Abstract
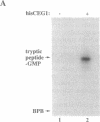
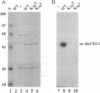
Nascent mRNA chains are capped at the 5' end by the addition of a guanylyl residue to form a G(5')ppp(5')N ... structure. During the capping reaction, the guanylyltransferase (GTP:mRNA guanylyltransferase, EC 2.7.7.50) is reversibly and covalently guanylylated. In this enzyme-GMP (E-GMP) intermediate, GMP is linked to the epsilon-amino group of a lysine residue via a phosphoamide bond. Lys-70 was identified as the GMP attachment site of the Saccharomyces cerevisiae guanylyltransferase (encoded by the CEG1 gene) by guanylylpeptide sequencing. CEG1 genes with substitutions at Lys-70 were unable to support viability in yeast and produced proteins that were not guanylylated in vitro. The CEG1 active site exhibits sequence similarity to the active sites of viral guanylyltransferases and polynucleotide ligases, suggesting similarity in the mechanisms of nucleotidyl transfer catalyzed by these enzymes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J., Brown R. S., Tsugita A. Primary structure and genetic organization of phage T4 DNA ligase. Nucleic Acids Res. 1983 Oct 25;11(20):7145–7156. doi: 10.1093/nar/11.20.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. Molecular characterisation of the DNA ligase gene, CDC17, from the fission yeast Schizosaccharomyces pombe. Eur J Biochem. 1987 Feb 2;162(3):659–667. doi: 10.1111/j.1432-1033.1987.tb10688.x. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. E., Johnston L. H., Kodama K., Tomkinson A. E., Lasko D. D., Lindahl T. Human DNA ligase I cDNA: cloning and functional expression in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. R., Zarbl H., Millward S. Reovirus guanylyltransferase is L2 gene product lambda 2. J Virol. 1986 Oct;60(1):307–311. doi: 10.1128/jvi.60.1.307-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong P., Shuman S. Covalent catalysis in nucleotidyl transfer. A KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J Biol Chem. 1993 Apr 5;268(10):7256–7260. [PubMed] [Google Scholar]
- Dadd C. A., Cook R. G., Allis C. D. Fractionation of small tryptic phosphopeptides by alkaline PAGE followed by amino acid sequencing. Biotechniques. 1993 Feb;14(2):266–273. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol. 1981 Jun 5;148(4):303–330. doi: 10.1016/0022-2836(81)90178-9. [DOI] [PubMed] [Google Scholar]
- Ensinger M. J., Moss B. Modification of the 5' terminus of mRNA by an RNA (guanine-7-)-methyltransferase from HeLa cells. J Biol Chem. 1976 Sep 10;251(17):5283–5291. [PubMed] [Google Scholar]
- Fausnaugh J., Shatkin A. J. Active site localization in a viral mRNA capping enzyme. J Biol Chem. 1990 May 5;265(13):7669–7672. [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A. J. Characterization of cap structures. Methods Enzymol. 1989;180:164–176. doi: 10.1016/0076-6879(89)80100-4. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Hagler J., Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992 Feb 21;255(5047):983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- Hammond J. M., Kerr S. M., Smith G. L., Dixon L. K. An African swine fever virus gene with homology to DNA ligases. Nucleic Acids Res. 1992 Jun 11;20(11):2667–2671. doi: 10.1093/nar/20.11.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy S., Singh M., Gait M. J. Effect of single amino acid changes in the region of the adenylylation site of T4 RNA ligase. Biochemistry. 1987 Mar 24;26(6):1688–1696. doi: 10.1021/bi00380a030. [DOI] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Tsunasawa S., Sakiyama F., Nakata A. Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol Gen Genet. 1986 Jul;204(1):1–7. doi: 10.1007/BF00330179. [DOI] [PubMed] [Google Scholar]
- Itoh N., Yamada H., Kaziro Y., Mizumoto K. Messenger RNA guanylyltransferase from Saccharomyces cerevisiae. Large scale purification, subunit functions, and subcellular localization. J Biol Chem. 1987 Feb 15;262(5):1989–1995. [PubMed] [Google Scholar]
- Kaliman A. V., Zimin A. A., Nazipova N. N., Kriukov V. M., Taniashin V. I. Sravnitel'nyi analiz genov DNK-ligaz fagov T6 i T4. Dokl Akad Nauk SSSR. 1988;299(3):737–742. [PubMed] [Google Scholar]
- Kletzin A. Molecular characterisation of a DNA ligase gene of the extremely thermophilic archaeon Desulfurolobus ambivalens shows close phylogenetic relationship to eukaryotic ligases. Nucleic Acids Res. 1992 Oct 25;20(20):5389–5396. doi: 10.1093/nar/20.20.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama K., Barnes D. E., Lindahl T. In vitro mutagenesis and functional expression in Escherichia coli of a cDNA encoding the catalytic domain of human DNA ligase I. Nucleic Acids Res. 1991 Nov 25;19(22):6093–6099. doi: 10.1093/nar/19.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Gorbalenya A. E. Related domains in yeast tRNA ligase, bacteriophage T4 polynucleotide kinase and RNA ligase, and mammalian myelin 2',3'-cyclic nucleotide phosphohydrolase revealed by amino acid sequence comparison. FEBS Lett. 1990 Jul 30;268(1):231–234. doi: 10.1016/0014-5793(90)81015-g. [DOI] [PubMed] [Google Scholar]
- Langberg S. R., Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2'-)-methyltransferases from HeLa cells. J Biol Chem. 1981 Oct 10;256(19):10054–10060. [PubMed] [Google Scholar]
- Lauer G., Rudd E. A., McKay D. L., Ally A., Ally D., Backman K. C. Cloning, nucleotide sequence, and engineered expression of Thermus thermophilus DNA ligase, a homolog of Escherichia coli DNA ligase. J Bacteriol. 1991 Aug;173(16):5047–5053. doi: 10.1128/jb.173.16.5047-5053.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blois H., French T., Mertens P. P., Burroughs J. N., Roy P. The expressed VP4 protein of bluetongue virus binds GTP and is the candidate guanylyl transferase of the virus. Virology. 1992 Aug;189(2):757–761. doi: 10.1016/0042-6822(92)90600-t. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Barnes D. E. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- Mao Z. X., Joklik W. K. Isolation and enzymatic characterization of protein lambda 2, the reovirus guanylyltransferase. Virology. 1991 Nov;185(1):377–386. doi: 10.1016/0042-6822(91)90785-a. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kaziro Y., Lipmann F. Reaction mechanism of mRNA guanylyltransferase from rat liver: isolation and characterization of a guanylyl-enzyme intermediate. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1693–1697. doi: 10.1073/pnas.79.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Kaziro Y. Messenger RNA capping enzymes from eukaryotic cells. Prog Nucleic Acid Res Mol Biol. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Lipmann F. Transmethylation and transguanylylation in 5'-RNA capping system isolated from rat liver nuclei. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4961–4965. doi: 10.1073/pnas.76.10.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles E. G., Christen L. Identification of the vaccinia virus mRNA guanyltransferase active site lysine. J Biol Chem. 1993 Nov 25;268(33):24986–24989. [PubMed] [Google Scholar]
- Niles E. G., Condit R. C., Caro P., Davidson K., Matusick L., Seto J. Nucleotide sequence and genetic map of the 16-kb vaccinia virus HindIII D fragment. Virology. 1986 Aug;153(1):96–112. doi: 10.1016/0042-6822(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Pena L., Yáez R. J., Revilla Y., Viñuela E., Salas M. L. African swine fever virus guanylyltransferase. Virology. 1993 Mar;193(1):319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- Pizarro J. L., Sandino A. M., Pizarro J. M., Fernández J., Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991 Feb;72(Pt 2):325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- Rand K. N., Gait M. J. Sequence and cloning of bacteriophage T4 gene 63 encoding RNA ligase and tail fibre attachment activities. EMBO J. 1984 Feb;3(2):397–402. doi: 10.1002/j.1460-2075.1984.tb01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen E. B., Lis J. T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Hurwitz J. RNA capping by the vaccinia virus guanylyltransferase. Structure of enzyme-guanylate intermediate. J Biol Chem. 1984 Nov 10;259(21):13488–13494. [PubMed] [Google Scholar]
- Schmitt M. P., Beck P. J., Kearney C. A., Spence J. L., DiGiovanni D., Condreay J. P., Molineux I. J. Sequence of a conditionally essential region of bacteriophage T3, including the primary origin of DNA replication. J Mol Biol. 1987 Feb 5;193(3):479–495. doi: 10.1016/0022-2836(87)90261-0. [DOI] [PubMed] [Google Scholar]
- Seliger L. S., Zheng K., Shatkin A. J. Complete nucleotide sequence of reovirus L2 gene and deduced amino acid sequence of viral mRNA guanylyltransferase. J Biol Chem. 1987 Dec 5;262(34):16289–16293. [PubMed] [Google Scholar]
- Shabarova Z. A. Synthetic nucleotide-peptides. Prog Nucleic Acid Res Mol Biol. 1970;10:145–182. doi: 10.1016/s0079-6603(08)60564-4. [DOI] [PubMed] [Google Scholar]
- Shark K. B., Conway T. Cloning and molecular characterization of the DNA ligase gene (lig) from Zymomonas mobilis. FEMS Microbiol Lett. 1992 Sep 1;75(1):19–26. doi: 10.1016/0378-1097(92)90450-3. [DOI] [PubMed] [Google Scholar]
- Shibagaki Y., Itoh N., Yamada H., Nagata S., Mizumoto K. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992 May 15;267(14):9521–9528. [PubMed] [Google Scholar]
- Shuman S., Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme--guanylate intermediate. Proc Natl Acad Sci U S A. 1981 Jan;78(1):187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Moss B. Purification and use of vaccinia virus messenger RNA capping enzyme. Methods Enzymol. 1990;181:170–180. doi: 10.1016/0076-6879(90)81119-f. [DOI] [PubMed] [Google Scholar]
- Shuman S. RNA capping by HeLa cell RNA guanylyltransferase. Characterization of a covalent protein-guanylate intermediate. J Biol Chem. 1982 Jun 25;257(12):7237–7245. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. L., Chan Y. S., Kerr S. M. Transcriptional mapping and nucleotide sequence of a vaccinia virus gene encoding a polypeptide with extensive homology to DNA ligases. Nucleic Acids Res. 1989 Nov 25;17(22):9051–9062. doi: 10.1093/nar/17.22.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thøgersen H. C., Morris H. R., Rand K. N., Gait M. J. Location of the adenylylation site in T4 RNA ligase. Eur J Biochem. 1985 Mar 1;147(2):325–329. doi: 10.1111/j.1432-1033.1985.tb08753.x. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Totty N. F., Ginsburg M., Lindahl T. Location of the active site for enzyme-adenylate formation in DNA ligases. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):400–404. doi: 10.1073/pnas.88.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama R., Mizumoto K., Nakahara Y., Tatsuno T., Kaziro Y. Mechanism of the mRNA guanylyltransferase reaction: isolation of N epsilon-phospholysine and GMP (5' leads to N epsilon) lysine from the guanylyl-enzyme intermediate. EMBO J. 1983;2(12):2195–2201. doi: 10.1002/j.1460-2075.1983.tb01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C., Stuart D., McFadden G. Identification and DNA sequence of the large subunit of the capping enzyme from Shope fibroma virus. Virology. 1991 Aug;183(2):773–777. doi: 10.1016/0042-6822(91)91009-6. [DOI] [PubMed] [Google Scholar]
- Venkatesan S., Moss B. Eukaryotic mRNA capping enzyme-guanylate covalent intermediate. Proc Natl Acad Sci U S A. 1982 Jan;79(2):340–344. doi: 10.1073/pnas.79.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Shatkin A. J. Synthesis of Gp4N and Gp3N compounds by guanylyltransferase purified from yeast. Nucleic Acids Res. 1984 Mar 12;12(5):2303–2315. doi: 10.1093/nar/12.5.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway S. K., Phizicky E. M., Abelson J. Structure and function of the yeast tRNA ligase gene. J Biol Chem. 1988 Mar 5;263(7):3171–3176. [PubMed] [Google Scholar]
- Xu Q., Teplow D., Lee T. D., Abelson J. Domain structure in yeast tRNA ligase. Biochemistry. 1990 Jul 3;29(26):6132–6138. doi: 10.1021/bi00478a004. [DOI] [PubMed] [Google Scholar]
- Yáez R. J., Viñuela E. African swine fever virus encodes a DNA ligase. Virology. 1993 Mar;193(1):531–536. doi: 10.1006/viro.1993.1161. [DOI] [PubMed] [Google Scholar]