Summary
Archaea employ the archaellum, a type IV pilus-like nanomachine, for swimming motility. In the crenarchaeon Sulfolobus acidocaldarius, the archaellum consists of seven proteins: FlaB/X/G/F/H/I/J. FlaF is conserved and essential for archaellum assembly but no FlaF structures exist. Here, we truncated the FlaF N terminus and solved 1.5-Å and 1.65-Å resolution crystal structures of this monotopic membrane protein. Structures revealed an N-terminal α-helix and an eight-strand β-sandwich, immunoglobulin-like fold with striking similarity to S-layer proteins. Crystal structures, X-ray scattering, and mutational analyses suggest dimer assembly is needed for in vivo function. The sole cell envelope component of S. acidocaldarius is a paracrystalline S-layer, and FlaF specifically bound to S-layer protein, suggesting that its interaction domain is located in the pseudoperiplasm with its N-terminal helix in the membrane. From these data, FlaF may act as the previously unknown archaellum stator protein that anchors the rotating archaellum to the archaeal cell envelope.
Graphical Abstract
Highlights
-
•
This is the first structural and functional study of an archaellum stator component
-
•
sFlaF is a β-sandwich, immunoglobulin-like dimeric protein
-
•
FlaF resembles and binds to the S-layer protein
-
•
FlaF exerts its function in the pseudoperiplasm
Banerjee et al. determined the crystal structure of soluble domain FlaF, an essential archaellum assembly component, at 1.5-Å resolution. The biochemical data suggest that FlaF may function as a stator protein in the archaeal pseudoperiplasm and interacts with the S-layer protein.
Introduction
Whereas bacteria use flagella for swimming motility, archaea employ archaella (formerly archaeal flagella) to move in liquid media or on surfaces. Archaella are found in all motile archaeal species and were recently also identified in the new archaeal phylum Thaumarchaeota (Jarrell and Albers, 2012; Spang et al., 2012). Although archaella functionally resemble bacterial flagella, they are assembled via a mechanism reminiscent of type IV pili (Craig et al., 2004). Archaea and bacteria possess many type IV pili structures that serve very diverse functions in adhesion to surfaces, DNA uptake, as well as virulence and twitching motility in bacteria (Burrows, 2012; Hamilton and Dillard, 2006; Henche et al., 2012). Importantly, the archaellum is the first type IV pilus-like structure that confers motility by rotary movement of the filament, and it is unknown how this rotation is achieved by its motor complex or how the rotating component may be attached to the archaeal cell envelope (Marwan et al., 1991; Shahapure et al., 2014).
In all known motile archaeal species, archaella subunits are encoded in one gene cluster that contains 7–15 different genes (Bayley et al., 1998; Lassak et al., 2012a), which comprise all the components needed for filament formation, rotation, and attachment to the cell envelope. Physiological and genetic analyses show that archaella act in motility by rotating the partially flexible structure (Alam and Oesterhelt, 1984; Lassak et al., 2012a; Shahapure et al., 2014) and that all of the genes present in the archaella gene cluster are essential for assembly and swimming motility (Bayley et al., 1998; Lassak et al., 2012b). The number of filament FlaB proteins varies from one copy in many crenarchaea to up to five different paralogs in euryarchaeota, and phylogenetic analysis revealed that flaHIJ are present in the same order in all archaella clusters. Hence, it was proposed that the polytopic membrane protein, FlaJ, the bifunctional ATPase, FlaI, and the RecA family protein, FlaH, form the conserved core complex for archaella assembly (Ghosh and Albers, 2011). The ATPase FlaI forms an ATP-dependent hexameric species, which shows enhanced ATP hydrolysis in the presence of native lipids (Ghosh et al., 2011). Recently, combined crystal structures and X-ray scattering solution analyses of hexameric FlaI showed that FlaI's first 29 structurally flexible amino acids are important for the switch between its assembly mode to the motility mode (Reindl et al., 2013). In agreement with FlaI's function in assembly and rotation, it was shown in haloarchaea that archaellum rotation is ATP dependent (Streif et al., 2008). In Sulfolobus acidocaldarius, FlaI interacts with FlaX, an archaellum component that is specific for crenarchaea. FlaX forms an oligomeric ring structure in the cytoplasm, and this ring structure was predicted to be the scaffold for the assembly of the motor complex of the archaellum (Banerjee et al., 2012, 2013). Euryarchaeota lack FlaX. Instead, FlaCDE, which are thought to interact with proteins that connect the archaellum to the chemotaxis cascade, promote directed movement along concentration gradients (Schlesner et al., 2009). The structure and function of the monotopic membrane proteins, FlaF and FlaG, are unknown but their gene order was found to be specific for crenarchaeota (flaG-flaF) and euryarchaeota (flaF-flaG) (Desmond et al., 2007). They are the top candidates for connecting the moving and stationary components of the filament as they possess transmembrane domains and might therefore localize extracellularly.
For other microbial nanomachines, the structures of the components and their architectural placement have led to key insights on their activities and mechanisms, such as for the type II secretion machine (Johnson et al., 2013; Korotkov et al., 2012; Lu et al., 2014; McLaughlin et al., 2012), and the structures of pilin filament subunits and the type IV pilus (Giltner et al., 2012; Melville and Craig, 2013; Takhar et al., 2013). However, for the archaellum, only ATPase FlaI structures are known (Reindl et al., 2013). Furthermore, there is no structural information for the archaellum on any proteins on the pseudoperiplasmic side of the membrane, which is the expected location for a stator that must both anchor the archaellum and ensure that the membrane does not rupture once the filament starts to rotate. We reasoned that FlaF is the likely candidate for this function. Yet, no structures exist for any FlaF homologs. To better understand the critical function of FlaF, which is conserved in all archaellum operons and essential for archaellum assembly, we characterized FlaF from S. acidocaldarius. To reduce aggregation of this monotopic membrane protein, we partially removed the predicted transmembrane N-terminal α-helix and then solved 1.5-Å and 1.65-Å resolution crystal structures of the native FlaF (sFlaF). Our structures revealed the end of the N-terminal α-helix protruding from a β-sandwich fold formed by an eight-stranded anti-parallel β-sheet domain that positions hydrophobic surfaces in a dimer interface. Moreover, FlaF was shown to bind the S-layer protein, the sole cell envelop forming protein in S. acidocaldarius (Albers and Meyer, 2011). To this end, we find that the FlaF soluble domain has an extracellular localization and its structure and position appear appropriate to anchor the flexible archaellum filament in the paracrystalline S-layer.
Results
FlaF, a Long Protruding N-Terminal α-Helix and β-Sheet Globular Domain Protein
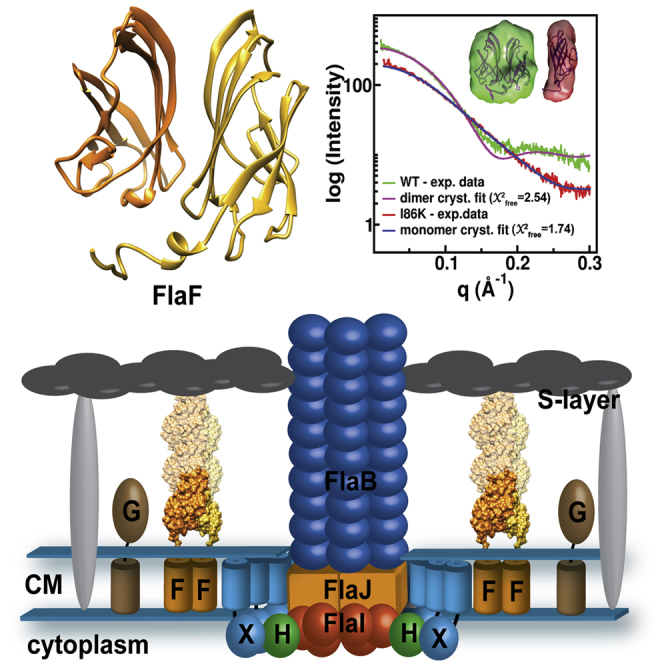
As FlaF is a conserved archaella assembly-associated protein and deletion or disruption of the flaF gene resulted in non-archaellated cells in Methanococcus and Sulfolobus species (Chaban et al., 2007; Lassak et al., 2012b), we sought to examine its structure to help understand its function. In silico analyses with secondary structure prediction servers suggested that FlaF is a monotopic transmembrane protein with an extended N-terminal α-helix (1–32 amino acids) that is connected to a globular domain with eight β-sheets (Figure 1). FlaF contains an incomplete archaellin (archaeal flagellin) domain, missing the class III signal peptide at the N terminus. Archaellins are like type IV pilin (T4P) proteins and form the archaellum filament. The heterologous expression of full-length FlaF in Escherichia coli was not successful. However, we reasoned that if this N-terminal sequence formed a protruding α-helix like other T4P pilin proteins (Parge et al., 1995), then this segment could be truncated without altering the subunit structure as shown for multiple T4P proteins (Craig et al., 2003; Hartung et al., 2011). We therefore constructed a 34-residue N-terminal deletion of FlaF and this resulted in highly soluble FlaF (sFlaF) lacking only the N-terminal transmembrane segment. N-terminal His6-tagged sFlaF was overproduced in Rosetta pLysS strain and purified by Ni2+ affinity chromatography (Figure S1A). To test the oligomeric state of sFlaF, purified sFlaF was analyzed by analytical size-exclusion chromatography (SEC) on a Superose 12 10/300 GL column and eluted as a dimer with a molecular weight of about 32 kDa. A less significant tetrameric species of 64 kDa was also observed in the elution profile (Figure S1B).
Figure 1.
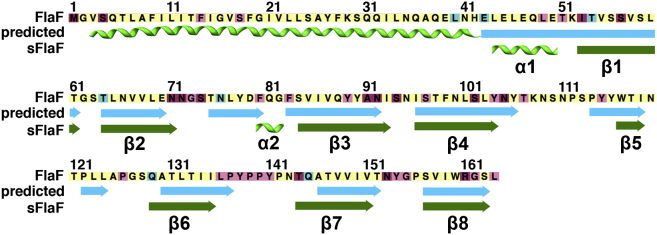
Secondary Structure Prediction and Comparison with the Crystal Structure of sFlaF
The full-length FlaF secondary structure was predicted by the Phyre 2 server. N-terminal residues 3–42 were predicted to be present as a long helical region. The actual secondary structure obtained from the crystal structure of sFlaF chain B (PDB, 4P94) is shown for comparison below the prediction. The sequence was color coded from cyan (most variable) to dark purple (most conserved) as conservation shown in Figure 6A.
Crystal Structure Determination
The native crystal structures of recombinant sFlaF (native 1, residues 46–164; PDB, 4ZBH; and native 2, residues 49–164 in chain A and 35–164 in chain B; PDB, 4P94) were determined at 1.50-Å and 1.65-Å resolution, respectively. The native 1 sFlaF structure was phased with a platinum derivative by single isomorphous replacement with anomalous scattering (SIRAS) (Table 1). The native 2 sFlaF structure was phased by molecular replacement with native 1 sFlaF as a search model. Both crystal structures were from the same construct (residues 35–164 with an N-terminal His-tag) but crystallized in different forms. The sFlaF native 1 crystal (Figure S2A) in space group P43212 contained one molecule per asymmetric unit with a solvent content of 43%; native 2 crystal (Figure S2B) in space group I23 contained two molecules (chain A and B) per asymmetric unit with a solvent content of 58%. The generated crystallographic symmetry mate of native 1 shows the nearly identical dimer structure to native 2 sFlaF. The root-mean-square deviation (rmsd) for both dimer crystal structures is about 0.50 Å for 207 aligned Cα atoms. Since chain B of the native 2 sFlaF structure (PDB, 4P94) contained the complete sequence, it was used for the analyses and figures unless otherwise specified.
Table 1.
X-Ray Diffraction Data Collection and Refinement Statistics of sFlaF
| sFlaF native 1 | sFlaF native 2 | sFlaF + Pt | |
|---|---|---|---|
| Data Collection | |||
| Synchrotron beamline | ALS 12.3.1 | ALS 12.3.1 | ALS 12.3.1 |
| PDB code | 4ZBH | 4P94 | |
| Wavelength (Å) | 0.9677 | 1.0332 | 0.9677 |
| Space group | P43212 | I 2 3 | P43212 |
| Cell dimensions | |||
| a, b, c (Å) | 52.41, 52.41, 100.24 | 127.26, 127.26, 127.26 | 52.71, 52.71, 99.30 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å)a | 50.00–1.50 (1.55–1.50) |
44.99–1.65 (1.68–1.65) |
50.00–2.50 (2.59–2.50) |
| Observations | 127,605 | 770,110 | 71,034 |
| Unique observation | 22,839 | 41,166 | 5,275 |
| Rsyma,b | 0.051 (0.516) | 0.054 (0.591) | 0.080 (0.409) |
| Mean I/σIa | 62.6 (3.7) | 30.4 (4.0) | 51.2 (8.0) |
| Completeness (%)a | 98.4 (97.6) | 100 (100) | 100 (100) |
| Redundancy | 5.6 | 18.7 | 13.5 |
| Refinement | |||
| Rwork/Rfree (%)c | 15.8/19.6 | 17.3/20.0 | |
| Rmsd bond length (Å) | 0.005 | 0.007 | |
| Rmsd bond angles (°) | 1.052 | 1.062 | |
| Protein molecules in asymmetric unit | 1 | 2 | |
| Residues not in model | 35–45 | Chain A: 35–48 | |
| Average B-factor | |||
| Protein | 26.10 | 23.50 | |
| Solvent | 45.60 | 37.60 | |
| Ramachandran (%) | |||
| Most favored | 97.44 | 97 | |
| Additional allowed | 2.56 | 3 | |
| Generous allowed | 0 | 0 | |
| Disallowed | 0 | 0 | |
Values in parentheses are the statistics for the highest-resolution shell of data.
Rsym = S |Ihkl − <I>|/S<I>, where <I> is the average individual measurement of Ihkl.
Rwork = (S|Fobs − Fcalc|)/S|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is calculated the same as Rwork but from the data (5%) that were excluded from the refinement.
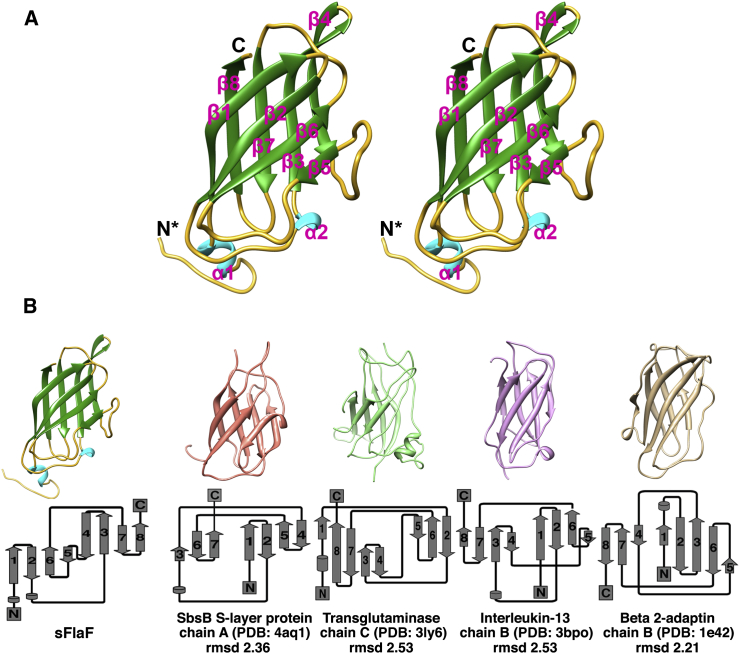
Crystal Structure of sFlaF, an Immunoglobulin-like β-Sandwich Protein
These two sFlaF crystal structures, determined independently in two space groups, provide the first structure determined among all the FlaF homologs. The sFlaF structure revealed a β-sandwich fold, which consisted of eight anti-parallel β-strands in two sheets (Figure 2). sFlaF, furthermore, is an anti-parallel Greek key β-barrel, where the β2/β3 and β6/β7 loops form +4 β-strand or Greek key connections as opposed to nearest-neighbor interactions (Figure 2B). This Greek key β-barrel fold with its cross-domain covalent connections imparts high stability and is utilized by stable proteins, such as immunoglobulins (Getzoff et al., 1988) and superoxide dismutase (Getzoff et al., 1989; Perry et al., 2010). The structure is more β-sandwich than barrel-like as four anti-parallel β1 (residues 53–61), β2 (residues 64–71), β6 (residues 129–135), and β5 (resides 117–119) strands form one β-sheet with the other four anti-parallel β4 (residues 96–104), β3 (residues 84–93), β7 (residues 145–151), and β8 (residues 157–163) strands forming another β-sheet that packs against the other (Figures 1 and 2). Structural alignments of the sFlaF structure against similar fold structures from the PDB using the DALI server (Holm and Rosenstrom, 2010) showed significant similarities with SbsB S-layer protein (chain A, residues 202–296), transglutaminase (chain C, residues 4–141), interleukin-13 (chain B, residues 1–94), and beta-2-adaptin (chain B, residues 704–820) (Figure 2B). All four protein structures have similar topologies to sFlaF and the rmsd of aligned structures are ∼2.2–2.5 Å even though the sequence identity is only about 8%–14%. The similar fold to the S-layer protein SbsB is intriguing as S-layer proteins can be part of the archaeal or bacterial cell envelope or even constitute their only component (Albers and Meyer, 2011).
Figure 2.
Crystal Structure of sFlaF
(A) Stereoview of the eight-strand β-sandwich fold of sFlaF with the N-terminal helix. The sFlaF structure reveals an immunoglobulin-like β-sandwich fold.
(B) Comparison of the structure fold similarity based on the DALI server (upper). The corresponding secondary structure topology (lower) is shown for each structure using the Pro-origami server (Stivala et al., 2011). The rmsd was calculated by aligning each structure with the sFlaF structure using Chimera. SbsB S-layer protein chain A (domain II), residues 202–296; transglutaminase chain C, residues 4–141; interleukin-13 chain B, residues 1–94; and beta 2-adaptin chain B, residues 704–820.
Secondary structure prediction of full-length FlaF predicts a long-extended N-terminal α-helix that contains a transmembrane region (residues 1–32) (Figure 1) reminiscent of T4P (Craig et al., 2003; Parge et al., 1995; Hartung et al., 2011). In the crystal structure of truncated sFlaF (residues 35–164), the N-terminal region is an 11-residue long-extended loop with a short helical region (Figure 3A, chain B, and Figure S3). Residues 35–48 from chain A of sFlaF native 2 structure were directed toward the bulk solvent space and were not visible in the electron density map. Similarly, residues 35–45 were also disordered in the other native 1 sFlaF crystal structure (PDB, 4ZBH) (Figure S5A). Contrarily, the N-terminal helix (residues 35–51) from chain B of the native 2 sFlaF structure was stabilized through electrostatic and hydrophobic interactions by the neighboring β-sheet in the chain A′ subunit that potentially can form a tetramer or filament of sFlaF (Figures S5B and S5C). The interactions of this N-terminal α-helix and neighboring β-sheet behave similarly to the T4P structure in which the N-terminal α-helix interacts with the β-sheet and forms an αβ roll fold (Craig et al., 2003), however, the orientation of the N-terminal α-helix from the T4P structure is protruding from the head domain, which is the opposite direction to the sFlaF N-terminal α-helix from the crystal structure. The lack of the N-terminal transmembrane domain may lead to the formation of the flexible loop/helix of the sFlaF structure (residues 35–51), which could impart a flexible connection into the N-terminal transmembrane helix or be stabilized by interactions to become part of this helix.
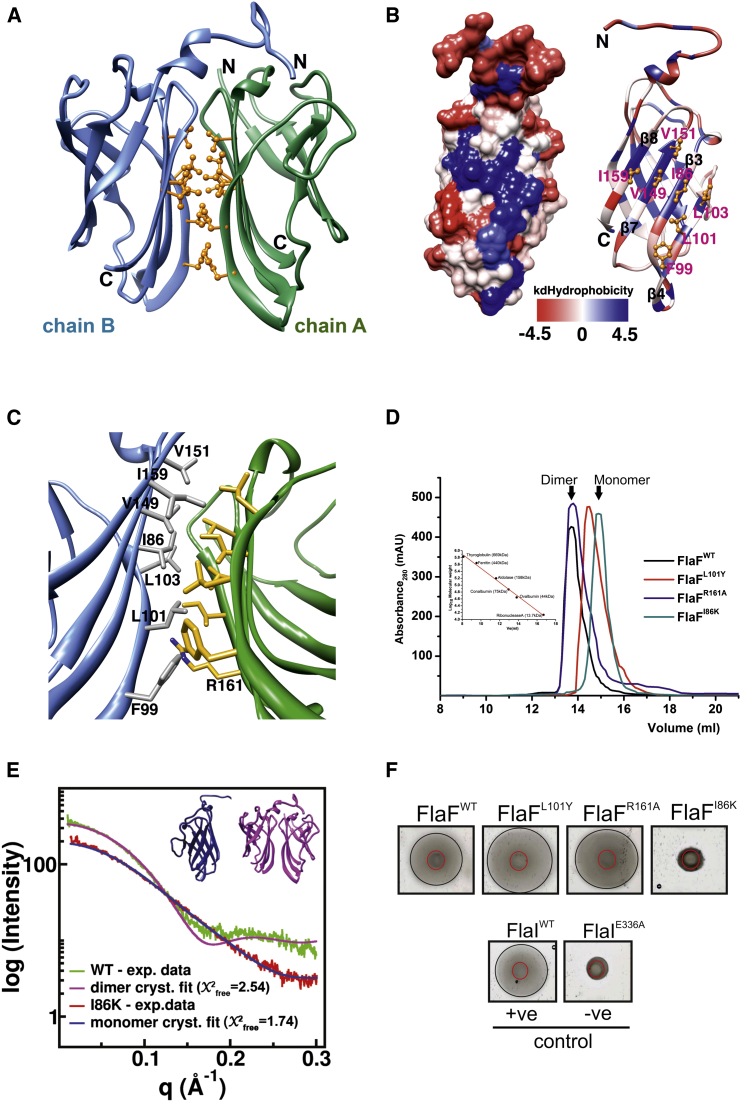
Figure 3.
sFlaF Dimer Formation through Hydrophobic Interactions
(A) The dimeric form of the sFlaF crystal structure. The hydrophobic residues are shown (gold, ball and stick). The dimer interface was between two β-sheets (β3, β4, β7, and β8) from each sFlaF subunit (chain A and B). The two subunits are in a two-fold symmetry.
(B) Left: the hydrophobicity surface presentation was prepared by Chimera. The color gradient ranges from blue (most hydrophobic), through white, to red (most hydrophilic) using the Kyte-DooLittle hydrophobicity scale (Kyte and Doolittle, 1982); right: the ribbon presentation with the corresponding color gradient. The hydrophobic residues (gold, ball and stick) that form a hydrophobic patch (the bluest region) on strand β3, β4, β7, and β8 are labeled in pink.
(C) The close-up view of the sFlaF dimer interface. The interface residues shown in stick are colored in dark gray and gold, respectively. Residue R161 has H-bonding with the main chain carbonyl oxygen of residue L101 (not shown).
(D) Analytical size-exclusion chromatogram of sFlaF dimer interface mutants. sFlaFWT and sFlaF-R161A elute as a homogeneous dimeric population, whereas sFlaF-L101Y contains a mixture of dimeric and monomeric sFlaF in solution. Interestingly, sFlaF-I86K elutes solely as a monomeric population. The size standard is shown in the inset.
(E) The SAXS results show that wild-type (WT)- and I86K-sFlaF exist in solution as dimer and monomer, respectively. The calculated scattering curves of the WT-sFlaF dimer (pink) and monomer (chain B, blue) were fit to the SAXS data of WT-sFlaF (green) and I86K-sFlaF (red) with X2free fit of 2.54 and 1.74, respectively.
(F) Motility assay of in-trans complemented ΔaapFΔFlaF strain using FlaF dimer interface mutants. The results clearly show that monomeric FlaF is not able to complement the deletion defect. The positive (+ve) and negative (−ve) controls used in this study were FlaIWT and FlaI-E336A complemented ΔaapFΔFlaI strains, respectively.
FlaF Dimerization Is Essential In Vivo
sFlaF crystallized in a dimeric conformation and the dimer interface was formed through hydrophobic interactions between two β-sheets (β4, β3, β7, and β8 strands) in two sFlaF molecules (Figure 3A). The hydrophobic residues (I86, F99, L101, L103, V149, V151, and I159) from two sFlaF molecules interacted with each other in a two-fold symmetry to form the dimer interface (Figure 3B). To help elucidate the physiological relevance of the dimerization of FlaF, we performed mutagenesis experiments and chose two residues at the dimer interface (I86 and L101) and one surface residue (R161) that forms an H-bond with the carbonyl oxygen of L101 (Figure 3C). I86 was substituted with lysine (I86K); for L101, we introduced a tyrosine (L101Y) to hinder dimerization and R161 was substituted with alanine (R161A). The mutants were analyzed by an in vitro dimerization assay using SEC and the results revealed that the sFlaF-I86K is monomeric in vitro (Figure 3D). To test if the dimer in the crystal exists in solution, we used small-angle X-ray scattering (SAXS), which allows conformations and assembly states to be objectively defined in solution (Hura et al., 2009, 2013; Rambo and Tainer, 2013). The SAXS data, including Chifree fits of dimer and monomer models to the SAXS data, indicate that the dimer assembly and interface in the crystal structures also exist in solution (Figures 3E and S5D–F). Except for residues L103 and V151, the rest of the hydrophobic residues in the dimer interface of the crystal structure and solution assembly are conserved in FlaF homologs. The solvation free energy gain upon the formation of this dimer is −15.2 kcal/mol based on the calculation by PISA (Krissinel and Henrick, 2007). The buried surface area is about 1,330 Å2, which is about 10% of the total surface area of 12,230 Å2. These results suggest that sFlaF favors dimer formation. We further tested these mutants by in-trans complementation of a flaF deleted strain to observe the in vivo effect. The results showed that I86K mutation cannot complement the ΔflaF strain (Figure 3F), which suggests that the dimer interface identified from the crystal structures is physiologically relevant and essential for its function in vivo.
Soluble Domain of FlaF Binds to S-layer Proteins
Most archaea produce a proteinaceous surface layer (S-layer) that surrounds the entire cell and gives it structural rigidity (Albers and Meyer, 2011). In Sulfolobales, the S-layer is composed of two different proteins: the small membrane-integrated protein SlaB and the outer large crystalline protein SlaA (Albers and Meyer, 2011; Veith et al., 2009). The space between the cell membrane and the pseudo cell wall is termed the pseudoperiplasmic space (Albers and Meyer, 2011); we reasoned that to hold the rotating assembly and help keep the membrane from rupture by a rotating filament would require a stator protein assembly positioned in this pseudoperiplasmic space. As analysis of FlaF for a class III signal peptide by the program Flafind was negative (Szabo et al., 2007), which shows that FlaF lacks an archaellin signal sequence. We suspected that FlaF would not be part of the archaellum filament but might have a different function in the extracellular pseudoperiplasmic space.
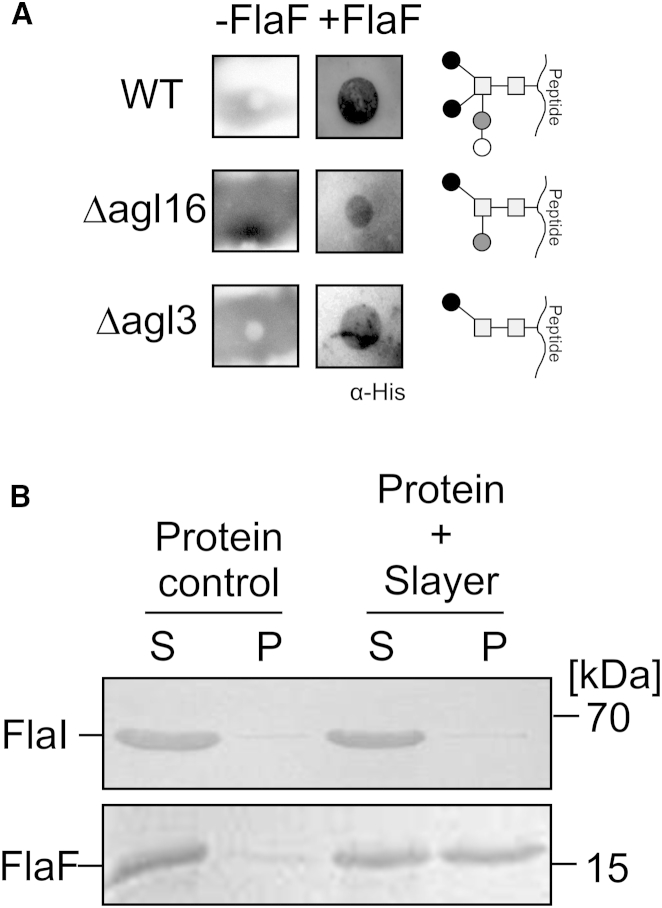
FlaF, which has an incomplete archaellin domain, is thus a logical candidate for this stator function. Hence, we tested whether FlaF can interact with the S-layer protein, the sole cell envelope component in the pseudoperiplasmic space. For this purpose, the S-layer was isolated and a dot far-western blot (Chan et al., 2008) was performed using purified sFlaF and S-layer protein (see Experimental Procedures). To that end, the S-layer was spotted on a polyvinylidene fluoride (PVDF) membrane and incubated with His-tagged sFlaF. Binding of FlaF to the S-layer protein was detected by incubation with an α-His-tag antibody and subsequent chemiluminescent detection. The results (Figure 4A) suggested that sFlaF binds to the isolated S-layer of S. acidocaldarius strain MW001 (Wagner et al., 2012). To further test the ability of sFlaF to bind the S-layer, we also used a sedimentation assay (see Experimental Procedures for details). The isolated S-layer proteins were incubated with either sFlaF or FlaI, a cytoplasmic archaellum subunit, and subsequently pelleted by centrifugation. Whereas FlaI did not co-pellet with the S-layer protein, FlaF was present in the S-layer pellet fraction (Figure 4B).
Figure 4.
Pseudoperiplasmic Localization of FlaF
(A) Dot far-western blot analysis of His6-tagged sFlaF. Surface layer proteins (S-layer) isolated from S. acidocaldarius MW001 (WT), Δagl16, and Δagl3 strains were spotted on to an activated PVDF membrane and incubated with either sFlaF or with buffer. Binding of sFlaF with the S-layer was visualized using α-His antibodies. No specific signal was detected in the negative control. The panel on the right shows the N-glycan tree (Meyer and Albers, 2013) of the different strains.  , N-acetyl glucosamine;
, N-acetyl glucosamine;  , sulfoquinovose; ○, glucose; ●, mannose.
, sulfoquinovose; ○, glucose; ●, mannose.
(B) Sedimentations assays were performed to analyze the interaction of sFlaF and the S-layer. FlaI was used as a negative control of the sedimentation assay. On the left, FlaI or FlaF were incubated alone. S and P indicate supernatant and pellet, respectively. In the two right lanes, FlaI and FlaF were incubated with the S-layer. Whereas FlaI shows no interaction with the S-layer, 50% of the FlaF was bound to the S-layer protein.
Archaeal S-layer proteins are highly glycosylated with N-glycans. In S. acidocaldarius, the N-glycan is tribranched and contains GlcNAc2 (Qui1-Glu1)(Man1)(Man1) (Figure 4A). To dissect whether the interaction of sFlaF with the S-layer proteins was mediated via sugar binding, S-layer proteins from two different mutant strains were isolated. The first strain was the S-layer from the Δagl3 strain missing the sulfoquinovose synthase that is essential for the synthesis of sulfoquinovose (Qui) (Meyer et al., 2011) and which therefore has a reduced glycan tree with Man1GlcNAc2 on its S-layer proteins. The second strain was purified from the Δagl16 strain that was missing only one terminal hexose of the N-glycan (Meyer et al., 2013). Importantly, the dot far-western blot analysis showed that FlaF bound to the S-layer proteins irrespective of their different N-glycan composition (Figure 4A), consistent with an interaction not requiring specific carbohydrate binding. As N-glycosylation is essential in S. acidocaldarius, we cannot test mutants void of N-glycans. Therefore N-glycans might still play a role in FlaF binding to the S-layer but the data suggest that the interaction may be through a protein contact, consistent with mimicking assemblies predicted from crystal lattice interactions (Figure S5C).
Conserved Residues and FlaF Function
The β-sandwich protein, FlaF, is essential for archaellum assembly. The crystal structure of sFlaF indicates that the β-sheets are forming a stable core and may primarily act in the protein's stable structural framework. To further identify regions implicated in the function of FlaF, we analyzed the sFlaF structure and identified two conserved loops (highlighted in Figure 5A and in Figures S5A and B) that are not involved in forming the β-sandwich fold (Figure 2A). As shown in the structure, the longest loop 136–143 (β6/β7) exposes conserved tyrosine side chains in a surface patch at the domain end distal to the N terminus. As there are no structural reasons to expect conservation of exposed and protruding regions, we reasoned that this conserved proline-rich loop (L136PYPPY141) and the serine (S74) loop between the β2-strand and the α2-helix, which lie along one face of the structure, may be important for FlaF's function and might therefore act in binding to the S-layer. So we chose the tyrosine residues (Y138 and Y141) in the proline loop as well as S74 and asparagine (N72) from the serine loop. We substituted all the chosen residues to alanine (A) to test the effect of these mutations in complementing the FlaF deletion defect. Individual Y138A and Y141A mutants in the proline-rich region do not show a significant effect in vivo (Figure 5B). However, the S74A mutant shows a 37% reduction in motility compared with the positive control supporting a role for this β2 α2 loop in function. S74 is flanked on one side by P126 in the β5-β6 loop and on the other side by L49 in the α1-β1 loop and by Y154 in the β7-β8 loop to form a possible binding surface (Figure S6). Interestingly, the comparison of chain A and chain B of sFlaF shows that S74 in chain B forms hydrogen bonds with the N-terminal α-helix residue E50 backbone carbonyl oxygen and with the E50 side chain through water-mediated interaction (Figure S7). Substitution of S74 to alanine results in a reduction of cell motility; this suggests the importance of this residue and possibly these interactions. We postulate that S74 might guard the N-terminal helix to loop switch for a function in motility.
Figure 5.
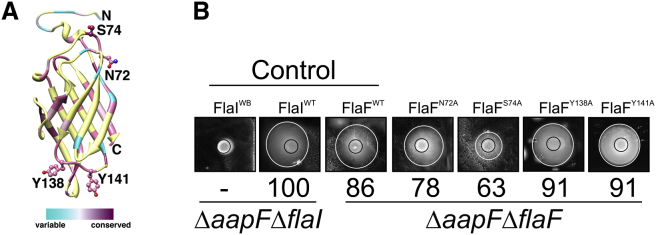
sFlaF Conserved Residues and Function
(A) The conserved residues chosen for further mutation analysis are depicted in a ball and stick model.
(B) Motility assay of complemented S. acidocaldarius strains. To check the importance of the conserved residues, the residues were mutated to alanine on a complementation plasmid and ΔaapFΔflaF strains were transformed and spotted on motility plates. The S. acidocaldarius ΔaapFΔflaI deletion mutant was complemented using either wild-type flaI (FlaIWT) as a positive control and E336A flaI (FlaIWB) as a negative control. The figure represents one of the three biological replicas of the motility assay and the numbers below each strain depict the percentage of the relative diameter of the swimming halo compared with the positive control from three individual biological samples. The black ring indicates the growth zone, whereas the white ring indicates the motility zone.
Discussion
Without understanding their component molecular structures, we can have only limited understanding of microbial nanomachines. One of the least structurally characterized microbial assembles is the archaellum, which is a rotary type IV pilus-like structure employed by all motile archaea to achieve swimming motility. In particular, no structural data have been available for any homolog of the critical, conserved FlaF protein of the archaellum. Our solved sFlaF structures revealed eight anti-parallel β-strands forming an immunoglobulin-like β-sandwich fold and a flexible N terminus connecting to the membrane-spanning helix. The observed binding of FlaF to the S-layer implies a pseudoperiplasmic localization of the soluble domain of FlaF. Interestingly, we observed that sFlaF shares structural similarity with the recently published S-layer protein, SbsB from Geobacillus stearothermophilus (Baranova et al., 2012).
S-layer proteins have ∼40%–60% of hydrophobic amino acids and self-assemble by an entropy-driven process (Sleytr and Beveridge 1999). The S-layer outer surface is usually hydrophobic with net neutral charge and the inner surface is hydrophilic and charged (Sára and Sleytr, 2000). The SbsB crystal structure shows six successive immunoglobulin-like β-sandwich domains, stabilized by Ca2+, and ∼50% hydrophobic residues. SbsB domain II, which is most structurally similar to sFlaF (Figure 2B), is suggested to orient toward the peptidoglycan layer. In Sulfolobales, the SlaB S-layer protein is predicted to contain 2–3 β-sandwich domains and an extended coil-coil region that anchors it to the cytoplasmic membrane (Veith et al., 2009). SlaA S-layer protein is also predicted to possess 67% β-strands and 6% α-helix. The FlaF-S-layer protein structural similarity and observed S-layer interaction support an extracellular localization for FlaF and imply possible hydrophobic interactions with S-layer proteins. We find that monomeric sFlaF-I86K shows impaired function in motility in vivo. Our results on FlaF and the known data on the S-layer suggest that FlaF may interact with the β-sandwich domains of the S-layer protein either through hydrophobic interactions similar to the FlaF dimer structure shown here or the β2-α2 Ser74 loop, which may be mimicking intermolecular interactions in the crystal packing contacts in our structures.
Regarding FlaF function in archaellum assembly and rotation, two possibilities exist, which are not mutually exclusive. First, as FlaF possesses an incomplete archaellin domain, it may act in recruiting FlaB at the base of the newly growing archaellum filament at an S-layer pore. In T4P and type II secretion systems, pseudopilins are not part of the main filament but are important for filament assembly and substrate secretion (Cisneros et al., 2012a, 2012b). Second, these results suggest that FlaF is the top candidate to act as a stator to hold the rotating assembly. It is essential that the archaellum is anchored in the cell envelope to ensure that the membrane does not rupture when the filament rotates. In the bacterial flagellum, this function is achieved by MotB in the MotAB stator complex (Berg, 2003). MotB binds to the peptidoglycan layers with a specific domain and thereby anchors the rotor complex of the flagellum in the cell envelope (Kojima et al., 2009; O'Neill et al., 2011; Reboul et al., 2011). Most archaea lack peptidoglycan (Albers and Meyer, 2011; Baumeister et al., 1989) but their extracellular proteins are heavily N-glycosylated. We propose that the data presented here on FlaF support an analogous function to MotB; its structure, location, and orientation appear suitable to anchor the rotor complex by binding to the S-layer via the FlaF soluble domain structure determined here and attaching to the membrane via its protruding N-terminal helix component (see Figure 6).
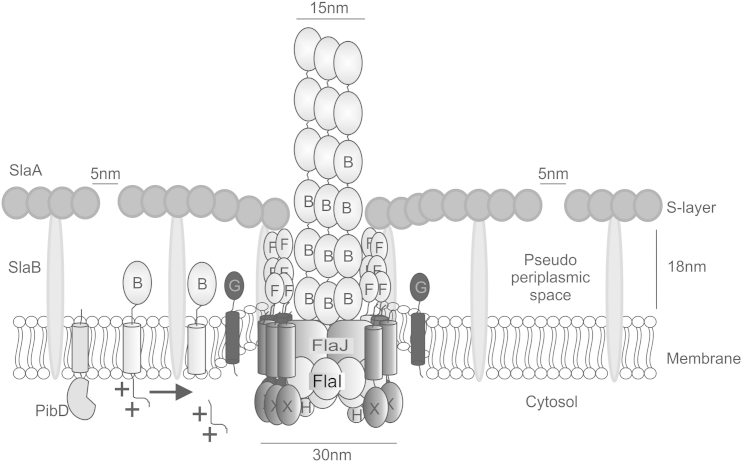
Figure 6.
Resulting Model of S. acidocaldarius Archaellum
S. acidocaldarius archaellum is composed of seven proteins, where FlaB (B) is the filament forming subunit (Desmond et al., 2007). FlaB expresses as a pre-protein with a class III signal sequence. PibD, a membrane-bound aspartic acid protease, specifically cleaves the signal sequence of FlaB (Albers et al., 2003; Szabo et al., 2007). FlaI, the dual function ATPase is involved in both archaellum assembly and rotation (Ghosh et al., 2011; Reindl et al., 2013). FlaH (H) is a unknown function RecA family protein. FlaX (X), a specific crenarchaeal protein, and the cytoplasmic domain form a 30-nm ring complex (Banerjee et al., 2012). The FlaX ring interacts strongly with FlaI and FlaH and together these three proteins form the archaellum motor complex (Banerjee et al., 2013). FlaG (G) function is so far unknown but it is predicted that its soluble domain is in the pseudoperiplasm (Ghosh and Albers, 2011). The S-layer of S. acidocaldarius is the cell wall structure and forms a crystalline array on the cell surface. It consists of the membrane-bound SlaB and the surface-covering SlaA. The pores of the rigid crystalline S-layer are about 5 nm in diameter (Baumeister et al., 1989) and the 18-nm space between the S-layer and the cell membrane is known as the pseudoperiplasmic space (Baumeister et al., 1989). We propose that FlaF (F) binds to the S-layer protein in its dimeric conformation and thereby anchors the archaellum in the cell envelop like stator protein in bacterial flagellum.
The bacterial flagellum stator complex must also act as a proton pump to generate the proton motif force for flagella rotation. Yet, archaellum rotation is driven by ATP hydrolysis, so we hypothesize that FlaF acts only as the static anchor of the archaellum in the cell envelope. FlaF may collaborate in this with FlaG; the function of FlaG is unknown but it is also predicted to be in the pseudoperiplasm (Ghosh and Albers, 2011). Furthermore, the S-layer in S. acidocaldarius is crystalline and rigid at a temperature of 80°C. The pores present between the hexagonal S-layer crystals are 4–5 nm in diameter, which is too small for proper protrusion of an archaellum filament (15 nm) (Figure 6) (Albers and Meyer, 2011; Baumeister et al., 1989). There may therefore need to be a mechanism by which the S-layer pore is suitably broadened; proteins that interact with the S-layer, such as FlaF, are chief candidates for this activity.
In general, immunoglobulin-like β-sandwich proteins such as FlaF can bind stably both to protein and to sugar moieties. So we propose that FlaF, possibly aided by FlaG, binds to the S-layer protein in its dimeric conformation and thereby anchors the archaellum in the cell envelope like the stator protein in bacterial flagellum. Going forward, it will be interesting to further investigate FlaF binding to S-layer protein and sugar moieties, its relationships with FlaG, and the stator complex architecture in relation to S-layer pores and archaellum rotatory motion.
Experimental Procedures
Bioinformatics Analysis
In silico analyses were carried out using available online tools as described in the Supplemental Information.
Site-Directed Mutagenesis
Mutagenesis of flaF was performed by round PCR on vectors pSVA1921 and pSVA2831. The respective amino acids were mutated to alanine and the resultant plasmids were named by the mutation as pSVA2832 (N72A), pSVA2833 (S74A), pSVA2834 (Y138A), pSVA2835 (Y141A), pSVA2816 (I86K), pSVA2809 (L101Y), and pSVA2813 (R161A). Oligonucleotide sequences used for mutagenesis are listed in Table S3.
Expression of Recombinant Δ34FlaF Protein (sFlaF) in E. coli
pET-based expression constructs for wild-type and different FlaF variants (N72A, S74A, Y138A, I86K, L101Y, R161A, and Y141A) were transformed into E. coli Rosetta (pLysS) strain and grown in Luria-Bertani-ampicilin-chloramphenicol medium at 37°C overnight as preculture for expression. 10 ml of preculture was inoculated into 1 l of Luria-Bertani medium supplemented with ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml) and was grown at 37°C until an optical density (OD600) of 0.6–0.7 was reached. 0.5–1 mM of IPTG (isopropyl β-D-thiogalactopyranoside) was added to induce protein expression. The induction was carried out at 37°C for 3–4 hr. The cells were collected by centrifugation, frozen in liquid nitrogen, and stored at −80°C.
Processing of the Cell Pellet and Purification of Recombinant Proteins
E. coli cell pellets were thawed on ice and resuspended in lysis buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 5% glycerol, 0.5% Triton X-100, and 20 mM imidazole. Cells were lysed using a Sonoplus HD3100 sonicator (Bandelin Sonorex Biotechnique) with the HD3100 probe. Cell debris was removed by centrifugation at 10,000 × g for 25 min at 4°C in a Beckman Coulter Avanti J-26 XP with JA 25.50 rotor. Membrane-associated proteins were removed from the cell-free lysate by ultracentrifugation at 150,000 × g for 45 min at 4°C in an Optima MAX-XP ultracentrifuge (rotor MLA55; Beckman Coulter). The soluble supernatant was collected for further analysis.
S. acidocaldarius cells expressing recombinant full-length FlaF (C-terminal His6-StrepII tagged) were lysed in 50 mM Tris-HCl (pH 8), and 150 mM NaCl buffer using Sonoplus HD3100 sonicator (Bandelin Sonorex Biotechnique) with an HD3100 probe and cell debris was removed by centrifugation (10,000 × g for 30 min in a Beckman Coulter Avanti J-26 XP with JA 25.50 rotor). To obtain the membrane-associated recombinant FlaF, the cell-free lysate was further ultracentrifuged at 150,000 × g for 45 min at 4°C in an Optima MAX-XP ultracentrifuge (Rotor MLA55; Beckman Coulter). The membrane fraction was collected and resuspended in 50 mM Tris-HCl (pH 8), 150 mM NaCl, 5% glycerol, 20 mM imidazole, and 2% (w/v) n-dodecyl-β-D-maltoside (DDM) followed by an incubation at 50°C for 1–2 hr for solubilization of the membrane proteins.
To purify His6-tagged (heterologous) or His6-StrepII-tagged (homologous) overexpressed FlaF protein, Ni-NTA affinity chromatography was used. Either soluble lysate or the solubilized membrane fraction was loaded onto an IMAC, Ni-NTA affinity column. The protein was eluted using a step gradient of imidazole from10 to 500 mM in 50 mM Tris-HCl (pH 8) and 150 mM NaCl. 0.05% (w/v) DDM was added to all buffers for the purification of FlaF from the solubilized membrane fraction. The eluted protein was desalted using a Profinia-desalting cartridge (Bio-Rad) and concentrated. The purity of the protein was assessed on Coomassie-stained SDS-PAGE and α-His western blot analysis.
To ensure the purity of heterologous overexpressed N-terminal His6-tagged FlaF, the concentrated protein was further loaded on to a Superose 12 10/300 GL gel-filtration column on an ÄKTA purifier system (GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 8) and 150 mM NaCl buffer. Elution fractions from several independent experiments were collected and concentrated using an Amicon filter (MWCO 10 kDa). The purity was monitored on SDS-PAGE and α-His western blot analysis. 95% pure protein was frozen in liquid N2 and stored at −80°C until further use.
Crystallization, Data Collection, and Structure Determination
sFlaF protein (residues 35–164 with an N-terminal His-tag [MGSSHHHHHHSQDPNS]), 6 mg/ml in 50 mM Tris (pH 8.0), 150 mM NaCl, and 3% glycerol, was mixed with an equal volume of the reservoir using the hanging drop or sitting drop vapor diffusion method. sFlaF was crystallized in three different conditions. For sFlaF native 1, the reservoir contained 3 M potassium chloride and 50 mM HEPES (pH 7.0); for sFlaF native 2, the reservoir contained 35% Tacsimate (pH 7.0); and for sFlaF derivative, protein was co-crystallized with 2 mM potassium tetrachloroplatinate(II) hydrate and the reservoir contained 3 M sodium acetate (pH 7.0). The crystals grew in 2–3 days at 15°C in all the conditions. Native 1 and derivative crystals were transferred to a reservoir including 15% glycerol as a cryoprotectant; native 2 crystals were transferred to a reservoir including 30% sucrose as a cryoprotectant for about 30 s and then flash cooled in liquid nitrogen for data collection.
The X-ray diffraction data for sFlaF were collected at the SIBYLS beamline 12.3.1 at Advanced Light Source (ALS) (Classen et al., 2013). Both sFlaF native 1 and derivative datasets were integrated and scaled using the HKL2000 program package (Otwinowski and Minor, 1997). sFlaF native 2 was integrated using XDS (Kabsch, 2010) and scaled using AIMLESS (Evans and Murshudov, 2013) in the CCP4 suite (Winn et al., 2011). The dataset for the sFlaF derivative with K2PtCl4 was used for SIRAS phasing. The native 1 structure of sFlaF was solved using the phenix.autosol program in the Phenix suite (Adams et al., 2010). The sFlaF native 2 structure was solved by molecular replacement using the native 1 sFlaF structure as a search model using Phaser in the Phenix suite. Both sFlaF native 1 and 2 structures were fitted manually using Coot (Emsley et al., 2010) and refined using the Phenix refine program. Rwork/Rfree were refined to 0.158/0.196 and 0.173/0.200 for the native 1 and native 2 sFlaF structures, respectively (Table 1). The His-tag was not visible in the electron density map in both native structures. Molecular graphics and analyses were performed with the UCSF Chimera package (Pettersen et al., 2004).
Surface Layer Protein Isolation
S-layer proteins were isolated using the protocol described by Meyer et al. (2011). Briefly, fresh culture was grown until the stationary phase, and the cell pellet was collected by centrifugation at 4,600 rpm for 20 min. Cell pellets were resuspended in 40 ml of buffer A, containing 10 mM NaCl and 0.4% sodium lauroylsarcosine. Resuspended cell suspension was incubated at 42°C for 2 hr under shaking conditions and a brownish pellet was obtained using a centrifugation step at 21,000 × g on an Optima Max-XU Ultracentrifuge (Beckman Coulter) for 50 min at 4°C. The upper white layer of the pellet was carefully resuspended in 1.5 ml of buffer A and incubated at 42°C for 20 min. Pure S-layer protein was isolated from the pellet fraction using a centrifugation step with maximum speed at 13,000 × g for 10 min in a table-top centrifuge. To ensure the purity, the S-layer protein was repeatedly washed with buffer B (10 mM NaCl, 0.5 mM MgSO4, and 0.5% SDS) and the white fluffy translucent pellet of pure S-layer protein was kept in water at 4°C until further use.
Dot Far-Western Blot Analysis
Dot blot far-western analysis was performed as described (Chan et al., 2008). Dots of varying concentrations of S-layer proteins were spotted onto a charged PVDF membrane (5–10 μl, equal concentration) and kept at either room temperature or 37°C until all the spots were dried. Immediately after a short washing step using PBST (1× PBS with 0.1% Tween 20), the membrane was incubated with ∼0.1 mg/ml His6-sFlaF in its native buffer (50 mM Tris-HCl [pH 8] and 150 mM NaCl). Bound sFlaF was chemiluminescently detected by a α-His mouseHRP antibody.
Sedimentation Assay and S. acidocaldarius Motility Assay
S-layer proteins are present in a paracrystalline form. To test the interaction between sFlaF and S-layer, freshly purified S-layer was mixed with either pure sFlaF as an experimental set or with pure FlaI, a soluble dual function ATPase (Reindl et al., 2013) as a negative control. Proteins were mixed in 20 mM Tris-HCl (pH 8) and 150 mM NaCl buffer for 2 hr at room temperature. After the incubation step, the reaction mixtures were centrifuged in a table-top centrifuge at 13,000 × g for 10 min. The supernatant (S) and pellet (P) fractions were collected and analyzed on SDS-PAGE to check binding with the S-layer. Swimming assays on semi-solid plates were performed following the protocols described by Lassak et al. (2012b).
Authors Contributions
A.B. and P.T. purified the protein, performed SEC, functional studies and designed the experiments, wrote the manuscript and created the figures. C.L.T. crystallized the FlaF native protein, collected crystal diffraction data, wrote the paper and created the figures. A.S.A. helped to solve the native FlaF crystal structure. P.C. prepared the constructs, performed motility assays, the sedimentation assay, and purified the proteins. J.P.I. co-crystallized the FlaF with and without heavy metal. J.A.T. and S.V.A. wrote and revised the manuscript and designed the experiments.
Acknowledgments
A.B., P.T., P.C., and S.V.A. were supported by intramural funds from the Max Planck Society and an ERC starting grant, ARCHAELLUM (no. 311523). We thank the Lawrence Berkeley National Laboratory Advanced Light Source and SIBYLS beamline staff, Scott Classen and Greg Hura, at 12.3.1 for assistance. This study was supported in part by NIH grant AI22160 to J.A.T. The SIBYLS beamline (BL12.3.1) is supported by the United States Department of Energy program (IDAT) and by NIH MINOS grant GM105404.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
John A. Tainer, Email: jatainer@lbl.gov.
Sonja-Verena Albers, Email: sonja.albers@biologie.uni-freiburg.de.
Accession Numbers
The native crystal structures of recombinant sFlaF have been deposited in the PDB under codes 4ZBH and 4P94.
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Oesterhelt D. Morphology, function and isolation of halobacterial flagella. J. Mol. Biol. 1984;176:459–475. doi: 10.1016/0022-2836(84)90172-4. [DOI] [PubMed] [Google Scholar]
- Albers S.V., Meyer B.H. The archaeal cell envelope. Nat. Rev. Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Albers S.V., Szabo Z., Driessen A.J. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 2003;185:3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Ghosh A., Mills D.J., Kahnt J., Vonck J., Albers S.V. FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI. J. Biol. Chem. 2012;287:43322–43330. doi: 10.1074/jbc.M112.414383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Neiner T., Tripp P., Albers S.V. Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J. 2013;280:6141–6149. doi: 10.1111/febs.12534. [DOI] [PubMed] [Google Scholar]
- Baranova E., Fronzes R., Garcia-Pino A., Van Gerven N., Papapostolou D., Pehau-Arnaudet G., Pardon E., Steyaert J., Howorka S., Remaut H. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature. 2012;487:119–122. doi: 10.1038/nature11155. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Wildhaber I., Phipps B.M. Principles of organization in eubacterial and archaebacterial surface proteins. Can. J. Microbiol. 1989;35:215–227. doi: 10.1139/m89-034. [DOI] [PubMed] [Google Scholar]
- Bayley D.P., Florian V., Klein A., Jarrell K.F. Flagellin genes of Methanococcus vannielii: amplification by the polymerase chain reaction, demonstration of signal peptides and identification of major components of the flagellar filament. Mol. Gen. Genet. 1998;258:639–645. doi: 10.1007/s004380050777. [DOI] [PubMed] [Google Scholar]
- Berg H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Burrows L.L. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- Chaban B., Ng S.Y., Kanbe M., Saltzman I., Nimmo G., Aizawa S., Jarrell K.F. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Chan C.S., Winstone T.M., Turner R.J. Investigating protein-protein interactions by far-Westerns. Adv. Biochem. Eng. Biotechnol. 2008;110:195–214. doi: 10.1007/10_2007_090. [DOI] [PubMed] [Google Scholar]
- Cisneros D.A., Bond P.J., Pugsley A.P., Campos M., Francetic O. Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 2012;31:1041–1053. doi: 10.1038/emboj.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros D.A., Pehau-Arnaudet G., Francetic O. Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol. Microbiol. 2012;86:805–818. doi: 10.1111/mmi.12033. [DOI] [PubMed] [Google Scholar]
- Classen S., Hura G.L., Holton J.M., Rambo R.P., Rodic I., McGuire P.J., Dyer K., Hammel M., Meigs G., Frankel K.A. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L., Taylor R.K., Pique M.E., Adair B.D., Arvai A.S., Singh M., Lloyd S.J., Shin D.S., Getzoff E.D., Yeager M. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- Craig L., Pique M.E., Tainer J.A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Desmond E., Brochier-Armanet C., Gribaldo S. Phylogenomics of the archaeal flagellum: rare horizontal gene transfer in a unique motility structure. BMC Evol. Biol. 2007;7:106. doi: 10.1186/1471-2148-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzoff E.D., Tainer J.A., Lerner R.A., Geysen H.M. The chemistry and mechanism of antibody binding to protein antigens. Adv. Immunol. 1988;43:1–98. doi: 10.1016/s0065-2776(08)60363-6. [DOI] [PubMed] [Google Scholar]
- Getzoff E.D., Tainer J.A., Stempien M.M., Bell G.I., Hallewell R.A. Evolution of CuZn superoxide dismutase and the Greek key beta-barrel structural motif. Proteins. 1989;5:322–336. doi: 10.1002/prot.340050408. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Albers S.V. Assembly and function of the archaeal flagellum. Biochem. Soc. T. 2011;39:64–69. doi: 10.1042/BST0390064. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Hartung S., van der Does C., Tainer J.A., Albers S.V. Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem. J. 2011;437:43–52. doi: 10.1042/BJ20110410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner C.L., Nguyen Y., Burrows L.L. Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. R. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton H.L., Dillard J.P. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 2006;59:376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- Hartung S., Arvai A.S., Wood T., Kolappan S., Shin D.S., Craig L., Tainer J.A. Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J. Biol. Chem. 2011;286:44254–44265. doi: 10.1074/jbc.M111.297242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henche A.L., Ghosh A., Yu X., Jeske T., Egelman E., Albers S.V. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ. Microbiol. 2012;14:3188–3202. doi: 10.1111/j.1462-2920.2012.02898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura G.L., Menon A.L., Hammel M., Rambo R.P., Poole F.L., Tsutakawa S.E., Jenney F.E., Classen S., Frankel K.A., Hopkins R.C. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–U683. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura G.L., Budworth H., Dyer K.N., Rambo R.P., Hammel M., McMurray C.T., Tainer J.A. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat. Methods. 2013;10:453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K.F., Albers S.V. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Johnson T., Sikora A., Zielke R., Sandkvist M. Fluorescence microscopy and proteomics to investigate subcellular localization, assembly, and function of the type II secretion system. Methods Mol. Biol. 2013;966:157–172. doi: 10.1007/978-1-62703-245-2_10. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Imada K., Sakuma M., Sudo Y., Kojima C., Minamino T., Homma M., Namba K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009;73:710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- Korotkov K.V., Sandkvist M., Hol W.G. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lassak K., Ghosh A., Albers S.V. Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res. Microbiol. 2012;163:630–644. doi: 10.1016/j.resmic.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Lassak K., Neiner T., Ghosh A., Klingl A., Wirth R., Albers S.V. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 2012;83:110–124. doi: 10.1111/j.1365-2958.2011.07916.x. [DOI] [PubMed] [Google Scholar]
- Lu C., Korotkov K.V., Hol W.G.J. Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL. J. Struct. Biol. 2014;187:223–235. doi: 10.1016/j.jsb.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwan W., Alam M., Oesterhelt D. Rotation and switching of the flagellar motor assembly in Halobacterium halobium. J. Bacteriol. 1991;173:1971–1977. doi: 10.1128/jb.173.6.1971-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin L.S., Haft R.J.F., Forest K.T. Structural insights into the type II secretion nanomachine. Curr. Opin. Struct. Biol. 2012;22:208–216. doi: 10.1016/j.sbi.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville S., Craig L. Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. R. 2013;77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B.H., Albers S.V. Hot and sweet: protein glycosylation in Crenarchaeota. Biochem. Soc. Trans. 2013;41:384–392. doi: 10.1042/BST20120296. [DOI] [PubMed] [Google Scholar]
- Meyer B.H., Zolghadr B., Peyfoon E., Pabst M., Panico M., Morris H.R., Haslam S.M., Messner P., Schaffer C., Dell A. Sulfoquinovose synthase - an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 2011;82:1150–1163. doi: 10.1111/j.1365-2958.2011.07875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B.H., Peyfoon E., Dietrich C., Hitchen P., Panico M., Morris H.R., Dell A., Albers S.-V. Agl16, a thermophilic glycosyltransferase mediating the last step of N-Glycan biosynthesis in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bacteriol. 2013;195:2177–2186. doi: 10.1128/JB.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill J., Xie M., Hijnen M., Roujeinikova A. Role of the MotB linker in the assembly and activation of the bacterial flagellar motor. Acta Crystallogr. D Biol. Crystallogr. 2011;67:1009–1016. doi: 10.1107/S0907444911041102. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parge H.E., Forest K.T., Hickey M.J., Christensen D.A., Getzoff E.D., Tainer J.A. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- Perry J.J.P., Shin D.S., Getzoff E.D., Tainer J.A. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta. 2010;1804:245–262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rambo R.P., Tainer J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul C.F., Andrews D.A., Nahar M.F., Buckle A.M., Roujeinikova A. Crystallographic and molecular dynamics analysis of loop motions unmasking the peptidoglycan-binding site in stator protein MotB of flagellar motor. PLoS One. 2011;6:e18981. doi: 10.1371/journal.pone.0018981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl S., Ghosh A., Williams G.J., Lassak K., Neiner T., Henche A.L., Albers S.V., Tainer J.A. Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol. Cell. 2013;49:1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sára M., Sleytr U.B. S-layer proteins. J. Bacteriol. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesner M., Miller A., Streif S., Staudinger W.F., Muller J., Scheffer B., Siedler F., Oesterhelt D. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 2009;9:56. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahapure R., Driessen R.P.C., Haurat M.F., Albers S.-V., Dame R.T. The archaellum: a rotating type IV pilus. Mol. Microbiol. 2014;91:716–723. doi: 10.1111/mmi.12486. [DOI] [PubMed] [Google Scholar]
- Sleytr U.B., Beveridge T.J. Bacterial S-layers. Trends Microbiol. 1999;7:253–260. doi: 10.1016/s0966-842x(99)01513-9. [DOI] [PubMed] [Google Scholar]
- Spang A., Poehlein A., Offre P., Zumbragel S., Haider S., Rychlik N., Nowka B., Schmeisser C., Lebedeva E.V., Rattei T. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 2012;14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- Stivala A., Wybrow M., Wirth A., Whisstock J.C., Stuckey P.J. Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics. 2011;27:3315–3316. doi: 10.1093/bioinformatics/btr575. [DOI] [PubMed] [Google Scholar]
- Streif S., Staudinger W.F., Marwan W., Oesterhelt D. Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J. Mol. Biol. 2008;384:1–8. doi: 10.1016/j.jmb.2008.08.057. [DOI] [PubMed] [Google Scholar]
- Szabo Z., Stahl A.O., Albers S.V., Kissinger J.C., Driessen A.J., Pohlschroder M. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 2007;189:772–778. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhar H.K., Kemp K., Kim M., Howell P.L., Burrows L.L. The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 2013;288:9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith A., Klingl A., Zolghadr B., Lauber K., Mentele R., Lottspeich F., Rachel R., Albers S.V., Kletzin A. Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol. Microbiol. 2009;73:58–72. doi: 10.1111/j.1365-2958.2009.06746.x. [DOI] [PubMed] [Google Scholar]
- Wagner M., van Wolferen M., Wagner A., Lassak K., Meyer B.H., Reimann J., Albers S.V. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 2012;3:214. doi: 10.3389/fmicb.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.