Abstract
Genome wide association studies (GWAS) have identified multiple loci associated with cross-sectional eGFR, but a systematic genetic analysis of kidney function decline over time is missing. Here we conducted a GWAS meta-analysis among 63,558 participants of European descent, initially from 16 cohorts with serial kidney function measurements within the CKDGen Consortium, followed by independent replication among additional participants from 13 cohorts. In stage 1 GWAS meta-analysis, SNPs at MEOX2, GALNT11, IL1RAP, NPPA, HPCAL1 and CDH23 showed the strongest associations for at least one trait, in addition to the known UMOD locus which showed genome-wide significance with an annual change in eGFR. In stage 2 meta-analysis, the significant association at UMOD was replicated. Associations at GALNT11 with Rapid Decline (annual eGFRdecline of 3ml/min/1.73m2 or more), and CDH23 with eGFR change among those with CKD showed significant suggestive evidence of replication. Combined stage 1 and 2 meta-analyses showed significance for UMOD, GALNT11 and CDH23. Morpholino knockdowns of galnt11 and cdh23 in zebrafish embryos each had signs of severe edema 72 hours after gentamicin treatment compared to controls, but no gross morphological renal abnormalities before gentamicin administration. Thus, our results suggest a role in the deterioration of kidney function for the loci GALNT11 and CDH23, and show that the UMOD locus is significantly associated with kidney function decline.
Keywords: chronic kidney disease, kidney development
Introduction
Chronic kidney disease (CKD) is an important public health problem affecting up to 10% of adults world-wide [1–3]. Faster rates of decline in estimated glomerular filtration rate (eGFR), and entry into CKD stages of increasing severity are associated with an increased risk of cardiovascular and all-cause mortality [4–9]. Thus, recently issued guidelines on the evaluation and management of patients with CKD have highlighted the importance of evaluating longitudinal measures of renal function in addition to determining eGFR and urinary albumin excretion at discrete time points [3].
Traditional risk factors for CKD include diabetes and hypertension, but these do not fully account for CKD risk [10]. There is evidence for considerable clustering of CKD within families [11] and the heritability of eGFR has been estimated at up to 36–75% in population-based studies [12]. Using genome-wide association studies (GWAS), multiple loci have been identified in association with eGFR and CKD in both European [13–16] and non-European populations [17,18] using data from one time point. However, multiple lines of evidence suggest that there may be unique genetic contributions to renal function decline above and beyond baseline renal function. First, there is substantial variability in the rate of eGFR decline in studies of healthy persons as well as among those with CKD [3,4,19,20]. Second, we have previously shown that some genetic loci associated with cross-sectional eGFR are also associated with incident CKD even after accounting for baseline eGFR [21]. Finally, genetic background has been shown to affect CKD progression in animal models [22,23].
Taken together, these data suggest that unique loci may exist for renal function decline in addition to those identified for a one-time measure of eGFR. Thus, we conducted a genome-wide association study (GWAS) meta-analysis among participants from 16 cohorts with serial kidney function measurements within the CKDGen Consortium, followed by independent replication among additional participants from 13 cohorts.
Results
Study participants
Changes in renal function over time were derived from 45,530 individuals who participated in stage 1 meta-analysis of study-specific GWAS, and an additional 18,028 independent individuals who participated in stage 2 meta-analysis (Table 1). Details on study design and genotyping are provided in Supplementary Tables 1 and 2 respectively.
Table 1.
Stage 1 and Stage 2 cohort characteristics.
| n | Age at baseline, years |
% women, n |
%HTN at baseline, n |
%DM at baseline, n |
%CKD at baseline, n |
eGFR baseline, ml/min/1.73m2 |
eGFR follow- up, ml/min/1.73m2 |
Duration between baseline and follow-up (Years) mean, SD |
|
|---|---|---|---|---|---|---|---|---|---|
| Stage 1 discovery cohorts | |||||||||
| AGES | 3219 | 54.2 (8.98) | 58.0, 1867 | 21.6, 694 | 3.2, 103 | 3.2, 104 | 89.6 (19.3) | 73.0 (20.0) | 22.2 (6.7) |
| Amish | 458 | 46.9 (14.3) | 54.4, 249 | 8.7, 40 | 4.2, 19 | NA | 95.9 (24.3) | 89.2 (19.4) | 5.2 (2.6) |
| ARIC | 9049 | 54.5 (5.7) | 52.9, 4793 | 26.9, 2427 | 8.6, 780 | 2.9, 260 | 89.7 (17.0) | 83.5 (17.1)A 80.6 (17.1)B |
8.0 (2.2) |
| ASPS | 848 | 65.8 (8.1) | 56.8, 482 | 72.5, 615 | 9.8, 83 | 12.7, 108 | 80.2 (20.2) | 74.6 (15.1) | 5.5 (2.0) |
| CHS | 2820 | 71.9 (5.0) | 61.3, 1729 | 34.5, 966 | 11.0, 310 | 7.9, 224 | 77.3 (20.8) | 75.4 (20.2) | 5.9 (1.8) |
| CoLaus | 1918 | 53.9 (10.8) | 54.9, 1053 | 35.4, 679 | 6.31, 121 | 5.0, 95 | 91.5 (20.4) | 84.9 (18.2) | 5.58 (0.29) |
| FHS (Offspring and Cohort) | 2523 | 58.1 (8.6) | 55.6, 1405 | 36.8, 927 | 8.1, 206 | 8.3, 210 | 88.7 (25.5) | 79.5 (19.1) | 11.1 (3.6) |
| GENOA | 1041 | 54.7 (10.3) | 55.6, 579 | 71.7, 746 | 8.8, 92 | 4.3, 45 | 92.1 (20.7) | 89.8 (22.8) | 4.0 (1.1) |
| HABC | 888 | 73.4 (2.8) | 48.5, 431 | 33.1, 294 | 8.5, 75 | 21.4, 190 | 71.8 (13.2) | 72.9 (21.2) | 9.0 (0.3) |
| JUPITER | 8780 | 66.1 (7.8) | 32.2, 2826 | 63.8, 5602 | 0.6, 54 | 11.5, 1008 | 80.1 (18.1) | 78.2 (17.7) | 2.6 (1.0) |
| KORA3 | 1641 | 52.5 (10.1) | 49.5, 813 | 38.3, 629 | 4.3, 71 | 3.2, 53 | 91.3 (18.0) | 83.9 (21.0) | 10 (0) |
| KORA4 | 1807 | 53.8 (8.9) | 51.3, 927 | 33.7, 606 | 3.7, 66 | 3.9, 70 | 89.5 (17.5) | 85.1 (20.2) | 7.1 (0.4) |
| MESA | 2324 | 63.2 (10.1) | 51.7, 1201 | 37.3, 866 | 5.5, 135 | 13.3, 310 | 74.2 (13.9) | 70.7 (15.1) | 4.8 (0.5) |
| The Rotterdam Study (RS-I) | 2422 | 66.5 (7.0) | 60.2, 1459 | 50.8, 1230 | 7.5, 182 | 7.7, 186 | 79.9 (15.5) | 73.7 (15.8) | 6.5 (0.4) |
| SHIP | 3203 | 49.2 (15.3) | 51.8, 1659 | 24.3, 778 | 7.0, 225 | 3.7, 119 | 92.4 (19.8) | 90.6 (23.6) | 5.3 (0.7) |
| Three Cities (3C) | 2589 | 73.0 (4.5) | 61.9, 1602 | 76.7, 1986 | 8.6, 223 | 18.9, 489 | 73.1 (16.1) | 71.0 (16.8) | 3.8 (0.3) |
| Stage 2 replication cohorts | |||||||||
| ADVANCE | 2034 | 67.0 (6.6) | 31.9, 649 | 55.2, 1123 | 100, 2034 | 16.0, 325 | 84.1 (28.1) | 84.8 (34.5) | 4.9 (0.9) |
| BMES | 1304 | 62.9 (7.7) | 60.1, 784 | 67.1, 875 | 5.4, 71 | 23.9, 312 | 82.6 (31.7) | 75.5 (34.9) | 10.4 (0.6) |
| COLAUS | 2238 | 53.1 (10.4) | 53.9, 1207 | 24.0, 538 | 4.1, 91 | 3.5, 79 | 90.5 (19.5) | 88.7 (18.7) | 5.5 (0.3) |
| HYPERGENES | 651 | 53.4 (7.5) | 45.3, 295 | 13.9, 91 | 0 | 0.61, 4 | 107.4 (23.5) | 103.4 (35.1) | 5.6 ( 3.2) |
| KORA3 | 1494 | 51.6 (13.3) | 52.5, 785 | 29.4, 437 | 5.1, 76 | 2.6, 39 | 98.0 (20.1) | 92.4 (21.3) | 9.6 (0.6) |
| KORA4 | 1200 | 49.2 (15.4) | 52.4, 629 | 13.3, 159 | 4, 48 | 5.8, 70 | 97.4 (21.7) | 92.6 (22.4) | 7.2 (0.5) |
| NESDA | 1270 | 43.3 (12.3) | 67.2, 854 | 32.4, 411 | 5.4, 69 | 0.9, 11 | 97.8 (20.4) | 95.5 (18.5) | 2.0 (0.3) |
| popgen | 577 | 60.2 (9.4) | 42.1, 243 | 50.4, 288 | 5.2, 30 | 6.1, 35 | 84.5 (17.2) | 79.9 (18.4) | 4.8 (0.8) |
| PREVEND (4 year follow-up) | 791 | 53.0 (13.3) | 50.8, 402 | 40.8, 323 | 4.2, 33 | 6.0, 47 | 89.2 (19.5) | 106.4 (34.2) | 4.3 (0.6) |
| PREVEND (9 year follow-up) | 2169 | 48.0 (11.1) | 48.0, 1040 | 28.2, 612 | 3.1, 66 | 1.7, 37 | 93.7 (17.7) | 86.9 (18.6) | 9.4 (0.84) |
| RS-II | 1243 | 61.8 (5.2) | 54.6, 679 | 21.4, 266 | 7.8, 97 | 8.6, 186 | 81.2 (17.0) | 73.7 (15.8) | 10.6 (0.4) |
| SAPHIR | 1374 | 51.6 (6.0) | 39.0. 536 | 54.9, 754 | 2.6, 36 | 0.9, 13 | 91.5 (15.8) | 88.0 (16.0) | 4.6 (0.7) |
| YFS | 1683 | 31.9 (4.9) | 56.0, 943 | 8.3, 139 | 1.1, 18 | 0.2, 3 | 105.4 (16.4) | 100.4 (16.0) | 6.0 (0) |
Unless indicated otherwise, values are given as mean (SD)
A: eGFR at visit 2; B: eGFR at visit 4; DM: Diabetes; HTN: hypertension; CKD: Chronic kidney disease (eGFR<60ml/min/1.73m2); eGFR: estimated glomerular filtration rate; SD: standard deviation
At the baseline examination, the prevalence of CKD, defined as eGFR<60 ml/min/1.73m2, ranged from 3.2% to 21.4% in stage 1 cohorts and from 0.2% to 23.9% in stage 2 replication cohorts. As expected, cohorts with lower mean age at baseline tended to have a lower baseline prevalence of CKD. Four kidney function decline traits were derived from serial eGFR values in each study participant to model mechanisms underlying different rates of kidney function change over time: 1) annual decline of eGFR (eGFRchange, in ml/min/1.73m2 decline per year; a positive value represents a decline in eGFR, whilst a negative value represents a rise in eGFR over time), 2) incident CKD to select individuals with a decline in kidney function to the clinical outcome CKD stage 3 or higher (CKDi, cases defined as those free of CKD at baseline but eGFR<60ml/min/1.73m2 during follow-up), 3) incident CKD with additionally at least a 25% eGFR decline from baseline to select individuals reaching CKD stage 3 after a sizeable decline in kidney function (CKDi25,) [24], and 4) rapid eGFR-decline to select individuals with the highest risk of adverse outcomes (Rapid Decline, cases defined as those with annual eGFR-decline≥3ml/min/1.73m2) [5]. Most cohorts showed a decline in kidney function over time (Table 1). The distribution of all four traits in stage 1 and stage 2 cohorts can be found in Supplementary Table 3.
Heritability of eGFR change
The heritability of eGFR change in the Framingham Heart Study was estimated as 38%, after adjusting for age, sex, and baseline eGFR.
Stage 1 meta-analysis of GWAS of measures of kidney function change over time
Stage 1 GWAS meta-analysis was performed in all samples for all four traits. Two secondary association analyses were performed to account for potentially different rates of kidney function decline in those with and without CKD: 1) eGFRchange stratified by baseline CKD status and 2) Rapid Decline in only those without baseline CKD; too few individuals with CKD fulfilled Rapid Decline criteria to perform this analysis. Supplementary Figure 1 shows the Manhattan and QQ-plots of the stage 1 meta-analysis of each trait. The genomic control factor ranged from 1.007 – 1.05, suggesting negligible evidence for population stratification.
In GWAS meta-analysis of stage 1 cohorts, the minor T allele of rs12917707 at the UMOD locus, previously identified by GWAS to be associated with higher eGFR in cross-sectional analysis [14], was associated with an increase in eGFR over time at a genome-wide significant level (p=2.6×10−14, Table 2), and showed at least nominally significant, direction consistent association with all other analyzed phenotypes (Supplementary Table 4). In addition, SNPs at the novel CDH23, GALNTL5/GALNT11, MEOX2, IL1RAP/OSTN, C2orf48/HPCAL1 and NPPB/NPPA loci were associated with at least one of the analyzed traits at a significance level of p<10−6 (Table 2). Thus, a total of 7 SNPs were moved forward to stage 2 meta-analysis. These SNPs mostly showed high imputation quality in each cohort or were genotyped de-novo (Supplementary Table 5), and low between-study heterogeneity (I2<25%).
Table 2.
Genetic association results of SNPs identified in stage 1 meta-analysis
| discovery stage 1 | Replication stage 2 |
Stage 1 and stage 2 combined |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPID | trait | Chr | Position (b36) |
Locus | coded allele |
non- coded allele |
AF coded allele |
beta | PVal2G C |
beta | one-sided pval1GC |
beta | two-sided pval1GC |
total sample size |
| rs12917707 | eGFRchange overall | 16 | 20275191 | UMOD, PDILT | t | g | 0.18 | −0.15 | 2.6×10−14 | −0.12 | 4.7×10−5 | −0.14 | 1.8×10−17 | 59373 |
| rs11803049 | eGFRchange CKD | 1 | 11851482 | NPPB, NPPA, KIAA2013, CLCN6 | a | g | 0.07 | −0.57 | 3.6×10−7 | 0.02 | 0.43* | −0.27 | 6.2×10−4 | 4116 |
| rs875860 | eGFRchange CKD | 10 | 72979535 | CDH23 | t | c | 0.12 | −0.49 | 6.2×10−7 | −0.15 | 0.047* | −0.31 | 4.6×10−6 | 4116 |
| rs11764932 | Rapid Decline overall | 7 | 15699643 | MEOX2 | a | g | 0.36 | 0.12 | 6.8×10−8 | 0.04 | 0.14 | 0.09 | 3.6×10−7 | 61078 |
| rs1019173 | Rapid Decline overall | 7 | 151341480 | GALNTL5, GALNT11, MLL3, CCT8L1 | a | g | 0.63 | −0.12 | 3.0×10−7 | −0.06 | 0.04 | −0.10 | 2.1×10−7 | 61077 |
| rs9814367 | Rapid Decline noCKD | 3 | 192075180 | IL1RAP, OSTN | t | c | 0.92 | −0.20 | 4.1×10−7 | 0.02 | 0.39 | −0.13 | 7.3×10−5 | 56687 |
| rs759341 | CKDi25 | 2 | 10297660 | C2orf48, HPCAL1, RRM2 | a | g | 0.31 | 0.18 | 1.5×10−6 | 0.06 | 0.27$ | 0.16 | 2.7×10−6 | 41122 |
“Locus” is based on build 36, hg18. The gene closest to the SNP is listed first and is in boldface if the SNP is located within the gene.
studies included: ADVANCE, BMES, COLAUS, RS-II
studies included: ADVANCE, BMES, RS-II
Stage 2 meta-analysis
Of the seven loci moved forward for stage 2 meta-analysis, only rs12917707 at UMOD was significantly associated with the stage 1 trait after correcting for multiple testing (p=4.7*×10−5). Two further SNPs showed suggestive significance (one-sided p<0.05) with their respective stage 1 trait: rs875860 in CDH23 with eGFRchange in those with CKD at baseline, and rs1019173 at GALNTL5/GALNT11 with Rapid Decline (Table 2). There was no significant heterogeneity between studies for these two SNPs (rs875860: I2=9.7%, p=0.34; rs1019173: I2=32.4%, p=0.12) or for the other SNPs analyzed in stage 2 meta-analysis (I2 <30.0%).
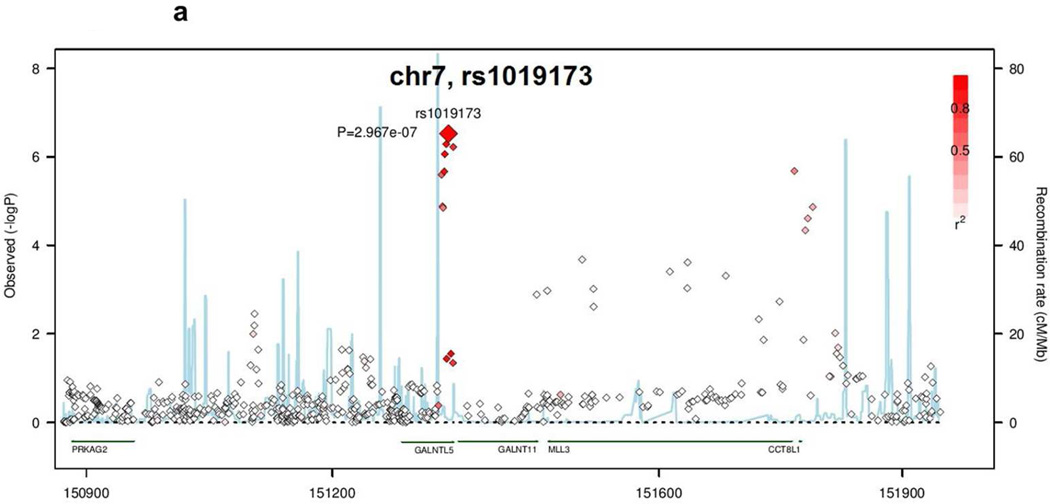
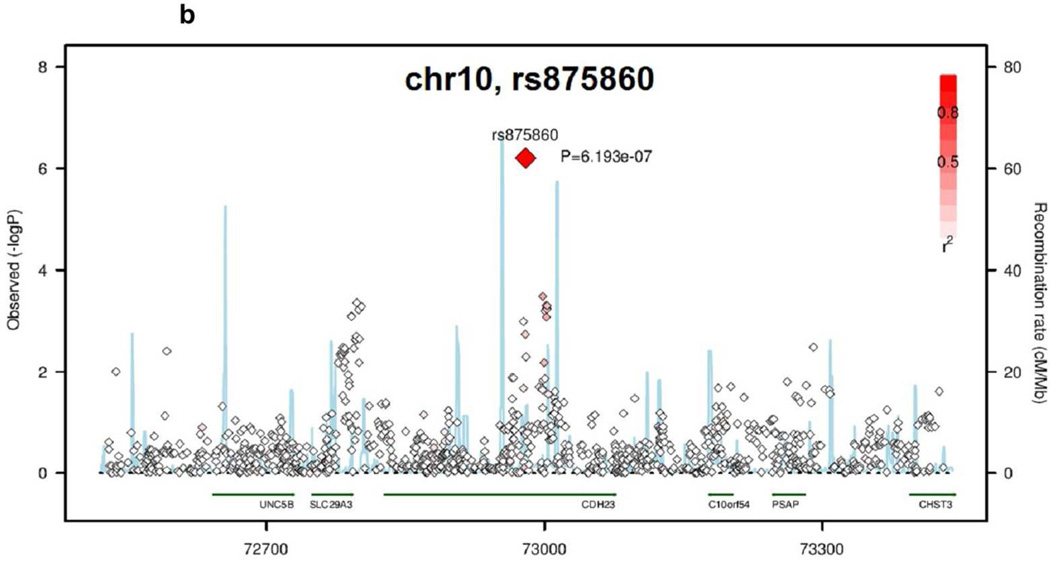
The SNP rs1019173 is located in an intron in the GALNTL5 gene, and lies in a linkage disequilibrium (LD) block spanning the genes GALNT11, MLL3, CCT8L, and part of the GALNTL5 gene (Figure 1a). The SNP in CDH23, rs875860, is an intronic SNP in an LD block whose boundaries lie within the coding region of the CDH23 gene (Figure 1b).
Figure 1.
Regional Association Plots of the novel loci identified by GWAS of kidney function decline traits. Negative log10 pvalues are plotted versus genomic position (build 36, hg18). The lead SNP in each region is labeled. Other SNPs in each region are color-coded based on their LD to the lead SNP. Light blue lines indicate recombination rate (cM/Mb). (A) GALNTL5/GALNT11 locus. (B) CDH23 locus.
In the combined meta-analysis of these three SNPs from both stage 1 and stage 2 cohorts, there was no evidence of between-study heterogeneity in the combined metaanalysis (I2<25%). Only the SNP at UMOD showed genome-wide significant association (rs12917707, p=1.2×10−16) in the combined stage 1 and stage 2 analysis, whereas there was suggestive evidence of significance for the two novel loci identified in stage 1 (rs875860 in CDH23: p=1.5×10−6 for the association with eGFRchange in those with CKD; rs1019173 at GALNTL5/GALNT11: OR=0.91 for the A allele, p=2.2×10−7 for the association with Rapid Decline).
Functional validation of novel loci in zebrafish
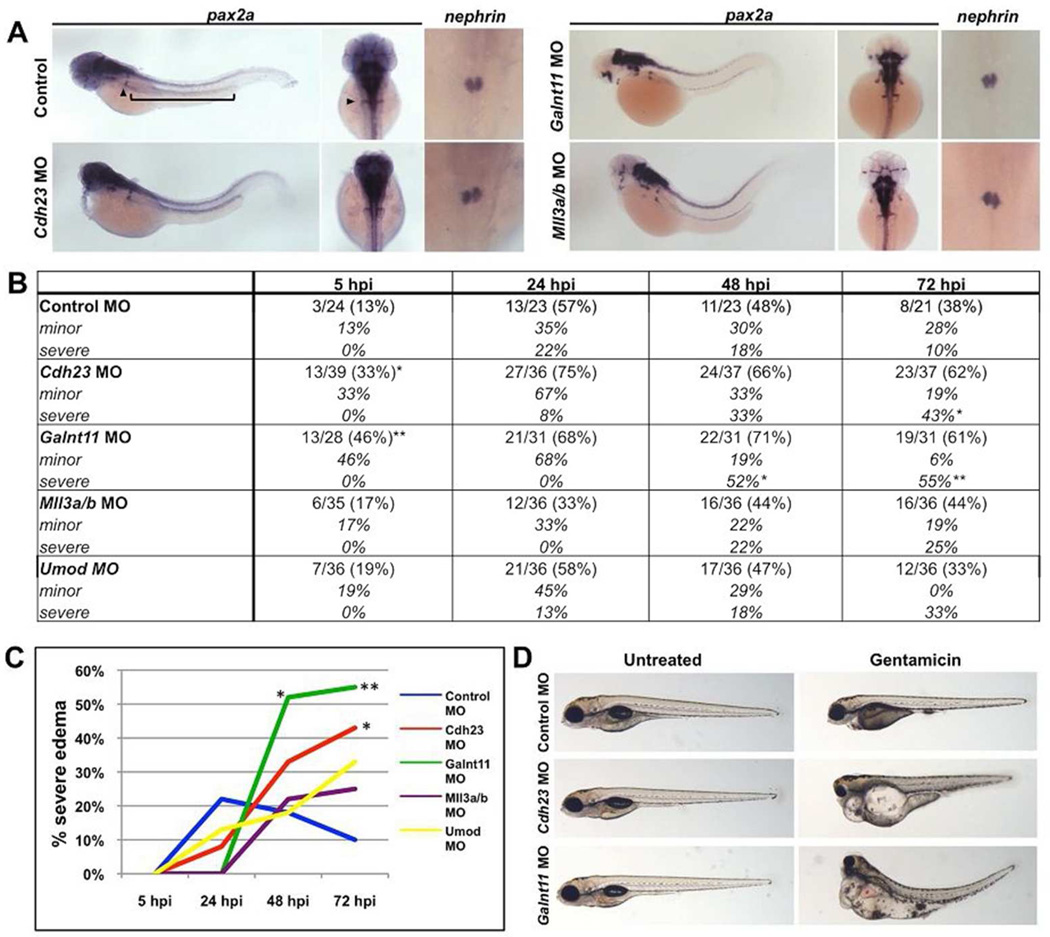
To investigate the role of the two suggestive novel loci in vertebrate kidney development and function and to bolster confidence in the nominally significant statistical associations in the replication studies, we knocked down the corresponding genes in the zebrafish using antisense morpholino (MO) technology. We focused on the CDH23 region and the block containing GALNTL5, GALNT11, MLL3 and CCT8L1. For the latter region, we focused on GALNT11 and MLL3, because there are no zebrafish GALNTL5 and CCT8L1 orthologs. Further, we investigated the effect of MO knockdown of umod. Following MO injection at the 1-cell stage, we performed in situ hybridization for the established renal markers pax2a (global kidney) and nephrin (podocytes) at 48 hours post-fertilization (hpf). Compared to control embryos, cdh23, galnt11, mll3a, mll3b and umod morphants did not display significant defects in glomerular or tubule gene expression (Figure 2A, n>25 embryos per MO injection).
Figure 2. Cdh23 and galnt11 knockdowns exacerbate nephrotoxic injury in zebrafish embryos.
(A) Whole mount in situ hybridization for the global kidney marker pax2a (arrowhead denotes the glomerulus, bracket denotes the tubule) and the podocyte marker nephrin demonstrates that morpholino (MO) knockdowns of cdh23, galnt11, mll3a, and mll3b do not result in changes in kidney gene expression compared to control embryos at 48 hours post-fertilization (hpf). Similar results were obtained for MO knockdowns of umod (images not shown). (B) Morpholino knockdown of cdh23 and galnt11 causes embryos to develop edema at a higher frequency than control embryos following gentamicin challenge. Data are presented as number of observed abnormalities per total number of embryos scored at 5, 24, 48, and 72 hours post-gentamicin injection (hpi), normalized to control experiments. *p < 0.05, **p < 0.005 by Fisher’s exact test. (C) Graphical representation of edema prevalence in embryos injected with gentamicin in (B). (D) Control embryos develop minor (cardiac) edema whereas cdh23 and galnt11 MO-injected embryos develop severe (cardiac, intestinal, ocular) edema 72 hours after gentamicin injection.
It is possible that morphant embryos develop a kidney function decline phenotype only after exposure to a nephrotoxin, despite observing no differences in renal marker expression at 48 hpf. Accordingly, after MO injection, we injected embryos with gentamicin at 48 hpf and observed edema prevalence and severity over the next three days. In control embryos, gentamicin injection predictably resulted in a majority of embryos developing minor (cardiac) edema by 24 hours post-injection (hpi) (Figure 2B–D). In comparison, cdh23 and galnt11 morphants developed significantly more severe (cardiac, intestinal, and ocular) and more frequent edema (Figure 2B–D). Specifically, whereas 10% of control embryos developed severe edema by 72 hpi, 43% of cdh23 morphants (p=0.009) and 55% of galnt11 morphants (p=0.001) developed severe edema at this time point. Additionally, a significant proportion of cdh23 (33%, p=0.035) and galnt11 morphant embryos (46%, p=0.005) injected with gentamicin developed edema earlier compared to controls at 5 hpi. In contrast, knockdown of mll3 or umod affected neither kidney development nor susceptibility to gentamicin (Figure 2B–C). Taken together, these data demonstrate that knockdown of cdh23 and galnt11 results in altered renal function after a nephrotoxic insult.
Interrogation of novel loci in eSNP databases and the CRIC Study
We interrogated eSNP data bases for evidence of SNPs at the CDH23 and GALNTL5/GALNT11 loci to evaluate an effect on gene expression [25] but found no relevant associations. Similarly, annotation information provided by ANNOVAR [26] did not yield genetic variants of potential functional interest within 500kb of and in linkage disequilibrium (r2 > 0.8 based on HapMap release 22) with the index SNPs.
In Caucasian participants of the Chronic Renal Insufficiency Cohort (CRIC) study, a prospective study of patients with CKD at baseline [27], neither SNPs in GALNTL5/GALNT11 or CDH23 were associated with eGFRchange (n=1476) or time to a composite renal event that consisted of incident end stage renal disease or halving of eGFR (n=1585, with a total of n=178 events; results not shown).
Discussion
Key findings
Our key findings are fourfold. First, we estimate the heritability of eGFR decline as being 38% in the general population of European descent, providing a rationale to search for genetic variants associated with kidney function decline. Second, we extend evidence of a known locus (UMOD) previously associated with incident CKD and ESRD [21,28] by showing genome-wide significant association with kidney function change. Third, we have identified two novel genetic loci (CDH23 and GALNTL5/GALNT11) with suggestive association with kidney function decline phenotypes. Finally, we show that knock-down of the two novel loci in zebrafish renders the nephron susceptible to a nephrotoxic insult.
Our findings in the context of the literature
We extend the current literature by performing the first large-scale GWAS of renal function decline traits in the general population. Previous studies analyzing progression of renal disease in African Americans [29−32], individuals of European descent [21], healthy nurses [33], and patients with diabetes [34,35], hypertension [31], IgA nephropathy [36,37] and ESRD [21] focused only on candidate genes.
The SNP in UMOD has previously been identified in a GWAS of eGFR measured at one time point [14], and was significantly associated with incident CKD and ESRD in a candidate gene study [21] and with salt-sensitive hypertension and kidney damage in rodents and humans [38]. Our data extend this knowledge base by providing strong evidence that genetic variation at the UMOD locus affects different definitions of kidney function decline.
For Rapid Decline, the associated region on chromosome 7 contains the genes GALNTL5, GALNT11, MLL3, and CCT8L1, with our zebrafish data suggesting GALNTL5 and GALNT11 as the genes of interest. GALNTL5 encodes the putative polypeptide N-acetylgalactosaminyltransferase-like protein 5, which by similarity has a presumed role in O-linked oligosaccharide biosynthesis. Polypeptide N-acetylgalactosaminyltransferase 11, encoded by GALNT11, is a glycosyl transferase that catalyzes the initial reaction in O-linked oligosaccharide biosynthesis. Studies in Xenopus support a role of the gene product in left-right patterning by modulating Notch1 signaling and thus establishing the crucial balance between motile and immotile cilia, and it is also expressed in the developing kidney of zebrafish [39,40]. Our data suggest that galnt11 is not essential for kidney development, but protects against susceptibility from nephrotoxins.
The region of chromosome 7 also contains a locus (rs7805747 in PRKAG2) that was previously identified in a GWAS meta-analysis of cross-sectional eGFR [15]. However, this SNP is independent of rs1019173 (r2=0.002, D’=0.061 in the 1000 Genomes Pilot Version 1, hg18); therefore, the novel locus identified in the present study is unlikely tagging the PRKAG2 locus. Moreover, conditional analysis using genotypes from both SNPs from individual level data from the ARIC study showed that the association between rs1019173 and Rapid Decline is unchanged when controlling for rs7805747 (data not shown).
The other locus identified from this study is an intronic SNP in CDH23 that is nominally associated with eGFR change in those with CKD at baseline. CDH23 encodes cadherin 23, a glycoprotein of the cadherin family. Cadherin 23 and protocadherin 15, encoded by PCDH15, form the tip-links spanning the stereocilia of the inner ear’s hair cells. These tip-links are key contributors to the mechanosensory transduction in hair cells required for hearing [41]. Rare mutations of CDH23 cause progressive, nonsyndromic deafness (DFNB12, MIM # 601386) [42−44] or Usher Syndrome 1D, characterized by profound deafness, vestibular dysfunction and retinitis pigmentosa (MIM # 601067). The transmembrane protein cadherin 23 is expressed in many tissues, including the kidney [44,45], where it is found predominantly in the tubulointerstitium [46]. While a kidney phenotype has not been reported for patients with DFNB12 or Usher syndrome, our zebrafish data provide evidence that cadherin 23 plays a role in protecting from susceptibility to nephrotoxins, while not being essential for nephrogenesis.
Implications
Our GWAS findings point towards two novel gene loci, CDH23 and GALNTL5/GALNT11, and one previously identified locus (UMOD) as being associated with kidney function decline. The zebrafish experiments support a role of the two newly identified loci in increasing renal susceptibility to nephrotoxic insults and may indicate that a perturbation model could serve as a model of longitudinal kidney function decline. In previous work, we have shown that knockdown of two genes identified by GWAS of cross-sectional eGFR, mpped2 and casp9, resulted in abnormal kidney development, with susceptibility to gentamicin only in casp9 knockdown [16]. Taken together, our current and previous data highlight the differential role of genes in affecting kidney development, function and susceptibility to damage.
Strengths and Limitations
Strengths of this study include the large sample size of renal function decline traits, follow-up in independent samples, analysis of several definitions of kidney function decline and validation in zebrafish. Some limitations warrant mention. Even though we addressed inter-assay differences of serum creatinine measurement by calibrating creatinine to representative NHANES standards, several other factors causing imprecision in defining kidney function decline phenotypes may have reduced our statistical power to identify genome-wide significant associations: 1) despite our use of different renal function decline definitions all featured in current guideline statements [3], there is no standard definition of renal function decline, 2) kidney function trajectories are less well-defined with two vs. several serum creatinine measurements given that renal function change may not be linear over time [3] and there may be day-to-day alterations in GFR, 3) GFR estimation equations are known to be imprecise especially at a GFR>60 ml/min/1.73m2, 4) we observed heterogeneity in design between studies including a wide range of length of follow-up. We cannot rule out that low statistical power also accounts for the negative finding in the CRIC study. Further, our findings, obtained mainly in general population cohorts, provide novel insights into mechanisms of kidney function decline, but may not be generalizable to cohorts enriched for CKD. This limitation deserves particular attention due to the unexpected observation that in most cohorts, the subgroup with baseline CKD (defined as eGFR<60 ml/min/1.73m2) showed a mean increase in eGFR over time irrespective of length of follow-up interval. This may indicate that in the CKD subgroup of these cohorts, a baseline eGFR<60 ml/min/1.73m2 may not represent progressive CKD with active disease but rather stable disease or imprecise GFR estimation. This highlights that more work with expanded datasets and functional models are necessary to further elucidate the genetics of CKD initiation and progression in population-based studies. Finally, the role of genes contributing to aging and chronic disease in humans may not be entirely modeled by transient morpholino knockdown and observation of a developmental phenotype: while zebrafish allows high throughput modeling of the effects of gene knockdown in a vertebrate organism, the developmental role of specific genes may well be different from homeostatic organ maintenance in the adult. Specifically, umod may not play a relevant role in zebrafish renal development or toxin susceptibility.
Conclusion
In a large GWAS of kidney function decline phenotypes in individuals of European descent, we showed that a SNP in UMOD is associated with kidney function decline phenotypes, and that there is suggestive statistical evidence for two novel loci (GALNTL5/GALNT11 and CDH23). Zebrafish experiments at the two novel loci suggest roles in the deterioration of kidney function after acute injury. Given the complexity of the kidney function decline phenotype, further interrogation of these regions is warranted.
Materials and Methods
Ethics Statement
In all studies, all participants gave informed consent. All studies were approved by their responsible Research Ethics Committees.
Phenotype definition
Serum creatinine was measured at a minimum of 2 time points spaced several years apart (2.0 – 22.2 years, median 5.6 years). In almost all studies, there were only two serum creatinine measurements in total. To be consistent across studies, we used each individual’s two creatinine measurements with the longest follow-up in between for phenotype creation in all cohorts (see below). Baseline and follow-up serum creatinine were calibrated to the US nationally representative National Health and Nutrition Examination Study (NHANES) data in all discovery and replication studies to account for between-laboratory variation [47]. In order to be consistent with our prior work, GFR based on serum creatinine (eGFRcrea) was estimated using the four-variable MDRD Study Equation. eGFRcrea values <15 ml/min/1.73m2 were set to 15, and those >200 were set to 200 ml/min/1.73m2.
Several phenotypes were used to model different mechanisms involved in change of renal function over time, using each individual’s two serum creatinine measurements with the longest follow-up. The continuous phenotype eGFRchange, modeling annual change in kidney function, was calculated by subtracting the eGFR at follow-up from the eGFR at baseline, and then dividing by the number of years of follow-up for each participant. Thus, a positive value of eGFRchange corresponds to a decline in kidney function over time, whereas a negative value of eGFRchange corresponds to an increase in kidney function over time. Three dichotomous phenotypes were calculated to model kidney function decline phenotypes with different clinical implications [5,24]: For Rapid Decline, cases were defined as individuals with a rapid decline in kidney function >= 3ml/min/1.73 m2 per year, and controls as those with a kidney function decline < 3ml/min/1.73 m2 per year [6]. For incident CKD (CKDi), cases were defined as participants with eGFR at baseline >= 60 ml/min/1.73m2 declining to an eGFR at follow-up < 60ml/min/1.73 m2; a more stringent definition of incident CKD (CKDi25) is restricted to incident CKD cases with a decline of eGFR >= 25% at follow-up. For both CKDi and CKDi25, controls were defined as those with an eGFR >= 60ml/min/1.73 m2 at baseline and follow-up.
Heritability of eGFR in the Framingham Heart Study
Heritability of eGFRchange was calculated with family data of the Framingham Heart Study using the variance components analysis implemented in SOLAR [48]. eGFRchange was calculated by taking follow-up eGFR (obtained between 2005−2008) and subtracting baseline eGFR (obtained in 1995−1998), divided by the number of years of follow-up. Residuals were created after adjusting for age, sex, baseline eGFR, and principal components as necessary. With residuals as response variable, a variance components model with an additive genetic and a random environmental variance components was fitted, where the correlation among relatives attributable to the genetic component is assumed proportional to the kinship coefficient matrix. Heritability is calculated as the ratio of the estimated genetic variance to the total phenotypic variance.
Definition of strata
Kidney function decline is known to differ depending on level of baseline eGFR. Thus, eGFRchange was analyzed (A) in the overall sample [eGFRchange overall], (B) in those with eGFR >= 60 ml/min/1.73m2 at baseline [eGFRchange noCKD], and (C) in those with eGFR < 60 ml/min/1.73m2 at baseline [eGFRchange withCKD]. Rapid Decline was analyzed in the overall sample [Rapid Decline overall] and in those with eGFR >= 60 ml/min/1.73m2 at baseline [Rapid Decline noCKD]. CKDi and CKDi25 were analyzed in the overall sample only.
Stage 1 genome-wide association analyses
All participating studies used a uniform analysis plan and each trait was created using standard programming commands that were provided to collaborating studies. The continuous trait (eGFRchange) was analyzed by linear regression, the dichotomous traits by logistic regression (Rapid Decline, CKDi, CKDi25). Models included the allelic dosage at each marker from imputed study data consisting of 2.5 million HapMap-II SNPs [49] on average, based on imputations with different programs and reference panels. Details of genotyping and imputation in each study are shown in Supplementary Table 2. We used the additive genetic model, adjusted for age and sex, baseline eGFR and, where applicable, for study site and principal components.
Stage 1 meta-analysis
For our stage 1 analysis, we used aggregated statistics of 16 population-based GWA studies of individuals of European ancestry for each of the longitudinal traits: eGFRchange overall, eGFRchange noCKD, eGFRchange with CKD, Rapid Decline overall, Rapid Decline noCKD, CKDi and CKDi25. All 16 stage 1 studies contributed data to every trait, except for the AMISH study, which provided data to eGFRchange overall and eGFRchange no CKD only due to low number of CKD cases at baseline and follow-up.
All input files underwent quality control using the GWAtoolbox package in R (www.eurac.edu/GWAtoolbox.html) [50], before including them into meta-analysis. Study data was meta-analyzed assuming fixed effects and using inverse-variance weighting. Thus the pooled effect βpooled is estimated as , where β and SE are the effect and standard error of the SNP on the outcome in the ith study. The meta-analyses were performed by METAL. We performed genomic control correction if the inflation factor λ in the study files was greater than 1 (1st GC correction) or if it was greater than 1 in the meta-analysis result (2nd GC correction) [51].
Next, we created a list of independent SNPs (pairwise r2<0.2, HAPMAP II release 22) that had a genomic control corrected p-value <10−6 and minor allele frequency > 5% in stage 1 meta-analysis and were present in at least 85% of the contributing studies.
Stage 2 meta-analysis
The stage 2 meta-analysis of SNPs identified in stage 1 was performed on the same phenotypes and using the same analysis plan as the stage 1 analysis, and was based on in silico genetic data or on de novo genotyped variants. Details on each stage 2 study’s genotyping and imputation platforms are shown in Supplementary Table 2. In addition, we also performed a combined inverse-variance weighted fixed-effects stage 1 and stage 2 meta-analysis using individual study files as input. Studies with less than 50 cases for a dichotomous trait or with an overall sample size of less than 50 for a continuous trait were excluded from the meta-analyses of the corresponding trait. SNPs with a stage 2 meta-analysis one-sided p-value <0.05 and effect direction consistency with the stage 1 meta-analysis effect direction were defined as showing nominally significant evidence of replication. The I2 statistic was computed to assess heterogeneity between studies.
Zebrafish functional experiments
Zebrafish were maintained according to established IACUC protocols. Zebrafish were injected at the 1-cell stage with 2 nl of 400 uM morpholinos (MO; GeneTools, Philomath, OR) designed to block the ATG start site or an exon-intron splice site of the target gene (Supplementary Table 6). Embryos were fixed in 4% PFA at the appropriate stages for in situ hybridization using well established protocols (http://zfin.org/ZFIN/Methods/ThisseProtocol.html). Renal gene expression was visualized using established markers for pax2a (global kidney) and nephrin (podocytes) [52,53]. The number of embryos displaying abnormal renal gene expression was compared to uninjected control embryos, and statistical significance was determined by Fisher’s exact test. For the gentamicin nephrotoxin experiment, embryos were injected with MO at the 1-cell stage and then injected with 5 nl of 10 mg/ml gentamicin prepared from one stock solution in the cardiac sinus venosus at 48 hpf after being anesthetized in a 1:20 dilution of 4 mg/ml Tricaine in embryo water. Live embryo development and edema prevalence was documented over the next three days.
Supplementary Material
Acknowledgements
Stage 1 cohorts:
AGES: This study has been funded by NIH contract N01-AG-1-2100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The study is approved by the Icelandic National Bioethics Committee, VSN: 00-063. The researchers are indebted to the participants for their willingness to participate in the study
AMISH: The Amish studies are supported by grants and contracts from the NIH including R01 AG18728 (Amish Longevity Study), R01 HL088119 (Amish Calcification Study), U01 GM074518-04 (PAPI Study), U01 HL072515-06 (HAPI Study), U01 HL084756 and NIH K12RR023250 (University of Maryland MCRDP), NIH P30 DK072488 (Clinical Nutrition Research Unit), the University of Maryland General Clinical Research Center, grant M01 RR 16500 and the Baltimore Veterans Administration Medical Center Geriatrics Research and Education Clinical Center. We thank our Amish research volunteers for their long-standing partnership in research, and the research staff at the Amish Research Clinic for their hard work and dedication.
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. A.K. was supported by the grant KO3598/2-1 (Emmy Noether Programme) of the German Research Foundation.
ASPS: The research reported in this article was funded by the Austrian Science Fond (FWF) grant number P20545-P05 and P13180. The Medical University of Graz supports the databank of the ASPS. The authors thank the staff and the participants of the ASPS for their valuable contributions. We thank Birgit Reinhart for her long-term administrative commitment and Ing Johann Semmler for the technical assistance at creating the DNA-bank.
The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
The CoLaus study received financial contributions from GlaxoSmithKline; the Faculty of Biology and Medicine of Lausanne; the Swiss National Science Foundation (33CSCO-122661; 3200BO-111361/2; 3100AO-116323/1;310000-112552). M.B is supported by the Swiss School of Public Health Plus.
FHS: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
GENOA: This research was partially supported by the National Heart Lung and Blood Institute of the National Institutes of Health R01 HL-87660.
The Health Aging and Body Composition Study (Health ABC) was funded by the National Institutes of Aging. This research was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
The JUPITER trial and the genotyping were supported by AstraZeneca
KORA studies: The genetic epidemiological work was funded by the NIH subcontract from the Children’s Hospital, Boston, US, (H.E.W., I.M.H, prime grant 1 R01 DK075787-01A1), the German National Genome Research Net NGFN2 and NGFNplus (H.E.W. 01GS0823; WK project A3, number 01GS0834), the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ, and by the Else Kröner-Fresenius-Stiftung (P48/08//A11/08 to C.A.B. and B.K.K.; 2012_A147 to CAB and IMH). The kidney parameter measurements in F3 were funded by the Else Kröner-Fresenius-Stiftung (C.A.B., B.K.K.) and the Regensburg University Medical Center, Germany; in F4 by the University of Ulm, Germany (W.K.). Genome wide genotyping costs in F3 and F4 were in part funded by the Else Kröner-Fresenius-Stiftung (C.A.B., B.K.K.). De novo genotyping in F3 and F4 were funded by the Else Kröner-Fresenius-Stiftung (C.A.B., IMH). The KORA research platform and the MONICA Augsburg studies were initiated and financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, by the German Federal Ministry of Education and Research and by the State of Bavaria. Geno-typing was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum München. The LINUX platform for computation was funded by the University of Regensburg for the Department of Epidemiology and Preventive Medicine at the Regensburg University Medical Center.
MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung and Blod Institute (NHLBI) in collaboration with MESA Investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and CTSA UL1-RR-024156
Rotterdam Study 1: The GWA study was funded by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Dr Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are very grateful to the participants and staff from the Rotterdam Study, the participating general practitioners and the pharmacists. We would like to thank Dr. Tobias A. Knoch, Luc V. de Zeeuw, Anis Abuseiris, and Rob de Graaf as well as their institutions the Erasmus Computing Grid, Rotterdam, The Netherlands, and especially the national German MediGRID and Services@MediGRID part of the German D-Grid, both funded by the German Bundes-ministerium für Forschung und Technology under grants #01 AK 803 A-H and #01 IG 07015 G, for access to their grid resources. Abbas Dehghan is supported by NWO grant (vici, 918-76-619).
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg- West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG.
Three Cities: The work was made possible by the generous participation of the control subjects, the patients, and their families. We thank Dr. Anne Boland (CNG) for her technical help in preparing the DNA samples for analyses. This work was supported by the National Foundation for Alzheimer’s disease and related disorders, the Institut Pasteur de Lille and the Centre National de Génotypage. The Three-City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (Inserm), the Victor Segalen Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Fondation de France and the joint French Ministry of Research/INSERM “Cohortes et collections de donnees biologiques” programme. Lille Génopôle received an unconditional grant from Eisai.
Stage 2 cohorts:
ADVANCE: The genetic epidemiological work was funded by Prognomix Inc. and by grants from Genome Quebec and Canadian Institutes for Health Research (CIHR). The clinical study was managed by the George Institute for International Health (Sydney, Australia) with grants received from Les Laboratoires Servier, France and from Medical Research Council of Australia. The genotyping was performed at the genomic platform of CRCHUM. The authors acknowledge the technical help of Carole Long and Mounsif Haloui and the bioinformatic analyses performed by Gilles Godefroid, François-Christophe Blanchet-Marois and François Harvey. The members of the genetic sub-study of ADVANCE, Stephen Harrap and Michel Marre are also acknowledged.
The Blue Mountains Eye Study (BMES) was supported by the Australian National Health & Medical Research Council (NHMRC), Canberra Australia (NHMRC project grant IDs 974159, 211069, 302068, and Centre for Clinical Research Excellence in Translational Clinical Research in Eye Diseases, CCRE in TCR-Eye, grant ID 529923). The BMES GWAS and genotyping costs was supported by Australian NHMRC, Canberra Australia (NHMRC project grant IDs 512423, 475604 and 529912), and the Wellcome Trust, UK as part of Wellcome Trust Case Control Consortium 2 (A Viswanathan, P McGuffin, P Mitchell, F Topouzis, P Foster, grant IDs 085475/B/08/Z and 085475/08/Z). EGH is supported by the NHMRC Fellowship scheme.
HYPERGENES (FP7 - HEALTH-F4-2007-201550); INTEROMICS (MIUR - CNR Italian Flagship Project); IC15-CT98-0329-EPOGH; LSHM-CT-2006-037093; HEALTH-2011-278249-EU-MASCARA; and ERC Advanced Grant-2011-294713-EPLORE and the Fonds voor Wetenschappelijk Onderzoek Vlaanderen; Ministry of the Flemish Community; Brussels; Belgium (grants G.0575.06 and G.0734.09)
KORA studies: The genetic epidemiological work was funded by the NIH subcontract from the Children’s Hospital, Boston, US, (H.E.W., I.M.H, prime grant 1 R01 DK075787-01A1), the German National Genome Research Net NGFN2 and NGFNplus (H.E.W. 01GS0823; WK project A3, number 01GS0834), the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ, and by the Else Kröner-Fresenius-Stiftung (P48/08//A11/08 to C.A.B. and B.K.K.; 2012_A147 to CAB and IMH). The kidney parameter measurements in F3 were funded by the Else Kröner-Fresenius-Stiftung (C.A.B., B.K.K.) and the Regensburg University Medical Center, Germany; in F4 by the University of Ulm, Germany (W.K.). Genome wide genotyping costs in F3 and F4 were in part funded by the Else Kröner-Fresenius-Stiftung (C.A.B., B.K.K.). De novo genotyping in F3 and F4 were funded by the Else Kröner-Fresenius-Stiftung (C.A.B., IMH). The KORA research platform and the MONICA Augsburg studies were initiated and financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, by the German Federal Ministry of Education and Research and by the State of Bavaria. Geno-typing was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum München. The LINUX platform for computation was funded by the University of Regensburg for the Department of Epidemiology and Preventive Medicine at the Regensburg University Medical Center.
NESDA was supported by the Geestkracht program of ZonMW [grant 10-000-1002]; matching funds from universities and mental health care institutes involved in NESDA. Funding support was also provided by the Netherlands Scientific Organization (904-61-090, 904-61-193, 480-04-004, 400-05-717), Centre for Medical Systems Biology (NWO Genomics), the Neuroscience Campus Amsterdam and the EMGO institute; the European Union (EU/WLRT-2001-01254), NIMH (RO1 MH059160). Genotyping was funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and analysis were supported by grants from GAIN and the NIMH (MH081802) and the Center for Molecular and Systems Biology (CMSB).
Genotype data were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/dbgap, accession number phs000020.v1.p1. Statistical analyses were partly conducted at the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation.
POPGEN: This study was funded by the German National Genome Research Network (NGFN; Federal Ministry of Education and Research, grant numbers 1GS0121, 01GS0171, 01GR0468) and by the DFG Excellence Cluster ‘Inflammation at Interfaces’ (EXC 306).
The PREVEND Study was financially supported by several grants from the Dutch Kidney Foundation.
The PREVEND Study was financially supported by several grants from the Dutch Kidney Foundation.
Rotterdam Study II: The GWA study was funded by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Dr Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are very grateful to the participants and staff from the Rotterdam Study, the participating general practitioners and the pharmacists. We would like to thank Dr. Tobias A. Knoch, Luc V. de Zeeuw, Anis Abuseiris, and Rob de Graaf as well as their institutions the Erasmus Computing Grid, Rotterdam, The Netherlands, and especially the national German MediGRID and Services@MediGRID part of the German D-Grid, both funded by the German Bundes-ministerium für Forschung und Technology under grants #01 AK 803 A-H and #01 IG 07015 G, for access to their grid resources. Abbas Dehghan is supported by NWO grant (vici, 918-76-619).
The SAPHIR-study was partially supported by a grant from the Kamillo Eisner Stiftung to B. Paulweber and by grants from the "Genomics of Lipid-associated Disorders – GOLD" of the "Austrian Genome Research Programme GEN-AU" to F. Kronenberg.
The Young Finns Study has been financially supported by the Academy of Finland: grants 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi), the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9M048 and 9N035 for TeLeht), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation and Emil Aaltonen Foundation (T.L). The expert technical assistance in the statistical analyses by Ville Aalto and Irina Lisinen is gratefully acknowledged.
Footnotes
Author contributions
Study design: ATi, ARSh, AHo, AGUi, AKö, BTa, BSt, BDMi, BWPe, CSFo, CHe, DSSi, GEi, HKr, IHdeB, JDi, JCh, JCo, KEn, LJLa, MWo, MdAn, ORa, PMRi, PHa, RRe, RSc, SERo, TBHa, THa, TLe, UNö, UVö, VGu, PVo, GWa, WHLKa, YLi,
Study Management: ARSh, AHo, AFr, AGUi, BPa, MBo, BDMi, BWPe, CSFo, CHe, DSSi, GEi, JJWa, JDi, JTr, JCh, MWo, MPi, NAu, ORa, OHFr, PMRi, PMi, PHa, RSc, SLRKa, STTu, THa, TLe, UNo, VGu, PVo, YLi,
Subject Recruitment: ARSh, AHo, BPa, BDMi, CSFo, CHe, CMe, CBa, DCu, DSSi, FKr, GEi, JCh, JASt, NAu, ORa, PMa, PMRi, PMi, PHa, RSc, STTu, THa, TLe, UNö, VGu, PVo, GWa
Zebra fish experiments: MGa, WGoe.
Interpretation of Results: ADe, ATi, APa, AVSm, AKö, AYCh, BTa, BPa, MBo, CSFo, CABö, CPa, DICh, GMMc, HKr, IHdeB, IMHe, JDi, JTr, JCh, KEn, MGa, MWo, MGo, MPi, NAu, ORa, PHa, PvdHa, RRe, RTGa, SSe, SJLBa, SERo, THa, TLe, TAs, UVö, WGoe, WHLKa, YLi
Drafting of manuscript: ATi, AKö, CSFo, CABö, GMMc, IMHe, MGa, MGo, WGoe, WHLKa.
Statistical Methods and Analysis: ADe, ATi, APa, AVSm, ATe, AKö, AYCh, BTa, BKo, BSt, BPa, CSFö, CABo, CPa, DICh, EHo, EGHo, ESa, FDGM, FKr, GMMc, GLi, HKr, IMNo, IMHe, JGu, JAn, LJROC, JDi, JCh, L-PLy, MLi, MWo, MMe, MM-Nu, MGo, MOl, MOl, PAKa, SSe, S-JHw, SCo, TCo, TAs, WHLKa, YLi, ZKo
Genotyping: ARSh, AVSm, ATe, AFr, AGUi, BKo, BDMi, CSFö, CABo, CBa, DICh, EJAt, FRi, FKr, GLi, HGr, HSc, JGu, J-CL, LJROC, JTr, L-PLy, MHa, MdAn, PFr, PHa, PAKa, PvdHa, STTu, SLRKa, TLe, UVö, YLi
Bioinformatics: AVSm, ATe, AFr, AJo, BTa, CABö, CPa, DICh, EHo, ESa, FRi, FKr, GMMc, GLi, HSc, HKr, IMHe, JGu, J-CL, JAn, LJROC, L-PLy, MWo, MGo, MOl, PFr, PAKa, QYa, S-JHw, SCo, VCh
Critical review and final approval of manuscript: all.
Financial disclosures
Johanne Tremblay: Consultant Servier; Pavel Hamet: Cconsultant Servier, John Chalmers: Grant Servier.
References
- 1.Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 2.Meguid El Nahas A, Bello AK. Chronic kidney disease: the global challenge. Lancet. 2005;365:331–340. doi: 10.1016/S0140-6736(05)17789-7. [DOI] [PubMed] [Google Scholar]
- 3.KDIGO. KDIGO Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int Suppl. 2013;3:1–150. doi: 10.1038/ki.2013.243. [DOI] [PubMed] [Google Scholar]
- 4.Matsushita K, Selvin E, Bash LD, et al. Change in estimated GFR associates with coronary heart disease and mortality. J Am Soc Nephrol. 2009;20:2617–2624. doi: 10.1681/ASN.2009010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rifkin DE, Shlipak MG, Katz R, et al. Rapid kidney function decline and mortality risk in older adults. Arch Intern Med. 2008;168:2212–2218. doi: 10.1001/archinte.168.20.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shlipak MG, Katz R, Kestenbaum B, et al. Rapid decline of kidney function increases cardiovascular risk in the elderly. J Am Soc Nephrol. 2009;20:2625–2630. doi: 10.1681/ASN.2009050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turin TC, Coresh J, Tonelli M, et al. One-year change in kidney function is associated with an increased mortality risk. Am J Nephrol. 2012;36:41–49. doi: 10.1159/000339289. [DOI] [PubMed] [Google Scholar]
- 8.Turin TC, Coresh J, Tonelli M, et al. Short-term change in kidney function and risk of end-stage renal disease. Nephrol Dial Transplant. 2012;27:3835–3843. doi: 10.1093/ndt/gfs263. [DOI] [PubMed] [Google Scholar]
- 9.Turin TC, Coresh J, Tonelli M, et al. Change in the estimated glomerular filtration rate over time and risk of all-cause mortality. Kidney Int. 2013;83:684–691. doi: 10.1038/ki.2012.443. [DOI] [PubMed] [Google Scholar]
- 10.Fox CS, Larson MG, Leip EP, et al. Predictors of new-onset kidney disease in a community-based population. JAMA. 2004;291:844–850. doi: 10.1001/jama.291.7.844. [DOI] [PubMed] [Google Scholar]
- 11.Satko SG, Sedor JR, Iyengar SK, Freedman BI. Familial clustering of chronic kidney disease. Semin Dial. 2007;20:229–236. doi: 10.1111/j.1525-139X.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 12.O'Seaghdha CM, Fox CS. Genome-wide association studies of chronic kidney disease: what have we learned? Nat Rev Nephrol. 2012;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers JC, Zhang W, Lord GM, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42:373–375. doi: 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köttgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köttgen A, Pattaro C, Böger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattaro C, Köttgen A, Teumer A, et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012;8:e1002584. doi: 10.1371/journal.pgen.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu CT, Garnaas MK, Tin A, et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y, Sim X, Go MJ, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44:904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng TY, Wen SF, Astor BC, et al. Mortality risks for all causes and cardiovascular diseases and reduced GFR in a middle-aged working population in Taiwan. Am J Kidney Dis. 2008;52:1051–1060. doi: 10.1053/j.ajkd.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Astor BC, Lewis J, et al. Longitudinal progression trajectory of GFR among patients with CKD. Am J Kidney Dis. 2012;59:504–512. doi: 10.1053/j.ajkd.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Böger CA, Gorski M, Li M, et al. Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD. PLoS Genet. 2011;7:e1002292. doi: 10.1371/journal.pgen.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regner KR, Harmon AC, Williams JM, et al. Increased susceptibility to kidney injury by transfer of genomic segment from SHR onto Dahl S genetic background. Physiol Genomics. 2012;44:629–637. doi: 10.1152/physiolgenomics.00015.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Su Y, Paueksakon P, et al. Integrin alpha1/Akita double-knockout mice on a Balb/c background develop advanced features of human diabetic nephropathy. Kidney Int. 2012;81:1086–1097. doi: 10.1038/ki.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bash LD, Coresh J, Kottgen A, et al. Defining incident chronic kidney disease in the research setting: The ARIC Study. Am J Epidemiol. 2009;170:414–424. doi: 10.1093/aje/kwp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lash JP, Go AS, Appel LJ, et al. Chronic Renal Insufficiency Cohort (CRIC) Study: baseline characteristics and associations with kidney function. Clin J Am Soc Nephrol. 2009;4:1302–1311. doi: 10.2215/CJN.00070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reznichenko A, Böger CA, Snieder H, et al. UMOD as a susceptibility gene for end-stage renal disease. BMC Med Genet. 2012;13:78. doi: 10.1186/1471-2350-13-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Lipkowitz MS, Salem RM, et al. Progression of chronic kidney disease: Adrenergic genetic influence on glomerular filtration rate decline in hypertensive nephrosclerosis. Am J Nephrol. 2010;32:23–30. doi: 10.1159/000313927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Mahata M, Rao F, et al. Chromogranin A regulates renal function by triggering Weibel-Palade body exocytosis. J Am Soc Nephrol. 2009;20:1623–1632. doi: 10.1681/ASN.2008111148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Salem RM, Rao F, et al. Common charge-shift mutation Glu65Lys in K+ channel beta(1)-Subunit KCNMB1: pleiotropic consequences for glomerular filtration rate and progressive renal disease. Am J Nephrol. 2010;32:414–424. doi: 10.1159/000320131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung MM, Chen Y, Lipkowitz MS, et al. Adrenergic beta-1 receptor genetic variation predicts longitudinal rate of GFR decline in hypertensive nephrosclerosis. Nephrol Dial Transplant. 2009;24:3677–3686. doi: 10.1093/ndt/gfp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper Worobey C, Fisher ND, Cox D, et al. Genetic polymorphisms and the risk of accelerated renal function decline in women. PLoS One. 2009;4:e4787. doi: 10.1371/journal.pone.0004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorsal A, Tarnow L, Lajer M, et al. The PPAR gamma 2 Pro12Ala variant predicts ESRD and mortality in patients with type 1 diabetes and diabetic nephropathy. Mol Genet Metab. 2008;94:347–351. doi: 10.1016/j.ymgme.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Sandholm N, Salem RM, McKnight AJ, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng W, Zhou X, Zhu L, et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with the progression of IgA nephropathy in Chinese. Nephrol Dial Transplant. 2011;26:2544–2549. doi: 10.1093/ndt/gfq768. [DOI] [PubMed] [Google Scholar]
- 37.Ferrandi M, Cusi D, Molinari I, et al. alpha- and beta-Adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy. J Mol Med (Berl) 2010;88:203–217. doi: 10.1007/s00109-009-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trudu M, Janas S, Lanzani C, et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fakhro KA, Choi M, Ware SM, et al. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci U S A. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boskovski MT, Yuan S, Pedersen NB, et al. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013 doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwander M, Xiong W, Tokita J, et al. A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells. Proc Natl Acad Sci U S A. 2009;106:5252–5257. doi: 10.1073/pnas.0900691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolz H, von Brederlow B, Ramirez A, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 43.Bork JM, Peters LM, Riazuddin S, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Palma F, Holme RH, Bryda EC, et al. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- 45.Siemens J, Kazmierczak P, Reynolds A, et al. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc Natl Acad Sci U S A. 2002;99:14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 47.Coresh J, Astor BC, McQuillan G, et al. Calibration and random variation of the serum creatinine assay as critical elements of using equations to estimate glomerular filtration rate. Am J Kidney Dis. 2002;39:920–929. doi: 10.1053/ajkd.2002.32765. [DOI] [PubMed] [Google Scholar]
- 48.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchsberger C, Taliun D, Pramstaller PP, Pattaro C. GWAtoolbox: an R package for fast quality control and handling of genome-wide association studies meta-analysis data. Bioinformatics. 2012;28:444–445. doi: 10.1093/bioinformatics/btr679. [DOI] [PubMed] [Google Scholar]
- 51.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 52.Drummond IA, Majumdar A, Hentschel H, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 53.Kramer-Zucker AG, Wiessner S, Jensen AM, Drummond IA. Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Dev Biol. 2005;285:316–329. doi: 10.1016/j.ydbio.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.