Abstract
The cellular and molecular characteristics of a cell line (BME26) derived from embryos of the cattle tick Rhipicephalus (Boophilus) microplus were studied. The cells contained glycogen inclusions, numerous mitochondria, and vesicles with heterogeneous electron densities dispersed throughout the cytoplasm. Vesicles contained lipids and sequestered palladium meso-porphyrin (Pd-mP) and rhodamine–hemoglobin, suggesting their involvement in the autophagic and endocytic pathways. The cells phagocytosed yeast and expressed genes encoding the antimicrobial peptides (microplusin and defensin). A cDNA library was made and 898 unique mRNA sequences were obtained. Among them, 556 sequences were not significantly similar to any sequence found in public databases. Annotation using Gene Ontology revealed transcripts related to several different functional classes. We identified transcripts involved in immune response such as ferritin, serine proteases, protease inhibitors, antimicrobial peptides, heat shock protein, glutathione S-transferase, peroxidase, and NADPH oxidase. BME26 cells transfected with a plasmid carrying a red fluorescent protein reporter gene (DsRed2) transiently expressed DsRed2 for up to 5 weeks. We conclude that BME26 can be used to experimentally analyze diverse biological processes that occur in R. (B.) microplus such as the innate immune response to tick-borne pathogens.
Keywords: Tick, Embryonic cells, Immunity, Transfection, Sleeping Beauty, DsRed2 gene
1. Introduction
Ticks are obligate ectoparasite arthropods that infest numerous species of terrestrial vertebrates. There are currently 866 described species of ticks (Horak et al., 2002). Ticks are more versatile vectors than mosquitoes, in that they can transmit a wide variety of pathogenic organisms, such as fungi, viruses, rickettsiae, bacteria, and protozoa, during the feeding process (Sonenshine, 1991). The tick Rhipicephalus (Boophilus) microplus is the most important cattle ectoparasite in the southern hemisphere. It thrives in regions of high humidity and elevated temperature found throughout Brazil. R. (B.) microplus is the vector of babesiosis and anaplasmosis caused by protozoan and rickettsial microorganisms, respectively, imposing serious difficulties to farmers and to the economy of tropical and subtropical countries. Such diseases, along with the direct parasitic action of ticks in cattle, make the infestation of bovine herds by R. (B.) microplus one of the main causes of low productivity of cattle grazing in these regions. New strategies for controlling tick populations to an acceptable level are needed to prevent enormous economic losses in cattle production (Sonenshine et al., 2006; Willadsen, 2006).
An effective immune response is essential for tick survival against microbial infections. We have purified four antimicrobial peptides from fully engorged R. (B.) microplus. One was obtained from intestinal contents (Fogaça et al., 1999) and the other three were purified from the hemolymph (Fogaça et al., 2004, 2006). Surprisingly, the peptide (Hb31–63) from the intestinal contents corresponded to fragment 33–61 of the α-chain of bovine hemoglobin that is active against Gram-positive bacteria, fungi, and the protozoan Leishmania amazonensis (Fogaça et al., 1999). We have shown that peptide cytotoxicity is due to permeabilization of the microbial membrane (Sforça et al., 2005). Microplusin, isolated from cell-free hemolymph (Fogaça et al., 2004), belongs to a new class of antimicrobial peptide and was also found in ovaries of fully engorged females and eggs (Esteves, 2003). The other two peptides, isolated directly from hemocytes, were a defensin (Fogaça et al., 2004) with sequence similarity with insect defensins and ixodidin (Fogaça et al., 2006), similar to some inhibitors of serine proteinases. We have shown that the hemocytes of R. (B.) microplus also produce reactive species of oxygen (ROS) when stimulated by both membrane components of bacteria and phorbol ester (PMA) (Pereira et al., 2001). The diverse primary structures and sites of synthesis and storage of these antimicrobial peptides, added to the phagocytic activity and ROS production by hemocytes, suggest that these defense mechanisms might work together, preventing infection of the vector and allowing these animals to survive.
Recently, we developed interest in the study of the immunological aspects of the interactions between R. (B.) microplus and pathogens transmitted by this tick. We are using tick cell culture as a model for analyzing the immunological interactions of R. (B.) microplus with tick-borne pathogens. Several cell lines isolated from embryonic tissues of R. (B.) microplus have been reported (Pudney et al., 1973; Holman and Ronald, 1980; Holman, 1981; Kurtti et al., 1988; Bell-Sakyi, 2004). We started these studies by characterizing one of these cell lines, BME26 (Kurtti et al., 1988), that had remained largely uncharacterized, except for its interaction with tick-associated spirochetes and rickettsiae (Kurtti et al., 1993, 2005). We carried out the cytological characterization of BME26 by light and transmission electron microscopy (TEM). The cell line identity was confirmed by partial sequencing of the mitochondrial 16S rRNA gene. A cDNA library was constructed and the analysis of 898 unique sequences revealed several abundant transcripts related to different functional classes including the immune system. In preparation for future immune gene silencing studies using RNAi to explore aspects of the immunological pathogen–vector interaction, a method for transfecting BME26 cells with exogenous nucleic acid was also studied in the present work.
2. Materials and methods
2.1. Establishment and maintenance of BME26 cells
Cell line BME26 was derived from embryos of R. (B.) microplus following protocols developed for isolating cell lines from tick embryos (Pudney et al., 1973; Bhat and Yunker, 1977; Holman and Ronald, 1980; Holman, 1981). The primary culture was made on August 1981 using an egg mass from a single engorged female 17 days after the onset of oviposition, but before larval eclosion. The line originated from ticks collected from cattle near the town of Ciudad Victoria, Tamaulipas, Mexico in 1964. The ticks were maintained under laboratory conditions at the USDA, ARS, Cattle Fever Tick Research Laboratory, Falcon Heights, TX from 1964 until 1981 when they were donated to TJK and UGM. Briefly, eggs were disinfected with 70% ethanol containing Tween 80 (1 drop of ~50 μl per 10 ml) for 5 min and rinsed with sterile water followed by Leibovitz’s L-15 medium (Gibco). The eggs were transferred in 200 μl of Leibovitz’s L-15 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 10% tryptose phosphate broth (TPB) to a 35 mm Petri dish and crushed with a glass rod. The embryonic tissues were transferred to a centrifuge tube and centrifuged at 300g for 5 min. The pellet was resuspended in 2.5 ml of the medium described above and transferred to a flat-bottom culture tube (Nunc). Early subcultures were kept in the culture medium described previously (L-15B medium supplemented with 5% heat-inactivated FBS, 5% TPB, and 0.1% lipoprotein concentrate) (Munderloh and Kurtti, 1989). Starting with the serial passage 6 (210 days post-isolation of the primary culture) and every 5–10 subsequent serial transfers, BME26 cells were cryopre-served in liquid nitrogen using a protocol routinely used to prepare tick cells for freezing (Munderloh et al., 1994). These frozen stocks made available for comparing cells that were at different passage levels.
For the studies reported herein, the line BME26 had been serially transferred approximately 40 times. Starting with frozen stock from the 27th passage the cell line has been in continuous culture, and typically 6–10 serial transfers were made annually. The cells, unless noted otherwise, were grown in L-15B300 (Munderloh et al., 1999) (prepared from L-15B medium diluted 3:1 with sterile tissue-culture-grade water) and supplemented with 20% heat-inactivated FBS (Gibco), 10% TPB (Difco), 100 units ml−1 penicillin (Gibco), and 100 μg ml−1 streptomycin (Gibco), denominated in the present work as complete medium. Cultures were incubated at 34 °C in 25 cm2 plastic flasks (Nunc) containing 5 ml of the complete medium, which was fully changed weekly. Monolayers were subcultured when they reached a density of approximately 107 cells ml−1. Attached cells were resuspended in fresh complete medium using a 14-gauge, 10 cm laboratory cannula (Becton Dickinson, USA) with tip fitted to a 5 ml plastic syringe. Approximately 8 × 105 cells ml−1 were transferred to new flasks. Culture density was determined using a Neubauer hemocytometer and the cell viability was evaluated by Trypan Blue exclusion (0.4%, Sigma). The cell doubling time was determined by counting viable cells every 15 days during 45 days of cell growth. Two independent experiments were done for determination of cell doubling time.
2.2. Light microscopy
Approximately 1000 of BME26 cells from the 57th passage in their late exponential phase were plated on glass-cover slips in 24-well plates and incubated in complete medium at 34 °C for 3 days. After this period each of the following fluorescent probes were added to a different aliquot of cells and incubated for 1 min: 10 mmol l−1 DAPI; 1 μg ml−1 rhodamine 123; (a specific fluorescent probe for mitochondria) 30 μg ml−1 orange acridine; or 1 μg ml−1 Nile Red (all obtained from Sigma Chemical). The cells were washed with 0.15 mol l−1 NaCl, 10 mmol l−1 sodium phosphate, pH 7.2 (PBS) and observed under differential interference contrast (DIC) using a Zeiss Axioplan 2 microscope. The fluorescence images were obtained using a filtered 100 W mercury lamp as the excitation light source with filter sets Zeiss-09 (BP450-490 nm/FT510 nm/LP515 nm), or Zeiss-15 (BP546-12 nm/FT580 nm/LP590 nm), or Zeiss-01 (BP365-12 nm/FT395nm/LP397 nm). In some experiments, the BME26 cells from the 61st passage in their late exponential phase were incubated in complete medium containing 20 μmol l−1 hemoglobin (Sigma) labeled with rhodamine isothiocyanate or 100 μmol l−1 fluorescent metalloporphyrin Pd-mP (Porphyrin Products Inc. Logan) for 24 h. Alternatively, the cells were incubated simultaneously with 2 mmol l−1 hemoglobin and 20 μmol l−1 hemoglobin–rhodamine. The labeled hemoglobin and Pd-mP were prepared as described earlier (Lara et al., 2005). After incubation, the cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min and observed on an Axioplan 2 microscope (Zeiss, USA). In the phagocytosis experiments 100 μl of yeast (Saccharomyces cerevisiae) suspension in PBS (3.7 × 106 cells ml−1) was incubated with 1 μl of Cytotracker (Sigma) for 15 min. After this time, yeast was centrifuged and washed twice in 500 μl of complete medium. The BME26 cells from the 61st passage in the late exponential phase at a density of 3.5 × 104 cells ml−1 were incubated with 10 μl of the labeled yeast with multiplicity of infection (MOI) of 1 for 24 h. After incubation, the cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min and observed in an Axioplan 2 microscope (Zeiss, USA).
2.3. Transmission electron microscopy (TEM)
The BME26 cells from the 52nd passage in their late exponential phase were fixed at 4 °C with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 mol l−1 cacodylate buffer, pH 7.4, post-fixed in 1% OsO4 and embedded in Spurr resin (Sigma). Ultra thin sections of 70 nm were gathered onto copper grids and stained with 2% uranyl acetate in distilled water for 1 h, then washed in distilled water and stained in 0.5% lead citrate in distilled water (Silva et al., 1999). Ultrastructure was examined in a Jeol 100 CX-II electron microscope. For phagocytosis experiments, the cells were incubated with yeast cells (S. cerevisiae) (MOI of 1) for 24 h and prepared for TEM as described above.
2.4. Karyology
Metaphase chromosomes were prepared from the BME26 cells of 20th and 83rd passage and incubated for 24 h in medium containing 0.1 μg l−1 demecolcine (Sigma). Treated cells were washed in PBS, suspended in a hypo-osmotic solution of 75 mmol l−1 KCl for 30 min and fixed in a 3:1 solution of methanol:acetic acid. Fixed cells were dropped onto cold slides, air dried, and stained with Giemsa. To determine chromosome number, 200 sets of well-separated metaphase chromosomes were examined and counted.
2.5. DNA extraction, amplification by polymerase chain reaction (PCR), and sequencing
DNA was extracted from 107 BME26 cells (48th passage in their late exponential phase) using the protocol described previously (Medina-Acosta and Cross, 1993). The primers used for the amplification of a fragment of the 16S rRNA gene and PCR conditions were the same as described earlier (Mangold et al., 1998). The PCR product was separated by 1% agarose gel electrophoresis with ethidium bromide staining and the amplified fragment was purified using the Wizard SV Gel kit (Promega), and cloned on pGEM®-T Easy Vector System (Promega). Sequencing of both strands was performed using the Big Dye Primer Cycle Terminator System and analyzed on PRISM® 310 Genetic Analyzer (reagents and equipment from Applied Biosystems). The 16S rRNA sequence obtained from the BME26 cell line was aligned with sequences of other hard-tick species available in GenBank using the CLUSTAL (W). The sequences used were: Boophilus microplus (L34310), Boophilus annulatus (Z97877), Rhipicephalus bursa (Z97878), Haemaphysalis cretica (L34308), Amblyomma hebraeum (L34316), and Ixodes ricinus (Z97882).
2.6. RNA extraction, cloning, expression, and sequencing of the antimicrobial peptides
Total RNA was isolated from a confluent culture of BME26 cells (50th passage in their late exponential phase) using Trizol reagent (Gibco/BRL), as recommended by the manufacturer, and used as model for the reverse-transcription using the SMART RACE cDNA Amplification Kit (Clontech). The oligonucleotides used to amplify microplusin and defensin sequences were derived from their sequences described previously (Fogaça et al., 2004): 5′-TGAAGGCCATCTTCGTGTCCG-3′ (microplusin sense oligonucleotide); 5′-AAGCCCACCATGAGCACGAC-CA-3′ (microplusin antisense oligonucleotide); 5′-GCA-GAAGGTTGCTAAA-3′ (defensin sense oligonucleotide); and 5′-TAGCGTTTGTGCGACACACTG-3′ (defensin antisense oligonucleotide). The PCR conditions were 5 min at 94 °C, 37 cycles of 30 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C, followed by an extension period of 7 min at 72 °C. The analysis of the products obtained by PCR, cloning, and sequencing were realized as described above.
2.7. cDNA library
The total RNA was isolated from BME26 cells (55th passage in their late exponential phase) as described above. The mRNA was purified from the total RNA using the PolyATract mRNA Isolation System III kit (Promega). The cDNA library was prepared using the CloneMinerTM cDNA Library Construction kit (Invitrogen). The 5′ end of the cDNA inserts from randomly selected clones were sequenced on an ABI PRISM® 3700 DNA sequencer (Applied Biosystems, USA) using the BigDye® Terminator cycle sequencing protocol (Applied Biosystems) and the M13 reverse sequencing primer (5′-AACAGCTATGAC-CATG-3′). Chromatograms from sequenced clones were automatically processed for sequence extraction, quality control, and assembly of redundant sequences as described previously (Lorenzini et al., 2006). The assembled sequences were submitted to similarity searches (BlastX-e-value cutoff 1 × 10−6) against public database sequences (Uniprot, http://www.pir.uniprot.org/). The Gene Ontology terms associated with Uniprot sequences found in the similarity searches were automatically annotated to the corresponding assemble sequences. The assemble sequences were also manually annotated, where possible, with only one Gene Ontology entry for molecular function and biological process. The manual annotation was based on the results of automatic annotations and on information from related sequences on other public databases (Interpro, PubMed). The identification of immune-related sequences was assisted by a list of Gene Ontology terms prepared previously (Lorenzini et al., 2006).
2.8. Transfection of BME26 cells
We evaluated transfection efficiency and possible stable gene transfer using a plasmid that included both Sleeping Beauty transposon and transposase (Ivics et al., 1997) functions on the same plasmid (Sauer et al., 2004) (generously provided by Drs. Blazar and Kren, University of Minnesota). In this construct the SB10 transposase gene, driven by the cytomegalovirus (CMV) promoter, resided outside the IR/DR of the transposable element containing the reporter gene, red fluorescent protein DsRed2 (Clontech). The DsRed2 was driven by the chicken β actin promoter and flanked by the IR/DR regions of the Sleeping Beauty transposon (Ivics et al., 1997). The CAGGS promoter was a chimera constructed from the chicken β actin promoter and CMV immediate early promoter sequences (Sauer et al., 2004). Subconfluent cultures (12.5 or 25 cm2 flasks with 1 × 105 to 5 × 105 cells ml−1) were seeded 7 days before transfection. The transfection was prepared using the Transfection Reagent Selector Kit following the protocol as given by the manufacturer (Qiagen) for transfection of adherent cells. Cells in passages 43–52 were grown in L15B supplemented with FBS (5%), TPB (5%), and lipoprotein concentrate (0.1%) (Munderloh and Kurtti, 1989). Endotoxin-free plasmid DNA (1–2 μg) was diluted in EC buffer (150 μl) and enhancer was added to condense plasmid DNA (8 μl enhancer per μg of DNA). It was then added to non-liposomal lipid Effectene (25 μl per μg of DNA) to form DNA micelles. This was further diluted in 1 ml of medium and added drop-wise onto “dry” cell layers that had been previously washed with PBS. As a control, vector DNA was added directly to 1.2 ml of complete medium without the use of Enhancer or Effectene. The flasks were incubated for 6 h at 34 °C and complete medium was added to each flask (2 ml to 12.5 cm2 flasks or 4 ml to 25 cm2 flasks). Transfected cultures were incubated at 34 °C for 1 week in medium containing the plasmid DNA complexes. Medium was changed weekly and examined periodically with a Nikon Diaphot inverted fluorescence microscope to determine the frequency of DsRed2 positive cells. At selected times after transfection, the number of DsRed expressing cells per field in 10 randomly selected microscope fields was determined. Data were expressed as the means of DsRed2 positive cells per mm2 of culture surface±standard deviations of the means.
3. Results
3.1. Growth characteristics of BME26 cell line
BME26 cells maintained in complete medium had a doubling time of approximately 15 days. The cells multiplied to form confluent cell layers within 2 months, reaching a density of approximately 1 × 107 cells ml−1. In the medium with 5% FBS, 10% TPB, and lipoprotein supplement the doubling time was approximately 7 days, closer to the 110–114 h doubling time observed previously (Munderloh and Kurtti, 1989).
3.2. BME26 cell line karyotype
The chromosome profile of line BME26 changed with increasing number of passages. In the 20th passage BME26 cells had the chromosome complement standard for R. (B.) microplus and most other Metastriata ticks (Hilburn et al., 1989; Gunn et al., 1993; Garcia et al., 2002). All chromosomes were acrocentric and the X chromosome was the largest. One pair of autosomes was distinguished by only one distinguishable pair of autosomes. There was only one distinguishable pair of autosomes that had a secondary constriction near the centromere. The predominant number of chromosomes per cell was 21 (79%) with one large X chromosome, the male complement of chromosomes. The remaining cells were aneuploid (20 or 23 chromosomes) with one X chromosome. The line in the 83rd passage was composed of diploid (69%) and tetraploid (31%) cells and most of the cells had the male complement of chromosomes (2n = 21, with one X chromosome, and 4n = 42, with two X chromosomes).
3.3. Confirmation of BME26 cell line species identity
We confirmed the R. (B.) microplus origin of the BME26 cell line by PCR amplification of the mitochondrial 16S rRNA gene. There was complete identity of the line BME26 partial 16S rRNA sequence with R. (B.) microplus (L34310), confirming that the BME26 cells originated from this tick species (Fig. 1). The position of the BME26 partial 16S rRNA sequence is 5-455 of the 16S rRNA gene (L34310). The percent identity of the line BME26 16S rRNA gene fragment with those from other ixodes ticks were: B. annulatus (Z97877), 97%; R. bursa (Z97878), 91%; H. cretica (L34308), 80%; A. hebraeum (L34316), 77%; and I. ricinus (Z97882), 76%.
Fig. 1.

Sequence alignment of the 16S rRNA fragment obtained from the Rhipicephalus (Boophilus) microplus (RBm) (L34310) and BME26 cell line (BME26). Dots represent identical nucleotides.
3.4. Cytological and functional characteristics
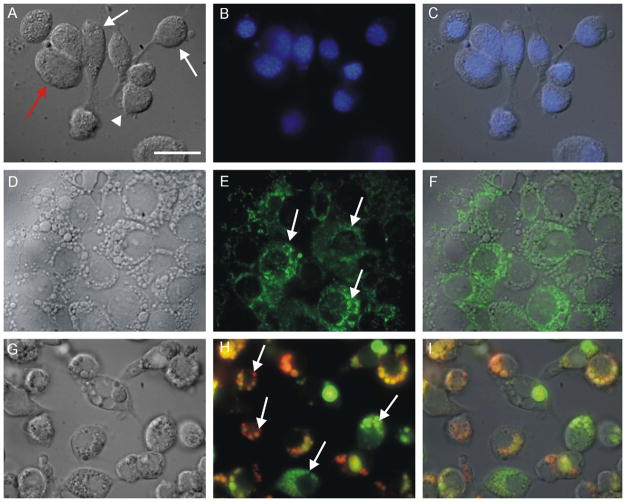
BME26 cells firmly adhered to the substrate and were morphologically heterogeneous at the beginning of the subculture. Cell cultures up to 3 days old had some cells with a fibroblast-like appearance (Fig. 2A, white arrow). In this initial stage, round cells were also observed having nuclei with areas extending over most of the cytoplasm (Fig. 2A–C, arrowhead) along with slightly larger cells with cytoplasmic vesicles (Fig. 2A and D, red arrow). Despite these morphological differences among cells soon after the subculture, when the monolayer reached confluence larger cells with cytoplasmic vesicles predominated.
Fig. 2.
General characterization of cellular morphology and structures of R. (B.) microplus embryonic cells (BME26), 3 days after subculture. (A, D, G) DIC microscopy of live cells; (B, E, H) fluorescence microscopy; (C, F, I) merged images. (A–C) DAPI staining demonstrating different morphologies (white arrow: cell with a fibroblast-like appearance, red arrow: cell with cytoplasmic vesicles, and arrowhead: round cell with nucleus occupying most of the cytoplasm); (D–F) rhodamine 123 fluorescence reveals a large and heterogeneous mitochondrion (arrows); (G–I) acridine orange staining showing large and small vesicles with different degrees of acidification (arrows). Scale bar = 20 μm.
The BME26 cells labeled with rhodamine-123 contained numerous mitochondria dispersed in the cytoplasm, indicating a high rate of respiratory metabolism (Fig. 2D–F). BME26 cells contained a high number of cytoplasmic vesicles that was independent of culture age. These vesicles displayed different degrees of acidification after staining with acridine orange (Fig. 2G–I). Cells incubated with Nile Red displayed fluorescence signals within large vesicles, indicating that they contained lipids (Fig. 3A and B). This feature was confirmed by a strong green emission after incubation of cells with palladium meso-porphyrin (Pd-mP) (Fig. 3C and D). As described earlier (Lara et al., 2005), Pd-mP shifts its emission from red, when bound to proteins that are in the cytoplasm of these cells, to green when exposed to an environment with low water content, as in lipid droplets and lipid bilayers. The lipid content of these large vesicles is shown in Fig. 3D.
Fig. 3.
Characterization of contents of large vesicles in BME26 by light microscopy. (A, C) DIC microscopy; (B, D) fluorescence microscopy; (A, B) Nile Red incorporation in large vesicles indicating high lipid contents (arrows); (C, D) cells pre-incubated with Palladium Mesoporphyrin IX (Pd-mP) show a strong green emission inside the large vesicles, another indicator of lipid content inside these organelles (arrows). Scale bar = 20 μm.
The ultrastructure of BME26 cells revealed a cytoplasm full of large electron-dense vesicles (Fig. 4A, asterisks) that in higher magnification (Fig. 4B) were shown to contain heterogeneous and most likely high lipid content. The number of these vesicles is variable in BME26 cells. In the interior of these structures we observed small vesicles (arrow), an indicator of autophagy. We also observed a high level of cytoplasmic glycogen content (Fig. 4B, asterisks).
Fig. 4.
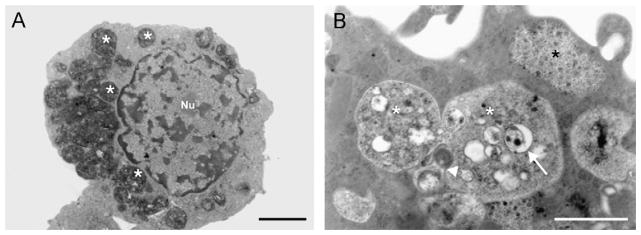
Characterization of BME26 cells by transmission electron microscopy. (A) Cell showing pronounced number of large electron-dense vesicles (white asterisks); and (B) large vesicles (white asterisks) with heterogeneous contents, rich in vesicles (white arrow), probably derived from autophagy (arrowhead). Black asterisk showing glycogen. Scale bar = 1 μm.
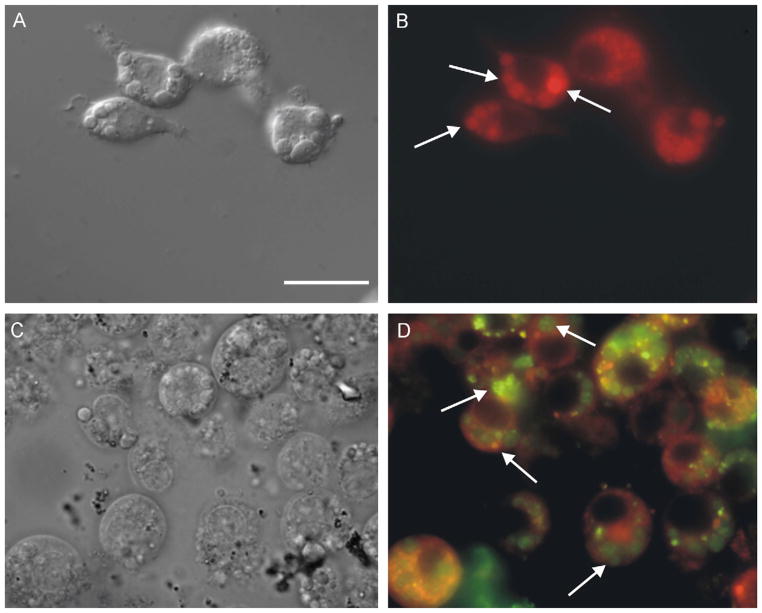
On the other hand, the uptake of rhodamine–hemoglobin by BME-26 cells indicated that these large vesicles were not involved in the endocytic pathway, and that the small vesicles were the final destination of endocytosed proteins (Fig. 5A–C). Addition of a 100-fold excess of unlabeled hemoglobin to the incubation medium did not block the uptake by hemoglobin–rhodamine in part of the cells, indicating that this endocytosis was not receptor mediated (Fig. 5D–F).
Fig. 5.
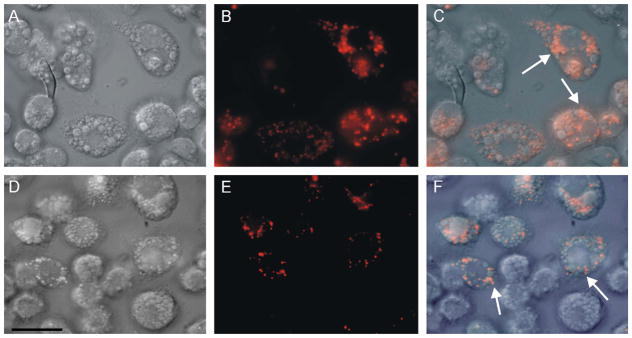
Light microscopy of live BME26 cells after uptake of hemoglobin conjugated to rhodamine. (A, D) DIC microscopy; (B, E) fluorescence microscopy; (C, F) merged images. Incubation conditions were: (A–C) cells exposed to 20 μmol l−1 rhodamine–hemoglobin, presenting labeling in small vesicles (arrows in merged image); (D–F) cells exposed to 20 μmol l−1 rhodamine–hemoglobin plus 2 mmol l−1 unlabeled hemoglobin, labeling is maintained in small vesicles (arrows in merged image). Scale bar = 20 μm.
Due to our interest in the immunological aspects of ticks, we investigated whether BME26 cells displayed any immune functions. The results presented in Fig. 6 demonstrated the ability of BME26 cells to phagocytose yeast cells. Fluorescent yeast cells appeared within the cytoplasm of the BME26 cells within 24 h of incubation (Fig. 6A) and TEM revealed that these yeast cells were inside vacuoles (Fig. 6B). In addition, sequences of two genes encoding antimicrobial peptides (defensin and microplusin) were amplified from RNA of BME26 cells by RT-PCR. They were 100% identical to the corresponding genes from R. (B.) microplus ticks (Fogaça et al., 2004).
Fig. 6.
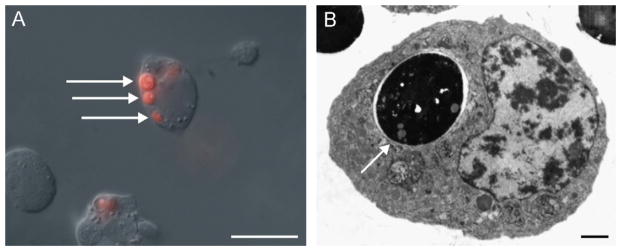
Light and transmission electron microscopy of BME26 cells phagocytosing S. cerevisiae. (A) Merged image of DIC of live cells showing phagocytosed yeast cells pre-stained with cytotracker (arrows). Scale bar = 20 μm. (B) Transmission electron microscopy showing phagocytosed yeast cell within vacuole (arrow). Nu: nucleus. Scale bar = 1 μm.
3.5. Molecular characterization
Our sequencing of BME26 cDNA library resulted in 898 unique sequences having an average length of 477 bp. The high-quality ESTs were deposited in the GenBank. Among them, 556 did not match any sequences in the Uniprot database and the remaining sequences were manually annotated with only one term of the molecular function ontology (Gene Ontology) (Fig. 7). This annotation revealed that several functional gene classes were abundantly expressed by the BM26 cell line. The sequences annotated with Gene Ontology terms as related to the immune response were individually analyzed (Table 1).
Fig. 7.
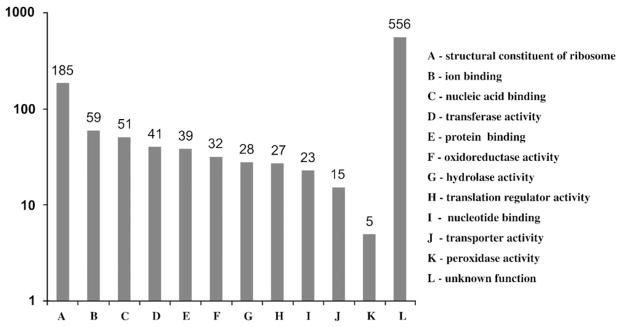
Gene expression profile of BME26 cell line using Gene Ontology. Values indicate the number of sequenced clones from the library grouped in categories of Molecular Function Ontology.
Table 1.
Putative immune-related transcripts of the BME26 cells’ cDNA library
| AS code | Acc. code | Description | Species | E-value |
|---|---|---|---|---|
| BMJFCE1001D06.g | Q6WNX4_BOOMI | Ferritin | Boophilus microplus | 1.0e–94 |
| BMJFCE1002H03.g | Q8MVB7JXOSC | Alpha-2-macroglobulin | Ixodes scapularis | 5.0e–22 |
| BMJFCE1001D02.g | Q86LE5_BOOMI | Microplusin preprotein | Boophilus microplus | 1.Oe–27 |
| Q64K37_9ACAR | Hebraein | Amblyomma hebraeum | 1.0e–20 | |
| BMJFCE1007B06.g | Q5I2A7_MYTGA | Heat shock 70 kDa protein | Mytilus galloprovincialis | 7.0e–82 |
| BMJFCE1010G05.g | Q6JVM9_RHIAP | Glutathione S-transferase | Rhipicephalus appendiculatus | 1.0e–121 |
| BMJFCE1018B10.b | Q7PTJ0_ANOGA | Peroxidase activity | Anopheles gambiae | 3.0e–55 |
| BMJFCE1004E12.b | Q9VQH2_DROME | NADPH oxidase | Drosophila melanogaster | 6.0e–29 |
| BMJFCE1006G02.g | Q8I862_DERVA | Factor D-like protein | Dermacentor variabilis | 5.0e–71 |
| BMJFCE1012H01.b | Q9NFY2_ANOGA | Serine protease | Anopheles gambiae | 3.0e–31 |
| BMJFCE1019E08.b | Q8WQX0_RHIAP | Serine proteinase inhibitor | Rhipicephalus appendiculatus | 3.0e–48 |
The BLASTX hits with the lowest E-values (implying the most significant similarities) are indicated.
3.6. Transfection of BME26 cells
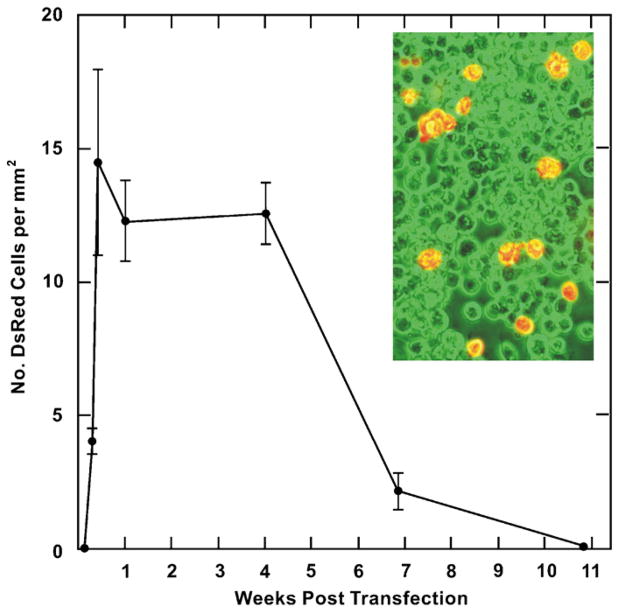
The expression of DsRed2 in transfected BME26 cells was followed over time by fluorescence microscopy to determine how rapidly transient expression arose and lasted. Cultures were also examined for evidence of colonies of stably transformed BME26 cells. Expression of DsRed2 was first noted 48 h after transfection (Fig. 8). The most rapid increase in red fluorescent cells occurred during the first week and the number of DsRed2 cells per mm2 remained stable for 4 weeks post-transfection (Fig. 8, inset). Cells expressing DsRed2 were initially seen as individual or paired cells and later as small colonies. The prevalence of colonies, indicative of transfected cell replication and possible insertion of the DsRed2 gene into the genome, was very low. By the end of 5 weeks, 8–10% of the cells expressed DsRed2 (approximated by counting the number of DsRed expressing cells in a field and dividing it by the total number of cells in that field) after which the number of DsRed2 positive cells declined. After 7 weeks there were very few DsRed2 expressing colonies of BME26 cells (5–10 colonies per flask, each containing 20–30 DsRed positive cells). At this point the cell layers were overgrown and detached from the flask. We have not developed a method for clonal selection of transformed BME26 cells and stably transformed BME26 sublines expressing DsRed2 were not obtained. Without the use of the DNA-condensing enhancer and Effectene to transfect the cells with the vector there was no DsRed2 expression.
Fig. 8.
Transient expression of red fluorescent protein DsRed2 by BME26 cells transfected with a cis-plasmid containing the Sleeping Beauty SB10 transposase and a transposon containing the DsRed2 gene. Data points are the means±standard deviations. Inset: DsRed2 expressing BME26 cells 3 weeks post-transfection. Fluorescent microscopic image (red cells) was superimposed over the phase contrast microscopic image (green filter used) of the same field.
4. Discussion
In this paper we describe the cytological characteristics of BME26 cells by light and TEM and report on the gene expression profile of these cells by cDNA sequencing.
Firstly, we confirmed the origin of the BME26 cells by comparison of the partial sequences of the mitochondrial 16S rRNA gene with those from different species of ixodid ticks. Our result shows clearly that BME26 cell line originated from R. (B.) microplus. However, as the line was isolated from embryonic tissue fragments from a single egg mass, the tissue of origin and level of differentiation of the cells comprising the line are unknown. Also, the line has never been cloned and as a result the number of different cell types that are present is also unknown.
All of the R. (B.) microplus cell lines that have been karyotyped to date, including BME26 in this study, are predominantly diploid with 21 (male complement) or 22 (female complement) chromosomes with a low proportion of aneuploid or tetraploid cells (Pudney et al., 1973; Holman and Ronald, 1980; Holman, 1981). Other than to identify the large sex chromosome and count the number of chromosomes present in a slide spreading, little attention has been paid to the autosomes. With the exception of an autosome having a secondary constriction (the third largest autosome noted by Garcia et al., 2002) the acrocentric autosomes are alike in shape, vary slightly in size from one another and are not clearly distinguishable by conventional staining methods. No effort has been made to determine which of the chromosomes are absent or duplicated in the aneuploid cells observed in the R. (B.) microplus cell lines. The technical difficulty to obtain good chromosome spreads for analysis without chromosome loss, overlap or breakage also needs to be considered. Cytogenetic studies similar to those carried out by Garcia et al. (2002) need to be done on cell line(s) being considered for R. (B.) microplus genomics research.
The BME26 cells had a relatively slow growth with doubling time of approximately 15 days. As with other tick cell lines, the doubling time of BME26 cells is strongly influenced by incubation temperature, medium supplementation, and passage history (e.g., frequency of feeding and transfer dilution) and can range from 5 (Munderloh and Kurtti, 1989) to 15 days. Other B. microplus line cells have a reported doubling time of 3.8 days (Holman, 1981) and 5.0 days (Cossio-Bayugar et al., 2002). However, in other lines of embryonic cells established from B. microplus, growth was also slow and subcultures were made only after 3–4 weeks (Pudney et al., 1973). The reported in vitro doubling times are in striking contrast to the rapid rate of cell multiplication that occurs during embryogenesis. The length of time from oviposition to hatching, that can occur in a laboratory growing R. (B.) microplus, is as short as 15 days (Nuñ ez et al., 1985). There is thus a clear need to learn more about the nutritional and physiological requirements of cultured R. (B.) microplus cells.
At the passage levels used in the present study, BME26 cells morphology was heterogeneous initially after the subculture, with attached cells having a fibroblast-like appearance and others being rounded up and small. After this initial growth period, and in cultures with confluent cell layers, most of the cells lost their fibroblast-like appearance and became rounded and enlarged.
The results obtained by light and TEM showed the BME26 cells having numerous mitochondria and glycogen inclusions dispersed throughout the cytoplasm. The experiments with a hydrophobic fluorescent probe, Nile Red, showed that the content of the large vesicles was mainly lipidic. These results were confirmed by TEM and Pd-mP uptake and subsequent fluorescence emission analysis. On the other hand, the cytosol showed a red Pd-mP emission, indicating a putative cytosolic heme-binding protein. The study of heme fate in the digest cells of R. (B.) microplus also suggested the presence of a cytosolic heme-binding protein involved in heme transport to hemosomes, where it was detoxified (Lara et al., 2005). It was already evidenced that heme transport and detoxification systems could be a useful target to new acaracides (Citelli et al., 2007), and BME26 cells could be an interesting powerful tool for drug discovery. After incubation with acridine orange, we observed different degrees of acidification in the vesicles, indicating that these vesicles were in different stages of maturation, or represented different structures, with some involved in autophagy and others in endocytosis.
Uptake of hemoglobin–rhodamine by the small vesicles indicated their involvement in the endocytic pathway. The early endosomes can function as a sorting station on the endocytic pathway. The regulation of the vesicular transport is dependent on the Rab proteins, which are members of the Ras superfamily of GTPases (Goody et al., 2005). The different degrees of acidification observed in the vesicles present in the cytoplasm of the BME26 cells and the lipid contents of these vesicles suggest that the BME26 cells possess high endocytic activity. Besides, we found in the cDNA library from BME26 cells one transcript with similarity to GTPase rab 11 (BMJFCE1001C10), indicating that the recycling endosomes are present in the endocytic pathway of BME26 cells (Clague, 1998).
The gene expression profile of BME26 cells was obtained through the sequencing of a cDNA library. The number of transcripts without match in the database was high, which could be the result either of novel genes or of sequencing problems that resulted in short sequences. Abundant transcripts were identified in functional classes including structural molecular activity, nucleic acid binding, ion binding, oxidoreductase activity, and transferase activity. Among them, the transcripts related with structural constituents of ribosome were the most abundant, indicating a high rate of the protein synthesis by BME26 cells.
Several transcripts that have been associated with immune functions were identified in the cDNA library. Among them, a ferritin sequence (BMJFCE1001D06) was the most abundant transcript in this category, which is well-known to be involved in defense functions and iron storage and transport in ticks and insects (Nichol et al., 2002; Mulenga et al., 2003, 2004).
In ovaries of the tick Dermacentor variabilis a partial cDNA fragment encoding an HC-like ferritin homolog (DVFER) was identified in a subtraction hybridization library. Interestingly, DVFER was up-regulated in response to Rickettsia montanensis infection (Mulenga et al., 2003). It has also been shown that both mechanical injury and Escherichia coli-challenge of D. variabilis induced significant increase of the mRNA level of DVFER. However, the mRNA level was 2-fold higher during bacterial challenge than during mechanical injury (Mulenga et al., 2004). Different results were obtained in Ixodes scapularis tick cells (IDE8) challenged with Anaplasma marginale where down-regulation of ferritin gene was detected (de la Fuente et al., 2007).
Other transcripts related to immune defense were serine proteases (BMJFCE1012H01.b) and protease inhibitors (BMJFCE1019E08.b). These molecules are involved in several invertebrate defense responses, such as hemolymph coagulation and melanization of pathogen surface (Kanost, 1999) as well as antimicrobial peptide production (Kambris et al., 2006; Wang et al., 2007).
We found one transcript similar to two antimicrobial peptides, microplusin and hebrain (BMJFCE1001D02). The gene expression of microplusin was detected in the ovaries, fat body, and hemocytes of the fully engorged females from R. (B.) microplus (Esteves, 2003; Fogaça et al., 2004). Microplusin is effective against several Gram-positive bacteria and filamentous fungi and its effect on Micrococcus luteus is bacteriostatic (Fogaça et al., 2004). Hebrain is an antimicrobial peptide isolated from the hemolymph of the tick A. hebraeum highly similar to microplusin (73%). Hebrain displays antimicrobial activity against Staphylococcus aureus, E. coli, and Candida albicans (Lai et al., 2004). In spite of the absence of defensin transcripts in the cDNA library, we have demonstrated the constitutive expression of defensin by sequencing the RT-PCR products obtained from non-challenged BME26 cells (data not shown), which showed 100% identity with the defensin sequence obtained from R. (B.) microplus (Fogaça et al., 2004).
Matilla and collaborators investigated the expression profile of two antimicrobial genes: lysozyme and defensin in I. scapularis and Dermacentor andersoni cell lines challenged by non-natural microorganisms transmitted by ticks (E. coli and M. luteus) and the endosymbiont Rickettsia peacockii (Mattila et al., 2007). Lysozyme gene was up-regulated in the two cell lines following challenge by E. coli and M. luteus, but not by R. peacockii. In addition, the up-regulation of defensin gene was observed only in I. scapularis cell line after bacterial stimulation. Interestingly, these results suggest that rickettsial endosymbionts may avoid activating the immune response of their host tick.
Other transcripts related to an immune response were also identified, including a heat shock protein (BMJFCE1007B06), glutathione S-transferase (BMJFCE-1010G05), peroxidase (BMJFCE1018B10), and NADPH oxidase (BMJFCE1004E12) (de Morais Guedes et al., 2005). Interestingly, heat shock protein and GST were identified as genes up-regulated in response to A. marginale infection of IDE8 tick cells (de la Fuente et al., 2007).
Transfection of cultured animal cells is widely used to analyze gene and promoter function and develop cultured cells of a desired phenotype. The most widely used transposition systems for the transformation of animal cells are those belonging to the Tc1mariner group. The Sleeping Beauty system is a broad host range transposon vector reconstructed from inactive mariner elements found in the genome of teleost fish (Ivics et al., 1997). It has also been used for the genetic transformation of the I. scapularis cell line ISE6 (Kurtti et al., 2006; Felsheim et al., 2006).
Our results indicated that the BME26 cells were transfected with a cis-plasmid carrying the Sleeping Beauty transposase and its transposon containing the DsRed2 gene. The use of transposon and transposase functions on the same plasmid proved to be convenient and effective. We also demonstrated that the CAGGS promoter was functional and that the heterologous DsRed2 gene was a suitable reporter gene for R. (B.) microplus cells. Our results also suggested that a non-liposomal lipid formulation (Effectene Reagent) could be used to introduce vector plasmids or interfering RNA into BME26 cells. The apparent decline in DsRed2 expression (number of DsRed2 cells per mm2) and in the intensity of cellular fluorescence that was seen after 4–5 weeks was taken as evidence of how long it took transiently transfected cells to eliminate or inactivate the DsRed2 gene introduced with the plasmid rather than their overgrowth by non-transfected cells. The presence of DsRed2 expressing colonies after 7 weeks could be taken as evidence of stable transformation. Nevertheless, further studies are needed to increase the overall level of transfection and select for stable transformants from a population of untransformed cells.
In conclusion, we performed a cellular and molecular characterization of the BME26 cells, which will certainly be useful for the investigation of aspects of the immunological pathogen–vector interaction as well as heme transport and detoxification. Currently, we are investigating the expression profile of the antimicrobial peptides, after challenge with different microorganisms, and in the near future we intend to silence their encoding genes by RNAi and evaluate the survival of BME26 cells upon challenge with pathogens. Interesting data obtained by RNAi gene silencing have been demonstrated in the tick D. variabilis and in the IDE8 cell line (de la Fuente et al., 2007). Four genes, which encode putative GST, salivary selenoprotein M, vATPase, and ubiquitin, affected A. marginale infection in the gut cells and the transmission from salivary glands. The A. marginale infection in IDE8 tick cells seems to be affected by six genes, which encode putative selenoprotein W2a, hematopoietic stem/progenitor cells protein-like, proteasome 26S subunit, ferritin, GST, and subolesin. Therefore the results of these studies demonstrated the successful use of RNAi in ticks or cultured cell ticks to evaluate the pathogen–vector interaction.
Acknowledgments
This work was supported by FAPESP (Fundaçã o de Amparo a Pesquisa do Estado de Sã o Paulo, Brazil) and CNPq (Conselho Nacional de Desenvolvimento Cientí fico e Tecnoló gico, Brazil). We are grateful to Susana P. Lima for technical assistance, Bruno Vellutini for microscopy backing, and Cassiano Pereira for figure preparation.
References
- Bell-Sakyi L. Ehrlichia ruminantium grows in cell lines from four ixodid tick genera. J Comp Pathol. 2004;130:285–293. doi: 10.1016/j.jcpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Bhat UK, Yunker CE. Establishment and characterization of a diploid cell line from the tick Dermacentor parumapertus Neumann (Acarina: Ixodidae) J Parasitol. 1977;63:1092–1098. [PubMed] [Google Scholar]
- Citelli M, Lara FA, Vaz JIdS, Oliveira PL. Oxidative stress impairs heme detoxification in the midgut of the cattle tick Rhipicephalus (Boophilus) microplus. Mol Biochem Parasitol. 2007;151:81–88. doi: 10.1016/j.molbiopara.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Clague MJ. Molecular aspects of the endocytic pathway. Biochem J. 1998;336(Part 2):271–282. doi: 10.1042/bj3360271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossio-Bayugar R, Wagner GG, Holman PJ. In vitro generation of organophosphate resistant Boophilus microplus (Acari: Ixodidae) cell lines. J Med Entomol. 2002;39:278–284. doi: 10.1603/0022-2585-39.2.278. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Blouin EF, Manzano-Roman R, Naranjo V, Almazan C, Perez de la Lastra JM, Zivkovic Z, Jongejan F, Kocan KM. Functional genomic studies of tick cells in response to infection with the cattle pathogen Anaplasma marginale. Genomics. 2007 doi: 10.1016/j.ygeno.2007.08.009. [DOI] [PubMed] [Google Scholar]
- de Morais Guedes S, Vitorino R, Domingues R, Tomer K, Correia AJ, Amado F, Domingues P. Proteomics of immune-challenged Drosophila melanogaster larvae hemolymph. Biochem Biophys Res Commun. 2005;328:106–115. doi: 10.1016/j.bbrc.2004.12.135. [DOI] [PubMed] [Google Scholar]
- Esteves E. MSc Thesis. Departamento de Parasitologia, ICB, Universidade de Sã o Paulo; Sã o Paulo: 2003. Biochemistry and molecular characterization of the antimicrobial peptide microplusin in ovaries and eggs of Boophilus microplus. [Google Scholar]
- Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 2006;6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogaça AC, Silva PI, Jr, Miranda MT, Bianchi AG, Miranda A, Ribolla PE, Daffre S. Antimicrobial activity of a bovine hemoglobin fragment in the tick Boophilus microplus. J Biol Chem. 1999;274:25330–25334. doi: 10.1074/jbc.274.36.25330. [DOI] [PubMed] [Google Scholar]
- Fogaça AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Dev Comp Immunol. 2004;28:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Fogaça AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2006;27:667–674. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Garcia RN, Garcia-Fernandez C, Garcia SML, Valente VLS. Mitotic and meiotic chromosomes of a southern Brazilian population of Boophilus microplus (Acari, Ixodidae) Iheringia Sé rie Zool. 2002;92:63–70. [Google Scholar]
- Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn SJ, Hilburn LR, Burbach BS. Homology within the X chromosomes of Boophilus microplus (Canestrini) and B. annulatus (Say) J Hered. 1993;84:232–235. doi: 10.1093/oxfordjournals.jhered.a111327. [DOI] [PubMed] [Google Scholar]
- Hilburn LR, Gunn SJ, Davet RB. The genetics of New World Boophilus microplus (Canestrini) and Boophilus annulatus (Say) in their possible control. Bull Soc Vector Ecol. 1989;14:222–231. [Google Scholar]
- Holman PJ. Partial characterization of a unique female diploid cell strain from the tick Boophilus microplus (Acari: Ixodidae) J Med Entomol. 1981;18:84–88. doi: 10.1093/jmedent/18.1.84. [DOI] [PubMed] [Google Scholar]
- Holman PJ, Ronald NC. A new tick cell line derived from Boophilus microplus. Res Vet Sci. 1980;29:383–387. [PubMed] [Google Scholar]
- Horak IG, Camicas JL, Keirans JE. The Argasidae, Ixodidae and Nuttalliellidae (Acari: Ixodida): a world list of valid tick names. Exp Appl Acarol. 2002;28:27–54. doi: 10.1023/a:1025381712339. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Kurtti TJ, Munderloh UG, Ahlstrand GG, Johnson RC. Borrelia burgdorferi in tick cell culture: growth and cellular adherence. J Med Entomol. 1988;25:256–261. doi: 10.1093/jmedent/25.4.256. [DOI] [PubMed] [Google Scholar]
- Kurtti TJ, Munderloh UG, Krueger DE, Johnson RC, Schwan TG. Adhesion to and invasion of cultured tick (Acarina: Ixodidae) cells by Borrelia burgdorferi (Spirochaetales: Spirochaetaceae) and maintenance of infectivity. J Med Entomol. 1993;30:586–596. doi: 10.1093/jmedent/30.3.586. [DOI] [PubMed] [Google Scholar]
- Kurtti TJ, Simser JA, Baldridge GD, Palmer AT, Munderloh UG. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick Dermacentor andersoni (Acari, Ixodidae) J Invertebr Pathol. 2005;90:177–186. doi: 10.1016/j.jip.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti TJ, Felsheim RF, Mattila JT, Baldridge GD, Burkhardt NY, Munderloh UG. Stable transformation of a tick (Ixodes scapularis) cell line with the Sleeping Beauty transposon system. Soc In Vitro Cell Dev Biol J. 2006;42:32-A. [Google Scholar]
- Lai R, Takeuchi H, Lomas LO, Jonczy J, Rigden DJ, Rees HH, Turner PC. A new type of antimicrobial protein with multiple histidines from the hard tick Amblyomma hebraeum. FASEB J. 2004;18:1447–1449. doi: 10.1096/fj.03-1154fje. [DOI] [PubMed] [Google Scholar]
- Lara FA, Lins U, Bechara GH, Oliveira PL. Tracing heme in a living cell: hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus. J Exp Biol. 2005;208:3093–3101. doi: 10.1242/jeb.01749. [DOI] [PubMed] [Google Scholar]
- Lorenzini DM, Silva PI, Jr, Soares MB, Arruda P, Setubal J, Daffre S. Discovery of immune-related genes expressed in hemocytes of the tarantula spider Acanthoscurria gomesiana. Dev Comp Immunol. 2006;30:545–556. doi: 10.1016/j.dci.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Mangold AJ, Bargues MD, Mas-Coma S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae) Parasitol Res. 1998;84:478–484. doi: 10.1007/s004360050433. [DOI] [PubMed] [Google Scholar]
- Mattila JT, Munderloh UG, Kurtti TJ. Rickettsia peacockii, an endosymbiont of Dermacentor andersoni, does not elicit or inhibit humoral immune responses from immunocompetent D. andersoni or Ixodes scapularis cell lines. Dev Comp Immunol. 2007;31:1095–1106. doi: 10.1016/j.dci.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Acosta E, Cross GA. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol Biochem Parasitol. 1993;59:327–329. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Macaluso KR, Simser JA, Azad AF. Dynamics of Rickettsia-tick interactions: identification and characterization of differentially expressed mRNAs in uninfected and infected Dermacentor variabilis. Insect Mol Biol. 2003;12:185–193. doi: 10.1046/j.1365-2583.2003.00400.x. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Simser JA, Macaluso KR, Azad AF. Stress and transcriptional regulation of tick ferritin HC. Insect Mol Biol. 2004;13:423–433. doi: 10.1111/j.0962-1075.2004.00502.x. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Kurtti TJ. Formulation of medium for tick cell culture. Exp Appl Acarol. 1989;7:219–229. doi: 10.1007/BF01194061. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J Parasitol. 1994;80:533–543. [PubMed] [Google Scholar]
- Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol. 1999;37:2518–2524. doi: 10.1128/jcm.37.8.2518-2524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol H, Law JH, Winzerling JJ. Iron metabolism in insects. Annu Rev Entomol. 2002;47:535–559. doi: 10.1146/annurev.ento.47.091201.145237. [DOI] [PubMed] [Google Scholar]
- Nuñez JL, Muñoz-Cobeñas ME, Moltedo HL. The Common Cattle Tick. Springer; Berlin: 1985. Boophilus microplus. [Google Scholar]
- Pereira LS, Oliveira PL, Barja-Fidalgo C, Daffre S. Production of reactive oxygen species by hemocytes from the cattle tick Boophilus microplus. Exp Parasitol. 2001;99:66–72. doi: 10.1006/expr.2001.4657. [DOI] [PubMed] [Google Scholar]
- Pudney M, Varma MG, Leake CJ. Culture of embryonic cells from the tick Boophilus microplus (Ixodidae) J Med Entomol. 1973;10:493–496. doi: 10.1093/jmedent/10.5.493. [DOI] [PubMed] [Google Scholar]
- Sauer MG, Ericson ME, Weigel BJ, Herron MJ, Panoskaltsis-Mortari A, Kren BT, Levine BL, Serody JS, June CH, Taylor PA, Blazar BR. A novel system for simultaneous in vivo tracking and biological assessment of leukemia cells and ex vivo generated leukemia-reactive cytotoxic T cells. Cancer Res. 2004;64:3914–3921. doi: 10.1158/0008-5472.CAN-03-3991. [DOI] [PubMed] [Google Scholar]
- Sforça ML, Machado A, Figueredo RC, Oyama S, Jr, Silva FD, Miranda A, Daffre S, Miranda MT, Spisni A, Pertinhez TA. The micelle-bound structure of an antimicrobial peptide derived from the alpha-chain of bovine hemoglobin isolated from the tick Boophilus microplus. Biochemistry. 2005;44:6440–6451. doi: 10.1021/bi0475323. [DOI] [PubMed] [Google Scholar]
- Silva JRMC, Stainess N, Hernandez-Blazquez FJ. Experimental studies on the response of the fish (Nothothenia corticeps, Richardson, 1844) to parasite (Pseudoterranova decipiens, Krabbe, 1878) and other irritant stimuli at Antarctic temperatures. Polar Biol. 1999;22:417–424. [Google Scholar]
- Sonenshine DE. Biology of Ticks. Oxford University Press; New York/Oxford: 1991. [Google Scholar]
- Sonenshine DE, Kocan KM, de la Fuente J. Tick control: further thoughts on a research agenda. Trends Parasitol. 2006;22:550–551. doi: 10.1016/j.pt.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng T, Rayaprolu S, Zou Z, Xia Q, Xiang Z, Jiang H. Proteolytic activation of pro-spatzle is required for the induced transcription of antimicrobial peptide genes in lepidopteran insects. Dev Comp Immunol. 2007;31:1002–1012. doi: 10.1016/j.dci.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willadsen P. Tick control: thoughts on a research agenda. Vet Parasitol. 2006;138:161–168. doi: 10.1016/j.vetpar.2006.01.050. [DOI] [PubMed] [Google Scholar]