Abstract
A vast diversity of microorganisms, including bacteria, fungi, viruses, and arthropods, colonize the human skin. Culture-independent genomic approaches for identifying and characterizing microbial communities have provided glimpses into the topographical, temporal, and interpersonal complexity that defines the skin microbiome. Identification of changes associated with cutaneous disease, including acne, atopic dermatitis, rosacea, and psoriasis, are being established. In this review, our current knowledge of the skin microbiome in health and disease is discussed, with particular attention to potential opportunities to leverage the skin microbiome as a diagnostic, prognostic, and/or therapeutic tool.
Keywords: Microbiome, genomics, metagenomics, microbiology, dysbiosis
In and on our bodies, microbial cells outnumber human cells by a factor of 10. Microbial genes outnumber human genes by a factor of 100. It is increasingly apparent that this collective set of microorganisms and their genetic material, the “microbiome”, contributes genetic diversity, modulates disease, influences metabolic processes, and is essential for immunity. The human microbiome is also dynamic, and changes associated with health and disease have been described and mechanistically investigated. Being a tractable entity, the microbiome is also a prime target for manipulation to influence health and disease processes. In most cases, but especially on the skin, the microbiota is an easily accessible target for therapeutic intervention and/or diagnostic testing.
Our awareness and investigation into human-associated microbial communities was once limited by culture-based techniques. These methods that rely upon cultivation and isolation of microorganisms are biased towards those <10% that are able to thrive in standard laboratory conditions. Importantly, culture-based methods exclude microbes that rely on microbe-microbe interactions to thrive. However, cutaneous microbiology has been an active area of research for decades, and early culture-based studies provided important insight into bacterial, fungal, and viral populations on the skin.1
The advent of next-generation sequencing technologies has revolutionized our view of human-associated microbial communities. Using DNA sequencing methodology, we are now able to characterize and analyze microbiomes with greater precision and accuracy, and less bias compared to culture-based approaches. A common approach used to identify bacterial populations is based on sequencing of the small subunit bacterial 16S ribosomal RNA (rRNA) gene. Hypervariable regions within this gene contain species-specific sequences, that when compared to reference databases, allow identification of the bacteria of origin.2 A similar approach for fungal identification, based on the sequence lying between the fungal 18S and 5.8S rRNA genes, the internal transcribed spacer region (ITS), has been used to sequence and survey fungal populations of the skin.3 Increasing throughput and decreasing costs associated with DNA sequencing, along with the development of computational tools for the analysis of resultant sequence data, have made these approaches feasible to query microbial communities in health and disease states. Importantly, these methods do not rely upon cultivation of the microorganism, thus eliminating biases associated with culture-based techniques.
Multiple studies have employed 16S rRNA gene sequencing to characterize bacterial communities colonizing the skin, in health and disease. Immediately following birth, our skin is colonized with microbiota, and delivery mode may in part contribute to differences in colonization. Analysis of neonate skin microbiota indicates that those babies delivered via vaginal birth had skin microbiota similar to their mother's vaginal microbiota, and babies delivered via Caesarean section had skin microbiota similar their mother's skin microbiota.4 It is unclear how long these colonization differences persist, as most vaginal bacteria are not typically found on the skin, and if the associated differences translate into functional consequences and/or disease risk. Other factors, including geography, lifestyle, and/or ethnicity may influence the skin microbiota. For example, Amerindians residing in the Venezuelan Amazon are colonized with distinctly different microbiota compared to subjects residing in the United States.5
Generally, the four dominant phyla of bacteria residing on the skin are the Actinobacteria, Proteobacteria, Firmicutes, and Bacteroidetes.6 A feature of the skin is the topographical diversity of bacterial populations, with composition and diversity of bacteria colonizing the skin depending on the microenvironment (Figure). Variability between individuals is high, similar to other sites of the human microbiome,7 as is temporal variability within the same individual.8 However, the dominant types of bacteria that reside on the skin appear to be relatively stable, with the rarer, less abundant types of bacteria accounting for the variability. These dominant types of bacteria, primarily Staphylococcus, Propionibacterium, and Corynebacterium, are differentially abundant depending on the skin site (Figure).8,9 Differentially distributed hair follicles, eccrine and apocrine glands, and sebaceous glands contribute to the variable cutaneous microenvironments, and likely select for subsets of bacteria that can thrive in those specialized conditions. Bacteria may also be present in subepidermal compartments of the skin,10 though it is unclear if these or any microorganisms identified by sequencing approaches are alive, as these techniques are unable to distinguish viable from non-viable microorganisms. In the following sections, each microenvironment of the skin (dry, moist, sebaceous) will be further discussed, with particular attention to skin disorders that may be influenced by the microbiome at those sites (Table).
Figure.
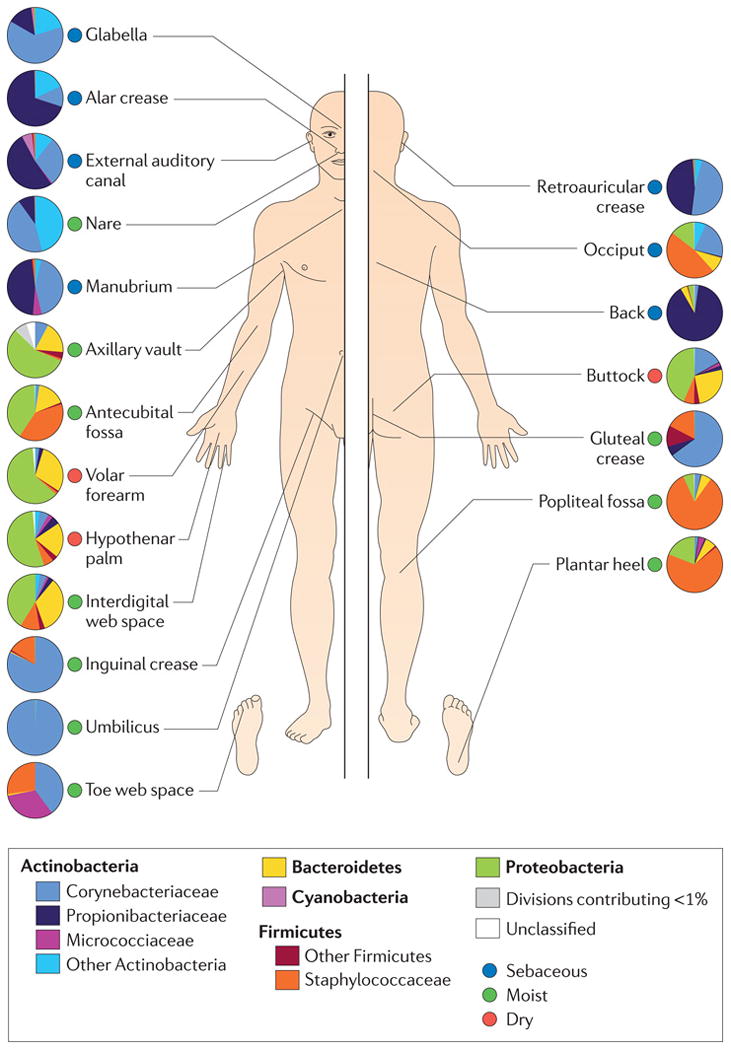
Topographical distribution of cutaneous microbiota and associated microenvironments (dry, sebaceous, moist). Family level classification of bacterial taxa is shown and pie charts represent relative abundance. Figure reproduced with permission of Nature Publishing Group.
Table.
Findings of select studies employing genomic approaches to characterize changes in skin microbiota associated with various dermatology disorders.
| Disease | Study | Sample size | Major disease-associated findings |
|---|---|---|---|
|
| |||
| Acne vulgaris | Fitz-Gibbon et al 201311 | 49 acne + 52 controls | Association of P. acnes strain with acne |
|
| |||
| Rosacea | Casas C et al 201212 | 50 rosacea + 48 controls | Increased density and frequency of Demodex follicularum |
|
| |||
| Atopic Dermatitis | Kong et al 201213 | 12 AD + 11 controls | Increased Staphylococcus and decreased diversity during flares |
|
| |||
| Psoriasis vulgaris | Alekseyenko et al 201314 | 51 psoriasis + 51 controls | Combined increase in Corynebacteria, Propionibacterium, Streptococcus, and Staphylococcus |
| Gao et al 200815 | 6 psoriasis | Increased Firmicutes and Actinobacteria | |
| Fahlen et al 201216 | 10 psoriasis + 12 controls | Decreased Staphylococcus and Propionibacterium | |
With growing recognition that the human-associated microbiota is a component of health and also many disease processes, strategies targeting the microbiota are on the horizon. Probiotics are live microorganisms with known health benefits, and are gaining increasing popularity for use in the gastrointestinal tract. Prebiotics are substrates that encourage the growth of beneficial bacteria. More research is needed to identify probiotic strains and prebiotic substrates that may benefit skin microbial communities, and thus skin health. In addition to therapeutic strategies, diagnostic tests based on the microbiota have the potential to provide more precision to the management and treatment of skin disorders. Microbial communities colonizing the human body are exquisitely sensitive to their underlying environment and changing conditions. Thus, microorganisms can rapidly change in number in response to perturbation, providing a sensitive readout of the environment in which they thrive. While these implementations of personalized and precision medicine are on the horizon, more research is needed to define the skin microbiota and its involvement in cutaneous disorders. Here we describe the healthy, skin microbiome as it is understood from recent genomic, culture-independent surveys (Figure), how it is perturbed in disease states (Table), and how microbiome research may one day inform precision and personalized medical approaches for the management and treatment of skin disease.
Sebaceous skin: acne and rosacea
Sebaceous areas, such as the back, face, behind the ears, tend to be colonized by high proportions of the lipophilic bacteria Propionibacterium (Figure).7-9 This finding is consistent with evidence that Propionibacterium hydrolyzes triglycerides found in sebum, thus releasing free fatty acids on the skin that then function to acidify and emolliate the skin.17 Further, sebaceous areas tend to be low in diversity and relatively stable temporally, compared to moist and dry sites of the skin,18 perhaps a reflection of the unique microenvironment provided by sebaceous secretions that selects for certain types of microbes.
The role of sebum in defining the skin microbiota may be reflected by age-associated changes in the composition and diversity of the skin microbiota. At birth, babies are colonized with higher relative abundance of Firmicutes than Actinobacteria.19 Generally, puberty-associated maturation of the sebaceous glands coincides with a shift in skin microbiota towards enrichment with Actinobacteria, including Corynebacterium and Propionibacterium.20 This age-related shift in skin microbiota may be a potential factor contributing to the decrease in incidence and severity of childhood skin diseases, for example atopic dermatitis.
The association between Propionibacterium acnes and acne vulgaris has been well-established.21 The effectiveness of antibiotics in acne treatment further supports a microbial role in disease pathogenesis. Based on 16S rRNA gene sequencing studies, it is now apparent that P. acnes is a member of the commensal microbiota even in those without acne. These findings raise the question of when and how does a commensal become a pathogen? Insight into this question may require strain-level analysis; a recent study demonstrated that even though P. acnes relative abundance did not differ between individuals with acne compared to healthy individuals, certain strains were highly associated with acne (Table).11 These strains carry unique genetic elements, not present in the strains associated with healthy individuals, which may contribute to virulence and pathogenicity.
Probiotic applications, where the skin microbiota is supplemented with a beneficial microorganism, may offer promise for treatment of conditions such as acne. For example, succinic acid, a fatty acid fermentation product of Staphylococcus epidermidis, can inhibit the growth of P. acnes.22 Similarly, P. acnes can function as a probiotic that suppresses growth of S. aureus, in particular USA300 community acquired methicillin resistant S. aureus (MRSA), through fermentation of glycerol.23 Replacing antibiotics with therapies that do not place selective pressures on microorganisms is desirable, as antibiotics encourage evolution of antibacterial resistance mechanisms. Long-term antibiotic treatment for acne has been associated with acquisition of antibiotic resistance.24,25 Gram-negative folliculitis26 and pharyngitis27 are also associated with antibiotic therapy of acne, and likely are opportunistic infections resulting from disturbed microbial community ecology on the skin and in the airways, respectively.
The fungal microbiota in sebaceous areas tends to be less diverse than bacterial communities, dominated by Malassezia species, specifically M. restricta and M. globosa, which requiring olive oil for culturing.3 In addition to Malassezia and Propionibacterium, the sebaceous areas of the skin support colonization by the arachnid Demodex skin mites, particularly D. folliculorum and D. brevis that colonize the pilosebaceous units, increasing in prevalence with age.28 This predilection may in part be explained by lipases produced by Demodex,29 allowing the mite to utilize sebum as a food source. Other nutritional sources may be cellular debris or bacteria such as P. acnes that reside in the pilosebaceous unit.1
Increased density of Demodex has been associated with rosacea of the subtypes erythematotelangiectatic rosacea (ETR) and papulopustular rosacea (PPR) (Table).12,30,31 In an additional layer of complexity, Demodex-associated microbiota, in particular Bacillus oleronius, a Gram-negative bacterium not typically found in the commensal skin microbiota, triggers inflammatory pathways in a manner that is mirrored in rosacea patients.32 Increased production of the antimicrobial peptide cathelicidin and aberrant processing are also associated with rosacea,33 which in part may contribute to dysbiotic cutaneous states. Because Demodex are present on healthy skin, and considered as part of the commensal microbiota, it is likely that host status contributes to their transition from commensal to pathogen. However, additional research is needed to provide insight into the mechanism in which microorganisms, including Demodex, influence the pathogenesis of rosacea.
Moist skin: body odor and atopic dermatitis
Skin areas that are moist and/or occluded tend to be colonized by bacteria such as Staphylococcus and Corynebacterium (Figure), organisms that prefer high humidity in their habitat. Culture-dependent analyses of the axilla microbiota has revealed microorganisms, namely the Corynebacterium, that process odorless eccrine, apocrine, and sebaceous gland secretions, thus producing volatile organic fatty acids and thioalcohols that are responsible for malodor associated with sweating of the axilla.34-37 Sequencing of 16S rRNA genes has confirmed that Corynebacterium and Staphylococcus are the predominant axilla bacteria, with higher proportions of Corynebacterium in males, and those using deodorants harboring greater bacterial diversity.38 Genetic variation in ABCC11, specifically a 538G→A single nucleotide polymorphism (SNP) prevalent in East Asian populations, results in reduced concentration of axillary apocrine secretions and decreased production of axillary odor.39 This genetic variant also impacts the composition of the axillary skin microbiota, where relative abundance of Staphylococcus is decreased and Corynebacterium increased in individuals carrying A/A genotype, as compared to those with G/A and G/G genotype.40 Deodorants containing probiotics and/or prebiotics may in the future be a feasible strategy to combat body odor, where odor-producing bacteria would be competed away with recolonization by non-odor producing bacteria.
Body odor also in part contributes to attractiveness to insects, and increased bacterial load and decreased diversity are associated with increased attractiveness to mosquitos.41 Further studies need to be completed, including identification of the volatile compounds produced, but engineering of the human skin microbiota to decrease attractiveness to insects, in particular those that are vectors of disease, could be a potential preventative measure for diseases such as malaria and dengue fever. Additionally, identification of the volatile compounds produced by the skin microbiota may provide the basis for new repellents or attractants.42
Several lines of the evidence point to microbial involvement in the pathogenesis of atopic dermatitis (AD), a chronic relapsing disorder affecting ∼15% of children in the US.43 The incidence of AD has increased markedly over the past 3 decades, suggesting an environmental component. Further, colonization with S. aureus is associated with AD and antibiotics and bleach baths are effective treatment strategies that target the microbiota.44 Profiling of the bacterial microbiota via 16S rRNA gene sequencing indeed confirmed increases in S. aureus relative abundance during AD flares, and decrease in bacterial diversity (Table).13 Whether S. aureus colonization is a cause or effect of changes in the host skin remains unclear, but could be addressed by fine-scale longitudinal study designs incorporating skin barrier testing and surveys of immune status and/or the use of animal models. Predicting the best treatment for AD can also be difficult, and information about the microbiome may be able to inform these decisions. Similar to fecal microbiota transplantation therapies to treat Clostridium difficile infections,45 one could envision the implementation of skin microbiota transplantations to ameliorate S. aureus colonization and AD symptoms without relying on antibiotic therapies that encourage acquisition of resistance.
Some evidence exists to suggest that gastrointestinal probiotics and prebiotics may offer benefit in preventing and reducing the severity of AD.46 In addition, low diversity in the gut microbiota during early infancy has been associated with development of atopic eczema later in life.47,48 These observations would suggest that there is a connection between the gut microbiota and the skin, potentially through stimulation and/or education of immune cell populations. Further exploration of this connection may provide insight into the mechanism of allergic and atopic disease, while providing an additional target, the gut microbiota, for manipulation (via probiotics, prebiotics, or microbiota transplant) in the treatment and/or prevention of skin disease.
Dry skin: psoriasis
Generally, dry areas of the skin harbor greater amounts of bacterial diversity and lesser bacterial load than sebaceous and moist sites, and seem to have less of a selective force, as those bacterial populations are not dominated by individual taxa.8 Instead, drier areas contain greater amounts of Proteobacteria and Bacteroidetes, in comparison to moist and sebaceous sites (Figure).8,9,49,50
Psoriasis vulgaris (plaque psoriasis) has a predilection for drier skin sites, such as the elbows, knees, and trunk. It has been hypothesized that similar to Crohn's disease, psoriasis results from failure in immune tolerance to microbiota.51 Guttate psoriasis, also forming lesions on the trunk and limbs, is associated with beta-hemolytic streptococcal infection, where streptococcal superantigens drive T-cell stimulation and expansion in the skin.52 The bacterial microbiota colonizing plaque psoriasis has been investigated using culture-independent techniques, and some changes have been identified, including decreased relative abundance of Propionibacterium in plaques (Table).15,16 In a larger study of 51 psoriasis patients and matched healthy controls, the psoriatic plaque microbiota was determined to contain an increased combined relative abundance of Corynebacterium, Propionibacterium, Staphylococcus, and Streptococcus, and clustered distinctly from controls.14 However, it is unclear if these changes are a cause or consequence of alteration of the skin barrier and the ecosystem. Longitudinal studies, profiling the dynamics of microbial populations during plaque resolution and relapsing could provide insight into the role of microbiota during the triggering, propagation, and maintenance of plaques. Should the previously mentioned hypothesis be correct regarding the similarity of psoriasis to Crohn's disease, microbiome-based therapies employed in conjunction with other therapies to prevent and/or treat the disease may be feasible. Further, complement activation is a known feature of psoriasis,53,54 and given previous findings connecting complement to skin microbiota composition and diversity,55 taken together would also suggest a microbial component to psoriasis. Complement therapeutics, which are under development and already used for some conditions resulting from complement activation, could potentially be employed to modify the skin microbiota and/or treat psoriasis.
Skin microbiome interactions with host immunity
Emerging evidence now suggests that host immunity influences the skin microbiota, and conversely, the skin microbiota in part modulates cutaneous immunity. Skin microbiota in patients with primary immunodeficiency syndromes (STAT-3 deficient hyper-IgE, Wiskott-Aldrich, and dedicator of cytokinesis 8 syndromes) is altered from a healthy state, colonized with different bacterial and fungal species, suggestive of increased ecological permissiveness.56 Primary immunodeficiency patients suffer from atopic dermatitis-like eczema and increased risk of skin infections. A separate study of STAT1 (chronic mucocutaneous candidiasis) and STAT3 primary immunodeficiency patients documented increased colonization with Gram-negative bacteria, in particular Acinetobacter, which also suppressed the cytokine response of primary leukocytes to S. aureus and Candida albicans, two pathogens that commonly cause skin infections in these patient populations.57
A study by our group recently demonstrated that antagonism of complement signaling (through the C5a receptor) in mice resulted in a shift in skin microbial composition and diversity.55 Notably, microbial diversity and richness decreased significantly when complement was blocked. Innate immunity, such as through complement signaling, may in part maintain microbial diversity at the skin barrier. Diversity can be advantageous in an ecosystem, and low diversity has been associated with some dysbiotic skin disorders (i.e. atopic dermatitis) and gastrointestinal disorders. Additionally, the commensal microbiota was found to induce expression of complement genes, suggesting that the skin microbiota in part regulates cutaneous immunity.55 Another study found that a molecule released by staphylococcal bacteria, lipoteichoic acid (LTA), inhibits inflammation following injury through a Toll-like receptor (TLR2) dependent pathway,58 suggesting that the microbiota modulates cutaneous inflammation and immunity. Regarding adaptive immunity, the commensal skin microbiota was found to autonomously direct T cell function and the local inflammatory milieu, in addition to conferring protective immunity to infection by Leishmania.59 These studies together support an emerging interactive relationship between the skin microbiota and cutaneous immunity.
Additional applications of microbiome science to precision cutaneous medicine
Another application of culture-independent profiling of microbial communities may lie in prognostic and diagnostic approaches to disease. For example, identifying variants of disease, based on the colonizing microbiota, may better inform the optimal treatment approach. In a study of 52 neuropathic diabetic foot ulcers, patients clustered into 3 groups based on their colonizing microbiota.60 While this study did not provide outcome information or association of specific microbiota with poor outcomes, it is possible that these clusters represent variants that may benefit from different management and treatment approaches. In this particular case, one cluster was characterized by high relative abundance of anaerobic bacteria, which were not identified by standard of care cultures the majority of the time, likely because anaerobes are notoriously difficult to culture. This information alone is useful in guiding decisions of what antibiotic regimens to implement. In a separate study of acute traumatic wounds, characteristic microbiota was identified that correlated with wound location, mechanism (blunt versus penetrating), and future complication.61 In this situation, microbiota may be useful in identifying those that will go on to future infectious complication. In both of these types of wounds, predicting those who will experience infectious complications is difficult and biomarkers to guide management and treatment are greatly needed.
The role of microbiota in skin aging, such as wrinkling and sagging, remains unclear and is an area where treatments based on the skin microbiota may offer promise. For example, its foreseeable that certain metabolites produced by skin microbiota may offer benefit by modulating cutaneous pro- and anti-inflammatory responses, similar to what has been shown in the gastrointestinal tract.62 Overall, more research is necessary to identify those microorganisms that may offer benefit, and their mechanism of action.
Conclusion
Advances in sequencing technology have enhanced our ability to identify and characterize microbial communities colonizing the skin. Our knowledge regarding the healthy cutaneous microbiota, and how it is altered in dermatology disease is steadily increasing. However, in order to apply this knowledge to diagnostic, prognostic, and therapeutic applications, further research is necessary to understand beneficial and harmful microorganisms and their mechanism. These applications are particularly promising in light of emerging antibiotic resistance across medically relevant bacterial strains. Further, microbiome therapeutics and diagnostics are highly applicable to precision and personalized medicine, and may in the future transform management and treatment of dermatological disease.
Acknowledgments
EAG gratefully acknowledges the support of the National Institutes of Health (AR060873) in this work.
Footnotes
Disclosures: Dr Grice received compensation as a consultant for Amway International, Inc. and GOJO Industries, Inc. Dr Grice's institution has grants/grants pending with Janssen Research and Development, LLC. She has been a paid lecturer for: TRI Princeton, Society for Advanced Wound Care, Personal Care Product Council and Society of Cosmetic Chemists.
References
- 1.Marples M. The Ecology of the Human Skin. Springfield, Ill: Charles C Thomas, Bannerstone House; 1965. [Google Scholar]
- 2.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Findley K, Oh J, Yang J, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498(7454):367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser MJ, Dominguez-Bello MG, Contreras M, et al. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013;7(1):85–95. doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannigan GD, Grice EA. Microbial ecology of the skin in the era of metagenomics and molecular microbiology. Cold Spring Harb Perspect Med. 2013;3(12):a015362. doi: 10.1101/cshperspect.a015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grice EA, Kong HK, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitz-Gibbon S, Tomida S, Chiu BH, et al. Propionibacterium Acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J Invest Dermatol. 2013;133(9):2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casas C, Paul C, Lahfa M, et al. Quantification of Demodex folliculorum by PCR in rosacea and its relationship to skin innate immune activation. Exp Dermatol. 2012;21(12):906–910. doi: 10.1111/exd.12030. [DOI] [PubMed] [Google Scholar]
- 13.Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alekseyenko AV, Perez-Perez GI, De Souza A, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1(1):31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One. 2008;3(7):e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahlen A, Engstrand L, Baker BS, Powles A, Fry L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res. 2012;304(1):15–22. doi: 10.1007/s00403-011-1189-x. [DOI] [PubMed] [Google Scholar]
- 17.Marples RR, Downing DT, Kligman AM. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J Invest Dermatol. 1971;56(2):127–131. doi: 10.1111/1523-1747.ep12260695. [DOI] [PubMed] [Google Scholar]
- 18.Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131(10):2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyden JJ, McGinley KJ, Mills OH, Kligman AM. Propionibacterium levels in patients with and without acne vulgaris. J Invest Dermatol. 1975;65(4):382–384. doi: 10.1111/1523-1747.ep12607634. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Kuo S, Shu M, et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol. 2014;98(1):411–424. doi: 10.1007/s00253-013-5394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu M, Wang Y, Yu J, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8(2):e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishijima S, Kurokawa I, Katoh N, Watanabe K. The bacteriology of acne vulgaris and antimicrobial susceptibility of Propionibacterium acnes and Staphylococcus epidermidis isolated from acne lesions. J Dermatol. 2000;27(5):318–323. doi: 10.1111/j.1346-8138.2000.tb02174.x. [DOI] [PubMed] [Google Scholar]
- 25.Levy RM, Huang EY, Roling D, Leyden JJ, Margolis DJ. Effect of antibiotics on the oropharyngeal flora in patients with acne. Arch Dermatol. 2003;139(4):467–471. doi: 10.1001/archderm.139.4.467. [DOI] [PubMed] [Google Scholar]
- 26.Fulton JE, Jr, McGinley K, Leyden J, Marples R. Gram-negative folliculitis in acne vulgaris. Arch Dermatol. 1968;98(4):349–353. [PubMed] [Google Scholar]
- 27.Margolis DJ, Fanelli M, Kupperman E, et al. Association of pharyngitis with oral antibiotic use for the treatment of acne: a cross-sectional and prospective cohort study. Arch Dermatol. 2012;148(3):326–332. doi: 10.1001/archdermatol.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elston DM. Demodex mites: facts and controversies. Clin Dermatol. 2010;28(5):502–504. doi: 10.1016/j.clindermatol.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez-Acosta F, Planas L, Penneys N. Demodex mites contain immunoreactive lipase. Arch Dermatol. 1989;125(10):1436–1437. doi: 10.1001/archderm.1989.01670220134028. [DOI] [PubMed] [Google Scholar]
- 30.Holmes AD. Potential role of microorganisms in the pathogenesis of rosacea. J Am Acad Dermatol. 2013;69(6):1025–1032. doi: 10.1016/j.jaad.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Roihu T, Kariniemi AL. Demodex mites in acne rosacea. J Cutan Pathol. 1998;25(10):550–552. doi: 10.1111/j.1600-0560.1998.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 32.O'Reilly N, Bergin D, Reeves EP, McElvaney NG, Kavanagh K. Demodex-associated bacterial proteins induce neutrophil activation. Br J Dermatol. 2012;166(4):753–760. doi: 10.1111/j.1365-2133.2011.10746.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamasaki K, Di Nardo A, Bardan A, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13(8):975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 34.Decreau RA, Marson CM, Smith KE, Behan JM. Production of malodorous steroids from androsta-5,16-dienes and androsta-4,16-dienes by Corynebacteria and other human axillary bacteria. J Steroid Biochem Mol Biol. 2003;87(4-5):327–336. doi: 10.1016/j.jsbmb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Emter R, Natsch A. The sequential action of a dipeptidase and a beta-lyase is required for the release of the human body odorant 3-methyl-3-sulfanylhexan-1-ol from a secreted Cys-Gly-(S) conjugate by Corynebacteria. J Biol Chem. 2008;283(30):20645–20652. doi: 10.1074/jbc.M800730200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leyden JJ, McGinley KJ, Holzle E, Labows JN, Kligman AM. The microbiology of the human axilla and its relationship to axillary odor. J Invest Dermatol. 1981;77(5):413–416. doi: 10.1111/1523-1747.ep12494624. [DOI] [PubMed] [Google Scholar]
- 37.James AG, Austin CJ, Cox DS, Taylor D, Calvert R. Microbiological and biochemical origins of human axillary odour. FEMS Microbiol Ecol. 2013;83(3):527–540. doi: 10.1111/1574-6941.12054. [DOI] [PubMed] [Google Scholar]
- 38.Callewaert C, Kerckhof FM, Granitsiotis MS, Van Gele M, Van de Wiele T, Boon N. Characterization of Staphylococcus and Corynebacterium clusters in the human axillary region. PLoS One. 2013;8(8):e70538. doi: 10.1371/journal.pone.0070538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin A, Saathoff M, Kuhn F, Max H, Terstegen L, Natsch A. A functional ABCC11 allele is essential in the biochemical formation of human axillary odor. J Invest Dermatol. 2010;130(2):529–540. doi: 10.1038/jid.2009.254. [DOI] [PubMed] [Google Scholar]
- 40.Harker M, Carvell AM, Marti VP, et al. Functional characterisation of a SNP in the ABCC11 allele-Effects on axillary skin metabolism, odour generation and associated behaviours. J Dermatol Sci. 2014;73(1):23–30. doi: 10.1016/j.jdermsci.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Verhulst NO, Qiu YT, Beijleveld H, et al. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS One. 2011;6(12):e28991. doi: 10.1371/journal.pone.0028991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhulst NO, Andriessen R, Groenhagen U, et al. Differential attraction of malaria mosquitoes to volatile blends produced by human skin bacteria. PLoS One. 2010;5(12):e15829. doi: 10.1371/journal.pone.0015829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw TE, Currie GP, Koudelka CW, Simpson EL. Eczema prevalence in the United States: data from the 2003 National Survey of Children's Health. J Invest Dermatol. 2011;131(1):67–73. doi: 10.1038/jid.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123(5):e808–814. doi: 10.1542/peds.2008-2217. [DOI] [PubMed] [Google Scholar]
- 45.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 46.Foolad N, Brezinski EA, Chase EP, Armstrong AW. Effect of nutrient supplementation on atopic dermatitis in children: a systematic review of probiotics, prebiotics, formula, and fatty acids. JAMA Dermatol. 2013;149(3):350–355. doi: 10.1001/jamadermatol.2013.1495. [DOI] [PubMed] [Google Scholar]
- 47.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129(2):434–440. 440 e431–432. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Karlsson C, Olsson C, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121(1):129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104(8):2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18(7):1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fry L, Baker BS, Powles AV, Fahlen A, Engstrand L. Is chronic plaque psoriasis triggered by microbiota in the skin? Br J Dermatol. 2013;169(1):47–52. doi: 10.1111/bjd.12322. [DOI] [PubMed] [Google Scholar]
- 52.Leung DY, Travers JB, Giorno R, et al. Evidence for a streptococcal superantigen-driven process in acute guttate psoriasis. J Clin Invest. 1995;96(5):2106–2112. doi: 10.1172/JCI118263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tagami H. The role of complement-derived mediators in inflammatory skin diseases. Arch Dermatol Res. 1992;284(Suppl 1):S2–9. doi: 10.1007/BF00638232. [DOI] [PubMed] [Google Scholar]
- 54.Kotnik V. Complement in skin diseases. Acta Dermatovenerol Alp Panonicat Adriat. 2011;20(1):3–11. [PubMed] [Google Scholar]
- 55.Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc Natl Acad Sci U S A. 2013;110(37):15061–15066. doi: 10.1073/pnas.1307855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh J, Freeman AF, Park M, et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013;23(12):2103–2114. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smeekens SP, Huttenhower C, Riza A, et al. Skin Microbiome Imbalance in Patients with STAT1/STAT3 Defects Impairs Innate Host Defense Responses. J Innate Immun. 2013 doi: 10.1159/000351912. ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai Y, Di Nardo A, Nakatsuji T, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15(12):1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naik S, Bouladoux N, Wilhelm C, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62(3):923–930. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hannigan GD, Hodkinson BP, Tyldsley AS, et al. Longitudinal culture-independent pilot study of microbiota colonizing open fractures and association with severity, mechanism, location, and complication from presentation to early outpatient follow up. J Orthop Res. 2014;32(4):597–605. doi: 10.1002/jor.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]


