Abstract
Cell wall formation and maintenance are crucial for hyphal morphogenesis. In many filamentous fungi, chitin is one of the main structural components of the cell wall. Aspergillus nidulans ChsB, a chitin synthase, and CsmA, a chitin synthase with a myosin motor-like domain (MMD) at its N-terminus, both localize predominantly at the hyphal tip regions and at forming septa. ChsB and CsmA play crucial roles in polarized hyphal growth in A. nidulans. In this study, we investigated the mechanism by which CsmA and ChsB accumulate at the hyphal tip in living hyphae. Deletion of kinA, a gene encoding conventional kinesin (kinesin-1), impaired the localization of GFP-CsmA and GFP-ChsB at the hyphal tips. The transport frequency of GFP-CsmA and GFP-ChsB in both anterograde and retrograde direction appeared lower in the kinA-deletion strain compared to wild type, although the velocities of the movements were comparable. Co-localization of GFP-ChsB and GFP-CsmA with mRFP1-KinArigor, a KinA mutant that binds to microtubules but does not move along them, was observed in the posterior of the hyphal tip regions. KinA co-immunoprecipitated with ChsB and CsmA. Co-localization and association of CsmA with KinA did not depend on the MMD. These findings indicate that ChsB and CsmA are transported along microtubules to the subapical region by KinA.
Introduction
Hyphae of filamentous fungi represent extremely polarized cells. Since “polarized growth” is a general growth mechanism common in the elongation of cells as distinct as neurons or pollen tubes, as well as in the differentiation of embryos [1], the mechanism of hyphal tip growth in filamentous fungi may provide a helpful basic model. This mechanism is thought to be highly elaborate and is supported by the continuous delivery of vesicles containing proteins required for polarized growth to the hyphal tips [2,3]. In some filamentous fungi, vesicles delivered to hyphal tips accumulate at apices prior to fusion with the membrane. These clusters of vesicles are called Spitzenkörper or apical bodies [4]. The Spitzenkörper is thought to act as a vesicle supply center, the place where cargo for the hyphal apex is sorted [5]. In the ascomycete filamentous fungus Aspergillus nidulans, some proteins involved in polarized growth are transported along microtubules and the actin cytoskeleton to the hyphal tips [6–9]. While there are 5~8 linear microtubules that extend to the hyphal tips in A. nidulans, actin cables are formed preferentially at the apical hyphal regions. The vesicles containing components of the tip growth machineries are thought to be transported along microtubules from posterior parts of hyphal tip regions to their apical regions, transferred to actin cables and delivered to the apical cortices of hyphae [10–12].
The cell surface of filamentous fungi is covered with a cell wall that consists mainly of chitin and glucan. Chitin is a β-1,4-linked homopolymer of N-acetylglucosamine and its synthesis is important for hyphal morphogenesis [13]. To date, fungal chitin synthases have been classified into seven classes and three divisions according to their structural characteristics [14]. In this report, we follow the classification proposed by Chigira et al. and Choquer et al. [15,16]. Fungal chitin synthases are membrane-bound proteins containing several transmembrane regions. Class I, II, and III chitin synthases belong to division I and class IV, V, and VI belong to division II. We previously isolated six chitin synthase genes from A. nidulans: chsA, chsB, chsC, chsD, csmA, and csmB; the products of these genes belong to classes II, III, I, IV, V, and VI, respectively [17–21](S1 Fig). The class V and VI chitin synthases, CsmA and CsmB, respectively, consist of a C-terminal chitin synthase domain (CSD) containing the amino acid sequences conserved in chitin synthases of other classes [22], and an N-terminal myosin motor-like domain (MMD). Myosins are motor proteins that move along actin filaments. In our previous studies, both csmA- and csmB-deletion mutations caused similar phenotypic abnormalities (e.g., formation of balloons and intrahyphal hyphae, and hyphal lysis especially under low osmotic conditions) [20,23], and these mutations were synthetically lethal [20]. The chsB-deletion mutant formed extremely small colonies and did not form conidiospores [24]. CsmA, CsmB, and ChsB localized near actin structures at the hyphal tips and at forming septa [20,24,25]. The MMDs of CsmA and CsmB bound to actin filaments in vitro [20,25] and the interaction between each MMD and actin was crucial for their proper localization and function [25,26]. However, the MMD of CsmB is not functionally equivalent to that of CsmA [27].
The orthologous genes encoding classes III, V, and VI chitin synthases have been isolated from filamentous fungi and some dimorphic yeasts, and their functions have been investigated [28–39]. The results obtained from these investigations indicate that the chitin synthases belonging to classes III, V, and VI play important roles in hyphal tip growth, maintenance of cell wall integrity, and pathogenicity.
Since the genomes of the yeasts Saccharomyces cerevisiae, Schizosaccharomyces pombe, and a dimorphic yeast Candida albicans do not possess classes III, V, and VI chitin synthases, the chitin synthases in these classes likely play critical roles in polarized growth, especially in filamentous fungi. In order to understand the dynamic aspects of polarized hyphal growth, it is necessary to clarify their transport and localization mechanisms in the hyphae.
Kinesins are motor proteins that move on microtubules in the plus end direction and are divided into 15 families according to their structural properties [40]. In these families, kinesin-1, kinesin-3, and kinesin-7 are involved in polarized growth of filamentous fungi [41]. In A. nidulans, there are 11 kinesin-encoding genes [42]. Among these genes, kinA, uncA, and kipA encode kinesins belonging to kinesin-1, kinesin-3, and kinesin-7, respectively. Kinesin-1 is suggested to function in the transport of secretory vesicles, dyneins, and dynactin, a microtubule minus end-directed motor and its regulator, while Kinesin-3 is suggested to be involved in the transport of secretory vesicles, early endosomes, peroxisomes, and mRNPs containing mRNA [11,43–53]. Zhang et al. [53] and Yao et al. [51] showed that dynein and dynactin localize along microtubules in the ΔkinA mutant, which provide good support for the idea that KinA transports dynein/dynactin along microtubules to the plus ends.
Although the localization of chitin synthases has already been investigated in some filamentous fungi, their localization mechanisms remain largely unknown. In N. crassa chitin synthases are thought to be transported on special vesicles called chitosomes [54,55]. In basidiomycete dimorphic yeast Ustilago maydis, it was reported that the proper localization of class V chitin synthase Msc1 depends on the presence of microtubules and/or actins, that its transport along microtubules does not depend on its MMD, and that the MMD is required for the tethering of Mcs1 at the hyphal tips to increase the efficiency of its exocytosis [36,50]. In this study, we report the transporting mechanisms of classes III and V chitin synthases, ChsB and CsmA, to hyphal tips in A. nidulans and the role of the MMD of CsmA in the transport process.
Materials and Methods
Strains, media, and bacterial and fungal transformations
The A. nidulans strains used in this study are listed in Table 1. Complete medium, YGuu medium (0.5% yeast extract, 1% glucose, 0.1% trace elements, uracil at 1.12 mg/ml, and uridine at 2.44 mg/ml) and minimal medium (MMGuu), minimal medium containing 2% glycerol (MMGlyuu), or minimal medium containing 100 mM threonine and 0.1% fructose instead of glucose (MMTFuu) for A. nidulans were used [56,57]. YGuu, MMGuu, and MMTFuu plates contained 1.5% agar. MMGuu and MMTFuu were supplemented with arginine at 0.2 μg/ml, biotin at 0.02 μg/ml, p-aminobenzoic acid at 1 μg/ml, and pyridoxine at 0.5 μg/ml, when necessary. Bacterial and fungal transformations were performed as described previously [57–59].
Table 1. A. nidulans strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| ABPU1 | biA1 pyrG89 argB2 pyroA4 wA3 | [19] |
| A1149 | pyrG89 pyroA4 ΔnkuA::argB | FGSC* |
| CsmA-HA/FLAG-ChsB1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::csmA-9xHA-pyrG ΔargB::argB-chsB(p)-3xFLAG-chsB | This study |
| EGFP-CsmA1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-egfp-csmA | This study |
| EB-5 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr4-alcA(p)-chsB ΔargB::argB-chsB(p)-egfp-chsB | [24] |
| SNR1 | ΔargB::trpΔB ΔkinA::pyr4 pyrG89 pyroA4 yA2 | [45] |
| SNZ9 | pyrG89 argB2 ΔnkuA::argB pyroA4 ΔuncA::pyroA | [48] |
| EGFP-CsmAΔkinA1, 2 | ΔargB::trpCΔB ΔkinA::pyr4 pyrG89 pyroA4 yA2 ΔcsmA::argB-alcA(p)-egfp-csmA | This study |
| EGFP-CsmAΔuncA1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-egfp-csmA ΔuncA::pyroA | This study |
| EGFP-ChsBΔkinA1, 2 | ΔargB::trpCΔB ΔkinA::pyr4 pyrG89 pyroA4 yA2 ΔargB::argB-chsB(p)-egfp-chsB | This study |
| EGFP-ChsBΔuncA1, 2 | argB2 pyroA4 wA3 ΔargB::argB-chsB(p)-egfp-chsB ΔuncA::pyroA | This study |
| EGFP-CsmA/mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-egfp-csmA ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| EGFP-ChsB/mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr4-alcA(p)-chsB ΔargB::argB-chsB(p)-egfp-chsB ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| ABPU1/mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| CA2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::csmA-9xHA-pyrG | [61] |
| FB-3 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr4-alcA(p)-chsB ΔargB::argB-chsB(p)-3XFLAG-chsB | [24] |
| CsmA-HA/mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::csmA-9HA-pyrG ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| FLAG-ChsB/ mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔchsB::pyr4-alcA(p)-chsB ΔargB::argB-chsB(p)-3XFLAG-chsB ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| EGFP-ΔMA1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-egfp-csmA(Δ1–860) | This study |
| EGFP-ΔMA/ mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-egfp-csmA(Δ1–860) ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| CΔMHA | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-csmA(Δ1–860)-9xHA-pyrG | [25] |
| D10HA | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-csmA(Δ347–356)-9xHA-pyrG | [25] |
| CΔMHA/ mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-csmA(Δ1–860)-9xHA-pyrG ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| D10HA/ mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::argB-alcA(p)-csmA(Δ347–356)-9xHA-pyrG ΔkinA::pyroA-alcA(p)-mrfp-kinA rigor | This study |
| ABPU1/GFP-KinA1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔkinA::pyroA-alcA(p)-gfp-kinA | This study |
| CsmA-HA/GFP-KinA1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔkinA::pyroA-alcA(p)-gfp-kinA ΔcsmA::pyrG-csmA-9HA | This study |
| CsmA-HA/FLAG-ChsB/ mRFP-KinArigor1, 2 | biA1 pyrG89 argB2 pyroA4 wA3 ΔcsmA::csmA-9xHA-pyrG argB::chsB(p)-3xFLAG-chsB ΔkinA::pyroA4-alcA(p)-mrfp-kinA rigor | This study |
*: Fungal Genetics Stock Center, Kansas City, Missouri
Constructions of plasmids and A. nidulans strains
To create the strains that produced CsmA with an EGFP tag at its N-terminus, we constructed the plasmid pMAEC as follows: The 0.8-kb KpnI-PstI fragment of pM-ALC2 [25] was cloned into the KpnI-PstI site of pUC19, yielding pUCCSM2. The 0.7-kb fragment encoding EGFP was amplified by PCR from pEGFP (Clontech) using primers 5′-GCGTTAACATGGTGAGCAAGGGCGAGG-3′ and 5′-GCCCATGGTCTTGTACAGCTCGTCCAT-3′, in which the underlined letters represent HincII and NcoI recognition sites, respectively, digested with HincII and NcoI, and cloned into the NcoI site of pUCCSM2, yielding pUCCSM. The 1.5-kb KpnI-PstI fragment of pUCCSM was cloned into the KpnI-PstI sites of pM-ALC2, yielding pMAEC. The 4.1-kb EcoRI and PstI fragment of pMAEC was used for the transformation of the ABPU1 and SNR1 (ΔkinA) strains (Table 1). Two transformants obtained from each strain, in which the DNA fragment encoding EGFP was integrated into the 5′-region of the csmA cording region, were selected and confirmed by Southern blot analysis using the 1.0-kb SpeI-XhoI fragment of pM-ALK-CHS5 [23] as a probe, and were designated as EGFP-CsmA1 and 2, and EGFP-CsmAΔkinA1 and 2, respectively (Table 1).
To obtain the strains that produced EGFP-CsmA in an uncA deletion mutant, a 4.0-kb fragment amplified from pNZ13 [48] using primers UncA-LB-fwd (5′-CGTCGATGGAAGGCATATACTACTCGC-3′) and UncA-RB-rev (5′-CATCCACGTCCCCATAACTAATACCACC-3′) was used for the transformation of EGFP-CsmA1. Two transformants in which uncA was replaced with pyroA were selected and designated as EGFP-CsmAΔuncA1 and 2.
EB-5 (chsB(p)- egfp-chsB) [24] was crossed with SNZ9 (ΔuncA) [48], selected and designated as EGFP-ChsBΔuncA1 and 2. SNR1 (ΔkinA) was transformed with the BglII fragment of pEB-argB [24]. Two transformants in which chsB(p)-egfp-chsB was integrated into the argB locus were selected and designated as EGFP-ChsBΔkinA1 and 2.
The strains that produced CsmA without its MMD (ΔMA) tagged with EGFP at its N-terminus under the control of the alcA promoter were constructed as follows: The 4.2-kb fragment containing the csmA promoter, argB, the alcA promoter, and the coding region of EGFP was amplified with AP1 (5′-ATAGTAACAGGTCAGGGTAT-3′) and 3-DMA-EGFP (5′-AGTTGTGAAACATATCGCCCCTTGTACAGCTCGTCCATGC-3′) as primers using pMAEC as a template and the 1.0-kb fragment encoding the CSD of CsmA (861 a.a.) was amplified from pMAEC using 5-EGFP-DMA (5′-GCATGGACGAGCTGTACAAGGGGCGATATGTTTCACAACT-3′) and 3-DMA (5′-CACATGGCCGACAATGAACA-3′) as primers. The amplified 4.2-kb and 1.0-kb fragments were fused by double-joint PCR [60] and the obtained fragment designated as EGFP-ΔMA was used for transformation. Two transformants in which a single copy of the EGFP-ΔMA producing fragment was integrated into the csmA locus were selected by Southern blot analysis using the same strategy as in the case of EGFP-CsmA1, and were designated EGFP-ΔMA1 and 2. The strains that produced KinArigor tagged with mRFP at their N-termini were obtained as follows: EGFP-CsmA1, EB-5, ABPU1, CA2, FB-3 [24,61], EGFP-ΔMA1, CΔMHA, and D10HA [25] were transformed with the 5.1-kb EcoRI-BglII fragment of pCS5-NZ [48]. The transformants in which the wild-type kinA was replaced with alcA(p)-mrfp-kinA rigor were selected and designated as EGFP-CsmA/mRFP-KinArigor1 and 2, EGFP-ChsB/mRFP-KinArigor1 and 2, ABPU1/mRFP-KinArigor1 and 2, CsmA-HA/mRFP-KinArigor1 and 2, FLAG-ChsB/mRFPKinArigor1 and 2, EGFP-ΔMA/mRFP-KinArigor1 and 2, CΔMHA/mRFP-KinArigor1 and 2, D10HA/mRFP-KinArigor1 and 2, respectively (Table 1). The strains that produced CsmA tagged with 9xHA at its C-terminus and ChsB tagged with 3xFLAG at its N-terminus were obtained by the transformation of CA2 with pFB-argB [24] and were designated as CsmA-HA/FLAG-ChsB1 and 2 (Table 1). CsmA-HA/FLAG-ChsB1 was transformed with the 5.1-kb EcoRI-BglII fragment, and the transformants in which the wild-type kinA was replaced with alcA(p)-mrfp-kinA rigor were designated as CsmA-HA/FLAG-ChsB/mRFPKinArigor1 and 2. To obtain the strain that produce KinA tagged with GFP at its N-terminus, the ABPU1 strain was transformed with the 5.1-kb EcoRI-BglII fragment of pCS2-NZ [48]. Two transformants in which the transformed fragment was integrated into the kinA locus were selected and designated as ABPU1/GFP-KinA1 and 2. ABPU1/GFP-KinA1 was transformed with the 3.0-kb BglII-EcoRI fragment of pHA9 [61]. Two transformants in which the fragment was integrated into the csmA locus were selected and designated as CsmA-HA/GFP-KinA1 and 2.
We obtained at least two transformants in each case after the confirmation of their relevant DNA structures by Southern blot analysis. They exhibited the same phenotypes with respect to growth and hyphal morphology. We used one of them for further experiments.
Light/fluorescence microscopy
Live-cell imaging of germlings and young hyphae: cells were grown on cover slips in 0.5 ml MMGly (de-repression of the alcA promoter) were used. Although GFP-ChsB was expressed under the native promoter, to compare the data of GFP-CsmA, which was expressed under the alcA promoter, MMGly was selected. Cells were incubated at 30°C overnight or 1 day. The cover slips were mounted on a microscope slide. Tempcontrol mini (Pepcon) was used as needed to control the temperature of the specimens during microscopy. Images were captured using an Axiophot microscope using a Planapochromatic 63 times oil immersion objective lens, the Zeiss AxioCam MRM camera (Zeiss, Jena, Germany), and the HBO103 mercury arc lamp (Osram) or HXP 120 (Zeiss, Jena, Germany) possessing faster speed wavelength switching. Images were collected and analyzed using the AxioVision and Zen system (Zeiss). Kymographs were made using ImageJ software (http://rsb.info.nih.gov/ij/) and Zen system (Zeiss).
Other methods
Western blot analysis was performed as described previously [62] with slight modifications. Cell lysates were prepared as follows: Conidia were inoculated in MMTFuu liquid medium and incubated for 14–16 h. The mycelia were frozen with liquid nitrogen and broken by grinding. The cell lysates were fractionated as described previously [24,61]. 10,000 x g pellet (P10) and the 100,000 x g pellet (P100) were suspended with 100 μl of TNE buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA) and then 500 μl of IP buffer (0.1% Triton X-100, 100 mM NaCl, 10 mM EDTA, 50 mM Tris-HCl pH 7.4) and 2 μl of Protease Inhibitor Cocktail (Sigma) were added. Co-immunoprecipitation of CsmA-HA, FLAG-ChsB, ΔM-HA, and d10-HA were performed as described previously [25] using the P10 and P100 fractions. mRFP-KinArigor was detected using a rabbit anti-DsRed-monomer (mDsRed) polyclonal antibody (Cat. No. 632496, Clontech), which recognizes mRFP, at a 1:1,000 dilution and an anti-rabbit immunoglobulin G (IgG), horseradish peroxidase-linked antibody (A0545, Sigma) at a 1:1,000 dilution. Twenty μg proteins were loaded on SDS-PAGE for the detection of proteins tagged with EGFP or mRFP.
Results
Localization of ChsB and CsmA
In previous work chitin synthase class III ChsB, expressed from the native promoter and fused to GFP, was observed at hyphal tips and forming septa in A. nidulans [24]. Here we analyzed the localization mechanism for ChsB and the class V chitin synthase CsmA (S1 Fig). To analyze the dynamic localization of CsmA in living hyphae, we replaced the csmA gene with a C-terminally GFP tagged version (CsmA-GFP). These strains, however, exhibited phenotypic abnormalities similar to those observed in csmA-deletion mutants and only weak fluorescence of CsmA-GFP (data not shown). This suggested that the GFP tag at the C-terminus interferes with the function of CsmA. Thus, we constructed a strain expressing CsmA tagged with GFP at its N-terminus (GFP-CsmA) under the control of the alcA promoter (Table 1). This strain grew well and did not exhibit phenotypic abnormalities when the alcA promoter was derepressed (medium containing glycerol as a carbon source) or when the alcA promoter was induced (medium containing threonine and fructose as carbon sources). The correct insertion of the construct in two strains expressing GFP-CsmA (EGFP-CsmA1 and 2) was confirmed by Southern blot analysis (S2 Fig). The production of the GFP-CsmA protein was confirmed by Western blot analysis using anti-GFP antibodies (S3A Fig).
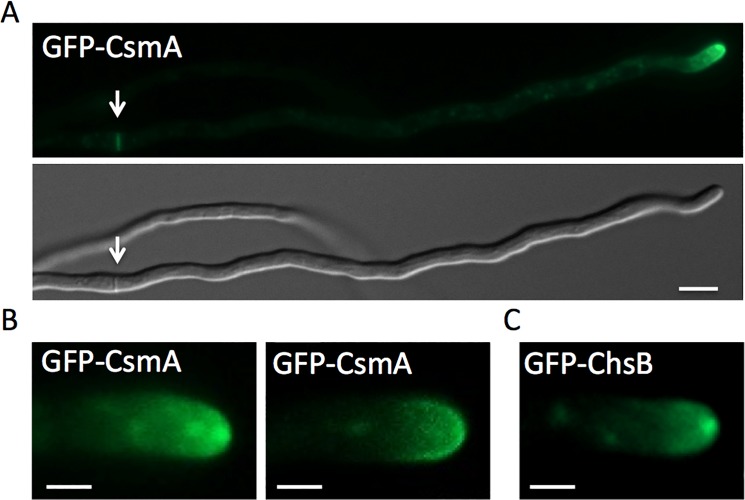
Fluorescence of GFP-CsmA was observed at hyphal tips and at forming septa in addition as moving spots in the cytoplasm (Fig 1A, and data not shown). At the apical membrane, GFP-CsmA was found at the Spitzenkörper and/or as a crescent along the apical membrane (Fig 1B, S1 and S2 Movies). Likewise, GFP-ChsB was observed at hyphal tips and forming septa [24]. However GFP-ChsB was found mainly at the Spitzenkörper at the hyphal tips under this condition (Fig 1C, S3 Movie).
Fig 1. Localization of GFP-CsmA.
(A) Strain EGFP-CsmA1 was grown in MMGlyuu overnight. GFP-CsmA was observed at the hyphal tips and forming septa (arrow). (B) GFP-CsmA was found at the Spitzenkörper (left) and/or as a crescent along apical membrane (right). (C) The strain producing GFP-ChsB (EB-5) was grown in MMGlyuu overnight. GFP-ChsB was found mainly at the Spitzenkörper at the hyphal tips. Scale bars represent 5 μm (A) and 2 μm (B,C).
Role of kinesin-1 (KinA) and kinesin-3 (UncA) for the localization of ChsB and CsmA
To investigate the roles of kinesins, KinA (kinesin-1) and UncA (kinesin-3), for the localization and transportation of ChsB and CsmA, we constructed strains expressing GFP-ChsB or GFP-CsmA in kinA- or uncA-deletion backgrounds, respectively. The production of GFP-ChsB and GFP-CsmA were confirmed by Western blot analysis using anti-GFP antibodies (S3A and S3B Fig).
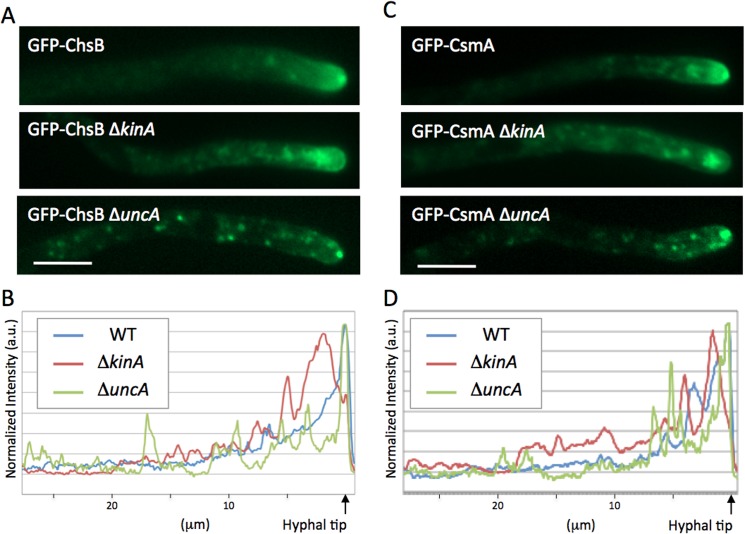
Typical localization patterns of GFP-ChsB or GFP-CsmA in the ΔkinA or ΔuncA are shown in Fig 2A and 2C. In the absence of KinA, both GFP-ChsB and GFP-CsmA showed large accumulations at subapical regions (Fig 2A and 2C). The profiles of GFP signal intensities along hyphae are shown in Fig 2B and 2D. In wild type hyphae, the maximum peaks of GFP-ChsB and GFP-CsmA were restricted sharply at the tip of hyphae. In contrast, the hyphae in kinA-deletion strains displayed wider peaks of GFP-ChsB and GFP-CsmA at approximately 1 μm behind the tip of hyphae. In the uncA-deletion strains, GFP-ChsB and GFP-CsmA localized at the apex of hyphal tips as in wild type stains (Fig 2A and 2C). However GFP-ChsB and GFP-CsmA appeared to be less diffused behind the apex. In addition, multiple punctate structures were observed along the hyphae. In the profiles of GFP signal intensities, several small peaks were often observed throughout the hyphae in the uncA-deletion strains (Fig 2B and 2D).
Fig 2. Localization of GFP-ChsB and GFP-CsmA in kinesin-deletion strains.
(A, C) The typical localization patterns of GFP-ChsB (A) and GFP-CsmA (C) in wild type (EB-5 and EGFP-CsmA1, respectively), ΔkinA (EGFP-ChsBΔkinA1 and EGFP-CsmAΔkinA1, respectively), and ΔuncA (EGFP-ChsBΔunkA1 and EGFP-CsmAΔunkA1, respectively) strains grown in MMGlyuu overnight. Scale bars represent 5 μm. (B, D) Signal intensity profiles of GFP-ChsB (B) and GFP-CsmA (D) along hyphae in wild type (blue line), ΔkinA (red line), and ΔuncA (green line) strains.
Role of KinA and UncA in the transportation of ChsB and CsmA
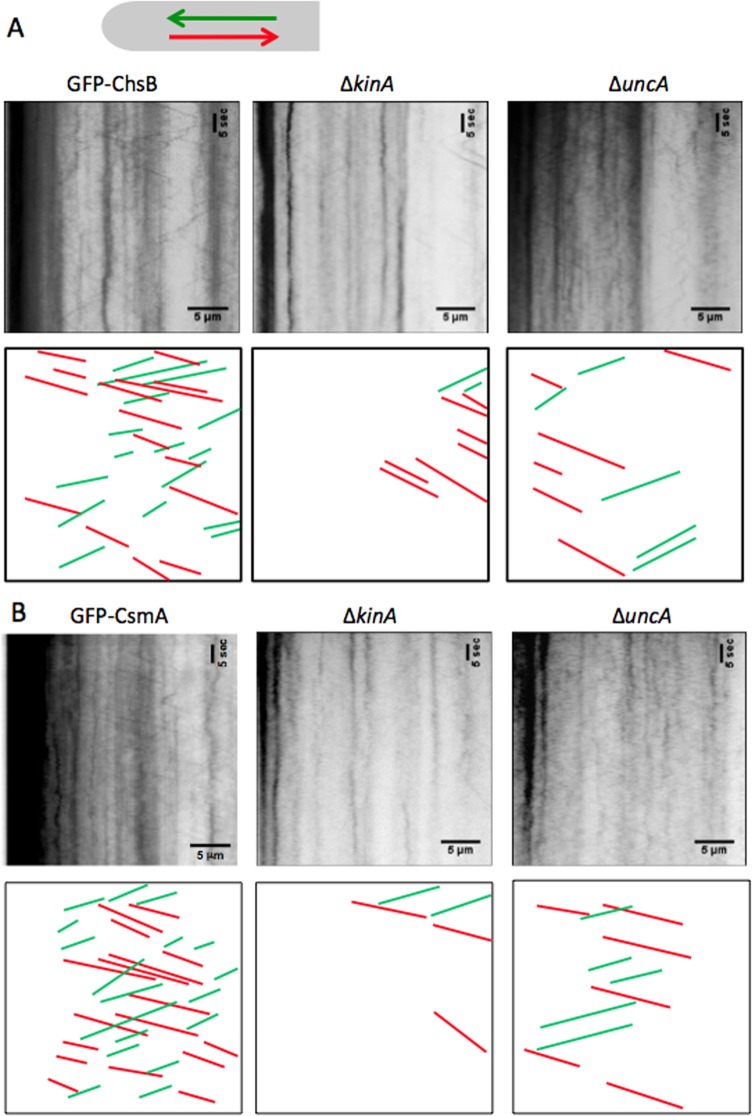
We investigated the roles of kinesins, KinA and UncA, in the transportation of ChsB and CsmA. To visualize the transportation events of GFP-ChsB and GFP-CsmA, kymographs were recorded at hyphal tips in wild type, the kinA-deletion, and the uncA-deletion strains (Fig 3)(S4–S9 Movies). The signals forming vertical lines in the kymographs represent immobile accumulations of GFP-ChsB (Fig 3A) and GFP-CsmA (Fig 3B). With the used microscopy setup it is impossible to detect single GFP molecules. The transport events shown in the kymographs are thought to represent events that multiple GFP-ChsB or GFP-CsmA molecules are transported together. It is not defined here whether the spots represent secretory vesicles, chitosomes, or early endosomes.
Fig 3. Kymographs GFP-ChsB and GFP-CsmA in kinesin-deletion strains.
(A) Kymographs of GFP-ChsB in the wild type, ΔkinA, and ΔuncA strains. (B) Kymographs of GFP-CsmA in wild type, ΔkinA, and ΔuncA strains. The kymographs were derived at hyphal tips (30 μm from the tips) from a 1 minute movie (200 ms intervals, total 300 frames). Horizontal scale bars represent 5 μm. Vertical scale bars represent 5 seconds. Anterograde movements are represented by green and retrograde movements are represented by red lines.
Transport frequency of ChsB and CsmA
The transport frequency of GFP-ChsB and GFP-CsmA in both anterograde and retrograde direction appeared higher in the wild type than in the kinA- or the uncA-deletion strains. To compare the transport frequencies, the transport events were quantified from the kymographs (Fig 4). In addition, the velocity of GFP spots was calculated from the kymographs and compared between wild type and the kinesin-deletion strains (Fig 5).
Fig 4. Transport frequency of GFP-ChsB and GFP-CsmA in kinesin-deletion strains.
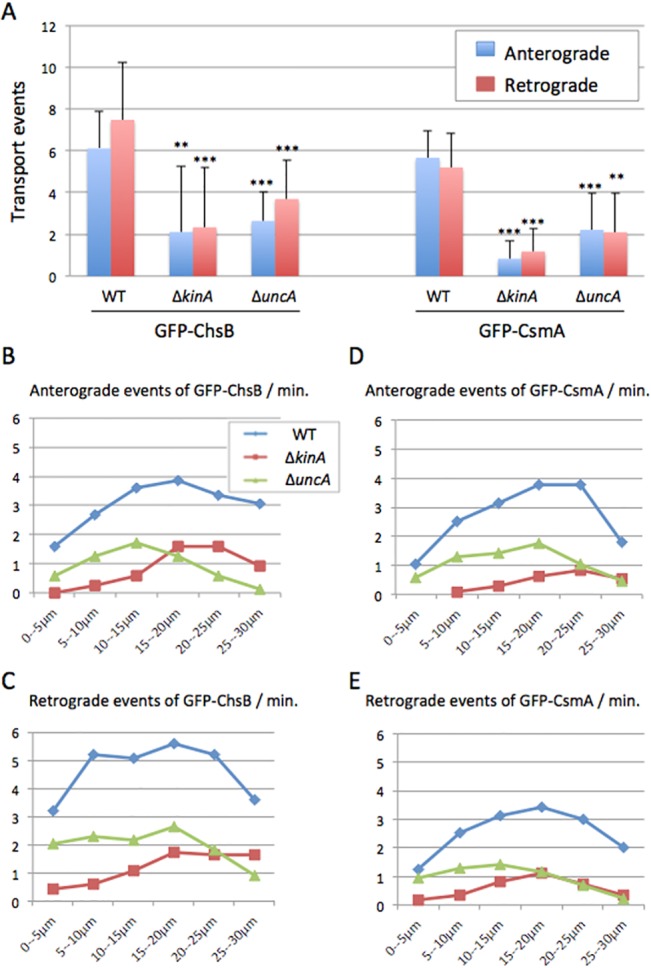
(A) The frequency of anterograde (blue) and retrograde (red) movements of GFP-ChsB (left) and GFP-CsmA (right) in the wild type, ΔkinA, and ΔuncA strains. Anterograde and retrograde movements were visualized in the kymographs. The numbers of motion events per minute are shown (mean ± S.D). **, P < 0.05 (statistically significant difference); ***, P < 0.001 (highly significant difference), compared to wild type strain. The p values were calculated by unpaired t-test. (B-E) Distribution diagrams of GFP-ChsB anterograde (B) and retrograde (C) events, and GFP-CsmA anterograde (D) and retrograde (E) events in each 5 μm-segment from hyphal tips in the wild type (blue line), ΔkinA (red line), and ΔuncA (green line) strains.
Fig 5. Velocity of GFP-ChsB and GFP-CsmA in kinesin-deletion strains.
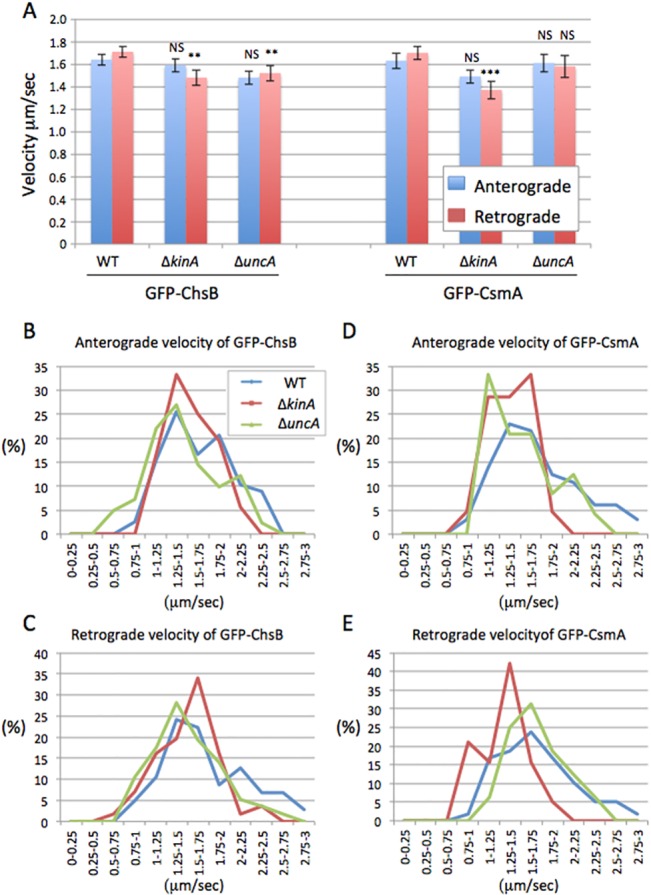
(A) The velocities of anterograde (blue) and retrograde (red) movements of GFP-ChsB (left) and GFP-CsmA (right) in wild type, ΔkinA, and ΔuncA strains are shown (mean ± S.E). NS, no significant difference; **, P < 0.05 (statistically significant difference); ***, P < 0.001 (highly significant difference), compared to wild type strain. The p values were calculated by unpaired t-test. (B-E) Distribution of velocities in the GFP-ChsB anterograde (B) and retrograde movements (C) and in the GFP-CsmA anterograde (D) and retrograde movements (E) are shown in wild type (blue line), ΔkinA (red line), and ΔuncA (green line) strains.
In wild type the frequency of anterograde and retrograde movements visualized by GFP-ChsB were 6.1 ± 1.8 and 7.5 ± 2.7 (mean ± S.D., n = 82 and 100) events per minute, respectively. In the kinA-deletion strain those (the frequencies of GFP-ChsB anterograde and retrograde movements) were reduced to 2.1 ± 3.1 and 2.3 ± 2.9 (mean ± S.D., n = 38 and 47) events per minute, respectively. In the uncA-deletion strain the frequencies were also reduced to 2.6 ± 1.4 and 3.7 ± 1.9 (mean ± S.D., n = 42 and 58) events per minute, respectively. The frequencies of transports visualized by GFP-ChsB in the kinA-deletion and the uncA-deletion strains were both significantly lower than those in wild type in anterograde (34% in ΔkinA and 46% in ΔuncA) and retrograde direction (31% in ΔkinA and 48% in ΔuncA) (Figs 3 and 4A).
In wild type the frequencies of GFP-CsmA anterograde and retrograde movements were 5.7 ± 1.3 and 5.2 ± 1.6 (mean ± S.D., n = 100 each) events per minute, respectively. In the kinA-deletion strain those (the frequencies of GFP-CsmA anterograde and retrograde movements) were significantly reduced to 0.8 ± 0.8 and 1.2 ± 1.1 (mean ± S.D., n = 40 and 45) events per minute, respectively. In the uncA-deletion strain the frequencies of GFP-CsmA anterograde and retrograde movement were reduced to 2.2 ± 1.8 and 2.1 ± 1.9 (mean ± S.D., n = 40 and 50) events per minute, respectively. Both frequencies of moving spots visualized by GFP-CsmA in the kinA-deletion and the uncA-deletion strains were significantly lower than those in wild type in both anterograde (15% in ΔkinA, 39% in ΔuncA) and retrograde (23% in ΔkinA, 40% in ΔuncA) direction (Figs 3 and 4A). Generally, the frequencies of GFP-ChsB movement were relatively higher than those of GFP-CsmA. The difference might represent different expression levels of GFP-ChsB and GFP-CsmA.
To analyze the distribution of transport events along the hyphae, the hyphae were divided into 5 μm-segments starting from hyphal tips and the frequencies of the movements of GFP-ChsB and GFP-CsmA were quantified in each segment in each strain (Fig 4B–4E). In cases where movements of GFP-ChsB and GFP-CsmA were observed in longer distances than 5 μm, they were counted in multiple segments. The frequencies of GFP-ChsB and GFP-CsmA in both anterograde and retrograde movements in the kinA-deletion strain were lower than those of the wild type strain. Especially in the region close to the tips (0–15 μm), movements of GFP-ChsB and GFP-CsmA were hardly observed in both anterograde and retrograde directions. The frequencies of GFP-ChsB and GFP-CsmA in both anterograde and retrograde direction in the uncA-deletion strain were also lower than those of the wild type strain along the hyphae, in contrast, especially behind the tip region (20–30 μm).
Transport speed of ChsB and CsmA
Although the frequency of GFP-ChsB and GFP-CsmA movements decreased in the kinA- and uncA-deletion strains, the velocities of the movements were comparable (Fig 5A). In wild type the velocities of GFP-ChsB anterograde and retrograde movements were 1.6 ± 0.1 and 1.7 ± 0.1 (μm/sec., mean ± S.E., n = 82 and 100), respectively. In the kinA-deletion strain those were 1.6 ± 0.1 and 1.5 ± 0.1 (μm/sec., mean ± S.E., n = 38 and 47), respectively, and 1.5 ± 0.1 and 1.5 ± 0.1 (μm/sec., mean ± S.E., n = 42 and 58), respectively, in the uncA-deletion strain. There were no significant differences in the velocities of anterograde movements in the kinesin-deletion strain, whereas the velocities of retrograde movements were slightly reduced (87% in ΔkinA, 89% in ΔuncA).
In the wild type strain the velocities of GFP-CsmA anterograde and retrograde movement were 1.6 ± 0.1 and 1.7 ± 0.1 (μm/sec., mean ± S.E., n = 100 each), respectively (Fig 5A). The velocities of GFP-CsmA showed no significant difference compared to the velocities of GFP-ChsB in both anterograde and retrograde movements, respectively. The velocities of anterograde and retrograde movements also showed no significant difference in both GFP-ChsB and GFP-CsmA. In the kinA-deletion strain the velocities of GFP-CsmA anterograde and retrograde movement were 1.5 ± 0.1 and 1.4 ± 0.1 (μm/sec., mean ± S.E., n = 40 and 45), respectively. In the uncA-deletion strain those were 1.6 ± 0.1 and 1.6 ± 0.1 (μm/sec., mean ± S.E., n = 40 and 50), respectively. There were no significant differences in the velocities of anterograde movements in the kinesin deletion strains, whereas the velocities of retrograde movements were slightly reduced in the kinA-deletion strain (81%) but not in the uncA-deletion strain.
The distribution of velocities in the anterograde and retrograde movements of GFP-ChsB and GFP-CsmA was analyzed in each strain (Fig 5B–5E). Velocities faster than 2 μm/sec were observed in wild type at all the anterograde and retrograde movements of GFP-ChsB and GFP-CsmA. However, the high speeds were not clearly found in the kinA-deletion strain. The average speed of early endosomes transport is slower than that of secretory vesicles (unpublished results). The higher speed peaks might indicate those signals derived from secretory vesicles.
Transport of ChsB and CsmA to the hyphal tips along microtubules by KinA
To further analyze the function of KinA in the transport of GFP-ChsB and GFP-CsmA, we constructed strains co-expressing either GFP-ChsB and mRFP-KinArigor (EGFP-ChsB/mRFP-KinArigor strain), or GFP-CsmA and mRFP-KinArigor (EGFP-CsmA/mRFP-KinArigor strain). The KinArigor protein, which has a point mutation in its ATPase domain, binds tightly to microtubules but does not move along them. Consequently, KinArigor decorates microtubules [48]. GFP-ChsB and GFP-CsmA localized to large accumulations at subapical regions similar to those in kinA-deletion strains. In addition, GFP-ChsB and GFP-CsmA were observed along microtubules decorated with mRFP-KinArigor throughout the hyphae (Fig 6A and 6B). These results suggest that ChsB and CsmA are transported to the hyphal tip regions along microtubules by KinA.
Fig 6. Colocalization of GFP-ChsB, GFP-CsmA, and GFP-ΔMA with mRFP1-KinArigor along microtubules.
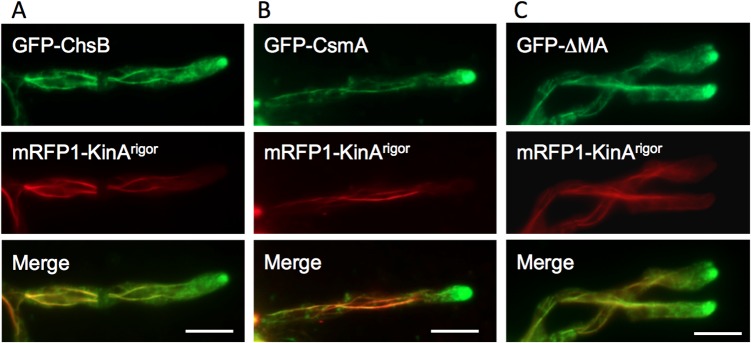
(A, B) GFP-ChsB (A) and GFP-CsmA (B) localized to large accumulations at the subapical tips similar to those in the ΔkinA strains, and were observed along microtubules decorated with mRFP-KinArigor throughout the hyphae. (C) GFP-ΔMA (CsmA without the MMD) also localized to large accumulations at the subapical tips and along microtubules decorated with mRFP-KinArigor. (A-C) These strains were grown in MMGlyuu overnight. Scale bars represent 5 μm.
Only the chitin synthase domain of CsmA without the MMD (GFP-ΔMA) did not show a clear accumulation at the hyphal tip membrane (data not shown)[25]. When the GFP-ΔMA was co-expressed with mRFP-KinArigor in the EGFP-ΔMA/ mRFP-KinArigor1 strain, the GFP-ΔMA also exhibited a similar localization pattern with GFP-CsmA co-expressed with mRFP-KinArigor, along microtubules and at subapical large accumulations (Fig 6C), suggesting that CsmA is transported to the hyphal tip regions by KinA independently of its MMD.
Interaction of ChsB and CsmA with KinA
To further examine the interaction between ChsB and KinArigor, or CsmA and KinArigor in vivo, we constructed strains co-expressing FLAG-ChsB and mRFP-KinArigor, or CsmA-HA and mRFP-KinArigor (FLAG-ChsB/mRFP-KinArigor strain or CsmA-HA/mRFP-KinArigor strain, respectively). The strains were subjected to a fractionation experiment (see Materials and methods). FLAG-ChsB and CsmA-HA were detected in a 10,000 x g pellet (P10) and in a 100,000 x g pellet (P100) that were obtained by cell fractionation of the crude cell extract of A. nidulans [24,61]. In S. cerevisiae, P100 fraction contains transport vesicles, endosomal membranes, and Golgi cisternae [63]. FLAG-ChsB and CsmA-HA were co-immunoprecipitated with mRFP-KinArigor from the P100 fractions of the FLAG-ChsB/mRFP-KinArigor strain and CsmA-HA/mRFP-KinArigor strain, respectively (Fig 7A and 7C). CsmA-HA was also co-immunoprecipitated with wild type KinA from the P100 fraction of the strain expressing CsmA-HA and GFP-KinA (Fig 7B). These results suggest that transport vesicles containing ChsB or CsmA are transported through the interaction with KinA. In addition, CsmA without the MMD (ΔMA) and CsmA truncated mutant protein lacking 10 amino acids from the actin binding domain of the MMD (D10M) also co-immunoprecipitated with mRFP-KinArigor (S4 Fig)[25], suggesting that the interaction of transport vesicles containing CsmA with KinA is independent of the MMD.
Fig 7. Co-immunoprecipitation of CsmA-HA with mRFP-KinArigor or GFP-KinA and FLAG-ChsB with mRFP-KinArigor in vivo.
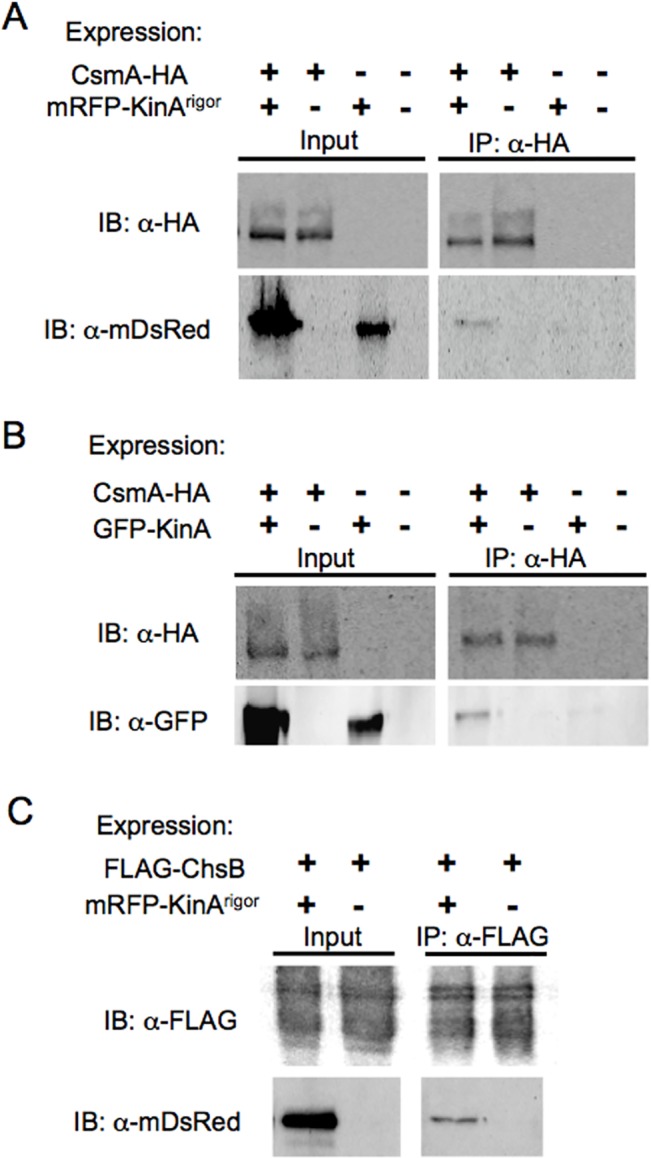
(A, B) 100,000 xg pellets (P100) of cell extracts of the strains expressing CsmA-HA, mRFP-KinArigor, or both of them (A), and the strains expressing CsmA-HA, GFP-KinA, or both of them (B) were immunoprecipitated with anti-HA antibody. P100 (Input) and immunoprecipitates with anti-HA antibody (IP: α-HA) were subjected to Western blot analysis using antibodies indicated by IB. (C) Those of the strains expressing FLAG-ChsB or FLAG-ChsB and mRFP-KinArigor were immunoprecipitated with anti-FLAG antibody and subjected to Western blot analysis.
We examined the interaction between ChsB and CsmA, but we did not detect co-immunoprecipitation of CsmA-HA or FLAG-ChsB from the P100 fractions of the strains that produce these tagged proteins together with or without mRFP-KinArigor (FLAG-ChsB/CsmA-HA/mRFP-KinArigor strain or FLAG-ChsB/CsmA-HA strain, respectively) (data not shown). These results suggest that ChsB and CsmA are transported on different vesicles by KinA.
Discussion
In filamentous fungi, protein machineries required for polarized growth, such as formin and regulators of polarity establishment, accumulate in the Spitzenkörper at the hyphal tips [3,5,7,10,12]. Chitin synthases of the classes I, III, and VII of Neurospora crassa (Chs3, Chs1, and Chs6) also localized at the Spitzenkörper and in subapical regions as punctate structures [55,64]. In the previous studies, we investigated the localization of class III and V chitin synthases in A. nidulans, ChsB and CsmA, respectively [24,25]. In this study, we investigated the transport mechanisms of ChsB and CsmA in living hyphae, especially the roles of kinesin-1 (KinA) and kinesin-3 (UncA) in the transportation of ChsB and CsmA. Our results suggest that these chitin synthases are delivered to hyphal tips along microtubules through the interaction with conventional kinesin (kinesin-1, KinA).
The localizations of GFP-CsmA and GFP-ChsB were disturbed especially around the hyphal tip region by the kinA deletion (Figs 2–5). In addition these chitin synthases co-immunoprecipitated with KinA. These results suggest that KinA transports CsmA and ChsB. Movements of ChsB and CsmA were hardly observed in kinA-deletion mutants and the number of movements with higher velocities were reduced in the mutants. Since it is suggested that smaller vesicles tend to move faster than larger vesicles [65], it is possible that these chitin synthases are mainly transported on small vesicles, such as secretory vesicles or chitosomes, at extreme hyphal tip regions by KinA. However, KinA is required for the transport of cytoplasmic dynein to the microtubule plus ends [52] and dynein itself is involved in membrane transport such as early endosomes in several fungi [11,47,66,67]. Since ChsB and CsmA accumulated at subapical regions of the hyphae in the kinA-deletion mutant and dynein is not transported to those regions in the kinA-deletion mutants [52], it is also possible that the defects of ChsB and CsmA transport in the kinA-deletion strains include the direct and/or indirect effect of mislocalization of dynein.
In U. maydis, Schuster et al. reported that class V chitin synthase Mcs1 was also transported by kinesin-1 [50]. These results indicate that the transport mechanism of class V chitin synthases is conserved between ascomycete and basidiomycete fungi. In contrast, the transport of the class III chitin synthase in N. crassa does not depend strictly on microtubules [64], suggesting that the transport mechanism of class III chitin synthase of A. nidulans is at least partially different from that of another ascomycete N. crassa. Since CsmA-HA and FLAG-ChsB did not co-immunoprecipitate, they are likely transported in different vesicle populations. In N. crassa, it is also suggested that classes I, III, and VII chitin synthases are contained in different vesicular subpopulations [64]. Thus chitin synthases of different classes are possibly transported in different vesicle subpopulations.
Kinesin-3, UncA, is involved in the transport of early endosome and peroxisome [11,48,68]. In U. maydis, kinesin-3 is also involved in the transport of early endosomes and the secretion of acid phosphatase [46,67,69], but is not involved in the transport of Mcs1 [50]. Although the transports of GFP-ChsB and GFP-CsmA were disturbed to some extent in the uncA-deletion strain, the localization at hyphal tips appeared nearly normal in the mutants. This suggests that UncA is involved in the transport of ChsB and CsmA, but not essential especially in hyphal tip regions. In A. nidulans, actin forms a ring structure at the subapical plasma membrane [70]. Endocytosis is suggested to occur nearby this structure, and the resultant endocytic vesicles fuse with each other in the cytoplasm to form early endosomes [7,70–73]. The early endosomes are transported along microtubules by kinesin-3 and dynein in the plus and minus end directions, respectively, in hyphae of A. nidulans and U. maydis [6,11,46,48,49] and the endosome system is thought to be involved in the recycling of membrane proteins at hyphal tips [74]. It is possible that kinesin-3, UncA, is involved in the transport of ChsB and CsmA through the recycling pathway. Alternatively the defects of early endosome transport might affect the transport of ChsB and CsmA indirectly. It is necessary to distinguish secretory vesicles and early endosomes in same hyphae in order to investigate the dependency of KinA and UncA on the transport.
It is well known that actin is also crucial for hyphal tip growth and that myosins are motor proteins that move on actin filaments. A. nidulans contains three genes, myoA, myoB, and myoE that encode myosin heavy chains belonging to the myosin families, myosin-1, myosin-2, and myosin-5, respectively [8,9,75]. Moreover, MyoE participates in vesicle transport in hyphae [8]. Mutagenesis of both, myosin-5 and kinesin-1 caused severe growth defects in A. nidulans and U. maydis [9,69], suggesting that the function of kinesin-1 could be partly substituted by myosin-5 and vice versa. Since myosin-5 functions in the transport of Mcs1 to the cell surface in a microtubule-independent manner [50], it is possible that a myosin-5, MyoE, is also involved in the transport of CsmA and ChsB. This idea is supported by the fact that the inhibition of actin polymerization by cytochalasin A severely disturbed the localization of these chitin synthases (data not shown)[25]. The MMDs of class V chitin synthases belong to the myosin-17 family. The binding of the MMD to actin filaments is required for the proper localization and function of CsmA [25], whereas no motor activity of the MMD of Mcs1 was detected in vitro [50]. The point mutation at the P-loop or Switch I in the MMD of CsmA does not affect its function and localization [25], suggesting that there is no motor activity or at least the motor activity is not essential for the transport mechanism. The idea is supported by our results that CsmA and CsmA without the MMD is transported to the hyphal tips by KinA.
If the MMD of CsmA is not necessary for the transport to hyphal tips, the role of MMD remains unrevealed. Schuster et al. suggested that the MMD of Mcs1 functions in the tethering of vesicles containing Mcs1 near the sites of exocytosis [50]. Our results showed that the deletion of kinesin-1 or kinesin-3 caused similar effects on the transport of ChsB and CsmA. The role of MMD may be important for the localization at hyphal tips after their transportation, rather than the transport towards hyphal tips. GFP-CsmA was found at the Spitzenkörper and/or as a crescent along the apical membrane (Fig 1B). In contrast, GFP-ChsB was found mainly at the Spitzenkörper at hyphal tips (Fig 1C). The difference in the localization between CsmA and ChsB was also observed in a significant population of hyphal tips by indirect immunofluorescence analysis [24,25]. ChsB and CsmA are likely to be transported to the Spitzenkörper in a similar way. Then, CsmA might be localized and/ or stay at special regions along the apical membrane depending on the interaction of MMD with the actin cytoskeleton.
Supporting Information
Alignment was done with the clustalW alignment program using the entire amino acid sequence of each chitin synthase. The tree was drawn using Njplot. Branch length indicates evolutionary distance. Chitin synthases belong to class III and V are shown in red and blue, respectively. Abbreviations: Nc, N. crassa; Sc, S. cerevisiae; Um, U. maydis.
(TIF)
(A) Strategy for constructing GFP-CsmA producing strains. (B) Southern blot analysis of EcoRV digested total DNAs of the EGFP-CsmA strains using the 1.0 kb SpeI-XhoI fragment of pM-ALK-CHS5 [23] as a probe. Lane 1, ABPU1; lane 2, EGFP-CsmA1; lane 3, EGFP-CsmA2.
(TIF)
(A) GFP-CsmA in the cell lysates of the EGFP-CsmA1 (WT), EGFP-CsmA/ΔkinA1 strain (ΔkinA), and EGFP-CsmA/ΔuncA1 (ΔuncA) strains were detected with anti-GFP antibody. (B) EGFP-ChsB in the cell lysates of the EB-5 strain (WT), EGFP-ChsB/ΔkinA1 (ΔkinA), and EGFP-ChsB/ΔuncA1 (ΔuncA) strains were detected using the same antibody. P10, 10,000 x g pellet; P100, 100,000 x g pellet; S100, 100,000 x g supernatant.
(TIF)
P100 fractions of the CΔMHA strain or the CΔMHA/mRFP-KinArigor1 strain (A), and the D10HA strain or the D10HA/mRFP-KinArigor1 strain (B) were immunoprecipitated with anti-HA antibody and subjected to Western blot analysis.
(TIF)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
Acknowledgments
MT was a research fellow of Japan Society for the Promotion of Science. The work was partly done the facilities of the Biotechnology Research Center of the University of Tokyo.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
NT was supported by German Research Foundation (DFG; http://www.dfg.de/) Grant No. TA819/2-1 and by the Baden-Württemberg Stiftung, Eliteprogramm für Postdocs (http://www.bwstiftung.de/startseite/). RF was supported by German Research Foundation Grant No. FOR1334. MT was supported by Grant-in-Aid for Japan Society for the Promotion of Science Grant No. 10J03260. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature 2003; 422: 766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rittenour WR, Si H, Harris SD. Hyphal morphogenesis in Aspergillus nidulans . Fungal Biol Rev. 2009; 23: 20–29. [Google Scholar]
- 3. Sudvery PE. Regulation of polarized growth in fungi. Fungal Biol Rev. 2008; 22: 44–55. [Google Scholar]
- 4. Harris SD. The Spitzenkörper: a signalling hub for the control of fungal development? Mol Microbiol 2009; 73: 733–736. 10.1111/j.1365-2958.2009.06803.x [DOI] [PubMed] [Google Scholar]
- 5. Riquelme M, Sánchez-León E. The Spitzenkörper: a choreographer of fungal growth and morphogenesis. Curr Opin Microbiol. 2014; 20: 27–33. 10.1016/j.mib.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 6. Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Peñalva MA. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 2009; 10: 57–75. 10.1111/j.1600-0854.2008.00848.x [DOI] [PubMed] [Google Scholar]
- 7. Taheri-Talesh N, Horio T, Araujo-Bazán L, Dou X, Espeso EA, Peñalva MA, et al. The tip growth apparatus of Aspergillus nidulans . Mol Biol Cell 2008; 19: 1439–1449. 10.1091/mbc.E07-05-0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taheri-Talesh N, Xiong Y, Oakley BR. The functions of myosin II and myosin V homologs in tip growth and septation in Aspergillus nidulans . PLoS One 2012; 7: e31218–e31218. 10.1371/journal.pone.0031218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J, Tan K, Wu X, Chen G, Sun J, Reck-Peterson SL, et al. Aspergillus myosin-V supports polarized growth in the absence of microtubule-based transport. PLoS One 2011; 6: e28575–e28575. 10.1371/journal.pone.0028575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fischer R, Zekert N, Takeshita N. Polarized growth in fungi—interplay between the cytoskeleton, positional markers and membrane domains. Mol Microbiol 2008; 68: 813–826. 10.1111/j.1365-2958.2008.06193.x [DOI] [PubMed] [Google Scholar]
- 11. Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012; 197: 971–982. 10.1083/jcb.201112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takeshita N, Manck R, Grun N, de Vega SH, Fischer R. Interdependence of the actin and the microtubule cytoskeleton during fungal growth. Curr Opin Microbiol. 2014; 20c: 34–41. [DOI] [PubMed] [Google Scholar]
- 13. Rogg LE, Fortwendel JR, Juvvadi PR, Steinbach WJ. Regulation of expression, activity and localization of fungal chitin synthases. Med Mycol. 2012; 50: 2–17. 10.3109/13693786.2011.577104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lenardon MD, Munro CA, Gow NAR. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 2010; 13: 416–423. 10.1016/j.mib.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chigira Y, Abe K, Gomi K, Nakajima T. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae . Curr Genet. 2002; 41: 261–267. [DOI] [PubMed] [Google Scholar]
- 16. Choquer M, Boccara M, Gonçalves IR, Soulié MC, Vidal-Cros A. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem. 2004; 271: 2153–2164. [DOI] [PubMed] [Google Scholar]
- 17. Fujiwara M, Horiuchi H, Ohta A, Takagi M. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem Biophys Res Commun. 1997; 236: 75–78. [DOI] [PubMed] [Google Scholar]
- 18. Motoyama T, Kojima N, Horiuchi H, Ohta A, Takagi M. Isolation and characterization of a chitin synthase gene (chsC) of Aspergillus nidulans . Biosci Biotechnol Biochem. 1994; 58: 2254–2257. [DOI] [PubMed] [Google Scholar]
- 19. Motoyama T, Fujiwara M, Kojima N, Horiuchi H, Ohta A, Takagi M. The Aspergillus nidulans chsD gene encodes a chitin synthase which has a redundant function with the chsA gene product in conidiophore formation. Mol Gen Genet. 1996; 251: 442–450 and 1997; 253: 520–528. 8709948 [Google Scholar]
- 20. Takeshita N, Yamashita S, Ohta A, Horiuchi H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol Microbiol. 2006; 59: 1380–1394. [DOI] [PubMed] [Google Scholar]
- 21. Yanai K, Kojima N, Takaya N, Horiuchi H, Ohta A, Takagi M. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans . Biosci Biotechnol Biochem. 1994; 58: 1828–1835. [DOI] [PubMed] [Google Scholar]
- 22. Yabe T, Yamada-Okabe T, Nakajima T, Sudoh M, Arisawa M, Yamada-Okabe H. Mutational analysis of chitin synthase 2 of Saccharomyces cerevisiae—Identification of additional amino acid residues involved in its catalytic activity. Eur J Biochem. 1998; 258: 941–947. [DOI] [PubMed] [Google Scholar]
- 23. Horiuchi H, Fujiwara M, Yamashita S, Ohta A, Takagi M. Proliferation of intrahyphal hyphae caused by disruption of csmA which encodes a class V-chitin synthase with a myosin motor-like domain in Aspergillus nidulans . J Bacteriol. 1999; 181: 3721–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fukuda K, Yamada K, Deoka K, Yamashita S, Ohta A, et al. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot cell 2009; 8: 945–956. 10.1128/EC.00326-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takeshita N, Ohta A, Horiuchi H. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans . Mol Biol Cell 2005; 16: 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsuizaki M, Takeshita N, Ohta A, Horiuchi H. Myosin motor-like domain of the class VI chitin synthase, CsmB, is essential for its functions in Aspergillus nidulans . Biosci Biotechnol Biochem. 2009; 73: 1163–1167. [DOI] [PubMed] [Google Scholar]
- 27. Tsuizaki M, Ohta A, Horiuchi H. Myosin motor-like domain of class VI chitin synthase CsmB of Aspergillus nidulans is not functionally equivalent to that of class V chitin synthase CsmA. Biosci Biotechnol Biochem. 2013; 77: 369–374. [DOI] [PubMed] [Google Scholar]
- 28. Aufauvre-Brown A, Mellado E, Gow NAR, Holden DW. Aspergillus fumigatus chsE: A gene related to CHS3 of Saccharomyces cerevisiae and important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet Biol. 1997; 21: 141–152. [DOI] [PubMed] [Google Scholar]
- 29. Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans . Eukaryot Cell 2005; 4: 1902–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim J-E, Lee H-J, Lee J, Kim KW, Yun S-H, Shim W-B, et al. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr Genet. 2009; 55: 449–459. 10.1007/s00294-009-0258-6 [DOI] [PubMed] [Google Scholar]
- 31. Kong L-A, Yang J, Li G-T, Qi L-L, Zhang Y-J, Wang C-F, et al. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae . PLoS Pathogens 2012; 8: e1002526–e1002526. 10.1371/journal.ppat.1002526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larson TM, Kendra DF, Busman M, Brown DW. Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr Genet. 2011; 57: 177–189. 10.1007/s00294-011-0334-6 [DOI] [PubMed] [Google Scholar]
- 33. Liu HB, Kauffman S, Becker JM, Szaniszlo PJ. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot Cell 2011; 3: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mellado E, Aufauvre-Brown A, Gow NAR, Holden DW. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol Microbiol. 1996; 20: 667–679. [DOI] [PubMed] [Google Scholar]
- 35. Pareja-Jaime Y, Martin-Urdíroz M, González Roncero MI, Antonio González-Reyes J, Ruiz Roldán MDC. Chitin synthase-deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild-type infection. Mol Plant Pathol. 2010; 11: 479–493. 10.1111/j.1364-3703.2010.00624.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Treitschke S, Doehlemann G, Schuster M, Steinberg G. The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis . Plant Cell 2010; 22: 2476–2494. 10.1105/tpc.110.075028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Werner S, Sugui JA, Steinberg G, Deising HB. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola . Mol Plant-Microbe Interact. 2007; 20: 1555–1567. [DOI] [PubMed] [Google Scholar]
- 38. Soulié M- C, Perino C, Piffeteau A, Choquer M, Malfatti P, Cimerman A, et al. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol. 2006; 8: 1310–1321. [DOI] [PubMed] [Google Scholar]
- 39. Sheng W, Yamashita S, Ohta A, Horiuchi H. Functional differentiation of chitin synthases in Yarrowia lipolytica . Biosci Biotechnol Biochem. 2013; 77: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 40. Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nature Rev Mol Cell Biol. 2009; 10: 682–696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- 41. Steinberg G. Motors in fungal morphogenesis: cooperation versus competition. Curr Opin Microbiol. 2011; 14: 660–667. 10.1016/j.mib.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 42. Schoch CL, Aist JR, Yoder OC, Turgeon BG. A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet Biol. 2003; 39: 1–15. [DOI] [PubMed] [Google Scholar]
- 43. Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbruegge M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci. 2012; 125: 2740–2752. 10.1242/jcs.101212 [DOI] [PubMed] [Google Scholar]
- 44. Konzack S, Rischitor PE, Enke C, Fischer R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans . Mol Biol Cell 2005;16: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Requena N, Alberti-Segui C, Winzenburg E, Horn C, Schliwa M, Philippsen P, et al. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans . Mol Microbiol. 2001; 42: 121–132. [DOI] [PubMed] [Google Scholar]
- 46. Schuster M, Kilaru S, Fink G, Collemare J, Roger Y, Steinberg G. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol Biol Cell 2011; 22: 3645–3657. 10.1091/mbc.E11-03-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seiler S, Plamann M, Schliwa M. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora . Curr Biol. 1999; 9: 779–785. [DOI] [PubMed] [Google Scholar]
- 48. Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol Biol Cell 2009; 20: 673–684. 10.1091/mbc.E08-07-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006; 25: 2275–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schuster M, Treitschke S, Kilaru S, Molloy J, Harmer NJ, Steinberg G. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J. 2012; 31: 214–227. 10.1038/emboj.2011.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yao XL, Zhang J, Zhou H, Wang E, Xiang X. In vivo roles of the basic domain of dynactin p150 in microtubule plus-end tracking and dynein function. Traffic 2012; 13: 375–387. 10.1111/j.1600-0854.2011.01312.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J, Li SH, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell 2003; 14: 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang J, Zhuang L, Lee Y, Abenza JF, Peñalva MA, Xiang X. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J Cell Sci. 2010; 123: 3596–3604. 10.1242/jcs.075259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bartnicki-García S. Chitosomes: past, present and future. FEMS Yeast Res. 2006; 6: 957–965. [DOI] [PubMed] [Google Scholar]
- 55. Riquelme M, Bartnicki-García S, González-Prieto JM, Sánchez-León E, Verdín-Ramos JA, Beltrán-Aguilar A, et al. Spitzenkörper localization and intracellular traffic of green fluorescent protein-labeled CHS-3 and CHS-6 chitin synthases in living hyphae of Neurospora crassa . Eukaryot Cell 2007; 6: 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ichinomiya M, Motoyama T, Fujiwara M, Takagi M, Horiuchi H, Ohta A. Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans . Microbiology 2002; 148: 1335–1347. [DOI] [PubMed] [Google Scholar]
- 57. Rowlands RT, Turner G. Nuclear and extranuclear inheritance of oligomycin resistance in Aspergillus nidulans . Mol Gen Genet. 1973; 126: 201–216. [DOI] [PubMed] [Google Scholar]
- 58. May G. Fungal technology In: Kinghorn JR, Turner G, editors. Applied molecular genetics of filamentous fungi. Glasgow, UK: Blackie Academic & Professional; 1992. pp. 1–27. [Google Scholar]
- 59. Sambrook J, Russell JW. Molecular Cloning: a laboratory manual, 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 60. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004; 41: 973–981. [DOI] [PubMed] [Google Scholar]
- 61. Takeshita N, Ohta A, Horiuchi H. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide, and regulated in response to osmotic conditions. Biochem Biophys Res Commun. 2002; 298: 103–109. [DOI] [PubMed] [Google Scholar]
- 62. Ichinomiya M, Yamada E, Yamashita S, Ohta A, Horiuchi H. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans . Eukaryot Cell 2005; 4: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rieder SE, Emr SD. Isolation of subcellular fractions from the yeast Saccharomyces cerevisiae. Curr Protoc Cell Biol. 2001; 2008/01/30 ed. pp. Unit 3.7. [DOI] [PubMed]
- 64. Sánchez-León E, Verdín J, Freitag M, Roberson RW, Bartnicki-Garcia S, Riquelme M. Traffic of chitin synthase 1 (CHS-1) to the Spitzenkörper and developing septa in hyphae of Neurospora crassa: Actin dependence and evidence of distinct microvesicle populations. Eukaryot Cell 2011; 10: 683–695. 10.1128/EC.00280-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abenza JF, Galindo A, Pantazopoulou A, Gil C, de los Ríos V, Peñalva MA. Aspergillus RabB (Rab5) integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol Biol Cell 2010; 21: 2756–2769. 10.1091/mbc.E10-02-0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schuster M, Kilaru S, Ashwin P, Lin C, Severs NJ, Steinberg G. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 2011; 30: 652–664. 10.1038/emboj.2010.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Steinberg G. Endocytosis and early endosome motility in filamentous fungi. Curr Opin Microbiol. 2014; 20: 10–18. 10.1016/j.mib.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Seidel C, Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 tail is necessary and sufficient to recognize modified microtubules. PLoS One 2012; 7: e30976–e30976. 10.1371/journal.pone.0030976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schuchardt I, Assmann D, Thines E, Schuberth C, Steinberg G. Myosin-V, kinesin-1, and kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis . Mol Biol Cell 2005; 16: 5191–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Araujo-Bazán L, Peñalva MA, Espeso EA. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans . Mol Microbiol. 2008; 67: 891–905. 10.1111/j.1365-2958.2007.06102.x [DOI] [PubMed] [Google Scholar]
- 71. Hervás-Aguilar A, Peñalva MA. Endocytic machinery protein SlaB is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans . Eukaryot Cell 2010; 9: 1504–1518. 10.1128/EC.00119-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peñalva MA. Endocytosis in filamentous fungi: Cinderella gets her reward. Curr Opin Microbiol. 2010; 13: 684–692. 10.1016/j.mib.2010.09.005 [DOI] [PubMed] [Google Scholar]
- 73. Upadhyay S, Shaw BD. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans . Mol Microbiol. 2008; 68: 690–705. 10.1111/j.1365-2958.2008.06178.x [DOI] [PubMed] [Google Scholar]
- 74. Peñalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, et al. Searching for gold beyond mitosis: Mining intracellular membrane traffic in Aspergillus nidulans . Cell Logist. 2012; 2: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McGoldrick CA, Gruver C, May GS. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995; 128: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment was done with the clustalW alignment program using the entire amino acid sequence of each chitin synthase. The tree was drawn using Njplot. Branch length indicates evolutionary distance. Chitin synthases belong to class III and V are shown in red and blue, respectively. Abbreviations: Nc, N. crassa; Sc, S. cerevisiae; Um, U. maydis.
(TIF)
(A) Strategy for constructing GFP-CsmA producing strains. (B) Southern blot analysis of EcoRV digested total DNAs of the EGFP-CsmA strains using the 1.0 kb SpeI-XhoI fragment of pM-ALK-CHS5 [23] as a probe. Lane 1, ABPU1; lane 2, EGFP-CsmA1; lane 3, EGFP-CsmA2.
(TIF)
(A) GFP-CsmA in the cell lysates of the EGFP-CsmA1 (WT), EGFP-CsmA/ΔkinA1 strain (ΔkinA), and EGFP-CsmA/ΔuncA1 (ΔuncA) strains were detected with anti-GFP antibody. (B) EGFP-ChsB in the cell lysates of the EB-5 strain (WT), EGFP-ChsB/ΔkinA1 (ΔkinA), and EGFP-ChsB/ΔuncA1 (ΔuncA) strains were detected using the same antibody. P10, 10,000 x g pellet; P100, 100,000 x g pellet; S100, 100,000 x g supernatant.
(TIF)
P100 fractions of the CΔMHA strain or the CΔMHA/mRFP-KinArigor1 strain (A), and the D10HA strain or the D10HA/mRFP-KinArigor1 strain (B) were immunoprecipitated with anti-HA antibody and subjected to Western blot analysis.
(TIF)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.