Abstract
Imaging mass spectrometry (IMS) has become a valuable tool for the production of molecular maps in samples ranging from solid inorganic materials to biologicals such as cells and tissues. The unique features of IMS are its ability to map a wide variety of different types of molecules, its superb molecular specificity, and its potential for discovery since no target specific reagents are needed. IMS has made significant contributions in biology and medicine and promises to be a next generation tool in anatomic pathology.
Keywords: imaging, MALDI MS, mass spectrometry, molecular microscopy
Throughout the ages of discovery even from early times of human existence, images have played a vital role in our efforts to understand the natural world and to educate future generations. This is clearly seen in cave paintings of early humans used to depict their environment and their desire to record their experiences to archive knowledge and culture. In looking back over the centuries, one can only marvel at the influence of the images produced by Galileo’s telescope and Van Leuwenhoek’s microscope and their impact on human scientific discovery. Fast forwarding to today, images have even become more important, playing critical roles in our personal lives as well as professional activities. It is nowhere more important than in science where imaging capabilities have provided the groundwork for quantum leaps in our understanding of the natural world in every field of science. Imaging technologies have enabled the molecular mapping of specimens in material science, geology, biology and so many other areas of human endeavor. Moreover, during the past decades, imaging technologies, from early X-ray images to advanced microscopy, MRI, and PET imaging technologies, have become absolutely essential in clinical settings.
As a result of extensive genomic information and the now rapidly growing proteomics and metabolomics era, we are well into a new age in biology and medicine based on the molecular expression of biomolecules in organs, tissues and cells. As we probe deeper into the molecular milieu of living cells, the complexity, including a vast array of molecular interactions occurring in time and space, is astounding even today. Mass spectrometry (MS) now finds itself in a critically important role of providing unparalleled molecular specificity and sensitivity to create molecular image maps in tissues. The mapping subjects are trace elements, low molecular weight organic molecules and endogenous metabolites, drugs, lipids, peptides, proteins, polynucleotides, saccharides, and polymers. Investigators can now produce a series of images, each presented at discrete molecular weights (measured as m/z values) over an entire sample. Perhaps the greatest challenge yet for MS lies in its capability of significantly helping us understand this complexity of life. An even greater reward for the future is being able to describe a detailed roadmap of biology and to enable the mitigation and perhaps even the cure of disease.
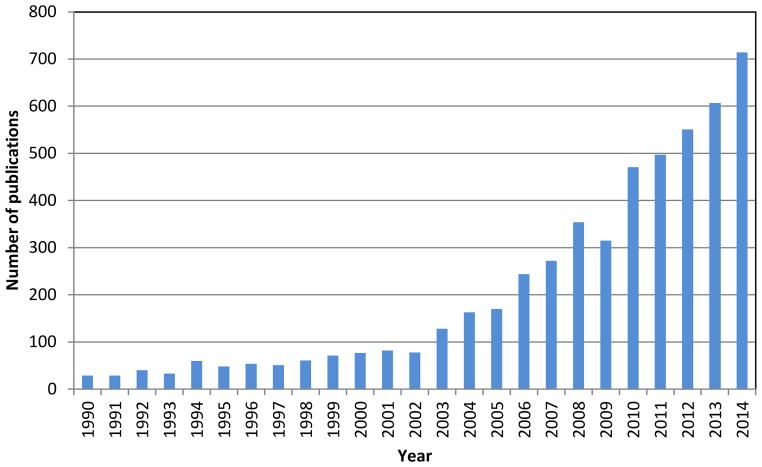
The interest and application of imaging MS technology has exploded over the past 10 years in different areas of science and has led to an astonishing number of papers published in this still rapidly growing field, as shown in Figure 1. Current commercial imaging mass spectrometers use a microprobe approach in which a laser or an atomic or molecular beam is employed to irradiate a sample spot that will then become pixels in an image.
Figure 1.
Number of publications by year from data obtained from a search with SciFinder Scholar for imaging mass spectrometry.
Ablation of these spots on the target forms an array of pixels across the area of interest, giving rise to a raw image dataset. Each pixel then has a mass spectrum associated with it that can represent the entire mass range of the instrument. The size of the laser beam on target and the pitch or distance between the ablation spots then define the size of the pixel and ultimately the spatial resolution of the image. The size of the pixel can be varied typically from 100 μm down to about 20 μm in current commercial instruments, and with some modified research instruments, down to 1 μm or less. A plot of any one of the specific m/z signals in the mass spectrum across the entire array of pixels then gives rise to an ion map or image of the sample for that value. Each m/z value recorded can give rise to a specific image by plotting the intensity of that m/z value over the entire pixel array. Typical MS images have 10,000 – 50,000 pixels, although some high resolution images can have more than a million pixels. In an alternative approach, the instrument acquisition mode can be targeted for just a specific mass or a very limited mass range encompassing a given compound or group of compounds.
Many different ionization modes have been used for imaging by MS, including SIMS [1], laser desorption ionization [2], MALDI [3], and desorption ESI of a number of varieties and variations [4], and all have unique capabilities and limitations. In the biological and medical arena, perhaps the most generally useful technology for imaging biomolecules in tissues and cells is MALDI MS. The first description of this imaging platform and its use for imaging cells and tissues was published in 1997 when it was demonstrated that signals for peptides and proteins could be obtained directly from tissues and blots of tissues [3]. This technology enjoys a wide applicability to biological and medical research owing to the balance among critical imaging parameters in terms of spatial resolution, molecular types amenable to analysis, molecular mass range, and sensitivity. Figure 2 shows a MALDI MS image for one lipid in a transverse section of a mouse brain as an example of the image quality attainable today.
Figure 2.
(left panel) – Partial microscopic image of a transverse section of mouse brain stained with hematoxylin/eosin. (right panel) – MALDI MS image of a partial brain section of the ion at m/z 885.5507 – [PI(38:4)] taken at a spatial resolution of 15 μm on a Bruker SolariX 15 T FTICR MS in the negative-ion mode and with >100,000 resolving power at m/z 400. The data were acquired at a laser repetition rate of 2 kHz and an acquisition speed of ~0.35s/pixel. The image dataset has a total of 364,740 pixels and a file size of ~800 GB.
One of the greatest opportunities and challenges for imaging MS lies in its clinical potential to aid diagnosis, prognosis, and effectiveness of therapy through the molecular assessment of tissues obtained from patients [5]. Through the unique combination of cell-specific sampling and the multiplex capabilities of the mass spectrometer, molecular signatures of specific diseased cells can be assayed where 10–20 or more proteins or metabolites can be measured virtually at the same time and at high sensitivity. For the first time, anatomical pathologists will have a tool to examine tissues directly without the limitations and expense of needing specific molecular probes. For clinical research, the potential for discovery is maximized because target-specific reagents such as antibodies are not required. It is clear that direct molecular mapping in disease will become increasingly important for molecular pathology in the coming years and will represent a technological paradigm shift in this field.
Although the vision of creating a functional molecular microscope goes back many years, the technology advances achieved today in MS make this vision a reality and enable its capability to provide a deeper understanding of molecular events ongoing in living systems. The technology is already advancing at a rapid rate, and some publications show images of biological tissue at the single cell level [6]. As our ability to desorb, collect and record ions improves, practical imaging at submicron levels will become routine and bring a wealth of information that details the molecular movement and compartmentalization of biomolecules within cellular substructures. In terms of the future of imaging MS, the picture is clear and quite bright.
References
- 1.Eccles AJ, Vickerman JC. The characterization of an imaging time-of-flight secondary ion mass spectrometry instrument. J Vac Sci Technol A. 1989;7(2):234–244. [Google Scholar]
- 2.Savina MR, Lykke KR. Chemical imaging of surfaces with laser desorption mass spectrometry. Trends Anal Chem. 1997;16(5):242–252. [Google Scholar]
- 3.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 4.Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32:218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norris JL, Caprioli RM. Imaging mass spectrometry: a new tool for pathology in a molecular age. Proteomics Clin Appl. 2013;7:733–738. doi: 10.1002/prca.201300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry. MALDI MS. J Mass Spectrom. 2012;11(47):1473–81. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]