Abstract
In microbiology, changes in specialized metabolite production (cell-to-cell signaling metabolites, virulence factors and natural products) are measured using phenotypic assays. However, advances in mass spectrometry based techniques including imaging mass spectrometry (IMS) now allow researchers to directly visualize the production of specialized metabolites from microbial colony biofilms. In this study, a combination of IMS and liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to visualize the effect of the macrolide antibiotic azithromycin (AZM) on colony biofilms of Pseudomonas aeruginosa. While previous research suggested that AZM may inhibit cell-to-cell signaling of P. aeruginosa and thereby reducing pathogenicity, we observed no clear decrease in specialized metabolite production.
Introduction
Pseudomonas aeruginosa is one of the most common and persistent opportunistic pathogens affecting patients with cystic fibrosis (CF). Chronic P. aeruginosa infections lead to progressive deterioration of patient lung function causing morbidity and mortality in CF patients.[1] P. aeruginosa is able to survive and thrive in these patients by growing as biofilms which provides tolerance to both inflammatory defense mechanisms of the host and antibiotic therapies.[2, 3] Several clinical studies have shown that long-term treatment of CF patients with chronic P. aeruginosa infections with the macrolide antibiotic azithromycin (AZM) leads to improved lung function and increased body weight.[4–7] The median concentration of AZM in the sputum of patients receiving high-dose therapy (250 mg AZM per day) is 9.5 g/mL, well below the minimum inhibitory concentration (MIC) for P. aeruginosa (128 to 512 g/mL).[4, 6]
It has been suggested that one way AZM treatment improves patient lung function is by inhibiting P. aeruginosa exchange of molecules involved in cell-to-cell interactions.[8, 9] This results in a reduction of exoproducts and pathogenicity. These exoproducts include specialized metabolites (quorum sensors, virulence factors and natural products) which are key components in the interactions between P. aeruginosa and the host. P. aeruginosa utilizes a hierarchical signaling pathway to control specialized metabolite production where the transcriptional factor pair RhlI-RhlR is subordinate to the LasI-LasR pair.[10, 11] LasI produces N-3-oxo-dodecanoyl-L-homoserine lactone (3-oxo-C12-HSL) while RhlI produces N-butanoyl-L-homoserine lactone (C4-HSL). These HSLs bind to the transcriptional activators LasR and RhlR, respectively, and activate target promoters. Both las and rhl have been implicated in regulating the production of a third signaling metabolite, 2-heptyl-hydroxy-4-quinolone (Pseudomonas quinolone signal (PQS)).[12–14] The HSLs and PQS have been shown to regulate the expression of genes required for specialized metabolite production including those for the siderophores pyochelin and pyoverdine, as well as, the phenazine, quinolone and rhamnolipid molecular families.[15] Herein, we describe the application of agar based microbial matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry (IMS) in combination with liquid chromatography-tandem mass spectrometry (LC-MS/MS) to investigate the effect of AZM on specialized metabolite production of three strains of P. aeruginosa: FLR01, a non-mucoid clinical isolate, and two common laboratory strains, PAO1 and PA14.
Methods
General
P. aeruginosa strain PA14 was provided by the D. Hung lab (Harvard Medical School, MA, USA) and originated in the lab of F. M. Ausubel (Massachusetts General Hospital, MA, USA).[16] Strain PAO1 was provided by the S. Noble lab (University of California, San Francisco, CA, USA) and originated in the lab of C. Manoil (University of Washington, Seattle, WA, USA).[17] Non-mucoid clinical isolate FLR01 was provided by the F. Rohwer lab (San Diego State University, CA, USA). All chemicals used for LB and ISP2 media were purchased from Sigma-Aldrich. Azithromycin was purchased from Sigma-Aldrich. LC-MS grade organic solvents were purchased from J.T. Baker.
P. aeruginosa culture
P. aeruginosa strain PA14, PAO1 and FLR01 were cultured to stationary phase (OD600 = 1.2) overnight in LB liquid medium from a single colony. The cultures were diluted to a 20% glycerol/water stock and stored at −80°C (~2.0×108 CFU/mL). P. aeruginosa (1 L) was inoculated directly from glycerol stocks on ISP2 agar medium (10 mL) containing 0, 2, 4, 6 or 8 g/mL azithromycin in 100 O.D.×25 mm Petri dishes (Fisherbrand). Cultures were incubated at 30°C for 48 hr.
MALDI IMS
MALDI IMS samples were prepared as previously described.[18, 19] Briefly, for each sample a region of agar including the colony biofilm was excised from the culture and laid on top of a Bruker MALDI MSP 96 anchor plate. To measure qualitative concentrations differences in metabolite production between samples, cultures of the same strain cultured under different AZM concentration were analyzed on the same MALDI plate. A photograph was taken of the agar sections transferred to the MALDI plates. Universal matrix (Sigma-Aldrich) was applied manually using a 53 m molecular sieve. Samples were dried at 37°C overnight. All colonies were subjected to MALDI-TOF IMS in positive reflectron mode using 500 m spatial resolution in both X and Y dimensions by a Bruker Daltonics Microflex.
Extraction procedure
General chemical extraction of the samples was performed. The agar from entire single plates including the colony biofilms was sliced into small pieces and extracted with 10 mL of ethyl acetate followed by 10 mL of methanol. The solvent was separated from agar pieces by filtration and concentrated in vacuo. Combined ethyl acetate-methanol extracts were resuspended in 1 mL of methanol and centrifuged prior to analysis.
LC-MS/MS analysis
Prior to LC-MS/MS analysis, samples were diluted 10-fold. Mass spectrometry was performed using a Bruker Daltronics Maxis qTOF mass spectrometer equipped with a standard electrospray ionization source. The mass spectrometer was tuned by infusion of Tuning Mix ES-TOF (Agilent Technologies) at a 3 L/min flow rate. For accurate mass measurements, a wick saturated with hexakis(1H,1H,3H-tetrafluoropropoxy)phophazene ions (Synquest Laboratories; m/z 922.0098) located within the source was used for lock mass internal calibration. Samples were introduced by a Thermo Scientific UltraMate 3000 Dionex ultraperformance liquid chromatograph (UPLC) using a 20-µL injection volume. Ethyl acetate-methanol extracts were separated using a Phenomenex Kinetex 2.6 m C18 (30×2.10 mm) UPLC column. A linear water-acetonitrile gradient (from 98:2 to 2:98 water/acetonitrile) containing 0.1% formic acid was utilized. The flow rate was 0.5 mL/min. The mass spectrometer was operated in data-dependent positive-ion mode, automatically switching between full-scan MS and MS/MS acquisitions. Full-scan MS spectra (m/z 50 to 2,000) were acquired, and the top 10 most intense ions in a particular scan were fragmented using collision-induced dissociation at 35 eV for +1 ions and 25 eV for +2 ions in the collision cell.
Data analysis
IMS data analysis was performed using Bruker Daltronics FlexImaging v3.0. Visualization of ion intensity was optimized to highlight differences between samples being compared. Structural verification of ions putatively identified in IMS was performed manually by comparing the exact mass from the LC-MS/MS data and the corresponding MS/MS spectra with previously reported structural characterization.[20, 21] LC-MS/MS data analysis was performed using Bruker Daltronics DataAnalysis v4.1 (Build 362.7). Lock mass internal calibration using hexakis(1H,1H,3H-tetrafluoropropoxy)phosphazene ions (Synquest Laboratories; m/z 922.0098) was applied. Extracted ion chromatograms (EICs) using the exact mass of a metabolite of interest were created. The MS/MS spectra from these EICs were manually compared with previously reported data. XCMS Online was used for peak matching, non-linear retention time alignment and quantification of mass spectral chromatographic peak areas across all samples.[22] To visualize these data, peak areas were normalized to culture growth (CFUs), fold-changes were calculated versus wild-type, log2 scaled and visualized as a heat map in R using a previously described script.[23]
Results and Discussion
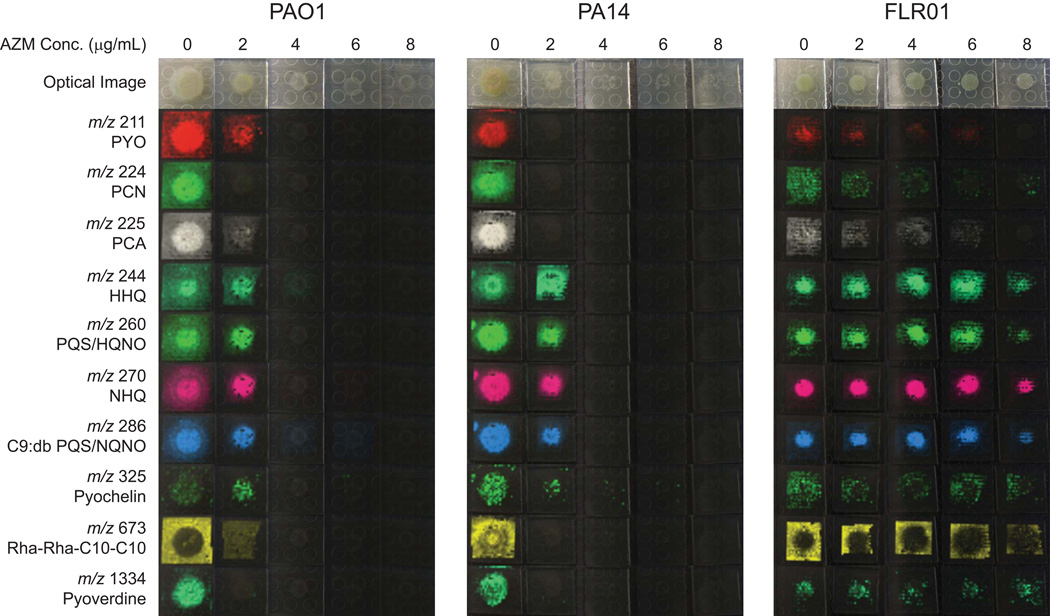
Macrolides, including AZM, have been reported to suppress the production of exoproducts including exopolysaccharide, exoenzymes and specialized metabolites through inhibition of P. aeruginosa’s signaling cascade.[8, 9] As interference with cell-to-cell signaling has been postulated as a potential beneficial effect of AZM treatment, we aimed to analyze the effect of AZM on specialized metabolite production of P. aeruginosa colony biofilms. Three P. aeruginosa strains including the laboratory strains PAO1 and PA14 and a clinical isolate from a CF patient treated with AZM, FLR01, were grown in the presence of 5 different concentrations of AZM (0, 2, 4, 6 and 8 g/mL) below the average level of AZM in the sputum of CF patients undergoing AZM therapy.[4] To visualize the effect of AZM on metabolite production, MALDI-IMS was used.[18, 19] In order to compare relative signal intensity between samples, IMS data for a single strain were collected in one experiment while data for the different strains were collected separately. As shown in Figure 1a wide variety of specialized metabolites were detected including the phenazines pyocyanin (PYO), phenazine-1-carboxamide (PCN) and phenazine-1-carboxylic acid (PCA); quinolones 2-heptyl-4-quinolone (HHQ), PQS, 4-hydroxy-2-heptylquinoline-N-oxide (HQNO), 2-nonyl-4-quinolone (NHQ) and 2-nonenyl-hydroxy-4-quinolone (C9:db PQS) and 4-hydroxy-2-nonenylquinoline-N-oxide (C9:db NQNO) among others; various rhamnolipids as exemplified by 3-[3-[(2R,3R,4R,5R,6S)-4,5-dihydroxy-6-methyl-3-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxyoxan-2-yl]oxydecanoyloxy]decanoic acid (Rha-Rha-C10-C10) and the siderophores pyochelin and pyoverdine.[20, 21]
Fig. 1.
Microbial IMS images of selected metabolites produced by P. aeruginosa strains PAO1, PA14 and FLR01 treated with increasing concentrations of AZM grown on ISP2 media for 48 h. Optical images are displayed in the top row. All other images are overlays of falsely colored m/z distributions over optical images. Both PAO1 and PA14 showed decreased metabolite production with increasing AZM concentration likely due to decreased growth. Strain FLR01 metabolite production was detected at all concentrations of AZM.
With increasing concentration of AZM, both PAO1 and PA14 showed decreased growth (Fig. 1) as measured by cell forming units (CFUs). Typically, antibiotic sensitivity is measured against exponentially growing P. aeruginosa with measured MIC values of AZM up to 512 g/mL.[6] The growth inhibition of the colony biofilms of AZM sensitive strains (PAO1 and PA14) supports previous finding that P. aeruginosa biofilms respond to AZM at much lower concentrations; similar to P. aeruginosa liquid cultures grown to stationary phase.[24–26] It is believed that macrolides exhibit antipseudomonal activity at clinically achievable levels in a time dependent manner against both biofilm and stationary phase P. aeruginosa by a combination of disruption of the cell membrane and inhibition of protein synthesis.[27, 28] Not surprisingly, in the IMS data for PAO1 and PA14, a decrease in observed metabolite production coincides with a reduction in CFUs. However, specialized metabolite production was detected for all concentrations of AZM administered to strain FLR01.
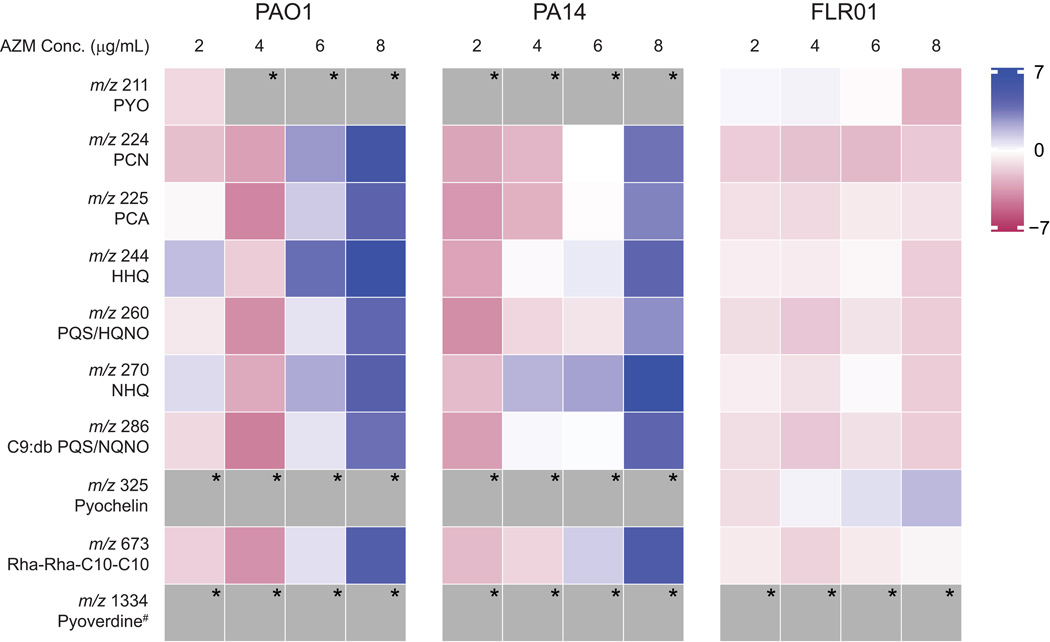
To more quantitatively assess specialized metabolite production, entire agar plates of replicate colony biofilms were extracted and subjected to LC-MS/MS. XCMS Online was used for peak matching, non-linear retention time alignment and quantification of mass spectral chromatographic peak areas across all samples.[22] To visualize specialized metabolite production as a heat map, the log2 scaling of the ratio of metabolite peak areas (AZM treated to not treated) normalized to cell growth was calculated. As shown in Fig. 2, the metabolite abundance normalized to growth shows an overall decrease for lower concentrations of AZM (2 and 4 g/mL) for PAO1 and PA14. However, in general, metabolite abundance increases for higher concentrations of AZM. This may be partially explained by the role of antibiotics as signaling metabolites.[29–31] Recent research suggests that low concentrations of antibiotics can induce specific changes in the behavior of bacteria susceptible to those antibiotics. For example, subMICs of the antibiotics tobramycin, ciprofloxacin and tetracycline led to increased transcription levels of genes involved in biofilm formation in strain PAO1.[29, 31] Interestingly, FLR01 does not respond to AZM in the same manner. This may be due to the fact that FLR01 shows some resistance to AZM and does not need to upregulate virulence genes to enhance survival.
Fig. 2.
Heat map for the LC-MS/MS profiles of selected specialized metabolites produced by P. aeruginosa strains PAO1, PA14 and FLR01 treated with increasing concentrations of AZM grown on ISP2 media for 48 h. Blue indicates increased metabolite production with respect to untreated biofilms, while red indicates decreased production. Both PAO1 and PA14 showed decreased metabolite production with lower concentrations of AZM, but increased metabolite production with higher concentrations. Metabolite production remained relatively consistent for strain FLR01 regardless of AZM concentration. * denotes not detected. # pyoverdine is not extracted under our conditions.
Conclusion
In this work, we described the application of microbial IMS in combination with LC-MS/MS to unravel the effect of AZM treatment on specialized metabolite production of P. aeruginosa biofilms. While previous research has suggested that AZM may act as a cell-to-cell signaling inhibitor at levels below the MIC, our results show that when corrected for a decrease in cell counts, specialized metabolite production increases with increasing concentration of AZM in AZM sensitive strains. Further investigation of the effect of common antibiotic classes at sub-inhibitory levels on P. aeruginosa specialized metabolites, both in single culture and co-culture, will help unravel the chemical response of pathogens affecting immunocompromised patients during treatment.
Acknowledgements
This work was supported by National Institutes of Health grant K01GM103809 (VVP). We also acknowledge the Bruker Therapeutic Discovery Mass Spectrometry Center at UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences partially funded byNIH Grant GMS10RR029121 (PCD). We gratefully acknowledge the Bugni and Currie laboratories at the University of Wisconsin-Madison for providing the R scripts and the Hung, Noble and Rohwer laboratories for providing the P. aeruginosa strains. Finally, the authors wish to congratulate Prof. Richard Caprioli on his recognition by ASMS for his achievements in imaging mass spectrometry.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15(2):194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi H. Biofilm disease: its clinical manifestation and therapeutic possibilities of macrolides. Am J Med. 1995;99(6A):26S–30S. doi: 10.1016/s0002-9343(99)80282-4. [DOI] [PubMed] [Google Scholar]
- 4.Baumann U, King M, App EM, Tai S, Konig A, Fischer JJ, Zimmermann T, Sextro W, von der Hardt H. Long term azithromycin therapy in cystic fibrosis patients: a study on drug levels and sputum properties. Can Respir J. 2004;11(2):151–155. doi: 10.1155/2004/747841. [DOI] [PubMed] [Google Scholar]
- 5.Equi A, Balfour-Lynn IM, Bush A, Rosenthal M. Long term azithromycin in children with cystic fibrosis: a randomised, placebo-controlled crossover trial. Lancet. 2002;360(9338):978–984. doi: 10.1016/s0140-6736(02)11081-6. [DOI] [PubMed] [Google Scholar]
- 6.Howe RA, Spencer RC. Macrolides for the treatment of Pseudomonas aeruginosa infections? J Antimicrob Chemother. 1997;40(2):153–155. doi: 10.1093/jac/40.2.153. [DOI] [PubMed] [Google Scholar]
- 7.Saiman L, Marshall BC, Mayer-Hamblett N, Burns JL, Quittner AL, Cibene DA, Coquillette S, Fieberg AY, Accurso FJ, Campbell PW., 3rd Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA. 2003;290(13):1749–1756. doi: 10.1001/jama.290.13.1749. [DOI] [PubMed] [Google Scholar]
- 8.Nalca Y, Jansch L, Bredenbruch F, Geffers R, Buer J, Haussler S. Quorumsensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob Agents Chemother. 2006;50(5):1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tateda K, Comte R, Pechere JC, Kohler T, Yamaguchi K, Van Delden C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45(6):1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181(4):1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiology and molecular biology reviews : MMBR. 2012;76(1):46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chemistry & biology. 2007;14(1):87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Molecular bioSystems. 2008;4(9):882–888. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 14.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M. Quinolones: from antibiotics to autoinducers. FEMS microbiology reviews. 2011;35(2):247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260(5111):1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 16.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, Fleming TC, Wenter R, Moore BS, Golden SS, Pogliano K, Dorrestein PC. Primer on agar-based microbial imaging mass spectrometry. J Bacteriol. 2012;194(22):6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5(12):885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moree WJ, Phelan VV, Wu CH, Bandeira N, Cornett DS, Duggan BM, Dorrestein PC. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci U S A. 2012;109(34):13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelan VV, Moree WJ, Aguilar J, Cornett DS, Koumoutsi A, Noble SM, Pogliano K, Guerrero CA, Dorrestein PC. Impact of a transposon insertion in phzF2 on the specialized metabolite production and interkingdom interactions of Pseudomonas aeruginosa. J Bacteriol. 2014;196(9):1683–1693. doi: 10.1128/JB.01258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Analytical chemistry. 2012;84(11):5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou Y, Tianero MD, Kwan JC, Wyche TP, Michel CR, Ellis GA, Vazquez- Rivera E, Braun DR, Rose WE, Schmidt EW, Bugni TS. Structure and biosynthesis of the antibiotic bottromycin D. Organic letters. 2012;14(19):5050–5053. doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillis RJ, Iglewski BH. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J Clin Microbiol. 2004;42(12):5842–5845. doi: 10.1128/JCM.42.12.5842-5845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichimiya T, Takeoka K, Hiramatsu K, Hirai K, Yamasaki T, Nasu M. The influence of azithromycin on the biofilm formation of Pseudomonas aeruginosa in vitro. Chemotherapy. 1996;42(3):186–191. doi: 10.1159/000239440. [DOI] [PubMed] [Google Scholar]
- 26.Mulet X, Macia MD, Mena A, Juan C, Perez JL, Oliver A. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob Agents Chemother. 2009;53(4):1552–1560. doi: 10.1128/AAC.01264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamura Y, Higashiyama Y, Tomono K, Izumikawa K, Yanagihara K, Ohno H, Miyazaki Y, Hirakata Y, Mizuta Y, Kadota J, Iglewski BH, Kohno S. Azithromycin exhibits bactericidal effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob Agents Chemother. 2005;49(4):1377–1380. doi: 10.1128/AAC.49.4.1377-1380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tateda K, Ishii Y, Matsumoto T, Furuya N, Nagashima M, Matsunaga T, Ohno A, Miyazaki S, Yamaguchi K. Direct evidence for antipseudomonal activity of macrolides: exposure-dependent bactericidal activity and inhibition of protein synthesis by erythromycin, clarithromycin, and azithromycin. Antimicrob Agents Chemother. 1996;40(10):2271–2275. doi: 10.1128/aac.40.10.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A. 2006;103(51):19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(26):17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Current opinion in microbiology. 2008;11(2):161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]