Summary
Adversity, particularly in early life, can cause illness. Clues to the responsible mechanisms may lie with the discovery of molecular signatures of stress, some of which include alterations to an individual’s somatic genome. Here, using genome sequences from 11,670 women, we observed a highly significant association between a stress-related disease, major depression, and the amount of mtDNA (p = 9.00 × 10−42, odds ratio 1.33 [95% confidence interval [CI] = 1.29–1.37]) and telomere length (p = 2.84 × 10−14, odds ratio 0.85 [95% CI = 0.81–0.89]). While both telomere length and mtDNA amount were associated with adverse life events, conditional regression analyses showed the molecular changes were contingent on the depressed state. We tested this hypothesis with experiments in mice, demonstrating that stress causes both molecular changes, which are partly reversible and can be elicited by the administration of corticosterone. Together, these results demonstrate that changes in the amount of mtDNA and telomere length are consequences of stress and entering a depressed state. These findings identify increased amounts of mtDNA as a molecular marker of MD and have important implications for understanding how stress causes the disease.
Highlights
-
•
Amount of mtDNA is increased, and telomeric DNA is shortened in major depression
-
•
Both changes can be induced with stress but are contingent on the depressed state
-
•
Changes are tissue specific and in part due to glucocorticoid secretion
-
•
Changes are in part reversible and represent switches in metabolic strategy
Cai et al. found increases in mtDNA and a reduction in telomeric DNA in cases of major depression using whole-genome sequencing. Both changes are depression state dependent. Mice exposed to chronic stress or glucorticoids showed that these changes reflect switches in metabolic strategy and are tissue specific and partial reversible.
Introduction
Adverse life experiences, particularly those in childhood, contribute to disease morbidity and mortality [1–7]. There is considerable interest in understanding the mechanisms through which they do so, as it remains unclear how illness becomes apparent decades after the presumed initiating event. Long-standing hypotheses include chronic activation of the hypothalamic-pituitary-adrenal axis [8–10] and alterations of neuroimmune function [11]. Molecular signatures of stressful life experiences and their relation to disease are therefore of special interest to clarify the causal relationship between signature, disease, and stress.
Causal associations between stressful life events and early adversities such as childhood sexual abuse and major depression (MD) are well documented [12–14], suggesting that molecular signatures of stress may be enriched in sufferers of MD. The China, Oxford and VCU Experimental Research on Genetic Epidemiology (CONVERGE) recruited 5,864 women with recurrent MD and 5,783 matched controls, from whom low-coverage genome sequences were obtained together with aggregate measures of lifetime adversities, including assessments of childhood sexual abuse [15, 16] and stressful life events [17, 18]. In CONVERGE, both childhood sexual abuse and stressful life events are strongly associated with risk for MD. More severe forms of abuse are more strongly associated with MD than milder forms, consistent with a causal relationship [16, 18–20].
We focused on two variable components of the somatic genome suspected to be associated with adverse life experiences: telomeric DNA and mtDNA. Accelerated shortening of telomeres, the sequence that caps the ends of chromosomes, has been associated with stress [21–23], anxiety [24], and MD [25] (although not all findings have been replicated [26, 27]). Abnormal mitochondrial morphology and altered metabolic activity has been reported in mood disorders [28]. Our aim was to establish whether telomere length and the amount of mtDNA represent markers of stress-related illness and to explore how such molecular signatures might arise.
Results
Shortened Telomeres and Increased mtDNA Are Associated with Adversity
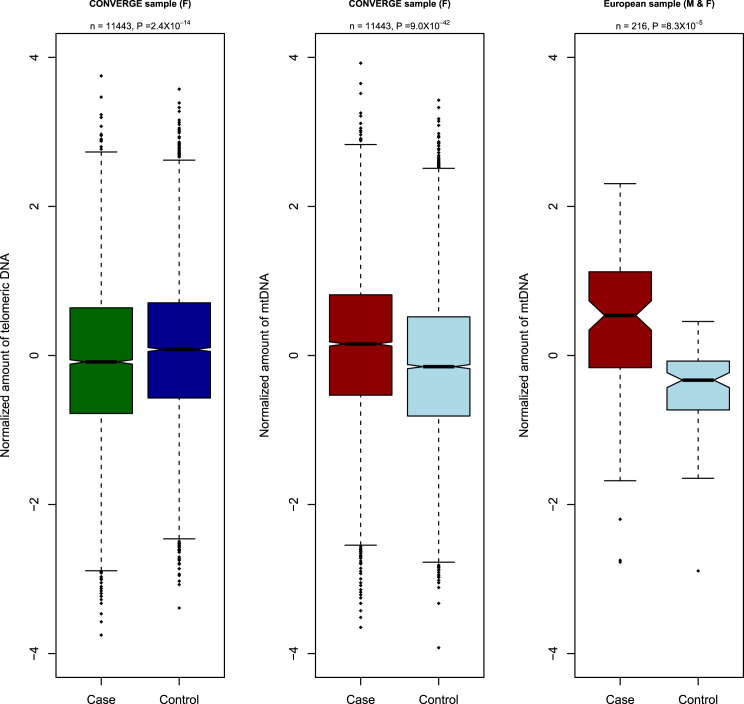
We first examined the relationship between mean telomere length and amount of mtDNA with MD. We assessed the mean length of telomeres (across all chromosomes) from low-coverage whole-genome sequencing (mean coverage of 1.7X) of saliva DNA samples of 11,670 subjects (Experimental Procedures). MD was associated with shorter mean telomere length: in a logistic regression model, the odds ratio for the contribution of normalized measure of mean telomere to the risk of MD is 0.85 (95% CI = 0.81–0.89, p = 2.84 × 10−14). Figure 1A shows the normalized distributions of mean telomere sequence length in cases and controls.
Figure 1.
Two Molecular Markers of Depression: Mitochondrial DNA and Telomere Length
Left: Boxplot of normalized measure of mean telomere length (vertical axis) for cases and controls in the CONVERGE study. Middle: Boxplot of the normalized amount of mtDNA (vertical axis) in cases and controls in the CONVERGE study. Right: Boxplot of the normalized amount of mtDNA (vertical axis) in cases and controls in the GENDEP/DECC studies (labeled IOP).
We obtained a mean coverage of 102X for the mitochondrial genome from which we estimated for each individual the amount of mtDNA. We observed a highly significant association between MD and the amount of mtDNA (p = 9.00 × 10−42 from logistic regression). Cases had more mtDNA than controls: the odds ratio for the contribution of normalized amount of mtDNA to the risk of MD was 1.33 (95% CI = 1.29–1.37). Note that the effect is in the opposite direction to that observed for telomeric DNA. Figure 1B shows the distributions of normalized amount of mtDNA coverage for the cases and controls.
We replicated the association between MD and increased amounts of mtDNA in a European case-control study [29, 30]. In contrast to the CONVERGE sample, the DNA was extracted from blood, and samples were of both sexes. We obtained quantitative PCR (qPCR) measures of mtDNA from 216 individuals (108 cases and 108 controls, 123 women and 93 men). In a logistic model, the odds ratio for the normalized measure of mtDNA’s contribution to the risk of MD was 1.35 (95% CI = 1.11–2.10, p = 8.3 × 10−5; Figure 1C).
We next explored the association in the CONVERGE data between stressful life events and both mean telomere length and amount of mtDNA. Telomere length was significantly shorter in those who had experienced more stressful life events (p = 0.0018, by linear regression) and in those reporting childhood sexual abuse (p = 0.043, by linear regression) (Table 1). The amount of mtDNA was significantly correlated with both the total number of stressful life events (linear regression p = 4.83 × 10−4) and childhood sexual abuse (linear regression p = 3.65 × 10−5). The association of both molecular markers with childhood sexual abuse was stronger with increasingly severe abuse (Table 1).
Table 1.
Relationship between Childhood Sexual Abuse, Telomere Length, and the Amount of Mitochondrial DNA
| CSA Type | Excess Telomeric DNAa | t Valueb | p Valuec | Excess mtDNAa | t Valueb | p Valuec | Number Casesd | Number Controlse | Totalf |
|---|---|---|---|---|---|---|---|---|---|
| Non-genital CSA | 0.02 | 0.35 | 0.73 | 0.08 | 1.37 | 0.169 | 186 | 81 | 267 |
| Genital CSA | −0.08 | −1.27 | 0.20 | 0.11 | 2.02 | 0.045 | 240 | 47 | 287 |
| Intercourse CSA | −0.20 | −2.45 | 0.01 | 0.38 | 4.67 | 3.05 × 10−6 | 159 | 17 | 176 |
Results for analysis of variance in which different forms of childhood sexual abuse (CSA) predict telomere length and the amount of mtDNA. Non-genital CSA refers to sexual invitation, sexual kissing, and exposing; genital CSA refers to fondling and sexual touching; and intercourse CSA refers to attempted or completed intercourse.
Estimated excess of telomeric or mtDNA over mean telomeric DNA or mtDNA in individuals with no CSA.
t statistic of tests of hypotheses that underlying excess is zero.
p value of tests of hypotheses that underlying excess is zero.
Number of MD cases.
Number of controls.
Number of total individuals.
Molecular Changes Are Not Due to Technical or Biological Artifacts
We explored a number of explanations for the association between molecular markers and MD (Figures 1, S1, and S2; Tables 1 and S1; Supplemental Experimental Procedures). First, we considered artifacts arising from incorrectly mapped reads. We found that the association between amount of mtDNA and MD could not be explained by contamination or mapping errors: none of the reads used for assessing the amount of mtDNA mapped to a set of all bacterial and plasmid genomes, and none mapped to nuclear copies of mtDNA.
Second, we considered whether the molecular changes might be due to medication. We could not explain the telomere length or mtDNA changes as a result of cases taking antidepressant medication: among the MD cases, 975 reported never having taken any antidepressants. Neither the amount of mtDNA nor telomeric length in these subjects differed significantly from that assayed in the 4,861 individuals reporting taking antidepressants (t test p = 0.96 and p = 0.88, respectively).
Third, we considered whether the effects might be explained by alterations in the cellular composition of the saliva between cases and controls (see Supplemental Experimental Procedures). Methylation of cytosine residues at cytosine-guanine (CpG) dinucleotides differs between cell types [31–35] and thus contains information about the cellular composition of the tissue from which it was extracted [36–38]. We assessed methylation in 156 individuals (78 cases and 78 controls), selected from the extremes of the distribution of amount of mtDNA, and matched for age and other potential confounds. The sites assayed are shown in Table S1, and the percentage of methylation at each CpG site is shown in Figure S1. MD case-control status remained highly significantly associated with the amount of mtDNA (t test p value = 5.14 × 10−18) and telomere length (p = 6.83 × 10−5) after accounting for the degree of methylation at each of the sites (Figure S2). Expressed as a change in effect size using Nagelkerke’s R2 measure, there is a 6% reduction in the R2 in a model including methylation and the amount of mtDNA to predict MD and a 9% reduction for telomere length. From this analysis, we concluded that the cellular composition of saliva collected from cases differed slightly from that of controls and explained less than 10% of the differences in the amount of mtDNA and telomere length between cases and controls.
Molecular Changes Are Contingent on the Depressed State
To investigate a causal relationship between stressful life events, MD, amount of mtDNA, and telomere length, we performed a series of conditional regression analyses, assuming that stressful life events preceded the onset of MD and the molecular changes (Supplemental Experimental Procedures). Table 2 shows the counts of individuals categorized by MD disease status and number of stressful life events, with the means and SEs for the amount of mtDNA (Table 2) and telomere length (Table 2) within each category.
Table 2.
Relationship between Stressful Life Events, mtDNA, Telomere Length, and Major Depression
| Difference in Normalized mtDNA Levels in Cases of MD and Controls per #SLE | |||
|---|---|---|---|
| #SLE | MD Control | MD Case | mtDNA Difference t Statistic, p Value |
| 0 | −0.132 (0.019), 2,487 | 0.142 (0.024), 1,689 | −8.86, 1.25 × 10−18 |
| 1 | −0.156 (0.026), 1,432 | 0.0987 (0.027), 1,441 | −6.83, 1.01 × 10−11 |
| 2 | −0.103 (0.034), 757 | 0.165 (0.033), 935 | −5.65, 1.84 × 10−08 |
| 3 | −0.068 (0.058), 334 | 0.085 (0.044), 507 | −2.12, 0.03 |
| 4+ | 0.062 (0.067), 221 | 0.132 (0.040), 666 | −0.89, 0.37 |
| Difference in Telomere Length in Cases of MD and Controls per #SLE | |||
| #SLE | MD Control | MD Case | Telomere Difference T Statistic, p Value |
| 0 | 0.078 (0.020), 2,542 | −0.053 (0.025), 1,722 | 4.10, 4.23 × 10−5 |
| 1 | 0.098 (0.026), 1,461 | −0.048 (0.027), 1,470 | 3.91, 9.42 × 10−5 |
| 2 | 0.093(0.035), 780 | −0.069 (0.032), 952 | 3.36, 8.05 × 10−4 |
| 3 | 0.042 (0.053), 342 | −0.085(0.045), 517 | 1.82, 0.069 |
| 4+ | 0.060 (0.060), 229 | −0.129 (0.038), 677 | 2.65, 0.0083 |
For each category of stressful life event (#SLE, ranging from none [0] to more than four [4+] reported events), Table 2 reports the means and SEs of the normalized mtDNA levels (top section of the table) and normalized telomere length measures (bottom section of the table), followed by the numbers of individuals, for MD cases and controls. The last column gives the t statistic and p value for the difference between cases and controls.
If stressful life events have independent causal effects on MD and the molecular measures, then the latter should become independent of MD after conditioning on the number of stressful events. Table 2 shows this is not the case because the mean differences in amount of mtDNA and telomere length between cases and controls, when stratified for the number of stressful life events, remained highly significant (t tests in third column of Table 2; mtDNA p values range from 1.25 × 10−18 to 0.37; telomere length p values range from 4.23 × 10−5 to 0.0083).
We next asked whether the effect of stressful life events on MD is entirely indirect, acting via changes in the amount of mtDNA or telomere length. We rejected this explanation because the association between MD and stressful life events remains highly significant after conditioning on either amount of mtDNA (p = 5.60 × 10−99; see Table S2, i) or telomere length (p = 2.x10−100; Table S3, i) in a logistic regression model. In contrast, the association between stress and amount of mtDNA or telomere length disappeared when conditioned on MD (p = 0.11 Table S4, i, and p = 0.11 Table S5, i, respectively). In other words, the predictive power of stress on amount of mtDNA and telomere length is mediated through a history of MD.
These conclusions also hold when the number of stressful life events is replaced by a history of childhood sexual abuse. In particular, there was no significant difference in the amount of mtDNA or telomere length when comparing controls who reported a history of childhood sexual abuse with those who did not. Mean values of normalized mtDNA for controls who reported any form of childhood sexual abuse was −0.136 (SE = −0.007) and −0.095 (SE = −0.001) for no such history; t test p value = 0.66. Comparable values for telomere length were 0.168 (SE = 0.0125) and 0.072 (SE = 0.001); t test p value = 0.27.
These analyses indicate that the molecular markers represent the current state of illness, regardless of the path by which it is reached, and predict that the most pronounced changes would be found in subjects currently reporting a severe mood disorder. Our analyses up to this point used subjects for whom we did not have a current state measure of mood. We therefore measured the amount of mtDNA in a separate Chinese case-control cohort of MD [39] where a state measure of mood was available (the Hamilton rating scale [40]). We selected 29 cases with scores greater than 25 (very severe) and 25 controls with scores of less than 5. Despite using such a small sample, we observed a highly significant difference (t test p value = 0.0008) and an odds ratio of 2.94 (95% CI 1.26–6.02), more than twice the odds ratio seen in the CONVERGE sample (odds ratio = 1.33).
Stress Increases the Amount of mtDNA and Shortens Telomeres
To gain a mechanistic understanding of the relationship between stress, amount of mtDNA, and telomere length, we undertook a mouse experiment. Sixteen C57BL/6J mice (eight males and eight females) were stressed for 4 weeks (for 5 days, a different stressor was administered: tail suspension, force-swim, foot shock, restraint, and sleep deprivation, followed by 2 days rest). After 0, 2, and 4 weeks of stress, amount of mtDNA and telomere length were assessed by qPCR and compared to age-matched non-stressed controls (eight males and eight females).
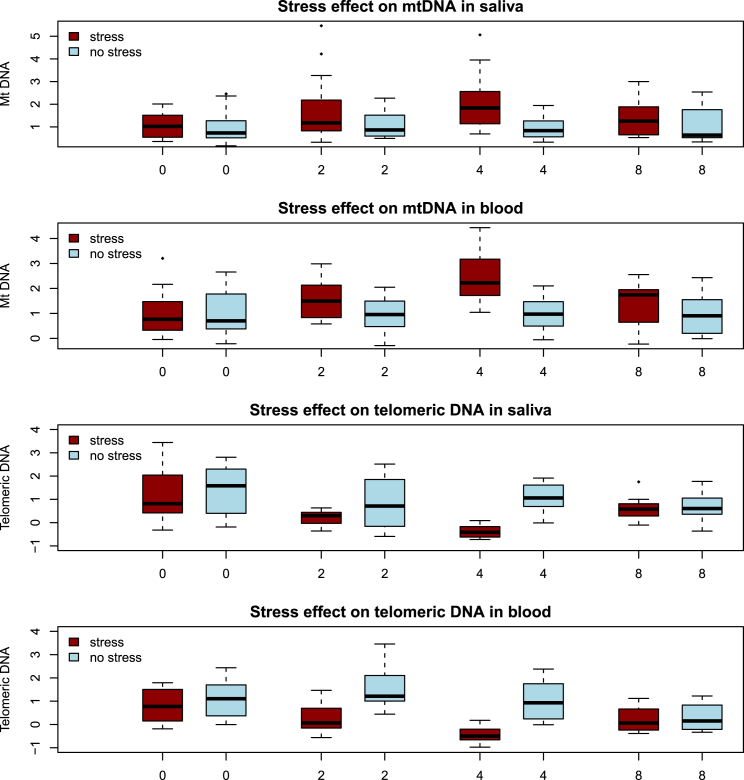
Consistent with our findings in humans, in mice, stress significantly increased the amount of mtDNA and decreased telomere length in saliva and in blood (Figure 2). After 4 weeks of stress, there was a mean increase in the amount of mtDNA of 210% compared to the unstressed animals in saliva (t test p = 0.0036) and 240% in blood (t test p = 6.1 × 10−5). At the same time, the length of telomeric DNA was reduced 28% in saliva (t test p = 0.0001) and 30% in blood (t test p = 0.0017) in stressed mice as compared to non-stressed. There were no significant differences in the white cell parameters between stressed and non-stressed animals (all p values > 0.05), indicating that this result is unlikely to be due to differences in the blood cellular composition.
Figure 2.
Effect of Chronic Stress on mtDNA in Saliva and Blood of Mice
Boxplot of relative mtDNA changes and relative mean telomere length over time in mice exposed to stress (red) and controls (blue). The vertical axis shows the amount of DNA, assessed by qPCR, relative to the mean of the values obtained before stress was imposed (week 0). The mean of week 0 is set to 1, so that results from subsequent weeks are fold changes relative to pre-stress levels. The horizontal axis is time in weeks from the beginning of the experiment. Stress was discontinued after week 4, so week 8 shows results for previously stressed animals after 4 weeks of living in a home cage. At the 4 week time point, the amount of mtDNA in blood and saliva was significantly greater in stressed animals (t test p = 6.1 × 10−5 and p = 0.0036, respectively). Also at the 4 week time point, relative mean telomere lengths in stressed mice were significantly lower in saliva (t test p = 0.0001) and blood (t test p = 0.0017) as compared to non-stressed mice. Differences between stressed and non-stressed mice in both measures were not significant at the start of the experiment or at the 8 week time point.
After 4 weeks of stress, half of the animals (eight stressed mice and eight controls) were kept in home cages without any intervention to model a recovery period of no stress. Molecular markers were again tested in blood and saliva, and results are shown as week 8 in Figure 2. Four weeks after the discontinuation of stress, there were no significant differences between control animals and those that had been previously exposed to stress (amount of mtDNA in saliva p = 0.50, in blood p = 0.38; telomere length in saliva p = 0.85, in blood p = 0.76; all p values from t tests). These results indicate that the molecular changes are, at least in part, reversible.
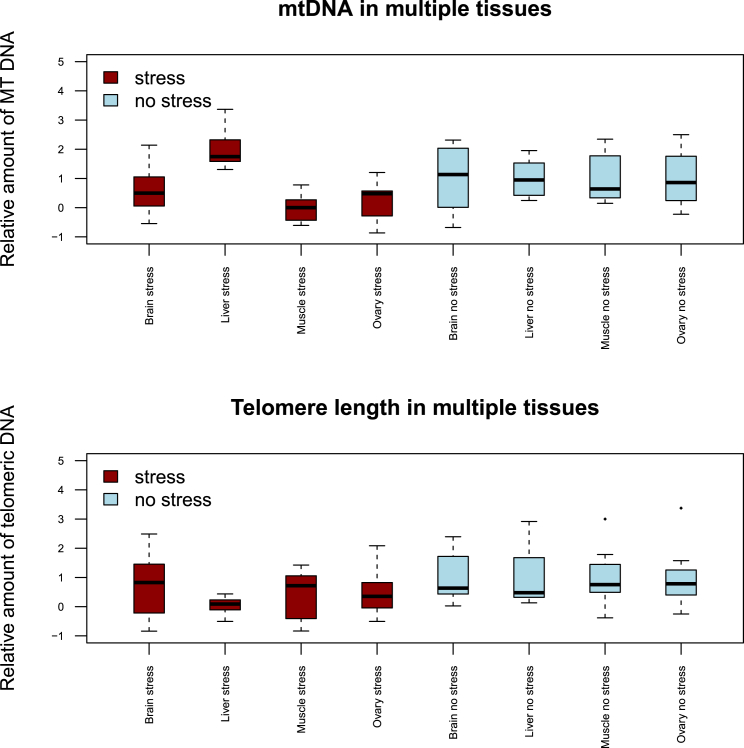
Immediately after the cessation of stress, multiple tissues from the other 16 animals were assayed for the amount of mtDNA and telomere length. Figure 3 shows results for four tissues: liver, muscle, brain (hippocampus), and ovary (ovary was chosen as we were interested to assess whether the changes might be transmitted to the next generation). For the amount of mtDNA, there was a significant increase in liver (p value from t test = 0.005), a significant decrease in muscle (p = 0.014), but no significant alterations in hippocampus (p = 0.50), and a suggestive change in ovary (p = 0.086). For telomere length, there was a significant 54% reduction in liver (p = 0.03), but no significant changes in other tissues (muscle p = 0.23, hippocampus p = 0.59, ovary p = 0.30). These results reveal tissue-specific changes in mtDNA, and possibly in telomere length, as a consequence of stress.
Figure 3.
Alterations of mtDNA in Different Tissues after 4 Weeks of Stress
Top: Assessment of mtDNA in four tissues. Bottom: Assessment of telomere length in four tissues. The vertical axis shows the amount of mtDNA or telomere length assessed by qPCR, relative to the mean of the values obtained for control animals (no stress). The horizontal axis gives the names of the tissues for the two conditions: stress (red) and no stress (blue).
Mitochondrial Function Is Altered in Tissues with Increased mtDNA
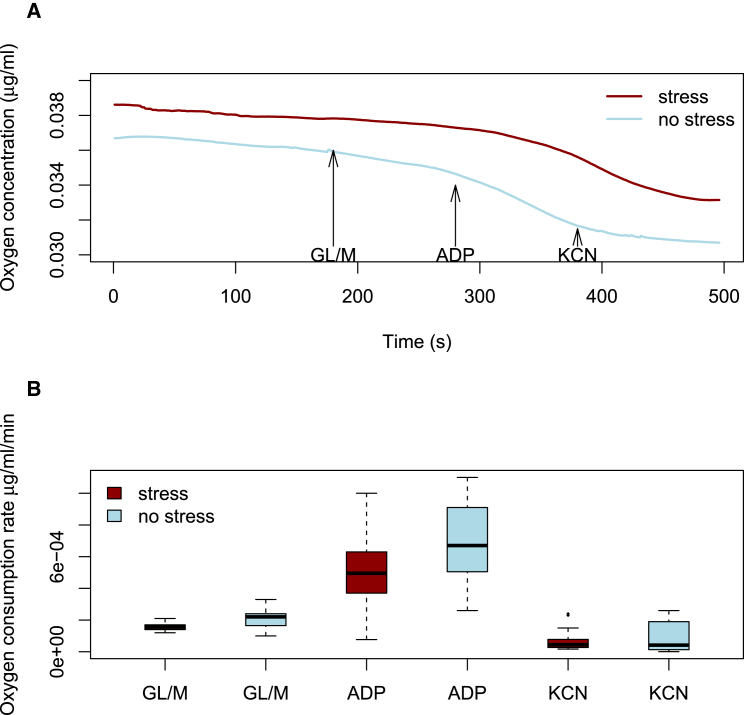
Altering the amount of mtDNA presumably reflects functional changes in mitochondria, a hypothesis we tested by measuring and comparing the oxidative phosphorylation (OXPHOS) capacity of mitochondria-enriched fractions from the liver of stressed and non-stressed mice. Figure 4A shows the mean values for the change in oxygen concentration over time for liver mitochondrial preparations taken from eight stressed and eight control (not stressed) animals. Addition of an equal amount of ADP (driving force for the electron transport chain after depletion of residual driving force in the fraction with excess glutatmate and malate) induced a greater increase in oxygen consumption in mitochondria from the non-stressed animals than stressed ones (p = 0.038, from a linear model; Figure 4B). The complete quenching of OXPHOS upon addition of electron transport chain inhibitor potassium cyanide in both stressed and non-stressed mice showed oxygen consumption during the experiment was solely due to OXPHOS. Results of this experiment showed that OXPHOS efficiency was reduced in the liver tissue of mice whose amount of mtDNA had increased in response to stress, suggesting either an adaptive switch to glycolysis or a compromise in mitochondrial function.
Figure 4.
The Oxygen Consumption of Mouse Liver after Stress Administration
(A) Oxygen concentration (vertical axis) detected per second (horizontal axis) per μg of mitochondria. The slope of the curve indicates the rate of oxygen consumption. Glutamate/malate (GL/MA) is added at 3 min after addition of isolated mitochondria, and oxygen consumption was assessed after substrate addition. The addition of ADP (100 s later) initiates active respiration while potassium cyanide (KCN) (100 s later) inhibits all mitochondrial function.
(B) Oxygen consumption rate per μg of mitochondria after the addition of the three compounds, comparing stressed and non-stressed animals.
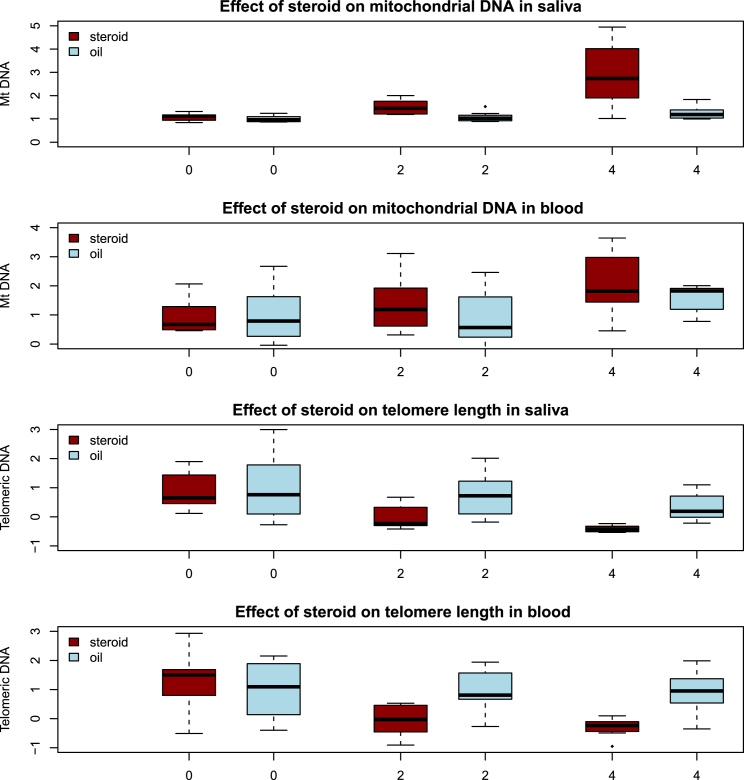
Glucocorticoid Administration Reproduces the Effects of Stress
What might be inducing the molecular changes? We considered one mechanism: activation of the hypothalamic pituitary adrenal (HPA) axis [41–48]. We administered corticosterone to eight C57BL/6J female mice over 4 weeks and oil vehicles of the same volume to eight control mice of the same strain. Figures 5 shows that after 4 weeks, there was significantly more mtDNA in the saliva (p = 0.011) and in the blood (p = 0.0013) of treated mice compared to controls and that telomere length had significantly reduced in both tissues (in saliva: p = 0.0023; in blood: p = 0.0016; all p values from t tests).
Figure 5.
Effect of Daily Subcutaneous Injection of Corticosterone on mtDNA and Telomere Length in Saliva and Blood in Mice
Boxplot of relative amount of mtDNA and relative mean telomere length over time in mice injected with corticosterone (red) and controls injected with the same volumes of oil vehicle (blue). The vertical axis shows the amount of mtDNA assessed by qPCR, relative to the mean of the values obtained before corticosterone was injected (week 0). The mean of week 0 is set to 1, so that results from subsequent weeks are fold changes relative to pre-stress levels. The horizontal axis is time in weeks from the beginning of the experiment. After 4 weeks, the amount of mtDNA levels in mice injected with corticosterone was significantly higher in saliva (t test p = 0.011) and blood (t test p = 0.0013) as compared to mice injected with oil vehicle; relative mean telomere lengths were significantly reduced in both tissues (in saliva: t test p = 0.0023; in blood: t test p = 0.0016) in mice injected with corticosterone as compared to mice injected with oil vehicle.
Discussion
We report here two important observations on the relationship between MD and two molecular signatures of adversity, the amount of mtDNA and mean telomere length. First, the changes in amount of mtDNA and telomere length are contingent on the presence of MD. We found no significant molecular changes in those who reported stressful life events, including childhood sexual abuse, but had never been depressed. Second, in a mouse model, while stress over a period of weeks did increase the amount of mtDNA and shorten telomere length, both changes were at least partly reversible. While early environmental adversity may result in permanent changes in physiology and risk of disease [49], our results indicate that it is important to recognize two trajectories, one leading to molecular signatures of stress and one to illness.
For the first trajectory leading from adversity to molecular changes, one possible pathway is through the endocrine system, particularly the activation of the hypothalamic pituitary axis, since changes in both molecular markers could be reproduced in mice by administration of corticosterone. Release of glucocorticoids is known to increase in response to stress. Severe stressors, such as childhood sexual abuse [42, 50], alter pituitary-adrenal and autonomic reactivity. In some circumstances, the consequences may be deleterious rather than adaptive: glucocorticoids have been implicated in the pathophysiology of posttraumatic stress disorder [51, 52], and it has been known for many years that some patients with MD exhibit hypersecretion of cortisol [41, 53, 54], in part due to corticotrophin releasing factor (CRF) hypersecretion [55].
For the second trajectory leading to illness, we hypothesize that while adversity may on its own have an effect on both the amount of mtDNA and mean telomere length, the extent and persistence of these molecular changes depend on an individual’s susceptibility to MD, either from genetic or additional environmental predisposing factors. In many individuals, the molecular signatures will be small and transitory, but in those with MD, the effects may be larger or last for a longer period of time. Subjects who have never been diagnosed with MD, yet suffered severe adversity, may have had detectable alterations in mtDNA levels and mean telomere length in particular tissues at the time they experienced stressful life events, but these changes would have reversed and no longer be detectable by the time they were interviewed. We emphasize that the molecular changes we observe are neither risk factors nor causes of MD. The correlation between stress, mtDNA, and telomere length is contingent upon MD; we could find no evidence that stressful life events act via changes in mtDNA or telomere length to increase the risk of MD. Thus, our data provide no support for a role of changes in the amount of mitochondrial DNA or length of telomeres in regulating mood.
The disease-state dependence of the measures is important when considering the potential use of the changes as biomarkers. It is noteworthy in this regard that in a sample when we assayed amount of mtDNA in currently severely ill subjects, a robust difference was detected in a comparison of just 29 cases and 25 controls. This suggests that, despite the relatively small effects and large variances seen in the saliva sample, there may be circumstances where the amount of mtDNA could serve as a useful biomarker. The relatively larger increases seen in the mouse experiment (up to 4-fold) suggest that controlling for inter-individual variation would improve the chances of the biomarkers having a clinical application.
Changes in mean telomere length and levels of mtDNA presumably reflect altered metabolic strategies in times of perceived or expected stress. Experiments assessing OXPHOS efficiency in mice showed a decrease in OXPHOS energy production in stressed mice with elevated mtDNA levels. The tissue-specific effects of stress on amount of mtDNA and mean telomere suggest different, or possibly sequential, pathways governing tissue-specific change. It is possible that these changes might in part explain changes in appetite and sleep occurring during the state of depression.
Experimental Procedures
The CONVERGE Study, Samples, DNA Preparation, and Sequencing
All 11,670 samples are drawn from the CONVERGE study of MD. The study protocol was approved centrally by the Ethical Review Board of Oxford University (Oxford Tropical Research Ethics Committee) and the ethics committees responsible for each hospital in China. The study posed minimal risk to the subjects (an interview and saliva sample). Stressful life events and childhood sexual abuse were assessed retrospectively. The stressful life events section of the CONVERGE interview was developed for the Virginia Adult Twin Study of Psychiatric and Use Disorders (VATSPUD) [17]. It assesses 16 traumatic lifetime events and the age at their occurrence. The childhood sexual abuse was a shortened version of the detailed module used in the VATSPSUD study, which was in turn based on the instrument developed by Martin et al. [56]. DNA was extracted from saliva samples using the Oragene protocol.
Sequencing libraries were constructed from DNA fragmented using the Covaris Adaptive Focused Acoustics (AFA) technology. QIAquick Gel Extraction kit was used to purify the DNA fragments. Each DNA sample was uniquely tagged with a sequencing index for multiplex library preparation. Insert sizes were on average 400 bp. Library quality was checked with an Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System. Libraries were sequenced on Illumina Hiseq 2000 machines. Sequencing reads for each of the 11,670 samples were aligned to Genome Reference Consortium Human Build 37 patch release 5 (GRCh37.p5) with Stampy (v.1.0.17) [57] and stored in BAM format [58].
The GENDEP and DeCC Studies, Samples, and qPCR
Cases and control samples were drawn from the United Kingdom Depression Case-Control (DeCC) study [29] and the Genome-Based Therapeutic Drugs for Depression (GENDEP) study [30]. mtDNA copy number was estimated from DNA extracted from blood samples by qPCR, using a TaqMan Universal PCR MasterMix on an ABI StepOnePlus Real-Time PCR System (Life Technologies). The pre-designed TaqMan assay Hs02596867_s1 was used to amplify a fragment of the MT-CYB gene on the mitochondrial chromosome in duplex with the TaqMan RNaseP Copy Number Reference Assay (Life Technologies, part number 4403326) as an internal control.
Extracting and Quality Control of Mitochondrial Reads from Low-Coverage Whole-Genome Sequencing Data
All reads mapped to the human mitochondrial genome NC_012920.1 were extracted from the whole-genome BAM files mapped to GRCh37.p5 using Samtools (v.0.1.18) [58]. The mitochondrial reads extracted were then converted to the FASTQ format using Picardtools (v.1.108, http://broadinstitute.github.io/picard/) and mapped to a combined reference containing 894 complete bacterial genomes, 2,024 complete bacterial chromosomes, 154 draft assemblies, and 4,373 complete plasmids sequences (in total, 7,390 unique bacterial DNA sequences) available on NCBI using BWA (v.0.5.6) [58]. All reads mapped to bacterial DNA sequences were filtered out using Samtools (v.0.1.18) [58] by imposing a mapping quality filter of 59 (Phred-scale probability of being wrongly mapped) and removing reads with FLAGs (-F 1804) that identify unmapped reads, unpaired reads, reads that do not pass quality control, reads that may be PCR or optical duplicates, and reads that are secondary alignments that also map to other areas of the reference. No reads from filtered BAM files of any sample map onto the combined bacterial reference.
Estimation of mtDNA Copy Number
Average read depth per 100 bp is calculated for the mtDNA reads mapped to NC_012920.1 both before and after filtering out poorly mapped reads including those potentially from bacterial genomes using SAMTOOLs (v.0.1.18) [58]. There are regions in the mitochondrial genome replicated in the nuclear genome commonly known as nuclear copies of mitochondrial DNA (NUMTs), which would most likely be present as secondary alignments. We calculated average read depth per segment of 100 bp in the mtDNA alignments both before and after filtering and compared the two sets of read depths. To reduce errors in estimation of coverage due to NUMTs, segments with big differences in read depth (>5% of the filtered read depth) between the filtered and unfiltered alignments that are more likely to span NUMTs were excluded from our calculation of mtDNA copy number. We arrived at a measure of mtDNA copy number by taking the mean read depth in the filtered alignments across all remaining 100 bp segments, then regressing it with sequencing batch, sample age, and average filtered read depth on chromosome 20, then transforming the residuals to normality using a quantile normal function in the R statistical software language [59].
Estimation of Mean Telomere Length
Mean telomere length was quantified from low-coverage whole-genome sequencing data mapped to Genome Reference Consortium Human Build 37 patch release 5 (GRCh37.p5) with Telseq v.0.0.1 [60]. The estimated mean telomere length output from Telseq was already corrected for whole-genome coverage and the GC content of DNA; it was then regressed with batch and sample age before the residuals were transformed to normality using a quantile normal function in the R statistical software language [59].
Association between Molecular Markers, MD, and Stress
We tested for association between MD and molecular markers using logistic regression in the R statistical software language [59]. All logistic regression models included as covariates the first three principal components (PCs) from a principal-component analysis (PCA) performed with Genome-wide Complex Trait Analysis (GCTA) v.1.24.4 [61] using a genetic relationship matrix (GRM). The GRM was generated with 561,819 common, tagging single nucleotide polymorphisms (SNPs) from all autosomes. All SNPs in this tagging set were polymorphic in 1,000G phase 1 Asian (ASN) panel, occur at greater than 5% minor allele frequency in CONVERGE study samples, and are out of linkage disequilibrium (LD) with each other (maximum pairwise LD = 0.8).
Mouse DNA Extraction
DNA was extracted from mouse tissues using a QIAamp DNA Investigator Kit (QIAGEN). Saliva was collected from mice by inserting a disposable inoculation loop into the animal’s mouth, allowing the animal to chew for a few seconds, before rinsing the loop in QIAamp DNA Investigator Kit ATL buffer. Blood was collected from a superficial tail vein.
Quantification of mtDNA Levels by qPCR
qPCR was carried out using the Bio-Rad iQ SYBR Green super mix supplied by Roche Molecular Biochemicals. A nuclear genomic fragment of 160 bp was amplified from the mouse Gapdh gene (forward 5′-TGACGTGCCGCCTGGAGAAAC-3′, reverse 5′-CCGGCATCGAAGGTGGAAGAG-3′. A 117 bp fragment of the mitochondrial genome (positions 13603–13719) was amplified with primers described in [62] (forward 5′- CCCAGCTACTACCATCATTCAAGT-3′, reverse 5′-GATGGTTTGGGAGATTGGTTGATGT-3′). qPCR was performed under the following conditions: denaturation 95°C for 10 min followed by 50 cycles of 15 s at 95°C and 1 min at 60°C. An estimate of the mtDNA copy number was calculated using the mean of Gapdh as a control [63]. All samples were duplicated at each time point. PCR efficiencies were between 90%–110% (average coefficient variance: 0.806). PCR runs were discarded if they failed to meet the following criteria: no template control (NTC) with a quantitation cycle (Cq) < 38 cycles; sample with a Cq > 30 cycles; PCR efficiency > 90% and < 110.0%; standard curve R2 < 0.980; replicate group Cq SD greater than 0.20. qPCRs were carried out at the end of each experiment and all time points were analyzed on a single plate, thus excluding batch effects.
Quantification of Telomere Length by Monochrome Multiplex qPCR
Average telomere length was measured from mouse DNA using a previously described monochrome multiplex qPCR (MMQPCR) method [64] with the following conditions: denaturation at 95°C for 15 min followed by 2 cycles of 15 s at 94°C and 60 s at 49°C, 4 cycles of 15 s at 94°C and 30 s at 59°C, 20 cycles of 15 s at 85°C and 30 s at 59°C, and 27 cycles of 15 s at 94°C, 10 s at 84°C, and 15 s at 85°C. Forward and reverse telomeric primers were 5′-ATACCAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTCATGG-3′ and 5′-GAGGCAATATCCCTATCCCTATCCCTATCCCTATCCCTAACC-3′. Average telomere length ratio was estimated from the ratio of telomere product to that of a single copy nuclear gene albumin, forward and reverse primers for which were 5′-CGGCGGCGGGCGGCGCGGGCTGGGCGGAAACGCTGCGCAGAATCCTTG-3′ and 5′-GCCCGGCCCGCCGCGCCCGTCCCGCCGCTGAAAAGTACGGTCGCCTG-3′.
Mitochondrial Oxygen Consumption
We measured oxygen consumption from mouse liver mitochondrial preparations over time using a Clark electrode. After the addition of respiratory substrates (glutamate and malate), oxygen consumption was monitored for 100 s, after which ADP was added and oxygen consumption measured for a further 100 s. Potassium cyanide (KCN) was added 100 s later to inhibit all mitochondrial oxygen consumption.
Animal Experiments
All experiments were carried out in strict accordance with the recommendations in the Guide for Laboratory Animals Facilities and Care as promulgated by the Council of Agriculture, Executive Yuan, ROC, Taiwan. The protocol was approved by the Institutional Animal Care and Use Committee of Chang Gung University (permit number: CGU13-067). Animals were group housed and randomly assigned to stress or non-stress experiments.
Mouse stress experiment: mice (strain C57BL/6J, female n = 8, male n = 8, aged 12 months) were stressed over 5 days followed by 2 days rest, repeated for 4 weeks. On the first day, animals were suspended from their tails for 10 min. This was repeated three times, with 5 min rest between tail suspensions. On the second day, animals were placed in a cylinder of deep water from which there was no escape for 10 min. The forced swim was repeated twice with a 10 min rest. On the third day, a foot shock was administered three times (0.75 mA for 10 s with 10 s rest). On the fourth day, animals were restrained in a cylindrical tube (12 cm in length and 3 cm in diameter) for 3 hr. On the fifth day, animals were sleep deprived for 24 hr (mice were put in water tank, containing multiple and visible platforms [4.5 cm in height and diameter] surrounded by water for 24 hr). For the glucocorticoid experiment, mice (strain C57BL/6J, female n = 8, aged 12 months) underwent daily subcutaneous injection of 30 mg/kg corticosterone (Sigma) or vehicle (oil) for 28 days. Association between mtDNA, telomere length, and stress was performed in a linear mixed model using the lme4 package in the statistical software language R [59]. The null model included only weight. Variation in the amount of mtDNA between different tissues was assessed by a t test, comparing values between controls and experimental animals.
Author Contributions
N.C., Yihan Li, S.C., K.K., R.M., and J.F. prepared the manuscript. Y.C., Yihan Li, H.D., B.D., Keqing Li, W.S., J.G., B.H., S.G., Jian Hu, C.H., G.H., G.J., Youhui Li, Kan Li, Yi Li, G.L., L. Liu, T.L., Ying Liu, L. Lv, H.M., H.S., J. Shen, J. Shi, J. Sun, M.T., Xumei Wang, Gang Wang, Xueyi Wang, J.Y., K.Z., N.S., J.Z., Z.Z., W.Z., H.Z., J.F., and K.K. handled sample collection. S.C., J.F., J.N., H.-Y.H., Y.-T.L., and G.-J.H. carried out animal experiments. N.C., Yihan Li, J.N., G.B., M.R., K.K., R.M., and J.F. handled human mtDNA and telomere analyses. Q.L., Jingchu Hu, W.K., W.J., Yihan Li, Guangbiao Wang, L.W., P.Q., Yuan Liu, T.J., Y. Lu, X.Z., Y.Y., Yingrui Li, H.Y., Jian Wang, X.G., R.M., J.M., J.F., Jun Wang, and X.X. carried out genome sequencing and analysis. N.C., Yihan Li, J.F., and R.M. carried out genetic analysis. N.C. identified the shortened telomeres and examined the relationship between both molecular markers with stress and depression. S.C. performed all animal experiments and analyzed the qPCR and OXPHOS output data. Yihan Li identified the excess of mtDNA in low-coverage sequencing data in cases of MD as compared to controls.
Acknowledgments
This work was funded by the Wellcome Trust (WT090532/Z/09/Z, WT083573/Z/07/Z, WT089269/Z/09/Z). All authors are part of the China, Oxford and VCU Experimental Research on Genetic Epidemiology (CONVERGE) consortium and gratefully acknowledge the support of all partners in hospitals across China. W.K. is funded by the Wellcome Trust (WT097307). N.C. is supported by the Agency of Science, Technology and Research (A∗STAR) Graduate Academy. Research was in part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London.
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Supplemental Information
References
- 1.Gluckman P.D., Hanson M.A., Cooper C., Thornburg K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rich-Edwards J.W., Spiegelman D., Lividoti Hibert E.N., Jun H.J., Todd T.J., Kawachi I., Wright R.J. Abuse in childhood and adolescence as a predictor of type 2 diabetes in adult women. Am. J. Prev. Med. 2010;39:529–536. doi: 10.1016/j.amepre.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nanni V., Uher R., Danese A. Childhood maltreatment predicts unfavorable course of illness and treatment outcome in depression: a meta-analysis. Am. J. Psychiatry. 2012;169:141–151. doi: 10.1176/appi.ajp.2011.11020335. [DOI] [PubMed] [Google Scholar]
- 4.Dong M., Giles W.H., Felitti V.J., Dube S.R., Williams J.E., Chapman D.P., Anda R.F. Insights into causal pathways for ischemic heart disease: adverse childhood experiences study. Circulation. 2004;110:1761–1766. doi: 10.1161/01.CIR.0000143074.54995.7F. [DOI] [PubMed] [Google Scholar]
- 5.Shonkoff J.P., Garner A.S., Committee on Psychosocial Aspects of Child and Family Health. Committee on Early Childhood, Adoption, and Dependent Care. Section on Developmental and Behavioral Pediatrics The lifelong effects of early childhood adversity and toxic stress. Pediatrics. 2012;129:e232–e246. doi: 10.1542/peds.2011-2663. [DOI] [PubMed] [Google Scholar]
- 6.Wegman H.L., Stetler C. A meta-analytic review of the effects of childhood abuse on medical outcomes in adulthood. Psychosom. Med. 2009;71:805–812. doi: 10.1097/PSY.0b013e3181bb2b46. [DOI] [PubMed] [Google Scholar]
- 7.Carroll J.E., Gruenewald T.L., Taylor S.E., Janicki-Deverts D., Matthews K.A., Seeman T.E. Childhood abuse, parental warmth, and adult multisystem biological risk in the Coronary Artery Risk Development in Young Adults study. Proc. Natl. Acad. Sci. USA. 2013;110:17149–17153. doi: 10.1073/pnas.1315458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McEwen B.S. Central effects of stress hormones in health and disease: understanding the protective and damaging effects of stress and stress mediators. Eur. J. Pharmacol. 2008;583:174–185. doi: 10.1016/j.ejphar.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEwen B.S. Protective and damaging effects of stress mediators. N. Engl. J. Med. 1998;338:171–179. doi: 10.1056/NEJM199801153380307. [DOI] [PubMed] [Google Scholar]
- 10.Miller G.E., Chen E., Parker K.J. Psychological stress in childhood and susceptibility to the chronic diseases of aging: moving toward a model of behavioral and biological mechanisms. Psychol. Bull. 2011;137:959–997. doi: 10.1037/a0024768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham J.E., Christian L.M., Kiecolt-Glaser J.K. Stress, age, and immune function: toward a lifespan approach. J. Behav. Med. 2006;29:389–400. doi: 10.1007/s10865-006-9057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendler K.S., Karkowski L.M., Prescott C.A. Causal relationship between stressful life events and the onset of major depression. Am. J. Psychiatry. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- 13.Fergusson D.M., Mullen P.E. Sage Publications, Inc; Thousand Oaks: 1999. Childhood Sexual Abuse: An Evidence Based Perspective. [Google Scholar]
- 14.Kendler K.S., Bulik C.M., Silberg J., Hettema J.M., Myers J., Prescott C.A. Childhood sexual abuse and adult psychiatric and substance use disorders in women: an epidemiological and cotwin control analysis. Arch. Gen. Psychiatry. 2000;57:953–959. doi: 10.1001/archpsyc.57.10.953. [DOI] [PubMed] [Google Scholar]
- 15.Martin J., Anderson J., Romans S., Mullen P., O’Shea M. Asking about child sexual abuse: methodological implications of a two stage survey. Child Abuse Negl. 1993;17:383–392. doi: 10.1016/0145-2134(93)90061-9. [DOI] [PubMed] [Google Scholar]
- 16.Chen J., Cai Y., Cong E., Liu Y., Gao J., Li Y., Tao M., Zhang K., Wang X., Gao C. Childhood sexual abuse and the development of recurrent major depression in Chinese women. PLoS ONE. 2014;9:e87569. doi: 10.1371/journal.pone.0087569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kendler K.S., Prescott C.A. Guildford Press; New York: 2006. Genes, Environment, and Psychopathology. [Google Scholar]
- 18.Tao M., Li Y., Xie D., Wang Z., Qiu J., Wu W., Sun J., Wang Z., Tao D., Zhao H. Examining the relationship between lifetime stressful life events and the onset of major depression in Chinese women. J. Affect. Disord. 2011;135:95–99. doi: 10.1016/j.jad.2011.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kendler K.S., Aggen S.H. Clarifying the causal relationship in women between childhood sexual abuse and lifetime major depression. Psychol. Med. 2014;44:1213–1221. doi: 10.1017/S0033291713001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong E., Li Y., Shao C., Chen J., Wu W., Shang X., Wang Z., Liu Y., Liu L., Gao C. Childhood sexual abuse and the risk for recurrent major depression in Chinese women. Psychol. Med. 2012;42:409–417. doi: 10.1017/S0033291711001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalev I., Moffitt T.E., Sugden K., Williams B., Houts R.M., Danese A., Mill J., Arseneault L., Caspi A. Exposure to violence during childhood is associated with telomere erosion from 5 to 10 years of age: a longitudinal study. Mol. Psychiatry. 2013;18:576–581. doi: 10.1038/mp.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epel E.S., Blackburn E.H., Lin J., Dhabhar F.S., Adler N.E., Morrow J.D., Cawthon R.M. Accelerated telomere shortening in response to life stress. Proc. Natl. Acad. Sci. USA. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahola K., Sirén I., Kivimäki M., Ripatti S., Aromaa A., Lönnqvist J., Hovatta I. Work-related exhaustion and telomere length: a population-based study. PLoS ONE. 2012;7:e40186. doi: 10.1371/journal.pone.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okereke O.I., Prescott J., Wong J.Y., Han J., Rexrode K.M., De Vivo I. High phobic anxiety is related to lower leukocyte telomere length in women. PLoS ONE. 2012;7:e40516. doi: 10.1371/journal.pone.0040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolkowitz O.M., Mellon S.H., Epel E.S., Lin J., Dhabhar F.S., Su Y., Reus V.I., Rosser R., Burke H.M., Kupferman E. Leukocyte telomere length in major depression: correlations with chronicity, inflammation and oxidative stress—preliminary findings. PLoS ONE. 2011;6:e17837. doi: 10.1371/journal.pone.0017837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jodczyk S., Fergusson D.M., Horwood L.J., Pearson J.F., Kennedy M.A. No association between mean telomere length and life stress observed in a 30 year birth cohort. PLoS ONE. 2014;9:e97102. doi: 10.1371/journal.pone.0097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glass D., Parts L., Knowles D., Aviv A., Spector T.D. No correlation between childhood maltreatment and telomere length. Biol. Psychiatry. 2010;68:e21–e22. doi: 10.1016/j.biopsych.2010.02.026. author reply e23–e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao L., Martin M.V., Watson S.J., Schatzberg A., Akil H., Myers R.M., Jones E.G., Bunney W.E., Vawter M.P. Mitochondrial involvement in psychiatric disorders. Ann. Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen-Woods S., Gaysina D., Craddock N., Farmer A., Gray J., Gunasinghe C., Hoda F., Jones L., Knight J., Korszun A. Depression Case Control (DeCC) Study fails to support involvement of the muscarinic acetylcholine receptor M2 (CHRM2) gene in recurrent major depressive disorder. Hum. Mol. Genet. 2009;18:1504–1509. doi: 10.1093/hmg/ddp051. [DOI] [PubMed] [Google Scholar]
- 30.Uher R., Huezo-Diaz P., Perroud N., Smith R., Rietschel M., Mors O., Hauser J., Maier W., Kozel D., Henigsberg N. Genetic predictors of response to antidepressants in the GENDEP project. Pharmacogenomics J. 2009;9:225–233. doi: 10.1038/tpj.2009.12. [DOI] [PubMed] [Google Scholar]
- 31.Wieczorek G., Asemissen A., Model F., Turbachova I., Floess S., Liebenberg V., Baron U., Stauch D., Kotsch K., Pratschke J. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69:599–608. doi: 10.1158/0008-5472.CAN-08-2361. [DOI] [PubMed] [Google Scholar]
- 32.Sehouli J., Loddenkemper C., Cornu T., Schwachula T., Hoffmüller U., Grützkau A., Lohneis P., Dickhaus T., Gröne J., Kruschewski M. Epigenetic quantification of tumor-infiltrating T-lymphocytes. Epigenetics. 2011;6:236–246. doi: 10.4161/epi.6.2.13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiencke J.K., Accomando W.P., Zheng S., Patoka J., Dou X., Phillips J.J., Hsuang G., Christensen B.C., Houseman E.A., Koestler D.C. Epigenetic biomarkers of T-cells in human glioma. Epigenetics. 2012;7:1391–1402. doi: 10.4161/epi.22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen B.C., Houseman E.A., Marsit C.J., Zheng S., Wrensch M.R., Wiemels J.L., Nelson H.H., Karagas M.R., Padbury J.F., Bueno R. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies M.N., Volta M., Pidsley R., Lunnon K., Dixit A., Lovestone S., Coarfa C., Harris R.A., Milosavljevic A., Troakes C. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Accomando W.P., Wiencke J.K., Houseman E.A., Nelson H.H., Kelsey K.T. Quantitative reconstruction of leukocyte subsets using DNA methylation. Genome Biol. 2014;15:R50. doi: 10.1186/gb-2014-15-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koestler D.C., Christensen B., Karagas M.R., Marsit C.J., Langevin S.M., Kelsey K.T., Wiencke J.K., Houseman E.A. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi C., Zhang K., Wang X., Shen Y., Xu Q. A study of the combined effects of the EHD3 and FREM3 genes in patients with major depressive disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2012;159B:336–342. doi: 10.1002/ajmg.b.32033. [DOI] [PubMed] [Google Scholar]
- 40.Hamilton M. A rating scale for depression. J. Neurol. Neurosurg. Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpenter W.T., Jr., Bunney W.E., Jr. Adrenal cortical activity in depressive illness. Am. J. Psychiatry. 1971;128:31–40. doi: 10.1176/ajp.128.1.31. [DOI] [PubMed] [Google Scholar]
- 42.Heim C., Newport D.J., Bonsall R., Miller A.H., Nemeroff C.B. Altered pituitary-adrenal axis responses to provocative challenge tests in adult survivors of childhood abuse. Am. J. Psychiatry. 2001;158:575–581. doi: 10.1176/appi.ajp.158.4.575. [DOI] [PubMed] [Google Scholar]
- 43.Di Giusto E.L., Cairncross K., King M.G. Hormonal influences on fear-motivated responses. Psychol. Bull. 1971;75:432–444. doi: 10.1037/h0031260. [DOI] [PubMed] [Google Scholar]
- 44.Du J., Wang Y., Hunter R., Wei Y., Blumenthal R., Falke C., Khairova R., Zhou R., Yuan P., Machado-Vieira R. Dynamic regulation of mitochondrial function by glucocorticoids. Proc. Natl. Acad. Sci. USA. 2009;106:3543–3548. doi: 10.1073/pnas.0812671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi J., Fauce S.R., Effros R.B. Reduced telomerase activity in human T lymphocytes exposed to cortisol. Brain Behav. Immun. 2008;22:600–605. doi: 10.1016/j.bbi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wikgren M., Maripuu M., Karlsson T., Nordfjäll K., Bergdahl J., Hultdin J., Del-Favero J., Roos G., Nilsson L.G., Adolfsson R., Norrback K.F. Short telomeres in depression and the general population are associated with a hypocortisolemic state. Biol. Psychiatry. 2012;71:294–300. doi: 10.1016/j.biopsych.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Tomiyama A.J., O’Donovan A., Lin J., Puterman E., Lazaro A., Chan J., Dhabhar F.S., Wolkowitz O., Kirschbaum C., Blackburn E., Epel E. Does cellular aging relate to patterns of allostasis? An examination of basal and stress reactive HPA axis activity and telomere length. Physiol. Behav. 2012;106:40–45. doi: 10.1016/j.physbeh.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Révész D., Verhoeven J.E., Milaneschi Y., de Geus E.J., Wolkowitz O.M., Penninx B.W. Dysregulated physiological stress systems and accelerated cellular aging. Neurobiol. Aging. 2014;35:1422–1430. doi: 10.1016/j.neurobiolaging.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 49.Gluckman P.D., Hanson M.A., Spencer H.G., Bateson P. Environmental influences during development and their later consequences for health and disease: implications for the interpretation of empirical studies. Proc. Biol. Sci. 2005;272:671–677. doi: 10.1098/rspb.2004.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heim C., Newport D.J., Heit S., Graham Y.P., Wilcox M., Bonsall R., Miller A.H., Nemeroff C.B. Pituitary-adrenal and autonomic responses to stress in women after sexual and physical abuse in childhood. JAMA. 2000;284:592–597. doi: 10.1001/jama.284.5.592. [DOI] [PubMed] [Google Scholar]
- 51.van Zuiden M., Geuze E., Willemen H.L., Vermetten E., Maas M., Amarouchi K., Kavelaars A., Heijnen C.J. Glucocorticoid receptor pathway components predict posttraumatic stress disorder symptom development: a prospective study. Biol. Psychiatry. 2012;71:309–316. doi: 10.1016/j.biopsych.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 52.Yehuda R. Biology of posttraumatic stress disorder. J. Clin. Psychiatry. 2001;62(17):41–46. [PubMed] [Google Scholar]
- 53.Gibbons J.L., McHUGH P.R. Plasma cortisol in depressive illness. J. Psychiatr. Res. 1962;1:162–171. doi: 10.1016/0022-3956(62)90006-7. [DOI] [PubMed] [Google Scholar]
- 54.Sachar E.J., Hellman L., Fukushima D.K., Gallagher T.F. Cortisol production in depressive illness. A clinical and biochemical clarification. Arch. Gen. Psychiatry. 1970;23:289–298. doi: 10.1001/archpsyc.1970.01750040001001. [DOI] [PubMed] [Google Scholar]
- 55.Arborelius L., Owens M.J., Plotsky P.M., Nemeroff C.B. The role of corticotropin-releasing factor in depression and anxiety disorders. J. Endocrinol. 1999;160:1–12. doi: 10.1677/joe.0.1600001. [DOI] [PubMed] [Google Scholar]
- 56.Zlotnick C., Kohn R., Keitner G., Della Grotta S.A. The relationship between quality of interpersonal relationships and major depressive disorder: findings from the National Comorbidity Survey. J. Affect. Disord. 2000;59:205–215. doi: 10.1016/s0165-0327(99)00153-6. [DOI] [PubMed] [Google Scholar]
- 57.Lunter G., Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R-Development-Core-Team . R Foundation for Statistical Computing; Vienna: 2004. A Language and Environment for Statistical Computing. [Google Scholar]
- 60.Ding Z., Mangino M., Aviv A., Spector T., Durbin R., UK10K Consortium Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014;42:e75. doi: 10.1093/nar/gku181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Furda A.M., Bess A.S., Meyer J.N., Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol. Biol. 2012;920:111–132. doi: 10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Jiao J., Kang J.X., Tan R., Wang J., Zhang Y. Multiplex time-reducing quantitative polymerase chain reaction assay for determination of telomere length in blood and tissue DNA. Anal. Bioanal. Chem. 2012;403:157–166. doi: 10.1007/s00216-012-5783-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.