Abstract
Activation-induced deaminase (AID) is a DNA cytosine deaminase that diversifies immunoglobulin genes in B cells. Recent work has shown that RNA Polymerase II (Pol II) accumulation correlates with AID recruitment. However, a direct link between Pol II and AID abundance has not been tested. We used the DT40 B-cell line to manipulate levels of Pol II by decreasing topoisomerase I (Top1), which relaxes DNA supercoiling in front of the transcription complex. Top1 was decreased by stable transfection of a short hairpin RNA against Top1, which produced an accumulation of Pol II in transcribed genes, compared to cells transfected with sh-control RNA. The increased Pol II density enhanced AID recruitment to variable genes in the λ light chain locus, and resulted in higher levels of somatic hypermutation and gene conversion. It has been proposed by another lab that AID itself might directly suppress Top1 to increase somatic hypermutation. However, we found that in both AID+/+ and AID−/− B cells from DT40 and mice, Top1 protein levels were identical, indicating that the presence or absence of AID did not decrease Top1 expression. Rather, our results suggest that the mechanism for increased diversity when Top1 is reduced is that Pol II accumulates and recruits AID to variable genes.
Keywords: DT40, Topoisomerase I, RNA Polymerase II, Activation-induced deaminase, Somatic hypermutation, Gene conversion
1. Introduction
B cells express activation induced-deaminase (AID) to generate diversity in antibody genes. AID converts cytosine to uracil in single-strand DNA, and the rogue uracil initiates a cascade of error-prone repair pathways to introduce mutations into the immunoglobulin loci (reviewed in (1)). Mutations which alter the variable (V) gene coding sequence on the heavy and light chain loci are then selected for increased affinity to antigen, producing a robust antibody response. Mutations also occur in switch regions preceding each constant (C) gene on the heavy chain locus, which initiate heavy chain class switching to further diversify antibodies. While AID primarily targets immunoglobulin genes, off-target events can occur in several oncogenes including Bcl6, Pax5, and Myc (2–4). This potential for AID-induced genomic instability warrants continuing investigation into the mechanisms of AID targeting.
One essential component of AID targeting is the need for transcription (5). In vitro studies have demonstrated that when RNA Polymerase II (Pol II) is paused, AID generates multiple mutations (6). Recently, we reported that AID mutagenesis correlated with Pol II accumulation in the switch and V regions of mice (7, 8). In switch regions, it has been proposed that the DNA sequence forms RNA:DNA hybrids, or R-loops (9), which inhibit transcriptional elongation and increase Pol II accumulation. The paused Pol II complexes then recruit Spt5, a transcription initiation factor, and the RNA exosome, which degrades RNA, to resume elongation (10, 11). Both of these factors have been shown to directly interact with AID, suggesting they play a role in recruiting AID. In V regions, however, there are no R-loops, and it is not known what directs AID to these regions. Nonetheless, Pol II accumulation appears to be involved, as we identified paused Pol II complexes that were associated with Spt5 in germinal center B cells (8). Furthermore, Pol II-Spt5 complexes correlated with AID accumulation, suggesting similar mechanisms of AID targeting in both switch and V regions. However, the hypothesis that increased Pol II density can actively promote AID mutagenesis has not been directly tested in vivo.
Topoisomerase I (Top1) is an essential enzyme which maintains proper helical tension in DNA. The maintenance of helical tension is especially important during the process of transcription. As Pol II separates the DNA strands, positive supercoils are generated ahead of the transcribing polymerase, followed by negative supercoils behind it (reviewed in (12)). It has been proposed that Top1 nicks positively supercoiled DNA to relieve the tension and enhance transcription elongation. In fact, it is noteworthy that Top1 cleavage sites are found throughout transcriptionally-active genes but not in silent genes (13, 14). Furthermore, inhibition of Top1 by camptothecin decreased Pol II elongation (15, 16), and increased Pol II density in actively transcribed genes (17). Thus, Top1 is an intricate regulator of Pol II function. To test the role of Pol II density in targeting AID to V genes, we artificially increased Pol II abundance by decreasing Top1 levels in the chicken DT40 B-cell line. We found that increased Pol II density elevated AID mutagenesis.
2. Materials and methods
2.1. DT40 cell lines and mice
Four engineered cell lines were used. For somatic hypermutation (SHM), a cell line was used that was surface IgM+ and lacked Vλ pseudogenes. We generated the ΦV− AIDR2 cell line using a DT40cre1 ΦV− AID−/− IgM+ progenitor cell line (18) that had AID reconstituted (AIDR2) using the vector pAidGpt. In this vector, an AID cDNA expression cassette was cloned downstream of the chicken β-actin promoter and upstream of an IRES-GPT sequence. For gene conversion (GC), a cell line was used that was surface IgM− and had Vλ pseudogenes. The DT40cre1 (called AID+/+ hereafter) cell line had a frameshift mutation in the rearranged V-joining (J) gene (19). For western blots of Top1 and AID, two additional cell lines were used: DT40cre1 AID−/− (called AID−/−), and DT40cre1 AIDR with AID reconstituted (called AIDR). For mouse studies, wild type C57BL/6 mice and Aid−/− mice (20) on a C57BL/6 background were bred in our mouse colony. All animal procedures were reviewed and approved by the Animal Care and Use Committee of the National Institute on Aging.
2.2. Top1 shRNA cloning, cell culture, transfection, and western blotting
Short hairpin (sh) RNA constructs were cloned into the pLKO.1 vector (Addgene) which contained a puromycin resistant selectable marker. Two shRNAs against Top1 were tried, using oligonucleotides confirmed by Sigma. The one listed in Supplemental Table S1 was selected, along with a control sequence. Cells were cultured in chicken medium (RPMI-1640 supplemented with 10% fetal bovine serum, 1% chicken serum, 2 mM L-glutamine, 0.1 μM β-mercaptoethanol, 100 I.U./mL penicillin, and 100 μg/mL streptomycin) at 41°C with 5% CO2. ΦV− AIDR2 and AID+/+ cell lines were stably transfected with 20–40 μg of linearized sh-control or sh-Top1 plasmids at 25 μF and 700 V with gene pulser Xcell (Bio-Rad). After electroporation, cells were incubated in chicken medium overnight, and the next day, puromycin (Sigma Aldrich) was added at a final concentration of 0.5 μg/ml. The cells were then incubated for another 6–8 days. Colonies were picked and cultured in fresh chicken medium for 2–3 days. For western blots, whole cell extracts from 5 × 106 cells were resuspended in 50 μl phosphate-buffered saline (PBS) solution, and lysed by addition of 50 μl SDS loading buffer followed by boiling. For Top1 and β-actin, 10 μl of lysate was separated by electrophoresis through a 12% gel. For AID, 40 μl of lysate was separated by electrophoresis through a 20% gel. Proteins were transferred onto polyvinylidene difluoride membranes (Biorad) and visualized with antibodies to Top1 (Bethyl Laboratories, Inc.), β-actin (Sigma-Aldrich), and AID (21).
2.3. Cell division and proliferation
For division, sh-control and sh-Top1 cells were treated with 5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen) and analyzed by flow cytometry on days 0, 1, and 2. For proliferation, 1000 cells/well were seeded into 4 individual wells containing 1 ml chicken media followed by incubation at 42°C. On each day, a single well per clone was counted for analysis of cell growth.
2.4. Chromatin immunoprecipitation
μChIP (22) analyses were performed on 0.5 million ΦV− AIDR2 sh-control or sh-Top1 cells. Briefly, 0.5 million cells were lysed in 120 μl of SDS-lysis buffer and sonicated to shear the genomic DNA to ~ 500 bp. Chromatin was diluted with 800 μl RIPA buffer and centrifuged to remove precipitates. 800 μl of supernatant was collected, and 100 μl used for input and pulldown experiments. Experiments were performed using 2 subclones from each of 3 independent clones for sh-control and 9 independent clones for sh-Top1. Antibodies against Pol II (Millipore, clone CTD4H8), AID, and non-specific rabbit IgG (Millipore) were used at 2.4 μg per reaction, and were incubated with protein-G dynabeads (Invitrogen). Chromatin samples were incubated with the antibody/bead complex, washed with RIPA buffer, and the bound fraction was eluted by incubation with 50 μg/ml proteinase K. Samples were extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 50 μl of nuclease free H2O. qPCR reactions were performed using 1 μl of ChIP DNA, Power SYBR green mastermix (Invitrogen), and primers listed in Supplemental Table S1. Calculation of % input was performed by deriving the % input value for the experimental antibody (2(Ct(input)-Ct(ChIP) x 100), and subtracting the % input value of the IgG control.
2.5. Somatic hypermutation
For SHM, 3 sh-control and 9 sh-Top1 independent clones were generated in the ΦV− AIDR2 parent cells. Cells were distributed by limiting dilution into 96-well plates. One week later, 24–36 subclones from each group were picked and put into 24-well plates containing chicken medium. On day 14, cells were washed and stained for surface IgM expression with FITC-labeled mouse anti-chicken IgM (Southern Biotech), and analyzed by flow cytometry. Ten sh-control and 11 sh-Top1 subclones, which represented the average percent of surface IgM loss (IgM+ to IgM−), were selected and cultured for 4 more weeks, for a total of 6 weeks incubation. DNA from all 21 subclones was prepared and cloned into plasmids. The primers shown in Supplemental Table S1 were used for sequencing.
2.6. Gene conversion
For GC, 2 sh-control and 4 sh-Top1 independent clones were generated in the AID+/+ parent cells. Cells were distributed by limiting dilution into 96-well plates. One week later, 24 subclones from each group were picked and put into 24-well plates containing chicken medium. On day 14, cells were stained for surface IgM, and analyzed by flow cytometry. Four sh-control and 8 sh-Top1 subclones, which represented the average percent of surface IgM gain (IgM− to IgM+), were selected and cultured for 4 more weeks, for a total of 6 weeks incubation. DNA from all 12 subclones was prepared and cloned into plasmids. The primers listed in Supplemental Table S1 were used for sequencing.
2.7. Western blot and mRNA analyses of AID, Top1, and β-actin in DT40 and mouse B cells
For DT40, cell extracts were prepared as described in 2.2, and membranes were stained with antibodies to AID, Top1, and β-actin. For mice, naïve splenic B cells were isolated by negative selection with anti-CD43 and anti-CD11b magnetic beads (Miltenyi Biotec). Cells were plated at a density of 0.5 million cells/ml and stimulated ex vivo with 5 μg/ml Escherichia coli lipopolysaccharide (LPS) serotype 0111:B4 (Sigma-Aldrich) and 5 ng/ml recombinant interleukin-4 (IL-4) (Biolegend) for 0, 6, or 24 h. Total RNA was isolated from cells using RNAeasy columns (Qiagen). cDNA was generated from 100 ng of RNA using oligo dT and M-MLV reverse transcriptase (Promega). Levels of AID and β-actin mRNA were determined by RT-PCR using rTaq polymerase (Takara) and primers described in Supplemental Table S1. For westerns, whole-cell extracts from 5 × 106 cells were prepared, separated by gel electrophoresis, and stained with antibodies to Top1 and β-actin.
3. Results and discussion
3.1. Cell division and proliferation are unaltered by Top1 deficiency
We previously showed a correlation between high levels of Pol II density and AID mutagenesis in V and switch regions from mice (7, 8). To test if Pol II accumulation directly affected AID targeting, we manipulated Top1 levels in chicken DT40 B cells, which undergo constitutive SHM and GC in the Igλ locus. We hypothesized that decreasing Top1 levels would increase positive supercoils ahead of transcribing Pol II, resulting in a pile up of Pol II, which may increase AID targeting. ΦV− AIDR2 and AID+/+ cells were stably transfected with shRNA to the enzyme (sh-Top1) and a control sequence (sh-control). For the ΦV− AIDR2 line used for SHM, 3 clones for sh-control, and 9 clones for sh-Top1 were generated. For the AID+/+ line used for GC, 2 clones for sh-control and 4 clones for sh-Top1 were made. Western blot analysis for Top1 and β-actin protein from both lines showed a decrease in Top1 in the sh-Top1 clones compared to sh-control clones (Supplemental Fig. S1). As summarized in Fig. 1A, Top1 protein was significantly reduced by 36% in ΦV− AIDR2 clones, and by 39% in AID+/+ clones. It has been recently shown that complete deletion of Top1 inhibits replication fork progression (23). To ensure that our partial Top1 deficiency did not affect replication, we examined cell division and proliferation. For cell division, CFSE dilution was measured for 2 days (Fig. 1B). When added to cells, CFSE covalently couples to intercellular molecules and is diluted by 50% into each daughter cell upon cell division. The number of cell divisions can then be monitored using flow cytometry to analyze mean fluorescence intensity. Comparison of sh-control and sh-Top1 cells from both ΦV− AIDR2 and AID+/+ clones showed no discernable difference in the decrease in fluorescence. For proliferation, growth curves were generated by plating 1000 cells into 1 ml media and counting the cells every 24 hrs (Fig. 1C). There was no effect on growth between sh-control and sh-Top1 cells. These results show that decreased levels of Top1 did not affect cell division and proliferation, and therefore the DT40 cells should be satisfactory to study SHM and GC processes.
Fig. 1.
Effects of Top1 knockdown on cell division and proliferation. For ΦV− AIDR2 cells, 3 sh-control and 9 sh-Top1 clones were tested, and for AID+/+ cells, 2 sh-control and 4 sh-Top1 clones were tested. (A) Western blot analysis comparing Top1 levels in sh-control and sh-Top1 clones. Representative blots are shown. Bar graphs represent the average Top1/β-actin ratio; error bars depict standard deviation (SD). P value, unpaired two-tailed Student’s t test. (B) Cell division experiments tracking the dilution of CFSE dye over time. Representative flow cytometry analysis of fluorescence intensity in a sh-control clone on days 0 (black), 1 (green), and 2 (orange). Bar graphs represent the average mean intensity; error bars indicate SD. Value for d 1 includes cells in both peaks. (C) Cell proliferation. Cells were counted on days 0–4.
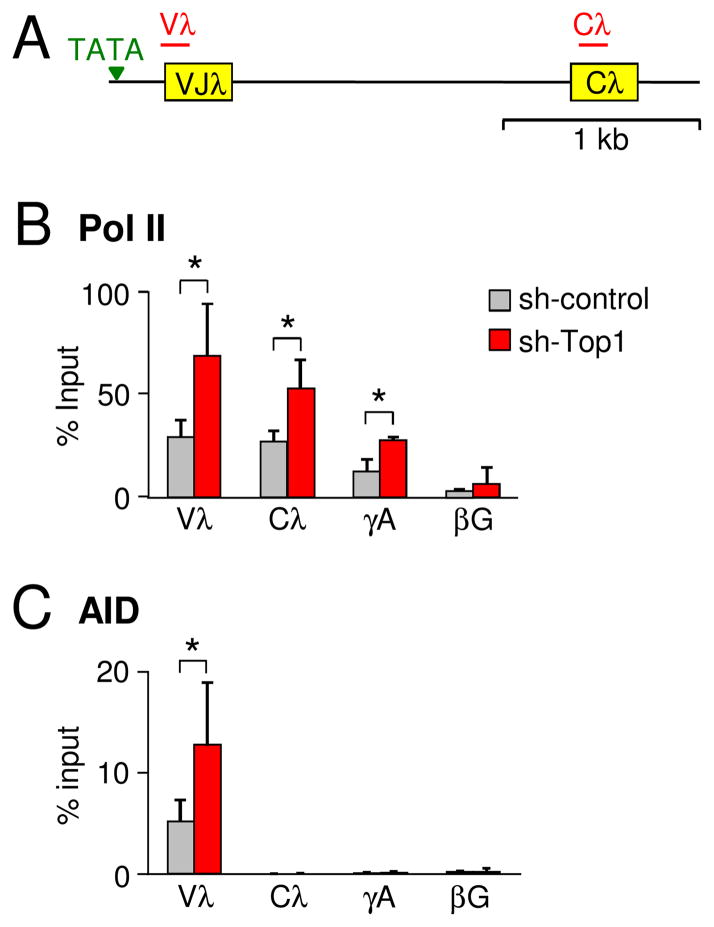
3.2. Top1 knockdown increases Pol II accumulation and AID recruitment to Vλ genes
Although cell division and proliferation were unaffected by Top1 knockdown, we predicted that transcription would be altered. Accordingly, Pol II density was measured in ΦV− AIDR2 cells by ChIP within the Igλ, γ-actin, and β-globin loci from sh-control and sh-Top1 cells. Cells with sh-Top1 had a significant accumulation of Pol II in Vλ and Cλ genes, compared to cells with the sh-control RNA (Fig. 2A and B). A significant increase was also seen in the promoter of the transcribed γ-actin gene, but not in the non-transcribed β-globin gene, which confirms previous reports that Top1 travels with elongating Pol II complexes (13, 14). To determine if increased Pol II density elevated recruitment of AID, we performed ChIP with anti-AID antibodies. The results showed a significant increase in AID in the Vλ gene in the Top1 knockdown cells compared to control cells (Fig. 2C). AID was absent in Cλ, γ-actin, and β-globin genes, consistent with the notion that additional factors are required to recruit AID to sites of paused Pol II. For example, we and others have reported that Pol II accumulation is associated with Spt5 (8, 10, 24); however we could not detect Spt5 in DT40 cells due to technical limitations, as commercial rabbit anti-Spt5 antibodies did not precipitate chicken Spt5. Taken together, Top1 deficiency promoted Pol II accumulation and increased AID targeting to Vλ genes.
Fig. 2.
ChIP analyses for RNA Pol II and AID. (A) Map of Igλ locus in ΦV− AIDR2 cells. TATA box, green triangle; VJ and C exons, yellow boxes. Red line indicates the area being amplified during ChIP assay. (B) RNA Pol II analysis on 2 subclones from each of 3 sh-control (gray) and 9 sh-Top1 (red) clones; error bars show SD. *, P ≤ 0.05 (unpaired two-tailed Student’s t test). (C) AID analysis on 2 subclones from 3 sh-control (gray) and 9 sh-Top1 (red) clones; error bars depict SD. *, P = 0.02 (unpaired two-tailed Student’s t test).
3.3. Pol II accumulation dictates levels of SHM and GC
V gene diversification in DT40 cells occurs by two mechanisms: SHM (25) and GC (26, 27). To determine if the elevated AID levels in shTop1 cells affected diversity, we analyzed point mutations and conversion tracks in VJλ genes. For SHM, we used a cell line with no pseudogenes (ΦV− AIDR2, surface IgM+), where AID deamination events are processed into single point mutations within the one functional, rearranged VJλ gene (Fig. 3A). Mutations were analyzed by two techniques (Fig. 3B). First, flow cytometry was used to assay cells that lost surface IgM after two weeks in culture due to mutations introducing stop codons in the V gene (IgM+ to IgM−). Analysis of sh-control and sh-Top1 clones revealed a significant increase in IgM− cells when Top1 levels were reduced. Second, DNA sequencing was used to assay the total population of cells (independent of IgM status) to detect all the mutations in VJλ genes. There was a significant increase in mutations from sh-Top1 clones compared to sh-control clones. Additionally, no mutation events were identified within the Cλ gene (data not shown) even with elevated Pol II levels, consistent with lack of AID localization by ChIP.
Fig. 3.
Somatic hypermutation and gene conversion. (A) Map of Igλ locus in ΦV− AIDR2 cells. TATA box, and VJ and C exons are shown. Double arrow represents the area amplified for sequencing. (B) SHM. Left graph; flow cytometry analysis of 3 sh-control (gray) or 9 sh-Top1 (red) independent clones, with 24–36 subclones analyzed per clone. P value, unpaired two-tailed Student’s t test. Right table; sequence analysis from 10 sh-control and 11 sh-Top1 subclones. P value, unpaired two-tailed Student’s t test. (C) Map of Igλ locus in AID+/+ cells. Yellow boxes represent the position of VJ and C exons, and black boxes show the position of V pseudogenes. Double arrow indicates the region sequenced. (D) GC. Left graph; flow cytometry analysis using 2 sh-control (gray) or 4 sh-Top1 (red) independent clones, with 24 subclones analyzed per clone. P value, unpaired two-tailed Student’s t test. Right table; sequence analysis from 4 sh-control and 8 sh-Top1 subclones. P value, unpaired two-tailed Student’s t test.
For GC, we used a cell line with pseudogenes (AID+/+, surface IgM−), where AID deamination events are processed into DNA strand breaks to initiate homologous recombination in the rearranged VJλ gene using any of the 25 ΦV genes for template repair (Fig. 3C). Similar to SHM, these events can be scored by flow cytometry and DNA sequencing (Fig. 3D). For flow cytometry, GC rescues surface IgM expression by reversing a frame shift mutation within the Vλ gene (IgM− to IgM+). Analysis of sh-control and sh-Top1 clones showed a significant increase in IgM+ revertants when Top1 was reduced. For sequencing, VJλ genes from the total population of cells (independent of IgM status) were analyzed. There was a significant increase in conversion frequency from sh-Top1 versus sh-control clones. Thus, Pol II accumulation, induced by Top1 deficiency, increased AID targeting to VJλ genes, which then escalated mutagenesis.
3.4. Top1 expression is unaffected by AID levels in DT40 and mouse cells
We have presented data here that Pol II abundance regulates AID accumulation and mutagenesis. In this scenario, anything that increases Pol II on V genes, such as Top1 reduction, will raise the mutation frequency. However, Honjo and colleagues proposed another mechanism to explain how Top1 knockdown increases mutagenesis (28, 29). They suggest that AID protein edits a microRNA that reduces the translation of Top1 mRNA. Diminished Top1 protein would amplify non-B structures in DNA, which undergo cleavage by the residual Top1 protein. DNA strand breaks then become substrates for SHM and class switch recombination. According to their model, AID+/+ cells will have less Top1 protein compared to AID−/− cells. We therefore tested this hypothesis to see if AID altered Top1 protein in DT40 cells. Western blot analyses for AID, Top1, and β-actin were performed on AID−/−, AID+/+, and AIDR DT40 cells (Fig. 4A). AID protein levels were different among the three strains, with cells reconstituted with AID (AIDR) expressing the most protein. However, Top1 expression was identical in all three lines. To repeat the experiments of Honjo and colleagues using mice (29), we then measured AID, Top1, and β-actin levels in Aid−/− and Aid+/+ primary mouse splenic B cells, before and after activation with LPS and IL-4 (Fig. 4B). AID and β-actin mRNA was assessed by RT-PCR, and Top1 and β-actin protein was quantified by western blots. At 0, 6, and 24 h after stimulation, AID mRNA was absent in Aid−/− cells, and increased in Aid+/+ cells. Top1 protein levels were unchanged by the presence or absence of AID, and were independent of AID upregulation after stimulation. Thus, in contrast to the results of Kobayashi et al. (29), AID had no detectable effect on Top1 protein levels. The reason for this discrepancy in data is not clear.
Fig. 4.
Top1 protein levels in DT40 and mouse B cells. (A) DT40 cells. Protein levels of AID, Top1, and β-actin are shown in representative western blots for AID−/−, AID+/+, or AIDR cell lines. Bar graph depicts the average Top1/β-actin ratio from 3 independent experiments; error bars represent SD. (B) Mouse cells. Splenic B cells were stimulated with LPS and IL-4 for 0, 6, and 24 h. mRNA levels by RT-PCR are shown in representative panels for AID and β-actin from Aid−/− and Aid+/+ cells. Similar results were seen in 2 additional mice per genotype (data not shown). Protein levels of Top1 and β-actin are shown in representative western blots for Aid−/− and Aid+/+ cells. Bar graph depicts the average Top1/β-actin ratio from 3 independent experiments with 1 mouse per experiment; error bars show SD.
4. Conclusion
Recent evidence suggests that Pol II and Spt5 are responsible for targeting AID to V and switch regions in mice (7, 8, 10, 30). The current theory is that when Pol II is paused, Spt5 accumulates and recruits AID. Attempts to identify what controls transcription and AID activity in the immunoglobulin loci have focused on the 3′ enhancer elements downstream of C genes. In mice, deletion of this region in the Igh locus diminishes Pol II and ablates Spt5 and AID recruitment (8, 31). In DT40 cells, deletions in this region in the Igλ locus modulate transcription and reduce mutagenesis (32–34). Thus, a definitive role for the 3′ enhancer region in transcription and mutation is unclear at this time. What is clear, however, is that Pol II is critical to initiate the assembly and activity of the mutation complex. For assembly, we demonstrate here that Top1 deficiency increased Pol II accumulation over transcribed Vλ, Cλ, and γ-actin genes, and magnified AID protein over the Vλ gene. For activity, Pol II abundance generates single-strand DNA (8, 9, 35, 36), which is the substrate for AID. Thus, Pol II has a dual role in both instigating recruitment of Spt5 and AID, and producing single-strand DNA for AID to act on. Pol II accumulation might be due to cis DNA sequences around the V gene that regulate transcription, perhaps by delaying promoter release of the initiating polymerases (8). The identification of such sequences may unlock the mystery of why V genes are a magnet for AID-induced mutagenesis.
Supplementary Material
Highlights.
Knockdown of Topoisomerase I increased RNA Polymerase II and AID in Vλ genes.
Pol II accumulation elevated somatic hypermutation and gene conversion in DT40 cells.
Topoisomerase I was unaffected by AID levels.
Acknowledgments
We thank William Yang for technical assistance, Jean Marie Buerstedde and Hiroshi Arakawa for generously providing DT40 cell lines, and Ranjan Sen and David Wilson III for critical reading of the manuscript. This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Appendix A. Supplementary data
Supplemental Table S1 lists the primers used for shRNA, ChIP, DNA sequencing, and RT-PCR.
Supplemental Fig. S1 shows western blots for Top1 and β-actin from all clones transfected with sh-control and sh-Top1.
Footnotes
Conflict of interest statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maul RW, Gearhart PJ. Refining the Neuberger model: Uracil processing by activated B cells. Eur J Immunol. 2014;44:1913–1916. doi: 10.1002/eji.201444813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 3.Chiarle R, Zhang Y, Frock R, Lewis S, Molinie B, Ho Y, Myers D, Choi V, Compagno M, Malkin D, Neuberg D, Monti S, Giallourakis C, Gostissa M, Alt FW. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein I, Resch W, Jankovic M, Oliveira TY, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, Casellas R, Nussenzweig M. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 6.Canugovi C, Samaranayake M, Bhagwat AS. Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase. FASEB J. 2009;23:34–44. doi: 10.1096/fj.08-115352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maul RW, Cao Z, Venkataraman L, Giorgetti CA, Press JL, Denizot Y, Du H, Sen R, Gearhart PJ. Spt5 accumulation at variable genes distinguishes somatic hypermutation in germinal center B cells from ex vivo-activated cells. J Exp Med. 2014;211:2297–2306. doi: 10.1084/jem.20131512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh CL, Lieber MR. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranello L, Kouzine F, Levens D. DNA topoisomerases beyond the standard role. Transcription. 2013;4:232–237. doi: 10.4161/trns.26598. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour DS, Pflugfelder G, Wang JC, Lis JT. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986;44:401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- 14.Stewart AF, Schutz G. Camptothecin-induced in vivo topoisomerase I cleavages in the transcriptionally active tyrosine aminotransferase gene. Cell. 1987;50:1109–1117. doi: 10.1016/0092-8674(87)90177-2. [DOI] [PubMed] [Google Scholar]
- 15.Stewart AF, Herrera RE, Nordheim A. Rapid induction of c-fos transcription reveals quantitative linkage of RNA polymerase II and DNA topoisomerase I enzyme activities. Cell. 1990;60:141–149. doi: 10.1016/0092-8674(90)90724-s. [DOI] [PubMed] [Google Scholar]
- 16.Ljungman M, Hanawalt PC. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis. 1996;17:31–35. doi: 10.1093/carcin/17.1.31. [DOI] [PubMed] [Google Scholar]
- 17.Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arakawa H, Saribasak H, Buerstedde JM. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 20.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 21.Kohli RM, Maul RW, Guminski AF, McClure RL, Gajula KS, Saribasak H, McMahon MA, Siliciano RF, Gearhart PJ, Stivers JT. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J Biol Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature protocols. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 23.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nature cell biology. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Humphries EH, Tjoelker L, Carlson L, Thompson CB. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol Cell Biol. 1990;10:3224–3231. doi: 10.1128/mcb.10.6.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buerstedde JM, Reynaud CA, Humphries EH, Olson W, Ewert DL, Weill JC. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi M, Sabouri Z, Sabouri S, Kitawaki Y, Pommier Y, Abe T, Kiyonari H, Honjo T. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc Natl Acad Sci U S A. 2011;108:19305–19310. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi M, Aida M, Nagaoka H, Begum NA, Kitawaki Y, Nakata M, Stanlie A, Doi T, Kato L, Okazaki IM, Shinkura R, Muramatsu M, Kinoshita K, Honjo T. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci U S A. 2009;106:22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogne M, Denizot Y. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210:1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kothapalli NR, Collura KM, Norton DD, Fugmann SD. Separation of mutational and transcriptional enhancers in Ig genes. J Immunol. 2011;187:3247–3255. doi: 10.4049/jimmunol.1101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buerstedde JM, Alinikula J, Arakawa H, McDonald JJ, Schatz DG. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 2014;12:e1001831. doi: 10.1371/journal.pbio.1001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohler KM, McDonald JJ, Duke JL, Arakawa H, Tan S, Kleinstein SH, Buerstedde JM, Schatz DG. Identification of core DNA elements that target somatic hypermutation. J Immunol. 2012;189:5314–5326. doi: 10.4049/jimmunol.1202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsa JY, Ramachandran S, Zaheen A, Nepal RM, Kapelnikov A, Belcheva A, Berru M, Ronai D, Martin A. Negative supercoiling creates single-stranded patches of DNA that are substrates for AID-mediated mutagenesis. PLoS Genet. 2012;8:e1002518. doi: 10.1371/journal.pgen.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.