymes as a trigger has a number of advantages because most enzymes catalyze chemical reactions under mild conditions (low temperature, neutral pH, and buffered aqueous solutions), where many conventional chemical reactions fail.25, 26 Furthermore, enzymes can also exhibit exceptional selectivity for their substrates, allowing for specific, sophisticated, biologically inspired chemical reactions.27
In the past years, a number of nanoscale materials have been employed for the design of enzyme-responsive drug delivery systems including polymer materials,28–30 phospholipids31, 32 and inorganic materials.33 The integration of nanomaterials with enzymatic responses can endow the formulations with bio-specificity and selectivity, allowing for the promising applications in diverse fields. For example, active tumor-targeting nanoparticles integrated with site-specific enzyme-triggered moieties can significantly achieve enhanced accumulation at the tumor site, reduced uptake by non-targeted tissue, as well as site-specific controlled drug release without a compromise in targeting efficiency and specificity.34 The nanomaterials can be rendered enzyme-responsive by containing moieties in their main chain or side groups which can be cleaved by enzyme. For instance, self-assembled nanoparticles are often incorporated with enzyme-responsive linkers which can be recognized by the biocatalyst or transformed by the product of the enzymatic reaction, in order to program the nanomaterials to release their cargo with spatial and temporal control.35 In other cases, especially for inorganic nanosystems, particles can be modified with active-targeting ligands that are sensitive to a certain enzyme.36, 37 This approach can further broaden the design flexibility and scope of applications by endowing the nanoparticle with enzyme responsive properties when the nanomaterial itself is not responsive to enzyme.
In this review, the most recent advances in the use of enzyme-responsive nanomaterials for a board range of controlled drug delivery applications are presented. First, the typical enzymes that can modulate drug delivery or offer a target for triggering drug release are summarized. Next, various nanomaterials, including polymers, lipid and inorganic materials that act as carriers or matrices for enzymatic-response drug delivery are surveyed. The opportunities and challenges of clinical applications are also discussed.
2. Typical enzymes for controlled drug delivery
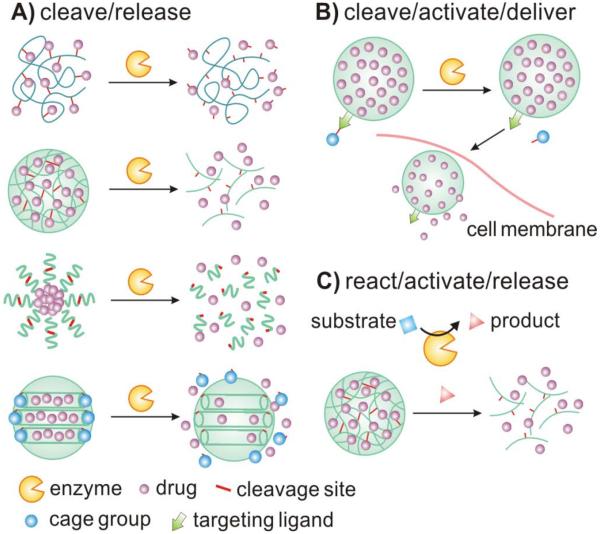
In this section, several typical classes of enzymes that serve as the triggers in enzyme-responsive controlled drug delivery are discussed. As summarized in Fig. 1, drugs can be triggered to release through a variety of enzymatic implementations. Drugs can be directly released from different nanocarriers via site-specific enzymatic cleavage. Loading of drugs into nanomaterials can be achieved through covalent attachment or physical encapsulation, involving cross-linked matrix, self-assembled system or caged porous structure. Drug carriers can also be activated by enzymes to expose the targeting ligand for the subsequent internalization into specific cells. Additionally, enzymes can facilitate the generation of specific products, such as acidic environment, promoting drug release from carriers.
Fig. 1.
A schematic illustrating typical implementations of enzyme-responsive nanomaterials for controlled drug delivery. (A) Drugs can be directly released from a variety of carriers upon site-specific cleavage by enzymes. (B) Drug carriers can be activated by enzymes to expose targeting ligands for the subsequent cellular delivery. (C) Enzymes can facilitate the generation of specific products that result in drug release from carriers.
2.1 Proteases
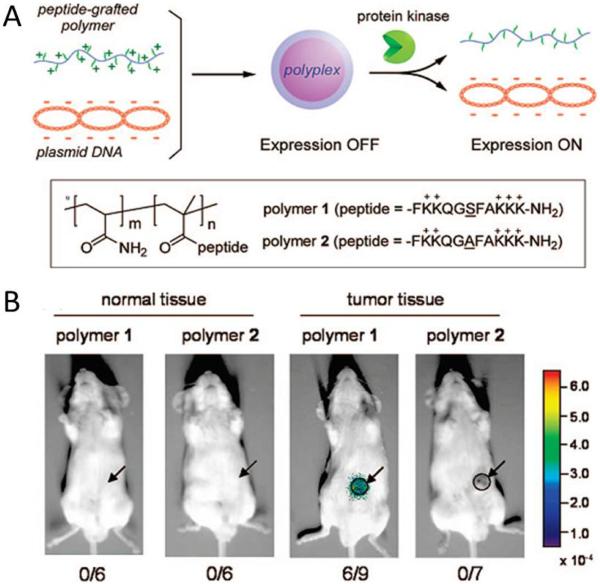
Proteases are known to be involved in many physiological processes such as tissue remodeling, wound healing and tumor invasion.38, 39 As many diseases are characterized by imbalances in the expression and activity of specific protease in the diseased tissue, protease overexpression could potentially be exploited to allow for selective activation of advanced drug delivery platforms. For example, Kang et al. synthesized peptide-grafted polymers (conjugates) that could activate gene expression in response to protein kinase or protease activity. Conjugates were able to preform cellular transfection in cultivated cells and also in an in vivo system.40 In the protease-responsive system, the peptide side chains were designed as a specific substrate of a target protease and could be phosphorylated with hyperactivated protease upon polyplex uptake by the target cancer cell (Fig. 2A). Results showed that mice (6/9) bearing xenograft tumors treated with peptide-grafted polymers exhibited luciferase expression while none of the six mice examined showed luciferase activity in normal subcutaneous tissue (Fig. 2B). Protein kinase Cα, including Caspase-3 protease, cleaved a specific sequence allowing the cationic portion of the polyplex to release and allowed for the activation of transgene transcription. In another case, Law et al. developed self-assembled peptide sequences which could release therapeutic payloads upon specific interaction with disease-associated proteases.41 The biologically inspired peptide sequence is composed of a protease cleavable region which self-assembled into a gel matrix in aqueous solutions. Upon addition of the targeted protease (uPA), the gel matrix could be digested at the substrate cleavage site, resulting in release of drugs and matrix fragments. Their results showed that the release of therapeutic peptide (B-r7-kla) fragment from the matrix was a result of the addition of enzyme and only can be observed in the presence of the enzyme. Furthermore, in HT-1080 cell lines, the encapsulated B-r7-kla was released slowly with no cytotoxic effects on cells lacking enzyme. However, enhanced drug release and increased cytotoxicity were observed under high uPA concentrations (500 nM) after 24 hours.
Fig. 2.
(A) Schematic illustration of development and enzyme-responsive mechanism of peptide-grafted polymers. (B) Luciferase activity in tumor site 24 h after intratumoral injection of peptide-grafted polymers 1 or 2 (C/A = 2.0). Arrows and circles indicate the site of xenografted-tumor tissue. Reproduced from ref. 40 with permission from American Chemical Society.
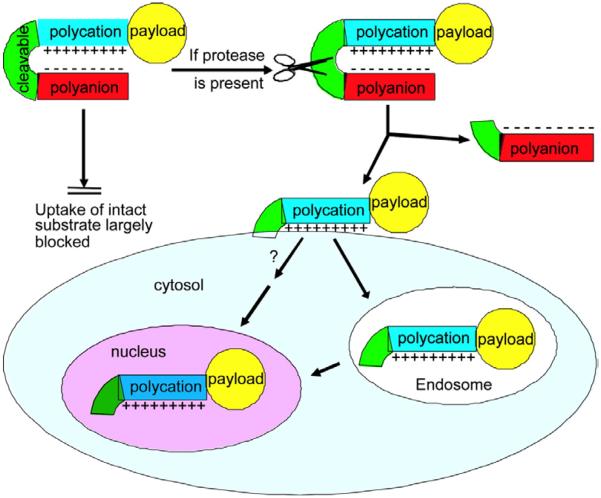
Another class of proteases which are often employed to serve as triggers of controlled drug delivery include matrix metalloproteinases (MMPs). MMPs have long been associated with cancer-cell invasion and metastasis.42, 43 MMPs are over expressed in tumor microenvironments in comparison to normal tissue in most cancers.44 MMPs are proteolytic enzymes and their basic mechanism of action — degradation of proteins — regulates various cell behaviors with relevance for cancer biology. Thus, MMPs have been widely employed to achieve enzyme-triggered site-specific drug release. Of note, Jiang et al. developed a proteolytically-activated cell-penetrating peptide-modified nanoparticle drug delivery system triggered by MMP-2 and MMP-9, labeled with Cy5, gadolinium, or both.45, 46 The reported ACPP was composed of a polycationic CPP sequence, a MMP-sensitive peptide linker (XPLGLAG) and a polyanionic inhibitory domain (poly Glu, E9), forming a hairpin structure (Fig. 3). ACPP surface decorated nanoparticles had increased tumor specificity. Upon addition of MMP, the inhibitory domain could be dissociated at cleavable site, resulting in releasing the polycationic peptides and their cargo to enter the cells. Residual tumor and metastases as small as 200 μm could be detected by fluorescence imaging. The as-prepared ACPP decorated nanoparticles exhibited more than 10-fold cellular association and showed high in vivo contrast ratio of images corresponding well to the MMP distribution.
Fig. 3.
Schematic illustration of activatable CPPs. The enzyme-responsive inhibitory domain can be dissociated at cleavable site, resulting in the internalization of nanoparticles. Reproduced from ref. 45 with permission from the National Academy of Sciences.
In addition to molecular imaging, the up-regulated expression of disease-associated MMP-2 in diseased tissues and the catalytic characteristic of proteolysis have made protease-activated drug delivery an attractive approach for tumor treatment. Recently, Gu et al. constructed an activatable LMWP (ALMWP, E10-PLGLAG-VSRRRRRRGGRRRR) modified PEG-PCL nanoparticulate drug delivery system which provided a new strategy for glioma treatment.47 A notable advantage of this approach is that the transport abillity of LMWP can be activated only in the presence of MMP-2 protease.
2.2 Phospholipases
In principle, phospholipase up-regulation has been a pathological indicator for multiple kinds of cancers and many other disease processes, including thrombosis, congestive heart failure, inflammation, neurodegeneration, and infectious pathogens.48–51 Phospholipase A2 (PLA2) is gaining much interest as therapeutic targets as they are known to be up-regulated in tumors. For example, in prostate cancers, Group IIa sPLA2 is reported to be expressed at levels 22-fold greater than disease-free paired controls.52 The upregulation of secreted phospholipase A2 in tumor microenvironment mediates carcinogenesis by two pathways: release of arachidonic acid, which produces carcinogenic metabolites; and release of lysophospholipids, including lysophosphatidic acids (LPA) that induce cell growth.53, 54 Microenvironment-upregulated PLA2s are being actively explored for the ability to produce environmentally responsive drug release in the tumor. Andresen et al. constructed an enzymatically activated liposome drug delivery system involving masked antitumor ether lipids (AELs) as prodrugs. AELs are activated by secretory phospholipase A2.55 AEL-forming liposomes offered the advantages of increased cytotoxity by utilizing the membrane permeability of AEL drugs. They synthesized (R)-1-O-hexadecyl-2-palmitoyl-sn-glycero-3-phosphocholine (1-O-DPPC) which was not only capable of forming liposomes but aslo carrying water-soluble prodrugs which could be activated by using PLA2 (proAELs) as the activating enzyme. Results showed that proAELs were successfully activated by PLA2 enzymes and converted to chemotherapeutic agents. The generated AELs and fatty acid hydrolysis products could perturb membranes and enhance the cellular uptake of drugs encapsulated in the liposomes. Additionally, severe side effects (hemolysis) were limited because PLA2 expression specifically in tumor tissue localized activation of proAELs.
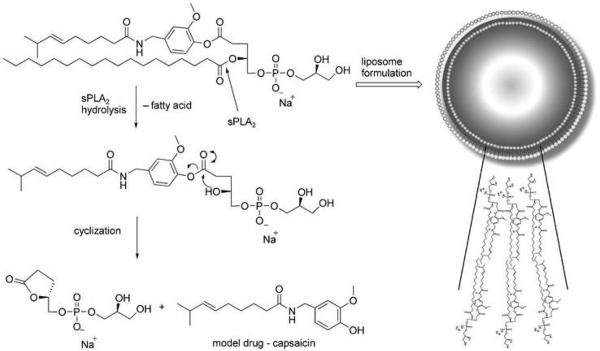
Linderoth et al. employed the overexpressed secretory phospholipase A2 as a trigger for activating liposomal drug-delivery systems (Fig. 4).56 PLA2 hydrolyzed the ester group in the sn-2 position of glycerophospholipids and facilitated the release the drugs at specific tumor sites. They used a capsaicin prodrug 8 as a model drug which could aggregate into small unilamellar vesicles (SUVs). Their results showed that capsaicin was released from the phospholipids after hydrolysis with amount was found to be (90 ± 11) % (n=3) after 24 h when the SUVs are exposed to human sPLA2-IIA, which indicated the prodrugs are good substrates for sPLA2. Additonally, the prodrug could stay in the buffer solution up to two weeks before the addition of snake-venom sPLA2. Attributed to the covalent attachment between the drug and glycerophospholipid derivative, lipid prodrug could react with an ester group at the sn-1 position, resulting in forming a five-membered lactone and releasing the carried drug.
Fig. 4.
Schematic illustration of the mechanism of prodrug based liposome which could be hydrolyzed by sPLA2 to release the drug after cyclization. Reproduced from ref. 56 with permission from WILEY.
2.3 Oxidoreductase
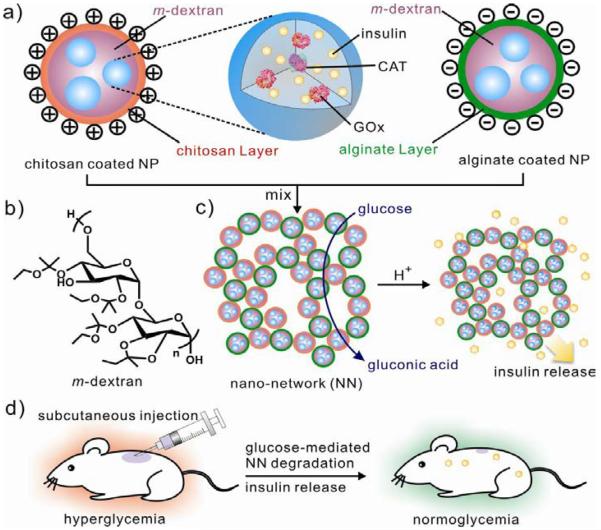
Oxidoreductases have been widely exploited to serve as a promising target for drug delivery systems or biosensing due to their fundamental role in oxidative environments which are generated by many diseases including diabetes and cancer.57, 58 For example, glucose oxidase (GOx) is extensively utilized as a gate in responsive, controlled release systems or a diagnostic tool in the detection of glucose values.4 For example, Gu et al. applied GOx as the trigger to develop a nano-network which can be responsive to blood-sugar levels for closed-loop insulin delivery (Fig. 5).5 In that case, the injectable nano-network, prepared via electrostatic interaction between positively charged nanoparticle (chitosan) and negatively charged nanoparticle (alginate), was composed of a mixture containing nanoparticles with a solid core of insulin, ketal modified dextran and enzymes (GOx and Catalase). Upon addition of hyperglycemic state, GOx encapsulated in nanoparticles began converting glucose into gluconic acid, resulting in dissociation of porous nano-network and subsequently release of insulin. A pulsatile release profile of insulin can be achieved in response to gluconse concentrations in vitro. To further examine the efficacy of the insulin-loaded nanonetwork for diabetes treatment, they utilized streptozotocin (STZ)-induced diabetic mice as the model to test the release of insulin in a glucose-responsive manner. In vivo results showed that the injectable nano-network was capable of stablizing blood glucose levels within the normoglycemic state for up to 10 days with a single injection. This glucose-responsive degradable nano-network allowed for self-regulated and long-term diabetes management. In another case, Gu et al. used glucose-responsive microgels for glucose-responsive release of insulin.59 One-step electrospray technology was utilized to encapsulate pH-responsive chitosan, enzyme nanocapsules and insulin into a monodisperse microgel. When exposed to a hyperglycemic condition, the microgel swelled as a result of the response of catalytic conversion of glucose into gluconic acid and subsequent protonation of the chitosan network. According to the in vivo studies, the microgel could reduce blood glucose levels in STZ-induced diabetic mice.
Fig. 5.
Injectable glucose-responsive nano-network for insulin delivery. (a) Ketal modified dextran-based nanoparticle encapsulated with insulin, Gox and catalase. (b) Decoration of nanoparticles with chitosan and alginate, respectively. (c) Formation of Nano-network (NN) via electrostatic interaction and dissociation via the catalytic conversion of glucose into gluconic acid (d) Schematic of glucose-responsive insulin delivery for type 1 diabetes treatment using the STZ-induced diabetic mice model. Reproduced from ref. 5 with permission from American Chemical Society.
2.4 Other enzymes
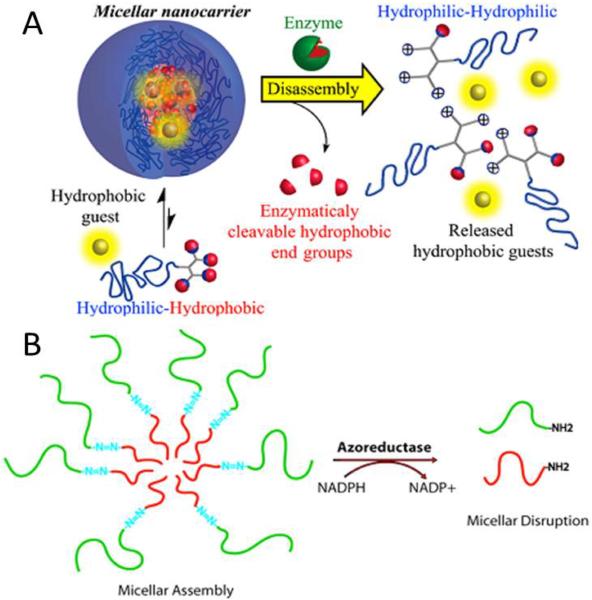
As many diseases are characterized by imbalances in the expression and activity of specific enzymes in the diseased tissue, dysregulatated enzymes can be exploited to permit the selective activation of advanced drug delivery platforms. Recently, the enzyme penicilin G amidase (PGA) was employed by Harnoy et al. to cleave the end groups of phenyl acetamide, which could disassemble the constructed micelles (Fig. 6A).60 The monomeric hybrid could be detected when enzymatic cleavage of the hydrophobic phenyl acetamide end groups happened. This result was supported by the change in the fluorescence of encapsulated Nile red dyes and fast disappearance of the amphiphilic hybrids in HPLC analysis in the presence of enzyme PGA. More interestingly, the formation of the fully degraded tetra-amine hybrids could not be observed at the relatively low enzyme PGA concentration (0.14 μM) after 24 h, while this formation was observed at the high enzyme PGA concentration of 1.4μM.
Fig. 6.
(A) Schematic of preparation and enzyme-responsive mechanism of a smart micellar nanocarrier. Reproduced from ref. 60 with permission from American Chemical Society. (B) Schematic representation of the disruption of PEG-N=N-PS block copolymer based micellar in the presence of azoreductase. Reproduced from ref. 61 with permission from American Chemical Society.
The azoreductase has received extensive attention in the field of the treatment of colon diseases due to the fact that azoreductase is produced by the microbialflora present in human colon. In a study by Rao et al, an azoreductase responsive assembly was prepared through the covalent coupling to azobenzene linkage and an amphiphilic diblock copolymer (Fig. 6B).61 The copolymer assembled into a micellar structure in water but dissociated in the presence of the azoreductase and coenzyme NADPH due to the azobenzene-based copolymer junction. The azobenzene-linked poly(ethylene glycol)-b-poly(styrene) (PEG-N=N-PS) amphiphilic copolymer was prepared through an ATRP-based macroinitiator approach. The assembled micellar disassembled into two segments (PEG segment and PS segment) when exposed to azoreductase. Due to the systems' azoreductase sensitivity and selectivity, the azoreductase responsive drug delivery system might have potential applicability in the treatment of colon diseases.
3. Enzyme responsive nanomaterials
In this section several nanomaterials that can be used as carriers for enzyme-responsive drug delivery systems are discussed. Popular strategies involve polymeric assemblies and liposomes that disintegrate, undergo structural reorganization, or cleavage to release functionalized ligands. Enzymes directly govern these changes or enzymatic products trigger advantageous changes in the physiochemical properties of the nanomaterial.
3.1 Polymeric nanoparticle
Nanotechnology, especially biodegradable polymeric nanoparticles, has emerged as a promising approach to augment delivery of chemical drugs and bioimaging agents.62, 63 Polymeric nanoparticles have drawn increasing interest for their ability to deliver drugs within an optimal dosage range, often resulting in increased therapeutic efficacy of the drug, mitigated side effects, and improved patient compliance.64–66 Polyethylene glycol (PEG)-surface modified nanoparticles, PEG is a relatively inert hydrophilic polymer, provide good steric hindrance preventing protein binding, reduce the rate of the reticuloendothelial system (RES) uptake, and increase circulation half-life of drugs.67, 68 Enzyme-responsive polymeric nanoparticles have been widely explored by taking advantage of 1) dysregulated enzymes in the disease tissue (e.g., overexpressed MMPs in most tumor tissue); 2) reversible or irreversible changes in chemical structures and/or properties of polymeric materials in response to specific enzymes. A lot of polymers have been synthetized to achieve enzyme-responsive function for disease treatment such as cancer and diabetes. For example, Lee et al. constructed enzyme-responsive linkers by using 4-hydroxymandelic acid as the framework.24 Then they attached the enzyme substrates to the 4-hydroxy group of the mandelate core, leading to spontaneous release of conjugated drugs when the substrate portion interacted with intracellular enzymes.
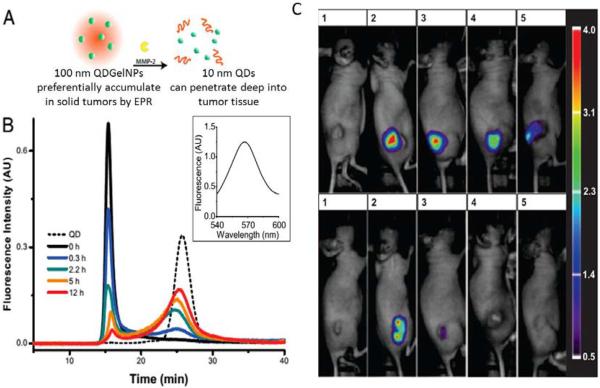
In another study, Wong et al. employed an enzyme trigger to shrink 100-nm nanoparticles to 10-nm nanoparticles after they were exposed to the tumor microenvironment with overexpressed enzymes such as MMP-2 (Fig. 7A).69 In the context, type A gelatin crosslinked with glutaraldehyde was used as the framework to form enzyme-degradable nanoparticles (~100 nm). PEG-stabilized quantum dots (QDs, ~10 nm) were conjugated to the nanoparticle surface. When these engineered nanoparticles preferentially extravasated from the leaky regions of the tumor vasculature by the enhanced permeation and retention (EPR) effect and were exposed to the upregulated MMP-2 in the tumor environment, the nanoparticles were degraded by the MMP enzyme and released 10 nm quantum dots, resulting in enhancement of diffusive transport and deep penetration in tumor tissue. (Fig. 7B). These enzyme-responsive, multistage nanoparticle systems allowed for additional tenability in the spatial control of drug delivery by changing in size, overcoming multiple physiological barriers thereby enhancing their penetration.
Fig. 7.
(A) Schematic illustration of size change of QDGelNPs and release of 10-nm QD NPs in the presence of MMP-2. (B) GFC chromatograms of QDGelNPs after incubation with MMP-2. Reproduced from ref. 69 with permission from the National Academy of Sciences. (C) Retention of enzyme-responsive nanoparticles in HT-1080 tumors after intratumoral injection. Reproduced from ref. 70 with permission from American Chemical Society.
Besides shrinking the size of polymeric nanoparticles, enzymes can also be used as the trigger to cleave substrates and increase the size of nanoparticle. Chien et al. reported a polymer–peptide hybrid block copolymers self-assembled micellar nanoparticle which exhibited enzyme-responsive characteristics.70 In their work, a hydrophobic block and a hydrophilic peptide were integrated into copolymers to prepare the micelles. Micelles passively diffused into the tumors and accessed to overexpressed enzymes. There micrometer-scale aggregates were formed via cleavage of peptides in the shell of the micelles. Essentially the aggregates were kinetically trapped within the tumor (Fig. 7C). By taking advantage of the formation of a new assembly in response to the enzymatic cleavage of the substrate, the aggregates were retained at the tumor site as long as one week, while non-responsive nanoparticles would be degraded within two days. This novel type of disintegrable nanoparticle which can be activated in vivo by MMP-2 exhibits potential applications for the effective delivery of chemotherapeutic drugs and imaging agents.
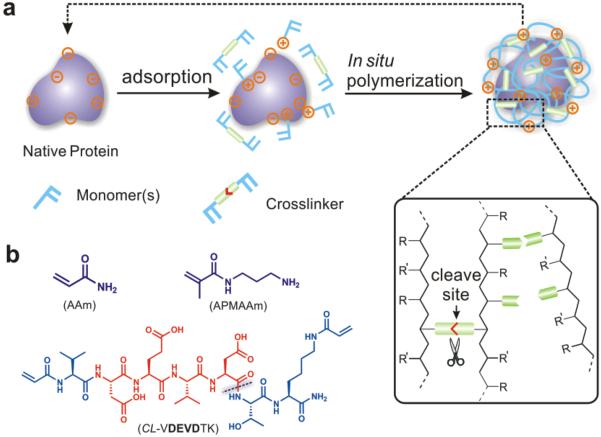
In addition to chemotherapeutical drugs, enzyme-responsive nanoparticles have also been harnessed to delivery therapeutic proteins. Gu et al. constructed an enzymatically degradable cocoon-like polymeric nanocapsule by using one-pot procedure (Fig. 8).71 The nanocapsule (CP3-NC) was composed of an apotosis-inducing target protein—Caspase-3 (CP3), acrylamide (AAm), N-(3-aminopropyl) methacrylamide (APMAAm) and a CP3-degradable VDEVDTK peptide as cross-linker (CL-VDEVDTK)72, 73. Upon proteolytic cleavage of the cross-linker at 37°C, the polymeric shell dissociated and the encapsulated protein CP3 released in functional form. In vitro results showed HeLa cells exhibited apoptotic hallmarks such as membrane blebbing and cell shrinkage when treated with 200 nM CP3-NC for 24 h, while cells treated with native CP3 or CP3-NCN (enzymatically nondegradable) showed no morphological changes. In another study, Biswas et al. exploited a protease-responsive nanocapsule for intracellular protein delivery.74 Nanocapsules were composed of monomers and an enzyme-degradable peptide cross-linker. Upon addition of furin (a ubiquitous intracellular protease), the peptide could be cleaved, leading to release of encapsulated protein. Their furin degradable protein delivery vehicle successfully delivered both cytosolic and nuclear proteins in active forms to a variety of cell lines.
Fig. 8.
Schematic illustration of preparation and mechanism of protein nanocapsules. (A) Preparation of protein nanocapsules via in situ polymerization. (B) Typical monomers and cross-linker used in enzyme-responsive nanocapsule system. Reproduced from ref. 71 with permission from American Chemical Society.
3.2 Liposome
Liposome development is rapidly evolving and dramatically changing the paradigms of drug delivery. The suitable sizes, unique chemical properties, large surface areas, structural diversity and multifunctionality of liposomes prove to be greatly advantageous for combating notoriously therapeutically evasive diseases such as cancer.75, 76 Because of their biocompatibility and ease of fabrication, liposomes are an excellent drug delivery platform and a number of smart liposomes have been designed to improve drug delivery efficiency and therapeutical efficacy.77 One of the emerging trends in smart liposome development is the integration of enzyme-responsive elements. As one of the earliest examples, Pak et al. demonstrated an enzymatically cleavable peptide–lipid N-Ac-AA-DOPE, resulted from covalent linkage of dioleoyl phosphatidylethanolamine DOPE and an elastase substrate. The liposome fusion could be achieved in the presence of human leukocyte elastase or proteinase K.78
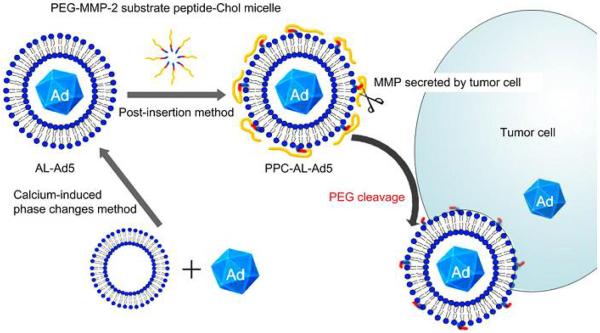
Liposomes can target the tissue of interest either passively (e.g. EPR effort in tumor tissue) or actively via the modification of targeting molecules that recognize receptors on the cell surface.79, 80 Several groups have taken an approach that involves the decoration of liposomes with enzyme-responsive ligands which allow for controlled drug release. Wan et al. reported an enzymatically cleavable PEG-lipid material which was composed of a PEG chain, matrix metalloproteinase (MMP)-substrate peptide, and cholesterol (PPC) (Fig. 9).81 As demonstrated, the linker peptide, Gly-Pro-Leu-Gly-Ile-Ala-Gly-Gln, between PEG and lipid was cleaved by MMPs including MMP-2, MMP-3, MMP-7 and MMP-9, resulting in the abandon of PEG chain and enhanced cellular uptake and transduction efficiency in a tumor specific manner. Their results indicated that PPC-modified enzyme-responsive anionic liposomal Ad vectors (PPC-AL-Ad) exhibited higher in vitro gene expression than non-cleavable PEG-lipid modified AL-Ad (PPC-AL-Ad). In another study, Terada et al. also used the PEGylated MMP-2 substrate peptide (Gly-Pro-Leu-Gly-Ile-Ala-Gly-Gln) to conjugate DOPE and prepared enzyme-responsive galactosylated liposome.82 Liposome could not enter into normal hepatocytes as a result of steric hindrance effect. Upon addition of upregulated MMPs, Liposome dissociated and subsequently was taken up. With this approach, it was possible to deliver a payload or increase the retention of liposomes within targeted cell populations via enzymatic catalyzed interactions.
Fig. 9.
Scheme of of the enzyme-responsive liposome for the tumor cell-specific delivery of adenoviral vectors. Reproduced from ref. 81 with permission from Elsevier.
3.3 Hybrid nanoparticle
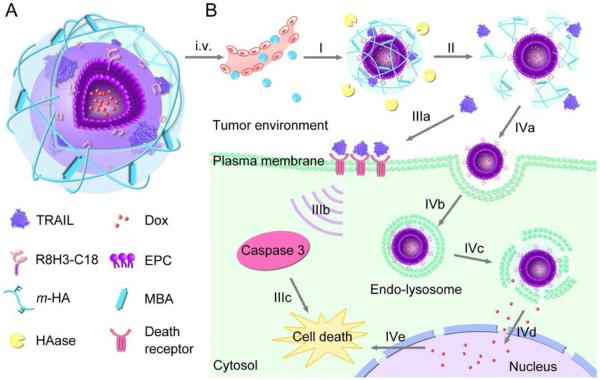
The preparation of organic/inorganic hybrid nanoparticles composed of organic polymers and/or inorganic colloids has been recently pursued as a route to combine the advantageous properties of both classes of macromolecules into one nanoparticle.83, 84 A hybrid design (core and shell structure) could take advantages of both organic/inorganic materials in the core and shell. For example, in lipid-polymer hybrid nanoparticles, solid cores act as supports that provide mechanical stability, controlled morphology, biodegradability, increased surface area-to-volume ratio and narrow size distribution. In addition, the core of these particles, often composed of PLA or PLGA polymer, can facilitate drug delivery and controlled drug release. At the same time, the lipid shell enveloping the core mimics biological membranes in interacting with the environment.85 Among the various emerging hybrid nanoparticles (e.g. lipid-polymer, gel-lipid, silica (SiO2)-polymer, iron oxides-polymer hybrid nanoparticle), enzyme-responsive hybrid nanoparticle are gaining increasing attraction as a new platform for drug delivery because their enzymatic properties can be engineered according to interest. Most recently, Jiang et al. developed a new programmed hybrid-nanoparticle drug-delivery system that could co-deliver different anticancer therapeutics (Fig. 10). The core–shell-based Gel–Liposome was composed of a R8H3 peptide decorated liposome core loaded with Dox and hyaluronic acid (HA) based shell encapsulated with trail.86 Besides, the crosslinked gel based outer shell degraded in concentrated hyaluronidase (HAase) that was overexpressed in tumor tissue. Degradation permitted the release of TRAIL and the exposure of targeted ligand R8H3-L. Subsequently, encapsulated Dox in the aqueous core of the modified liposome was taken up by tumor cells. In vivo biodistribution showed that the Dox amount in the tumor tissues using Gel-Liposome as delivery carrier was 5.72- and 2.70-fold higher than that of those delivered by the Dox solution and Dox-R8H3-Liposome after 48 h. The enzyme-responsive gel-liposome drug delivery system provided a potential delivery strategy to simultaneously deliver different antitumor drugs and achieve synergistic anticancer efficacy.
Fig. 10.
Schematic illustration of mechanism of core–shell-based Gel–Liposome for programmed and site-specific drug delivery. Reproduced from ref. 86 with permission from WILEY.
In another study, Bernardos et al. synthetized a hybrid silica mesoporous nanoparticle for on-command drug delivery.87 In the context, authors prepared the silica mesoporous nanoparticles by using an MCM-41 inorganic scaffold, containing mesopores (2–3 nm) to allow for the encapsulation of certain guests. The nanoscopic MCM-41-based nanoparticle were capped with different “saccharide” derivatives to form an enzyme-responsive hybrid silica mesoporous nanoparticle. This hybrid nanoparticle could open the gate and release cargoes when taken up by tumor cells and exposed to the specific enzymes (e.g. β-D-galactosidase). Mondragn et al. extended this enzyme-responsive hybrid nanoparticle platform by synthetizing capped hybrid systems. They used two different anchoring strategies to functionalize MCM-41 nanoparticles functionalized with polymer ε-poly-L-lysine (biodegradable by amidases).88 One anchoring strategy was the formation of a urea bonds via using lysine amino groups to decorate MCM41 nanoparticles with the ε-poly-L-lysine backbone (solid Ru-rLys-S1). The other strategy was specific attachment of polypeptide to MCM-41 nanoparticles via the carboxyl terminus. Upon addition of proteases, both hybrid silica mesoporous nanoparticles exhibited remarkable payload release as a result of the hydrolysis of the polymer's amide bonds.
3.4 Inorganic nanoparticles
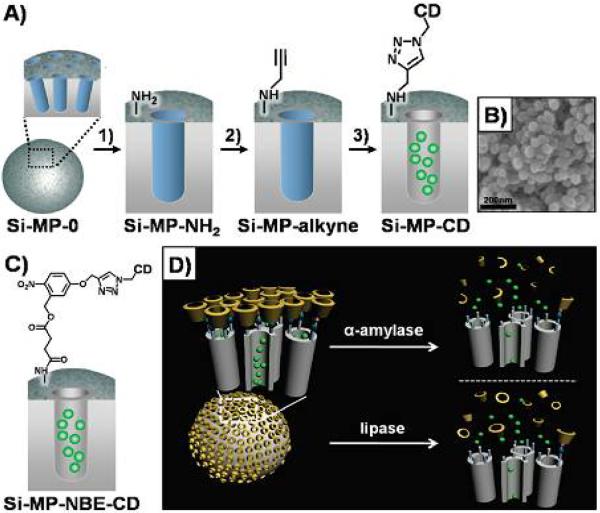
Inorganic nanoparticles, especially enzyme-responsive silica nanoparticles (Si-MPs), have great potential for useful applications in nanomedicine due to their unique responsiveness. In particular, enzyme-responsive Si-MPs equipped with gatekeepers are enormously appealing and are being implemented in controlled release systems. For instance, Park et al. reported an enzyme-responsive drug delivery system which could release cargoes in response to α-amylase and lipase (Fig. 11).89 Their enzyme-responsive Si-MPs were composed of the surface cyclodextrin (CD) gatekeepers, functional stalks, and fluorescence probes within porous channels. In the presence of α-amylase, the CD which initially block the the porous channel could be hydrolyzed. Subsequently stalk was degraded by lipase, resluting in release of crgoes.This strategy permitted the Si-MPs to exhibit enzyme-responsive characteristics.
Fig. 11.
Si-MPs-based enzyme-responsive drug delivery system. (A) Schematic illustration of Si-MP-CD. (B) FE-SEM image of Si-MP-CD. (C) Functional ligand on the surface of SiMP-NBE-CD. (D) α-amylase and lipase triggered release of guest molecules from Si-MP-CD. Reproduced from ref. 89 with permission from American Chemical Society.
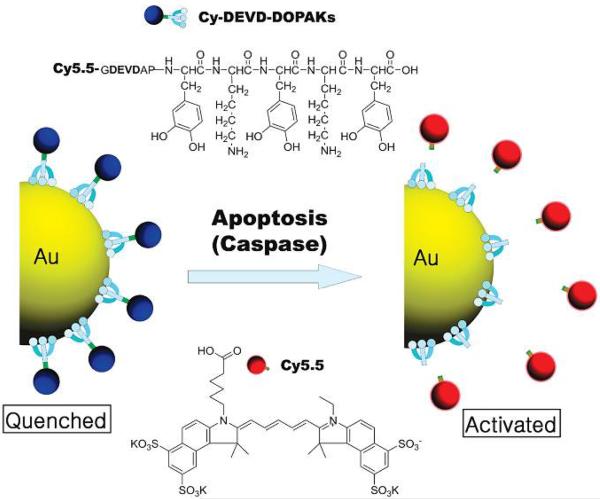
Another enzyme-responsive inorganic nanoparticle which has drawn great attention is gold (Au) nanoparticle due to its unique chemical and physical properties including ease of synthesis, versatile functionalization through thiol linkages and optical properties derived from the localized surface plasmon resonance (LSPR) phenomenon.90, 91 Sun et al. developed a caspase sensitive Au nanoparticle for apoptosis imaging in live cells (Fig. 12).92 Using caspase-3 degradable peptide substrate (DEVD) as a cross-linked bridge, a near-infrared fluorescence dye was decorated on the surface of AuNP. Without the presence of enzyme, the fluorescence was quenched. When the functionalized AuNP was taken up by live cell, the peptide substrate could be cleaved by caspase-3 and the quenched fluorescence was recovered. This new enzyme-responsive system provided a potential application for detecting the apoptosis process due to the involvement of caspase-3 in cellular apoptosis. Xia et al. developed an enzyme-sensitive multimodal gold nanocage probe that allowed for tumor localization with photoacoustic imaging and evaluation of metastasis with NIR fluorescence.93 They modified the surface of gold nanocages with dye-labeled peptides cleavable by metalloproteases (MMPs). Upon cleavage of the peptide in the presence of MMP-2, the dye was released from the gold nanocage and fluorescence emission was recovered. Furthermore, enzyme-responsive gold nanoparticles could be harnessed to detect and evaluate enzymatic activity. Using endonucleases as the trigger, Xu et al. reported a DNA-AuNP platform as colorimetric indicators to evaluate enzymatic activity and to screen enzyme inhibitors.94 Two functionalized, complmentary 13-nm AuNPs were hybridized to form a cross-linked network of nanoparticles, purple in color. When these aggregates were exposed to endonuclease, the DNA-duplex interconnecting the AuNPs was degraded and a red color was generated due to the separation of AuNPs.
Fig. 12.
Schematic illustration of enzyme-cleavable DEVD peptide-conjugated gold nanoparticles (DEVD-AuNPs). Reproduced from ref. 92 with permission from American Chemical Society.
4. Enzyme-based dual/multiple responsive systems
Over the past few decades enzyme-responsive nanomaterials have emerged as one promising and viable technology platform for controlled drug delivery. They have been actively designed to accomplish enhanced drug release at target sites (spatial control) and/or at the right time (temporal control). In an effort to further improve controlled drug release performances, enzyme-based dual/multiple responsive drug delivery systems that respond to a combination of two or more signals, i.e pH/enzyme and temperature/enzyme, have recently been developed. For example, Dong et al. described a pH/Enzyme-responsive tumor-specific delivery system for doxorubicin (Fig. 13A).95 Cationic gelatin sensitive to gelatinase (GA) and Dnase I was employed to form compact nanoscale complexes (CPX1) with polyGC-DOX intercalation. A pH-responsive pegylated alginate was utilized to form CPX2 when combined with CPX1. In the presence of GA and Dnase, CPX2 could be digested and released DOX when pH < 6.9. According to in vivo studies, pH/enzyme dual responsive CPX2 enhanced the drug accumulation in tumor tissue, resulting in enhanced anti-cancer activity (more than 2 times anti-cancer efficiency of DOX delivered by CPX2 when compared with free doxorubicin) and reduced the cardiotoxicity of the encapsulated DOX. In addition to pH/enzyme-responsive delivery systems, Chen et al. took another approach to design a temperature/enzyme-responsive polyvalent nucleic acid decorated mesoporous silica nanoparticle (MSP) intracellular drug delivery platform (Fig. 13B).96 They anchored a self-complementary duplex DNA to the channel of MSPs. This served as a cap which trapped the guest molecules within the MSP. The entrapped guest moleculars release was triggered either by thermal denaturation of the DNA duplex or by the presence of DNase I. Furthermore, their strategy extended the application of other functional nucleic acids to generate a multifunctional stimuli-responsive durg delivery system.
Fig. 13.
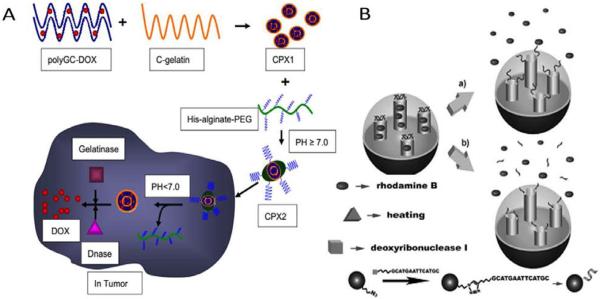
Enzyme-based dual responsive drug delivery system. (A) Schematic illustration of design of CPX1 and CPX2. Reproduced from ref. 95 with permission from Elsevier. (B) Release of cargoes from temperature/enzyme dual-responvie MSPs. Reproduced from ref. 96 with permission from WILEY.
5. Conclusion
The past several years have witnessed a rapid progress in the development of enzyme-responsive drug delivery system. These strategies exhibit tremendous therapeutic and detection potency for cancer or other diseases at both research and clinical levels. Nanomaterials, including polymers, lipids and inorganic materials, which are incorporated into an enzyme-responsive drug delivery system, hold incredible promise in overcoming the limitations and drawbacks of conventional drug delivery. Additionally, they offer unprecedented control over spatiotemporal drug release and delivery profiles leading to superior in vitro and/or in vivo theranostic efficiency62.
Despite emerging progress made in enzyme-responsive drug delivery research, there are still many challenges that need to be addressed. There is a tremendous variety of enzyme dysregulation activities in different diseases and even at different stages of one disease. Further fundamentally understanding of the spatial and temporal pattern of offers essential criteria for designing more effective and precise delivery vehicles. Second, there are many overlapping substrates between closely related enzyme families; more specific designs should be taken into account for enhancing delivery efficacy. Further, eventual translation of enzyme-responsive drug delivery requires comprehensive evaluation of biocompatibility of relevant formulations. Tailoring materials that present negligible cytotoxicity and systemic toxicity is highly desirable for clinical applications97–99.
Supplementary Material
Acknowledgements
This work was supported by the American Diabetes Association's Junior Faculty Award, the grant 550KR51307 from NC TraCS, the T32 GM008719 Medical Scientist Training Program grant from the National Institute for General Medical Sciences (NIGMS, NIH), NIH's Clinical and Translational Science Awards (CTSA) at UNC-CH, and the NC State Faculty Research and Professional Development Award to Z.G.
Reference
- 1.Farokhzad OC, Langer R. Adv Drug Deliv Rev. 2006;58:1456–1459. doi: 10.1016/j.addr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Farokhzad OC, Langer R. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 3.Brannon-Peppas L, Blanchette JO. Adv Drug Deliv Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Mo R, Jiang T, Di J, Tai W, Gu Z. Chem. Soc. Rev. 2014;43:3595–3629. doi: 10.1039/c3cs60436e. [DOI] [PubMed] [Google Scholar]
- 5.Gu Z, Aimetti AA, Wang Q, Dang TT, Zhang Y, Veiseh O, Cheng H, Langer RS, Anderson DG. ACS Nano. 2013;7:4194–4201. doi: 10.1021/nn400630x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaittanis C, Santra S, Perez JM. Adv Drug Deliv Rev. 2010;62:408–423. doi: 10.1016/j.addr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moghimi SM, Hunter AC, Murray JC. Pharmacol. Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 8.Wang H, Zhao Y, Wu Y, Hu YL, Nan K, Nie G, Chen H. Biomaterials. 2011;32:8281–8290. doi: 10.1016/j.biomaterials.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, Bimbo LM, Makila E, Villanova F, Kaasalainen M, Herranz-Blanco B, Caramella CM, Lehto VP, Salonen J, Herzig KH, Hirvonen J, Santos HA. J. Control. Release. 2013;170:268–278. doi: 10.1016/j.jconrel.2013.05.036. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X-X, Eden HS, Chen X. J. Control. Release. 2012;159:2–13. doi: 10.1016/j.jconrel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Adv Drug Deliv Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mura S, Nicolas J, Couvreur P. Nat. Mater. 2013;12:991–1003. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- 13.Lopes JR, Santos G, Barata P, Oliveira R, Lopes CM. Curr. Pharm. Des. 2013;19:7169–7184. doi: 10.2174/13816128113199990698. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Huang Y, Kumar A, Tan A, Jin S, Mozhi A, Liang XJ. Biotechnol. Adv. 2014;32:693–710. doi: 10.1016/j.biotechadv.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Schmaljohann D. Adv Drug Deliv Rev. 2006;58:1655–1670. doi: 10.1016/j.addr.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 16.Gao W, Chan JM, Farokhzad OC. Mol. Pharm. 2010;7:1913–1920. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ron ES, Bromberg LE. Adv Drug Deliv Rev. 1998;31:197–221. doi: 10.1016/s0169-409x(97)00121-x. [DOI] [PubMed] [Google Scholar]
- 18.Luo Z, Cai K, Hu Y, Zhao L, Liu P, Duan L, Yang W. Angew. Chem. Int. Ed. Engl. 2011;50:640–643. doi: 10.1002/anie.201005061. [DOI] [PubMed] [Google Scholar]
- 19.Markland P, Zhang Y, Amidon GL, Yang VC. J. Biomed. Mater. Res. 1999;47:595–602. doi: 10.1002/(sici)1097-4636(19991215)47:4<595::aid-jbm17>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 20.Murdan S. J. Control. Release. 2003;92:1–17. doi: 10.1016/s0168-3659(03)00303-1. [DOI] [PubMed] [Google Scholar]
- 21.Kumar CS, Mohammad F. Adv Drug Deliv Rev. 2011;63:789–808. doi: 10.1016/j.addr.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghadiali JE, Stevens MM. Adv. Mater. 2008;20:4359–4363. [Google Scholar]
- 23.Andresen TL, Thompson DH, Kaasgaard T. Mol. Membr. Biol. 2010;27:353–363. doi: 10.3109/09687688.2010.515950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MR, Baek KH, Jin HJ, Jung YG, Shin I. Angewandte Chemie International Edition. 2004;43:1675–1678. doi: 10.1002/anie.200353204. [DOI] [PubMed] [Google Scholar]
- 25.Hu J, Zhang G, Liu S. Chem. Soc. Rev. 2012;41:5933–5949. doi: 10.1039/c2cs35103j. [DOI] [PubMed] [Google Scholar]
- 26.Ulijn RV. J. Mater. Chem. 2006;16:2217–2225. [Google Scholar]
- 27.de la Rica R, Aili D, Stevens MM. Adv. Drug Delivery Rev. 2012;64:967–978. doi: 10.1016/j.addr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Chen Q, Wang Z, Zhang X. Angew. Chem. 2010;122:8794–8797. [Google Scholar]
- 29.de las Heras Alarcón C, Pennadam S, Alexander C. Chem. Soc. Rev. 2005;34:276–285. doi: 10.1039/b406727d. [DOI] [PubMed] [Google Scholar]
- 30.Bawa P, Pillay V, Choonara YE, Du Toit LC. Biomedical materials. 2009;4:022001. doi: 10.1088/1748-6041/4/2/022001. [DOI] [PubMed] [Google Scholar]
- 31.Wang S.-n., Deng Y.-h., Xu H, Wu H.-b., Qiu Y.-k., Chen D.-w. Eur. J. Pharm. Biopharm. 2006;62:32–38. doi: 10.1016/j.ejpb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Kawakami S, Munakata C, Fumoto S, Yamashita F, Hashida M. J. Pharm. Sci. 2001;90:105–113. doi: 10.1002/1520-6017(200102)90:2<105::aid-jps1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 33.Popat A, Ross BP, Liu J, Jambhrunkar S, Kleitz F, Qiao SZ. Angewandte Chemie International Edition. 2012;51:12486–12489. doi: 10.1002/anie.201206416. [DOI] [PubMed] [Google Scholar]
- 34.Allen TM. Nat. Rev. Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 35.Hahn ME, Gianneschi NC. Chem. Commun. 2011;47:11814–11821. doi: 10.1039/c1cc15220c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.HeeáKook Y, TaxáOh E, JooáPark H. J. Mater. Chem. 2009;19:2310–2315. [Google Scholar]
- 37.Liu J, Ong W, Román E, Lynn MJ, Kaifer AE. Langmuir. 2000;16:3000–3002. [Google Scholar]
- 38.Weissleder R, Tung C-H, Mahmood U, Bogdanov A. Nat. Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 39.Goel A, Chauhan S. Indian J. Exp. Biol. 1997;35:553–564. [PubMed] [Google Scholar]
- 40.Kang J-H, Asai D, Kim J-H, Mori T, Toita R, Tomiyama T, Asami Y, Oishi J, Sato YT, Niidome T. J. Am. Chem. Soc. 2008;130:14906–14907. doi: 10.1021/ja805364s. [DOI] [PubMed] [Google Scholar]
- 41.Law B, Weissleder R, Tung C-H. Biomacromolecules. 2006;7:1261–1265. doi: 10.1021/bm050920f. [DOI] [PubMed] [Google Scholar]
- 42.Crawford H, Matrisian L. Invasion Metastasis. 1993;14:234–245. [PubMed] [Google Scholar]
- 43.Yoon S-O, Park S-J, Yun C-H, Chung A-S. J. Biochem. Mol. Biol. 2003;36:128–137. doi: 10.5483/bmbrep.2003.36.1.128. [DOI] [PubMed] [Google Scholar]
- 44.Westermarck J, KÄHÄRI V-M. The FASEB Journal. 1999;13:781–792. [PubMed] [Google Scholar]
- 45.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson ES, Jiang T, Aguilera TA, Nguyen QT, Ellies LG, Scadeng M, Tsien RY. Proceedings of the National Academy of Sciences. 2010;107:4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu G, Xia H, Hu Q, Liu Z, Jiang M, Kang T, Miao D, Tu Y, Pang Z, Song Q, Yao L, Chen H, Gao X, Chen J. Biomaterials. 2013;34:196–208. doi: 10.1016/j.biomaterials.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 48.Scott KF, Sajinovic M, Hein J, Nixdorf S, Galettis P, Liauw W, de Souza P, Dong Q, Graham GG, Russell PJ. Biochimie. 2010;92:601–610. doi: 10.1016/j.biochi.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Wells A, Grandis JR. Clin. Exp. Metastasis. 2003;20:285–290. doi: 10.1023/a:1024088922957. [DOI] [PubMed] [Google Scholar]
- 50.Vadas P, Browning J, Edelson J, Pruzanski W. J. Lipid Mediat. 1993;8:1–30. [PubMed] [Google Scholar]
- 51.Schmiel DH, Miller VL. Microbes Infect. 1999;1:1103–1112. doi: 10.1016/s1286-4579(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 52.Graff JR, Konicek BW, Deddens JA, Chedid M, Hurst BM, Colligan B, Neubauer BL, Carter HW, Carter JH. Clin. Cancer Res. 2001;7:3857–3861. [PubMed] [Google Scholar]
- 53.Laye JP, Gill JH. Drug Discov. Today. 2003;8:710–716. doi: 10.1016/s1359-6446(03)02754-5. [DOI] [PubMed] [Google Scholar]
- 54.Cummings BS. Biochem. Pharmacol. 2007;74:949–959. doi: 10.1016/j.bcp.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 55.Andresen TL, Davidsen J, Begtrup M, Mouritsen OG, Jørgensen K. J. Med. Chem. 2004;47:1694–1703. doi: 10.1021/jm031029r. [DOI] [PubMed] [Google Scholar]
- 56.Linderoth L, Peters GH, Madsen R, Andresen TL. Angewandte Chemie International Edition. 2009;48:1823–1826. doi: 10.1002/anie.200805241. [DOI] [PubMed] [Google Scholar]
- 57.Butterfield DA, Hardas SS, Lange MLB. J. Alzheimer's Dis. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kundu JK, Surh Y-J. Pharm. Res. 2010;27:999–1013. doi: 10.1007/s11095-010-0096-8. [DOI] [PubMed] [Google Scholar]
- 59.Gu Z, Dang TT, Ma M, Tang BC, Cheng H, Jiang S, Dong Y, Zhang Y, Anderson DG. ACS Nano. 2013;7:6758–6766. doi: 10.1021/nn401617u. [DOI] [PubMed] [Google Scholar]
- 60.Harnoy AJ, Rosenbaum I, Tirosh E, Ebenstein Y, Shaharabani R, Beck R, Amir RJ. J. Am. Chem. Soc. 2014;136:7531–7534. doi: 10.1021/ja413036q. [DOI] [PubMed] [Google Scholar]
- 61.Rao J, Khan A. J. Am. Chem. Soc. 2013;135:14056–14059. doi: 10.1021/ja407514z. [DOI] [PubMed] [Google Scholar]
- 62.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 63.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 64.Davis ME. Nat. Rev. Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 65.Petros RA, DeSimone JM. Nat. Rev. Drug Discovery. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 66.Hu Q, Gu G, Liu Z, Jiang M, Kang T, Miao D, Tu Y, Pang Z, Song Q, Yao L, Xia H, Chen H, Jiang X, Gao X, Chen J. Biomaterials. 2013;34:1135–1145. doi: 10.1016/j.biomaterials.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 67.Mohanraj V, Chen Y. Tropical Journal of Pharmaceutical Research. 2007;5:561–573. [Google Scholar]
- 68.Soppimath KS, Aminabhavi TM, Kulkarni AR, Rudzinski WE. J. Control. Release. 2001;70:1–20. doi: 10.1016/s0168-3659(00)00339-4. [DOI] [PubMed] [Google Scholar]
- 69.Wong C, Stylianopoulos T, Cui J, Martin J, Chauhan VP, Jiang W, Popović Z, Jain RK, Bawendi MG, Fukumura D. Proceedings of the National Academy of Sciences. 2011;108:2426–2431. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chien M-P, Carlini AS, Hu D, Barback CV, Rush AM, Hall DJ, Orr G, Gianneschi NC. J. Am. Chem. Soc. 2013;135:18710–18713. doi: 10.1021/ja408182p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gu Z, Yan M, Hu B, Joo K-I, Biswas A, Huang Y, Lu Y, Wang P, Tang Y. Nano Lett. 2009;9:4533–4538. doi: 10.1021/nl902935b. [DOI] [PubMed] [Google Scholar]
- 72.Gu Z, Biswas A, Joo K-I, Hu B, Wang P, Tang Y. Chem. Commun. 2010;46:6467–6469. doi: 10.1039/c0cc01439g. [DOI] [PubMed] [Google Scholar]
- 73.Gu Z, Tang Y. Lab Chip. 2010;10:1946–1951. doi: 10.1039/c001335h. [DOI] [PubMed] [Google Scholar]
- 74.Biswas A, Joo K-I, Liu J, Zhao M, Fan G, Wang P, Gu Z, Tang Y. ACS Nano. 2011;5:1385–1394. doi: 10.1021/nn1031005. [DOI] [PubMed] [Google Scholar]
- 75.Torchilin VP. Nat. Rev. Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 76.Ramishetti S, Huang L. Therapeutic delivery. 2012;3:1429–1445. doi: 10.4155/tde.12.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samad A, Sultana Y, Aqil M. Curr. Drug Deliv. 2007;4:297–305. doi: 10.2174/156720107782151269. [DOI] [PubMed] [Google Scholar]
- 78.Pak CC, Ali S, Janoff AS, Meers P. Biochimica et Biophysica Acta (BBA)-Biomembranes. 1998;1372:13–27. doi: 10.1016/s0005-2736(98)00041-8. [DOI] [PubMed] [Google Scholar]
- 79.Gabizon AA. Adv. Drug Delivery Rev. 1995;16:285–294. [Google Scholar]
- 80.Torchilin V. Adv. Drug Delivery Rev. 2011;63:131–135. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 81.Wan Y, Han J, Fan G, Zhang Z, Gong T, Sun X. Biomaterials. 2013;34:3020–3030. doi: 10.1016/j.biomaterials.2012.12.051. [DOI] [PubMed] [Google Scholar]
- 82.Terada T, Iwai M, Kawakami S, Yamashita F, Hashida M. J. Control. Release. 2006;111:333–342. doi: 10.1016/j.jconrel.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 83.Schärtl W. Nanoscale. 2010;2:829–843. doi: 10.1039/c0nr00028k. [DOI] [PubMed] [Google Scholar]
- 84.Zhang L, Chan JM, Gu FX, Rhee J-W, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. ACS Nano. 2008;2:1696–1702. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu C-MJ, Fang RH, Luk BT, Zhang L. Nanoscale. 2014;6:65–75. doi: 10.1039/c3nr05444f. [DOI] [PubMed] [Google Scholar]
- 86.Jiang T, Mo R, Bellotti A, Zhou J, Gu Z. Adv. Funct. Mater. 2014;24:2295–2304. [Google Scholar]
- 87.Bernardos A, Mondragon L, Aznar E, Marcos MD, Martínez-Máñez R. n., Sancenón F. l., Soto J, Barat JM, Pérez-Payá E, Guillem C. ACS Nano. 2010;4:6353–6368. doi: 10.1021/nn101499d. [DOI] [PubMed] [Google Scholar]
- 88.Mondragon L, Mas N, Ferragud V, de la Torre C, Agostini A, Martinez-Manez R, Sancenon F, Amoros P, Perez-Paya E, Orzaez M. Chemistry (Weinheim an der Bergstrasse, Germany) 2014;20:5271–5281. doi: 10.1002/chem.201400148. [DOI] [PubMed] [Google Scholar]
- 89.Park C, Kim H, Kim S, Kim C. J. Am. Chem. Soc. 2009;131:16614–16615. doi: 10.1021/ja9061085. [DOI] [PubMed] [Google Scholar]
- 90.Daniel M-C, Astruc D. Chem. Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 91.Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv. Drug Delivery Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 92.Sun I-C, Lee S, Koo H, Kwon IC, Choi K, Ahn C-H, Kim K. Bioconjug. Chem. 2010;21:1939–1942. doi: 10.1021/bc1003026. [DOI] [PubMed] [Google Scholar]
- 93.Xia X, Yang M, Oetjen LK, Zhang Y, Chen Q. Li, J., Xia Y. Nanoscale. 2011;3:950–953. doi: 10.1039/c0nr00874e. [DOI] [PubMed] [Google Scholar]
- 94.Xu X, Han MS, Mirkin CA. Angew. Chem. 2007;119:3538–3540. [Google Scholar]
- 95.Dong L, Xia S, Wu K, Huang Z, Chen H, Chen J, Zhang J. Biomaterials. 2010;31:6309–6316. doi: 10.1016/j.biomaterials.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 96.Chen C, Geng J, Pu F, Yang X, Ren J, Qu X. Angewandte Chemie International Edition. 2011;50:882–886. doi: 10.1002/anie.201005471. [DOI] [PubMed] [Google Scholar]
- 97.Chow EK-H, Ho D. Sci. Transl. Med. 2013;5:216rv214–216rv214. doi: 10.1126/scitranslmed.3005872. [DOI] [PubMed] [Google Scholar]
- 98.Tai W, Mo R, Lu Y, Jiang T, Gu Z. Biomaterials. 2014 doi: 10.1016/j.biomaterials.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gu Z, Biswas A, Zhao M, Tang Y. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.