Abstract
In this model study, we explored the host’s contribution of breath volatiles to diagnostic secondary electrospray ionisation-mass spectrometry (SESI-MS) breathprints for acute bacterial lung infections, their correlation with the host’s immune response, and their use in identifying the lung pathogen.
Murine airways were exposed to Pseudomonas aeruginosa and Staphylococcus aureus bacterial cell lysates or to PBS (controls), and their breath and bronchoalveolar lavage fluid (BALF) were collected at six time points (from 6 to 120 h) after exposure. Five to six mice per treatment group and four to six mice per control group were sampled at each time. Breath volatiles were analysed using SESI-MS and the BALF total leukocytes, polymorphonuclear neutrophils, lactate dehydrogenase activity, and cytokine concentrations were quantified.
Lysate exposure breathprints contain host volatiles that persist for up to 120 h; are pathogen specific; are unique from breathprints of controls, active infections and cleared infections; and are correlated with the host’s immune response.
Bacterial lung infections induce changes to the host’s breath volatiles that are selective and specific predictors of the source of infection. Harnessing the pathogen-specific volatiles in the host’s breath may provide useful information for detecting latent bacterial lung infections and managing the spread of respiratory diseases.
Introduction
Recent developments in breath analysis have reinvigorated the quest for breath-based diagnostics, advancing the promise of noninvasive tests for diseases ranging from lung cancer to schizophrenia [1–8]. Breath contains hundreds of volatile organic compounds (VOCs) per individual, amounting to thousands in the human breath metabolome [9], potentially conveying a wealth of information about the human condition. The goal of our work is to use advanced mass spectrometry methods to capture the vast chemical information in breath for the development of rapid breath-based diagnostic tools. We have previously employed secondary electrospray ionisation-mass spectrometry (SESI-MS) to generate diagnostic fingerprints of acute bacterial lung infections from the VOCs of breath [10–14]. SESI-MS does not require any sample preparation; the mixture of breath volatiles is introduced directly into the mass spectrometer, where the VOCs are sorted by their mass-to-charge ratio (m/z) and counted, creating a breathprint that is used to identify the aetiology of lung infections without the need for compound identification [12, 14]. We have previously demonstrated that SESI-MS breathprints can be used to distinguish between seven different bacterial lung pathogens in vivo [12], and although the breathprint changes over time throughout the course of infection and clearance, portions of the breathprint are stable enough to identify the pathogen at any time from 6 to 120 h after initial pathogen exposure [11]. Even after the infections have been cleared (i.e. no viable bacteria recovered from the lung), components of the breathprint can be used to identify the pathogen with which the host had been previously infected [11]. This observation led us to speculate that the distinguishing features of each breathprint do not arise solely from pathogen metabolism but are also due to changes to host VOCs, possibly in conjunction with the immunological response.
In the murine model study described herein, we delve into the contribution of the host’s breath volatiles to acute lung infection breathprints. To isolate host-derived VOCs from the total volatile metabolome of an infection, we exposed mice airways to bacterial cell lysates of Pseudomonas aeruginosa and Staphylococcus aureus (two clinically important opportunistic pathogens) to obtain breathprints without the metabolic contributions from living bacterial cells. The lysates activate the host’s immune response, which we hypothesised would generate pathogen-specific changes in the breathprint. We found correlations between breathprint peaks and cytokine concentrations, neutrophils and leukocyte counts in bronchoalveolar lavage fluid (BALF), providing evidence that breath contains volatiles that are related to host immunology. In addition, we found that breathprints of lysate exposure differ to breathprints of the corresponding active infections as well as uninfected controls, and that they are predictive of the bacterial lysate source. These results have important implications for the detection of latent bacterial lung infections, for monitoring and managing the spread of respiratory diseases, and for assessing and monitoring new and existing bacterial vaccines.
Materials and methods
Bacterial strains and lysate preparation
Bacterial cell lysate preparation was adapted from the study by Allard et al. [15]. Briefly, P. aeruginosa strain PAO1-UW and S. aureus RN450 were grown aerobically in 100 mL of Dulbecco’s Modified Eagle Medium (Mediatech, Manassas, VA, USA) and tryptic soy broth, respectively. Cells were harvested by centrifugation, washed and resuspended in 100 mM phosphate buffer (pH 7). Bacterial cells were lysed by ballistic disintegration and heated (65°C; 10 min). Plate counts confirmed that no viable cells remained. Lysates were diluted in PBS to a final protein concentration of 1 mg·mL−1 and then stored frozen at −80°C until use.
Murine airway exposure, bronchoalveolar lavage, and breath collection and analysis
6–8-week-old male C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Treatment groups were exposed to 5 μg of protein in P. aeruginosa or S. aureus bacterial cell lysate in 40 μL of PBS by oropharyngeal aspiration [15], and their breath was collected 6, 12, 24, 48, 72 or 120 h after exposure and analysed by SESI-MS. Additional groups of mice were exposed to 40 μL of PBS by oropharyngeal aspiration as a negative control for each time point. After breath collection, bronchoalveolar lavage (BAL) was performed and the total leukocytes (or white blood cells (WBCs)) were counted and characterised as macrophages, eosinophils, polymorphonuclear neutrophils (PMNs) or lymphocytes based on characteristic morphology and staining. Lactate dehydrogenase (LDH) activity in the cell supernatant was quantified, as previously described [11]. Cytokine concentrations in the BALF were analysed using customised Milliplex assays (Millipore, Billerica, MA, USA). The cytokine panel is provided in the online supplementary material.
The protocols for animal infection and respiratory physiology measurements conformed to the Declaration of Helsinki and were approved by the Institutional Animal Care and Use Committee, in accordance with Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines. All mice were housed in an AAALAC-accredited animal facility at the University of Vermont (Burlington, VT, USA). Full protocols for animal care, airway exposure, breath collection and SESI-MS breath analysis used in this study can be found in previous publications [12, 14], and are summarised in the online supplementary material.
Sample size, data analysis and statistics
87 mice were used in this study; five mice per time point for S. aureus lysate (SAL) and P. aeruginosa lysate (PAL) and four mice per time point for PBS, with the exception of 24-h PAL and PBS, which included six mice each. Full details on sample size estimation are provided in the online supplementary material.
The statistical significance of BAL total leukocytes, PMN counts and LDH activity between lysate exposure and PBS control groups was determined using two-way ANOVA followed by Tukey’s Honestly Significant Difference test. To meet the assumption of normal distribution for the ANOVA and t-test, total leukocytes and PMN counts were log-transformed before the analysis. All values represent the mean±SEM of all replicates within each time point.
SESI-MS spectra collection and raw data processing were conducted using Analyst 1.4.2 software (Applied Biosystems, Foster City, CA, USA). Spectra are the averages of all sample replicates in each group, which were blank-subtracted and normalised to the peak of greatest intensity. The blank spectrum is humidified room air collected using the same procedure as for mouse breath. JMP version 10 (SAS Institute Inc., Cary, NC, USA) was used in this study for all statistical analyses. Spearman’s rank correlation coefficients (ρ) were calculated to compare the similarity of breathprint profiles from each treatment group at each time point. Two-sided non-parametric Mann–Whitney U-tests were carried out to identify breathprint peaks that were significantly different between pairs of test groups using a 5% significance level cut-off.
Spearman’s rank analyses were used to identify correlations between breathprint peaks and cytokine concentrations, total WBCs, PMNs and/or LDH activity at 6, 12, 24 and 48 h after exposure to PAL, SAL or PBS. The correlations reported in table 1 correspond to the average Spearman’s rank coefficients of all four time points. Partial least squares-discriminant analysis (PLS-DA) was conducted for group classification using spectral intensity data from all of the breathprints in each treatment group or using cytokine concentrations in BALF. Breathprints from active infections of P. aeruginosa and S. aureus that were used for the PLS-DA in this study were collected contemporaneously with the lysate breathprints, and have been previously published [11]. Details of the Spearman’s rank correlation and PLS-DA analyses are provided in the online supplementary material. To validate the utility of SESI-MS breathprints in classifying unknown breath samples, two rounds of 10-times PLS-DA cross-validation tests were performed. In the first round, 90% of the samples were used as a training set (randomly selected) and the remaining 10% of samples were used as a testing set. In the second round, 70% of the samples were used as a training set (randomly selected) and the remaining 30% of samples were used as a testing set. The correct classification rates for the training sets were 100% in both rounds. The correct classification rates for the testing sets were calculated as the number of correctly classified samples divided by the number of samples that were included in the testing sets over all 10 rounds of cross-validation.
TABLE 1.
Average Spearman’s rank correlation coefficients between the secondary electrospray ionisation-mass spectrometry peak intensities and host-derived parameters of lung inflammation up to 48 h post-exposure
| Positive correlations
|
Negative correlations
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| m/z 56 | m/z 70 | m/z 97 | m/z 115 | m/z 121 | m/z 41 | m/z 43 | m/z 44 | m/z 63 | m/z 94 | |
| G-CSF | 0.71 ± 0.13* | 0.73 ± 0.10# | 0.82 ± 0.02¶ | −0.74 ± 0.17* | −0.71 ± 0.16* | −0.82 ± 0.14¶ | −0.71 ± 0.18* | |||
| IL-6 | −0.80 ± 0.18* | −0.70 ± 0.14* | −0.78 ± 0.16* | |||||||
| KC | −0.81 ± 0.05¶ | −0.76 ± 0.06¶ | −0.84 ± 0.17¶ | |||||||
| MIP-1β | 0.78 ± 0.01¶ | −0.71 ± 0.18* | ||||||||
| RANTES | 0.79 ± 0.25# | 0.78 ± 0.15# | 0.81 ± 0.23# | 0.81 ± 0.16* | 0.73 ± 0.32* | −0.76 ± 0.01¶ | ||||
| TNF-α | −0.72 ± 0.11# | |||||||||
| PMNs | 0.71 ± 0.13# | 0.74 ± 0.13* | 0.79 ± 0.04# | −0.79 ± 0.18* | −0.72 ± 0.27* | −0.79 ± 0.19# | ||||
| WBCs | 0.74 ± 0.13* | 0.75 ± 0.09# | 0.79 ± 0.03¶ | −0.77 ± 0.22# | −0.79 ± 0.16# | |||||
Data are presented as mean ± SEM. m/z: mass-to-charge ratio; G-CSF: granulocyte-colony stimulating factor; IL-6: interleukin-6; KC: keratinocyte-derived chemokine; MIP-1β: macrophage inflammatory protein-1β; TNF: tumour necrosis factor; PMNs: polymorphonuclear neutrophils; WBCs: white blood cells.
p < 0.05;
p < 0.001;
p < 0.0001.
Results
Host VOCs and their correlation with the immune system response
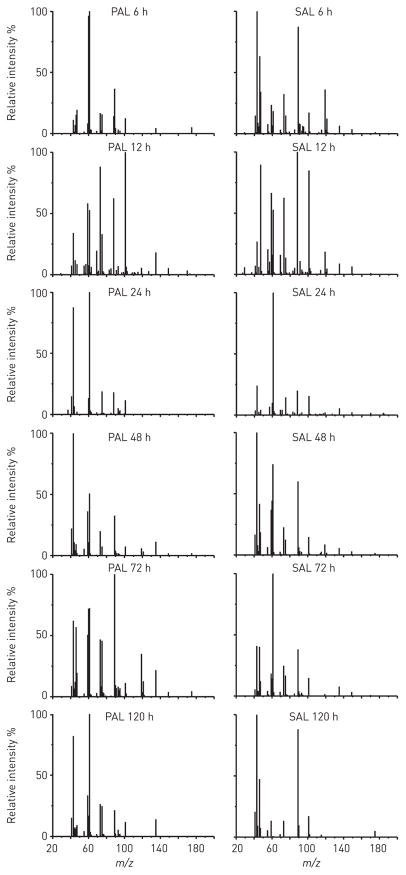
To capture the host-derived component of lung infection breathprints, we exposed mice airways to PAL and SAL, and analysed their breath volatiles using SESI-MS (m/z = 20–200 Da) at six time points from 6 to 120 h after exposure (fig. 1). The Spearman’s rank correlation coefficients (ρ), comparing the similarity of the five or six replicates within each time point and treatment group, were strongly correlated (all ρ≥0.85, SEM ≤0.06). Based on Mann–Whitney U-tests performed on a peak-by-peak basis at each time point, the breathprints of mice exposed to PAL or SAL differ from mice in the control group treated with PBS (table S1, PAL versus PBS and SAL versus PBS). We observed that 41 breathprint peaks increase with exposure to PAL or SAL (versus controls), and 24 peaks decrease, confirming that the host’s breath volatiles change in response to airway exposures to bacterial lysates.
FIGURE 1.
Secondary electrospray ionisation-mass spectrometry breathprints of mice 6–120 h after Pseudomonas aeruginosa lysate (PAL) or Staphylococcus aureus lysate (SAL) airway exposure. Each relative intensity spectrum is the average breathprint from at least five biological replicates. m/z: mass-to-change ratio.
The SESI-MS breathprints change over time after bacterial lysate exposure (fig. 1 and table S1) and we found that the immune system responds in kind. In comparison with control PBS exposures, the BALF of mice treated with PAL had greater numbers of WBCs 6–72 h after exposure (fig. 2a), and the same trend was observed for PMNs (fig. 2b). SAL airway exposure resulted in absolute WBC and PMN cell counts that were higher than in the PBS controls for time points up to 48 h (fig. 2a and b). In addition, we observed an increase in LDH activity (fig. 2c) and statistically significant increases in the concentrations of 10 cytokines in BALF up to 72 h after lysate exposure (table S2). While the PAL provokes a stronger immune and inflammatory response than the SAL in mouse airways, both lysates induced measurable immune responses that begin to abate after 48–72 h, with lung damage peaking during that same time period.
FIGURE 2.
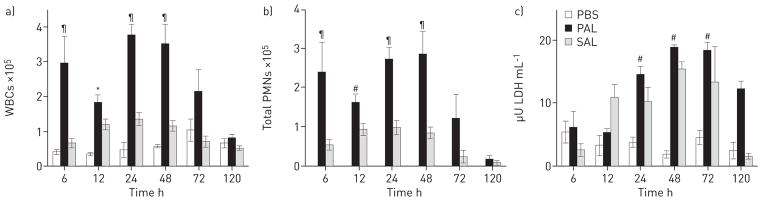
Host immune factors in bronchoalveolar lavage fluid (BALF), 6–120 h after airway exposure to Pseudomonas aeruginosa lysate (PAL), Staphylococcus aureus lysate (SAL) or to PBS control. a) White blood cells (WBCs). b) Total polymorphonuclear neutrophils (PMNs). The numbers of PMNs in BALF after PBS treatment are not visible on this scale. c) Lactate dehydrogenase (LDH) activity. Data represent the mean±SEM of at least five mice per treatment group, and at least four mice per control. Statistical significance was determined using two-way ANOVA by comparison with the appropriate PBS-treated mice at each time post-exposure (6, 12, 24, 48, 72 and 120 h). *: p<0.05; #: p<0.001; ¶: p<0.0001.
Airway exposure to bacterial lysates induces time-dependent changes in the host’s immune response as well as the host’s breath volatiles. We therefore postulated that these two phenomena are linked. We used the non-parametric Spearman’s rank correlation method to evaluate the relationship between the measurements of immunological response and the SESI-MS breathprints. The immune markers were only robustly different in the lysate-exposed groups versus PBS controls at 6–48 h (i.e. at least half of the immune markers measured at each time point were statistically different from the control) (fig. 2 and table S2); therefore, we restricted the correlation analysis to data collected at the first four time points. In table 1, we report the average Spearman’s rank correlation coefficients for the strongest positive and negative correlations (average >0.7 or < −0.7). All of the immunological markers trended together and had positive correlations with breathprint peaks (m/z) 56, 70, 97, 115 and 121, meaning that as the immune determinants increased, these breathprint peaks increased in magnitude. In contrast, negative correlations were observed with breathprint peaks (m/z) 41, 43, 44, 63 and 94, where peak magnitudes increased as the concentrations of host immune markers decreased (or vice versa). Corroborating these data, we found that peaks (m/z) 56, 70, 97 and 115 were also statistically significant indicators of PAL and/or SAL exposure (table S1, PAL versus PBS and SAL versus PBS), and peaks (m/z) 41, 43 and 44 decreased compared with the controls at early time points and increased at later times, in opposition to the response of WBCs and PMNs (fig. 2, and tables S1 and S2).
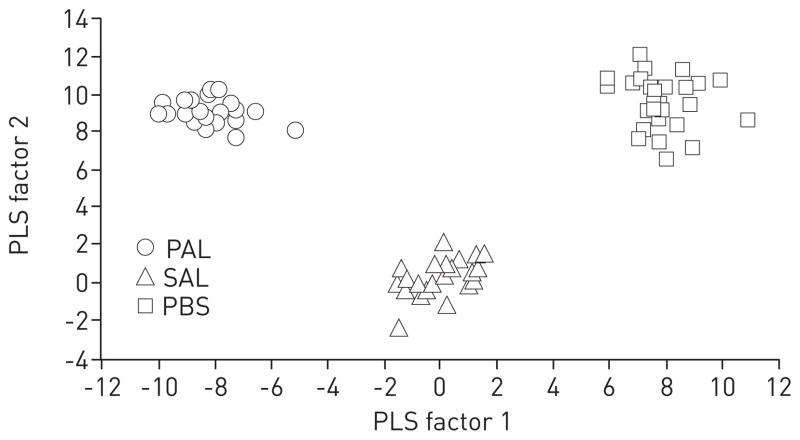
Host versus pathogen contributions to infection breathprints
We hypothesised that a portion of the time-independent SESI-MS breathprint peaks of active infections [11] arise from the host in response to the presence of the pathogen, while others are attributable to bacterial metabolites, making it possible to distinguish active infections from lysate exposure. Using PLS-DA, we aggregated all of the time points in each test group to identify breathprint peaks that were predictive of bacterial infection or lysate exposure and that endured for up to 120 h. The PLS-DA canonical plot of the P. aeruginosa treatment groups showed clear separations between the groups using the first two PLS factors (fig. 3a, p<0.0001). In addition, all of the replicates for any treatment group, regardless of the time point, could be clustered. Similar results were obtained with PLS-DA of the S. aureus treatment groups (fig. 3b).
FIGURE 3.
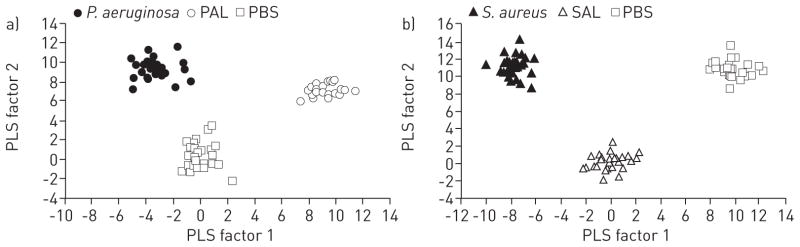
Discriminant analyses using prediction formulae from partial least squares (PLS) regressions for the separation of breathprints from mice with active infections, lysate exposure or untreated controls. a) Pseudomonas aeruginosa lung infection, P. aeruginosa lysate (PAL) exposure or the untreated PBS controls. The PLS algorithm used nine factors, with a minimum prediction sum of squares (PRESS) residual statistic of 0.45. b) Staphylococcus aureus lung infection, S. aureus lysate (SAL) exposure or the untreated PBS controls. The PLS algorithm used 13 factors, with a minimum PRESS residual statistic of 0.33. The first two PLS factors explain the largest percentage of the variation (P. aeruginosa 28.7%; S. aureus 27.0%) and provide a statistically significant separation of all three groups in each case (p<0.0001). All replicates for all six time points (6, 12, 24, 48, 72 and 120 h) for each group were included, resulting in 88 biological replicates for P. aeruginosa-PAL-PBS (a) and 86 for S. aureus-SAL-PBS (b).
The variable importance plot (VIP) showed 81 breathprint peaks strongly contributing to the separation of the P. aeruginosa-PAL-PBS treatment groups and 87 peaks for S. aureus-SAL-PBS (figs S1 and S2; VIP >0.8 [16]). To identify which of these peaks were significantly different between active infections, lysate exposure and controls, we performed Mann–Whitney U-tests on a peak-by-peak basis at each time point and compared these data to the VIP and the Spearman’s rank correlations (table S1). We found six peaks that strongly indicated the host had been exposed to live pathogens or their lysates (m/z 56, 59, 69, 75, 101 and 115), which were significantly increased in the treatment groups versus controls and had VIP >1.5 [17]. There were an additional 19 peaks that separated the controls from the infection and exposure groups, either by their presence only in the uninfected/unexposed controls, or their exclusive presence in the treatment groups. In addition, four of these 25 peaks were positively correlated with the host’s immune response (m/z 56, 70, 97 and 115) (table 1). Approximately two-thirds of the remaining peaks were selectively present or absent in one or two treatment groups, and these data can be used in aggregate in SESI-MS breathprinting to discriminate infection from exposure, and host versus pathogen contributions to breath volatiles.
Host breath VOCs are specific to the source of lysate exposure
We hypothesised that some of the host’s breath volatiles were changing in a pathogen-specific way and were contributing to the diagnostic power of the breathprint. In the Mann–Whitney data comparing P. aeruginosa versus PAL and S. aureus versus SAL (table S1), there were only two peaks that were shared exclusively by the lysate-exposed groups (m/z = 71 and 112), while there were many peaks that differed between SAL and PAL. To confirm that these differences were robust and predictive of the lysate source, we performed Mann–Whitney U-tests between the PAL and SAL breathprints and found 72 peaks that were significantly different between the two treatment groups at some time up to 120 h after lysate exposure (table S1; PAL versus SAL). The stable host breath volatiles that differentiated the exposure sources were identified by PLS-DA, which clearly separated the PAL, SAL and PBS groups (fig. 4; p<0.0001). The VIP indicated that there were 76 peaks from the lysate exposure breathprints driving the time-independent separation of the treatment groups (fig. S3), which included nine of the 10 breathprint peaks that were strongly correlated with host immune and inflammation markers (table 1).
FIGURE 4.
Discriminant analysis using prediction formulae from partial least squares (PLS) regression for the separation of breathprints from mice treated with Pseudomonas aeruginosa lysate (PAL) or Staphylococcus aureus lysate (SAL) versus the untreated PBS control. The PLS algorithm used 10 factors, with a minimum prediction sum of squares (PRESS) statistic of 0.39. The first two PLS factors explain the largest percentage of variation (22.6%), and provide statistically significant separation of all three groups (p<0.0001). All replicates for the six time points (6, 12, 24, 48, 72 and 120 h) were included for each group, resulting in 87 biological replicates in the analysis.
As a final verification that breathprints were a robust indicator of the source of bacterial lysate exposure, we used PLS-DA 10-times cross-validation to predict the classification of unknown samples (acquired at any of the six time points) as PAL-, SAL- or PBS-exposed breath (table 2). We used two testing set sizes, 10% and 30% of the total data, in order to compensate for the biases of holdout accuracy estimations using large or small testing sets [18]. When using 90% of the samples (randomly selected) as the training set and the remaining 10% as the testing set, we observed a 99% correct classification rate for the testing set, with just one false-negative classification of SAL as PBS. When using 70% of the data for training and 30% for testing, we still observed correct classification for 94% of the test samples (table 2). We also evaluated cytokines in the BALF for specificity and selectivity in predicting the lysate treatment groups in the study. Clustering all time points up to 120 h for each treatment group (PAL, SAL or PBS) and using all 10 cytokines for PLS-DA observation variables, the separation between groups using the first two PLS factors was not clear (fig. S4). The poorest separation was between SAL and PBS, which resulted in a misclassification rate between these groups that exceeded 35% (table S3).
TABLE 2.
Summary of the correct classification rate for the partial least squares-discriminant analysis (PLS-DA) 10-times cross-validation tests of breathprints arising from bacterial cell lysate exposures
| Predicted treatment | Actual treatment
|
||
|---|---|---|---|
| PBS | PAL | SAL | |
| Round 1# | |||
| PBS | 100% | 0 | 3% |
| PAL | 0 | 100% | 0 |
| SAL | 0 | 0 | 97% |
| Round 2¶ | |||
| PBS | 100% | 0 | 17% |
| PAL | 0 | 100% | 0 |
| SAL | 0 | 0 | 83% |
PLS-DA using 90% training data and 10% testing data.
PLS-DA using 70% training data and 30% testing data.
Discussion
In this model study, we probed the host’s contribution of volatile metabolites to the diagnostic SESI-MS breathprints for acute bacterial lung infections. We accomplished this by collecting the breathprints of mice that were airway-exposed to P. aeruginosa and S. aureus cell lysates, simulating an exposure to bacterial pathogens without an active infection and without bacterial metabolite contributions to breath volatiles. SESI-MS breathprints changed over time from 6–120 h after lysate exposure (fig. 1), and were correlated to the host immune response (table 1). In addition, the breathprints arising from exposures to bacterial lysates differed from breathprints of the concomitant active infections (fig. 3), and were specific to the lysate source (fig. 4). Notably, all three statistical methods used to evaluate the breathprint data corroborated the reproducibility and discriminating power of SESI-MS breathprints; nearly all of the peaks that were statistically different between treatment groups (indicated by coloured blocks in the Mann–Whitney U-test columns of table S1) also contributed significantly to the PLS-DA separations (VIP >0.8), and all 10 of the immune-correlated peaks had significant VIP and Mann–Whitney scores. Through PLS-DA 10-times cross-validation tests, we demonstrated that the host breath volatiles were selective and specific predictors of the source of lysate exposure (table 2). There have been efforts to use cytokines as biomarkers of lung disease and injury [19–22]. In this study, the cytokine concentrations in BALF were not nuanced enough to distinguish between exposures to P. aeruginosa and S. aureus cell lysates up to 120 h (fig. S4, table S3). However, breathprints were unique, reproducible and stable enough to be used for diagnosing P. aeruginosa and S. aureus lung infections [11], distinguishing active infections versus lysate exposure (fig. 3), and identifying the source of pathogen exposure (fig. 4) for up to 5 days after the initial event.
We postulated that the breathprints of cell lysate exposure and cleared infections (e.g. 72 and 120 h P. aeruginosa and S. aureus breathprints from mice that no longer harboured viable bacteria in the lung) may be undistinguishable, as the breathprints of cleared infections may arise from the host’s immune response to dead bacterial cells or cellular debris. Interestingly, there was a robust separation of cleared infections and lysate exposure (fig. 3), demonstrating that infection breathprints contain additional information, even after clearance. The distinction may arise from differences in the host immune response to live pathogens versus bacterial lysates, similar to a phenomenon that has been observed in human blood immune markers for hepatitis B and its corresponding vaccine [23]. If it is indeed possible to harness differentiating breath volatiles related to the host’s response to infection versus antigen exposure, breath testing could improve triage management for contagious respiratory diseases [24] and the identification of vaccinated persons in populations with high burdens of endemic disease (e.g. bacille Calmette–Guérin vaccination versus tuberculosis infection) [25, 26]. In addition, identifying immune-related breath volatiles may make it possible to differentiate between bacterial colonisation versus active infection, a key concern for ventilated patients in hospitals worldwide [27].
By using Gram-positive and Gram-negative bacterial lysates in this study, we have arguably induced immune responses that are more different than we might have achieved for two bacterial species that were more closely related. We also acknowledge that human immunology is different and more varied than the murine model used in this study, and that developing a diagnostic for bacterial lung infections in humans will require careful accounting of these variations. We would expect, for example, that comorbidities that suppress the immune system (e.g. HIV or diabetes) would be likely to decrease the sensitivity of breath-based diagnostics if the tests incorporated immune-dependent data [28]. Conversely, comorbidities related to hyperactive immune systems (e.g. autoimmune diseases or allergies) might generate false-positive diagnoses. However, breathprint markers associated with a dysfunctional immune response may provide clinicians and researchers with useful information; for example, they may facilitate the early detection and monitoring of acute respiratory distress syndrome [29] or allergic asthma [30], they may identify persons with latent infections who are at risk of reactivation due to immunosuppression [26], or they may detect lethal co-infection combinations such as H1N1 influenza virus and Streptococcus pneumoniae [31]. Indeed, while there is a large body of work that has begun to elucidate how the human immune system responds to bacterial, viral, fungal and environmental insults, we have very limited information about how these different immune triggers may translate to breath [2–5]. As far as we are aware, this is the first study to distinguish between host and metabolising pathogen contributions to the breathprint. We propose that the host information in the breathprint adds value to bacterial infection diagnostics by identifying the pathogen within the context of disease, and also has the potential to advance other important aspects of clinical care and disease management.
Supplementary Material
Table S1: Mann-Whitney U-tests comparing SESI-MS breathprints at 6 – 120 h after murine airway exposures to P. aeruginosa lysate (PAL), S. aureus lysate (SAL), live P. aeruginosa (PA) or S. aureus (SA) pathogens, or phosphate buffered saline (PBS) control (see Materials & Methods for calculation details). To facilitate comparisons to the other statistical methods employed for data analysis, the PLS-DA VIP scores (Figs. S1–S3) and immune correlations (Table 1) are summarized in the four left-most columns.
Table S2: Average cytokine concentrations (pg/mL) in bronchoalveolar lavage fluid at six time points 6 – 120 h after murine airway exposure to P. aeruginosa cell lysate, S. aureus cell lysate, or phosphate buffered saline (PBS) control.
Acknowledgments
Support statement: This project was financially supported by grants from the National Center for Research Resources (5P20RR021905-07), the National Institute of General Medical Sciences (8 P20 GM103496-07), the National Institutes of Health, NASA EPSCoR (NNH09ZNE002C), and by a postdoctoral fellowship from the Cystic Fibrosis Foundation (H.D. Bean).
The authors would like to thank the following faculty, staff and students of the University of Vermont (Burlington, VT, USA) (except where otherwise noted) for contributing to this study: Lennart Lundblad, Nirav Daphtary and Minara Aliyeva of the Vermont Lung Center ventilation facility and Jenna Allard for their technical assistance with the mouse work; Fiona Baird and Janice Bunn for their contribution to the experimental design; Beth Kirkpatrick, Matthew Poynter and Janet Lindow (Yale School of Medicine, New Haven, CT, USA) for helpful feedback on the data and manuscript; and Pierre Galea, Jackson Sengle and Frederick Naumann for their assistance with breath collection.
Footnotes
Conflict of interest: Disclosures can be found alongside the online version of this article at erj.erjournals.com
References
- 1.Broza YY, Haick H. Nanomaterial-based sensors for detection of disease by volatile organic compounds. Nanomedicine. 2013;8:785–806. doi: 10.2217/nnm.13.64. [DOI] [PubMed] [Google Scholar]
- 2.Bijland LR, Bomers MK, Smulders YM. Smelling the diagnosis: a review on the use of scent in diagnosing disease. Neth J Med. 2013;71:300–307. [PubMed] [Google Scholar]
- 3.Boots AW, van Berkel JJBN, Dallinga JW, et al. The versatile use of exhaled volatile organic compounds in human health and disease. J Breath Res. 2012;6:027108. doi: 10.1088/1752-7155/6/2/027108. [DOI] [PubMed] [Google Scholar]
- 4.Dummer J, Storer M, Swanney M, et al. Analysis of biogenic volatile organic compounds in human health and disease. Trends Anal Chem. 2011;30:960–967. [Google Scholar]
- 5.Pleil JD, Stiegel MA. Evolution of environmental exposure science: using breath-borne biomarkers for “discovery” of the human exposome. Anal Chem. 2013;85:9984–9990. doi: 10.1021/ac402306f. [DOI] [PubMed] [Google Scholar]
- 6.Amann A, Corradi M, Mazzone P, et al. Lung cancer biomarkers in exhaled breath. Expert Rev Mol Diagn. 2011;11:207–217. doi: 10.1586/erm.10.112. [DOI] [PubMed] [Google Scholar]
- 7.Haick H, Broza YY, Mochalski P, et al. Assessment, origin, and implementation of breath volatile cancer markers. Chem Soc Rev. 2014;43:1423–1449. doi: 10.1039/c3cs60329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buszewski B, Grzywinski D, Ligor T, et al. Detection of volatile organic compounds as biomarkers in breath analysis by different analytical techniques. Bioanalysis. 2013;5:2287–2306. doi: 10.4155/bio.13.183. [DOI] [PubMed] [Google Scholar]
- 9.Phillips M, Herrera J, Krishnan S, et al. Variation in volatile organic compounds in the breath of normal humans. J Chrom B. 1999;729:75–88. doi: 10.1016/s0378-4347(99)00127-9. [DOI] [PubMed] [Google Scholar]
- 10.Bean HD, Zhu J, Hill JE. Characterizing bacterial volatiles using secondary electrospray ionization mass spectrometry (SESI-MS) J Vis Exp. 2011;52:2664. doi: 10.3791/2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Jiménez-Díaz J, Bean HD, et al. Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: from initial infection to clearance. J Breath Res. 2013;7:037106. doi: 10.1088/1752-7155/7/3/037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Bean HD, Jiménez-Díaz J, et al. Secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting of multiple bacterial lung pathogens, a mouse model study. J Appl Physiol. 2013;114:1544–1549. doi: 10.1152/japplphysiol.00099.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Bean HD, Kuo YM, et al. Fast detection of volatile organic compounds from bacterial cultures by SESI-MS. J Clin Microbiol. 2010;48:4426–4431. doi: 10.1128/JCM.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, Bean HD, Wargo MJ, et al. Detecting bacterial lung infections: in vivo evaluation of in vitro volatile fingerprints. J Breath Res. 2013;7:016003. doi: 10.1088/1752-7155/7/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allard JB, Rinaldi L, Wargo MJ, et al. Th2 allergic immune response to inhaled fungal antigens is modulated by TLR-4-independent bacterial products. Eur J Immunol. 2009;39:776–788. doi: 10.1002/eji.200838932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometrics Intellig Lab Syst. 2001;58:109–130. [Google Scholar]
- 17.Jun CH, Sang-Ho L, Hae-Sang P, et al. Use of partial least squares regression for variable selection and quality prediction. International Conference on Computers & Industrial Engineering; New York: IEEE; 2009. pp. 1302–1307. [Google Scholar]
- 18.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the 14th International Joint Conference on Artificial Intelligence; Montreal: Morgan Kaufmann Publishers Inc; 1995. pp. 1137–1143. [Google Scholar]
- 19.Cross LJM, Matthay MA. Biomarkers in acute lung injury: insights into the pathogenesis of acute lung injury. Crit Care Clin. 2011;27:355–377. doi: 10.1016/j.ccc.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proudfoot AG, Hind M, Griffiths MJ. Biomarkers of acute lung injury: worth their salt? BMC Med. 2011;9:132. doi: 10.1186/1741-7015-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagel SD, Chmiel JF, Konstan MW. Sputum biomarkers of inflammation in cystic fibrosis lung disease. Proc Am Thorac Soc. 2007;4:406–417. doi: 10.1513/pats.200703-044BR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, Yeligar SM, Brown LA. Exhaled breath condensate: a promising source for biomarkers of lung disease. ScientificWorldJournal. 2012;2012:217518. doi: 10.1100/2012/217518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Centers for Disease Controland Prevention. Epidemiology and prevention of vaccine-preventable diseases. 12. Washington: Public Health Foundation; 2012. [Google Scholar]
- 24.Siegel JD, Rhinehart E, Jackson M, et al. 2007 guideline for isolation precautions: preventing transmission of infectious agents in health care settings. Am J Infect Control. 2007;35:S65–S164. doi: 10.1016/j.ajic.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walzl G, Ronacher K, Hanekom W, et al. Immunological biomarkers of tuberculosis. Nat Rev Immunol. 2011;11:343–354. doi: 10.1038/nri2960. [DOI] [PubMed] [Google Scholar]
- 26.Gideon HP, Flynn JL. Latent tuberculosis: what the host “sees”? Immunol Res. 2011;50:202–212. doi: 10.1007/s12026-011-8229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbs K, Holzman IR. Endotracheal tube: friend or foe? Bacteria, the endotracheal tube, and the impact of colonization and infection. Semin Perinatol. 2012;36:454–461. doi: 10.1053/j.semperi.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Mandalakas AM, van Wyk S, Kirchner HL, et al. Detecting tuberculosis infection in HIV-infected children: a study of diagnostic accuracy, confounding and interaction. Pediatr Infect Dis J. 2013;32:E111–E118. doi: 10.1097/INF.0b013e31827d77b7. [DOI] [PubMed] [Google Scholar]
- 29.Bhargava M, Wendt CH. Biomarkers in acute lung injury. Transl Res. 2012;159:205–217. doi: 10.1016/j.trsl.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robroeks CM, van Berkel JJ, Jobsis Q, et al. Exhaled volatile organic compounds predict exacerbations of childhood asthma in a 1-year prospective study. Eur Respir J. 2013;42:98–106. doi: 10.1183/09031936.00010712. [DOI] [PubMed] [Google Scholar]
- 31.Kash JC, Walters KA, Davis AS, et al. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. MBio. 2011;2:e00172–11. doi: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Mann-Whitney U-tests comparing SESI-MS breathprints at 6 – 120 h after murine airway exposures to P. aeruginosa lysate (PAL), S. aureus lysate (SAL), live P. aeruginosa (PA) or S. aureus (SA) pathogens, or phosphate buffered saline (PBS) control (see Materials & Methods for calculation details). To facilitate comparisons to the other statistical methods employed for data analysis, the PLS-DA VIP scores (Figs. S1–S3) and immune correlations (Table 1) are summarized in the four left-most columns.
Table S2: Average cytokine concentrations (pg/mL) in bronchoalveolar lavage fluid at six time points 6 – 120 h after murine airway exposure to P. aeruginosa cell lysate, S. aureus cell lysate, or phosphate buffered saline (PBS) control.