Abstract
Analyte signal in a laser desorption/postionization scheme such as infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) is strongly coupled to the degree of overlap between the desorbed plume of neutral material from a sample and an orthogonal electrospray. In this work, we systematically examine the effect of desorption conditions on IR-MALDESI response to pharmaceutical drugs and endogenous lipids in biological tissue using a design of experiments approach. Optimized desorption conditions have then been used to conduct an untargeted lipidomic analysis of whole body sagittal sections of neonate mouse. IR-MALDESI response to a wide range of lipid classes has been demonstrated, with enhanced lipid coverage received by varying the laser wavelength used for mass spectrometry imaging (MSI). Targeted MS2 imaging (MS2I) of an analyte, cocaine, deposited beneath whole body sections allowed determination of tissue-specific ion response factors, and CID fragments of cocaine were monitored to comment on wavelength-dependent internal energy deposition based on the “survival yield” method.
Keywords: IR-MALDESI, mass spectrometry imaging, laser desorption whole body imaging, lipids, xenobiotics
INTRODUCTION
Mass spectrometry imaging (MSI) is an analytical approach that provides the capability for simultaneously monitoring the spatial distribution within a biological sample of analytes ranging from small molecules, such as pharmaceutical drugs (1), to lipids (2, 3) to peptides and proteins (4). The utility of MSI has been demonstrated for a variety of fields, with particular emphasis on biomedical applications like drug distribution (5) and biomarker identification(6). Spatial scales of analysis can vary widely based on application from cellular (tumor margin identification) (7) to multi-organ (pathway analysis) to whole body (drug and metabolite distribution) imaging (8).
MSI sensitivity and selectivity toward different compound classes are dependent on sample preparation(9) and the methodology used to desorb and ionize an analyte from a sample. For the analysis of lipids and other labile small molecules, which have emerged as areas with relevance in drug studies and biomarker identification, the predominant MSI technique, matrix-assisted laser desorption/ionization (MALDI) has some limitations. This is largely due to the fact that MALDI requires the homogenous application of a non-native organic matrix which must be judiciously selected to target an analyte class of interest. For typical MALDI MSI using a time-of-flight (TOF) analyzer, the matrix and analytes must also be stable under high vacuum conditions. MALDI matrices also dominate the spectral response in the low mass range making the analysis of labile, small molecules and their metabolites challenging due to isobaric interference, particularly when coupled to relatively low resolving power TOF MS. Ambient ionization techniques such as desorption electrospray ionization (DESI) (10), liquid extraction surface analysis (LESA) (11), and matrix-assisted laser desorption electrospray ionization (MALDESI) (12) offer alternatives to MALDI MSI with minimal sample preparation and none of the sample stability limitations of high vacuum.
Unlike surface-based techniques that rely on liquid extraction of analytes, which can cause temporal response and selective extraction of analytes, desorption of materials by pulsed laser excitation is nearly instantaneous. At infrared wavelengths, incident radiation can excite and desorb a broad range of condensed phase biological targets (13–15). While the ionization yield of the resulting desorbed plume is low, limiting the applicability of IR-LDI, MALDESI was the first ambient ionization technique to overcome this issue by coupling resonant laser desorption with electrospray post-ionization (16). Like the subsequent and analogous methods LAESI (17) and LADESI (18), IR-MALDESI has utilized an incident wavelength of 2940 nm, corresponding to the absorption cross-section peak for the OH stretching band of liquid water. This wavelength excites species natively present in biological samples, such as endogenous water, thereby eliminating background interferences associated with non-native matrices. Pulsed infrared laser desorption of a water-rich biological target results in enhanced penetration depth relative to the UV excitation of a MALDI matrix, micrometers vs nanometers, respectively (14). Specimen sections of standard thickness (10–50 micrometers) are completely ablated in 2–3 IR laser pulses. This rapid ejection of material associated with the probed volumetric element, or voxel, means that IR-MALDESI is agnostic to the location of internal standards, which can be placed readily above or below a tissue sample(19).
The sensitivity of IR-MALDESI is contingent on a high degree of spatial overlap between the material ejected from the sample surface and the orthogonal electrospray. The vertical distance from a sample surface traveled by a laser desorbed plume front is sensitive to both the laser fluence and the degree to which the wavelength of laser emission overlaps spectral absorption features of the analyte or matrix (20). Recent IR-MALDESI results indicate significant gains in sensitivity toward drugs, drug metabolites, and lipids following the addition of an exogenous ice layer to the surface of a tissue sample (21, 22). Ice is deposited facilely and uniformly onto the surface of a tissue sample, and has an added benefit of maintaining cryopreservation of a sample throughout the imaging process. This is beneficial on multiple fronts since it preserves tissue structure and limits evaporative loss of volatile small molecules or their degradation due to enzymatic activity (23). While the role of ice as an IR-MALDI matrix remains uncertain (24, 25), we have recently investigated its effect on the IR-MALDESI desorption process (21). Using shadowgraphy, we showed that the dynamics of an IR-MALDESI plume of desorbed material ablated at an incident wavelength of 2940 nm from a tissue section with an exogenous ice layer are dramatically altered relative to material desorbed from tissue without an ice layer. Based on these observations, differences in plume dynamics with the ice layer were attributed to either a reduction in laser fluence or a more efficient transfer of energy to the sample, or a combination of these factors. Ions were shown to be accumulated over multiple ablation events, suggesting that the excitation matrix is likely a mixture of water and ice.
In this work, factors controlling ice-mediated IR-MALDESI laser desorption are systematically explored using the design of experiments (DOE) statistical approach (26, 27) and optimized desorption parameters for the detection of xenobiotics and lipids are identified. Previous IR-MALDESI method optimization using ice has fixed the incident excitation wavelength at 2940 nm, but the ice or ice/water matrix may be more resonantly excited at longer wavelengths. OH stretch absorption by a thin film of ice can be significantly red-shifted to a wavelength of 3100 nm relative to liquid due to structural and morphological changes that occur during freezing (28). Increased ion yields have been observed at 3100 nm from an IR-MALDI analysis of an ice thin film (29, 30). Both fluence and wavelength can influence IR-MALDI response to a biological analyte when using ice as a matrix (31). Here, we examine the role of excitation fluence and wavelength in the ice-mediated analysis of biological samples using IR-MALDESI.
The DOE-optimized desorption parameters have then been utilized to investigate IR-MALDESI lipid response across tissue types of a whole organism at two distinct incident wavelengths relevant to the water/ice matrix, 2940 nm and 3100 nm. Whole body imaging is critically important in ascertaining the localization and distribution of drug and its metabolites across an entire organism, or in discovery-based biomarker identification. It was anticipated that the attributes of IR-MALDESI MSI would complement efforts for whole body imaging by MALDI (32–35) or DESI (8). While the ultimate spatial resolution of IR-MALDESI cannot equal that of UV-MALDI due to fundamental diffraction limits of focused infrared light, its 100 µm voxel diameter is comparable to MALDI pixel dimensions selected for practical whole body analysis throughput(32) and equals that of DESI(8) and allows key anatomical features to be distinguished. The deposition and crystallization of an ice matrix is not expected to vary across different tissue types in the manner of MALDI matrices(36) (9) (37), which can lead to ion suppression that complicates interpretation of analyte response and localization across a tissue section encompassing multiple organs or a whole organism(33) (32). Further, the combination of the IR-MALDESI source with a high resolution, high mass accuracy mass spectrometer (22) is expected to allow more ion peaks of interest to be accurately assigned without the additional necessity of selected reaction monitoring (SRM) to ensure proper peak identification, which better preserves the multiplexed capability that is one of the hallmarks of MSI as first conceived by Caprioli and coworkers(38).
Finally, the response factors of a standard analyte across the tissues of a whole organism have been evaluated to investigate the potential for ion suppression/response factors for different tissue types, as has been observed with MALDI(39). Cocaine has been used as a standard, sprayed onto glass slides prior to mounting whole body cryosections. By imaging the tissue section using selective reaction monitoring (SRM) for cocaine and determining its fragmentation ratios in collision induced dissociation, we attempt to evaluate the internal energy distribution of IR-MALDESI at 2940 nm and 3100 nm.
EXPERIMENTAL
Chemicals and Materials
HPLC Grade methanol, water, and isopentane were purchased from Burdick and Jackson (Muskegon, MI) and formic acid was purchased from Sigma-Aldrich (St. Louis, MO). Emtricitabine and raltegravir were obtained from the NIH AIDS Reagent Program, directed by the Pathogenesis and Basic Research Branch, Basic Sciences Program, Division of AIDS (DAIDS), NIAID, NIH. Cocaine was purchased from Sigma-Aldrich (St. Louis, MO). All materials were used as purchased without further purification.
Cervical tissue was obtained from surgical waste via the University of North Carolina Tissue Procurement Facility through UNC IRB # 09–0921. Written informed consent was obtained from all patients. After harvest, cervical tissue was incubated in a solution of antiretroviral (ARV) therapies containing 100,000 ng/mL of both a nucleoside reverse transcriptase inhibitor emtricitabine (FTC) and an integrase strand transfer inhibitor raltegravir (RAL) for 24 hours at 37°C. Cervical tissue was then removed from drug solution and rinsed with fresh culture media before being frozen with dry ice vapor and stored at −80°C. Adult mouse liver and two-day old whole neonatal mouse pups were obtained according to Institutional Animal Care and Use Committee (IACUC) and North Carolina State University regulations approved for the Ghashghaei laboratory. Animals were sacrificed by hypothermia in an isopentane/dry ice bath to preserve tissue structure.
Sample Preparation
Each tissue was sectioned at −20°C using a Leica CM1950 cryomicrotome (Buffalo Grove, IL) into slices of 10 or 25 µm for incubated cervical tissue or whole body neonate mouse, respectively. The sections were then thaw-mounted directly onto glass microscope slides for imaging. For evaluation of desorbed internal energy distribution across whole body sections, cocaine was diluted as received in 50:50 methanol/water to a concentration of 411 µM, and then uniformly sprayed onto glass slides using a pneumatic sprayer (TM Sprayer™, LEAP Technologies) prior to mounting a tissue sample resulting in an on-slide analyte distribution of approximately 180 fmol/voxel. Once mounted, the sample was placed on a liquid cooled thermoelectric stage that was cooled to −10°C while under nitrogen purge, after which the sample was exposed to the ambient environment in order to deposit a thin layer of ice over the surface of the tissue. Preservation of the ice layer throughout imaging was ensured by maintaining a stable relative humidity of approximately 10% within the IR-MALDESI source enclosure through the addition of dry nitrogen gas.
Instrumentation
A more detailed description of the IR-MALDESI imaging source and its optimization for tissue imaging has previously been reported (21, 40). A tunable 20 Hz pulse rate, 7 ns pulse width mid-IR la-ser (IR-Opolette 2371, Opotek, Carlsbad, CA) is used to resonantly excite the ice matrix layer and sample, facilitating the desorption of neutral molecules from the tissue section. On tissue, the laser beam diameter exceeding the ablation threshold is 150 µm. Incident laser energy is controlled by a custom-built rutile polarizer-based attenuator, and measured using a thermopile detector (Nova 2; Ophir, Jerusalem, Israel). Each voxel is subjected to two laser pulses, completely ablating the probed sample volume. Desorbed neutrals then partition into the charged solvent droplets of an electrospray plume where ions are generated through an ESI-like process. For the imaging experiments, 50% (v/v) aqueous methanol with 0.2% formic acid was used for the positive electrospray solvent, which has been shown to work well for small molecules and lipids(22). All imaging experiments were performed with an over-sampled spot-to-spot distance of 100 µm (41, 42).
The IR-MALDESI imaging source has been coupled to a Thermo Fisher Scientific Q Exactive (Bremen, Germany) such that ion accumulation was triggered to temporally overlap with the pulsing of the laser, and resulting in a single orbitrap acquisition at each voxel. While this strategy precludes the use of automatic gain control (AGC) of the instrument due to the requirement for a fixed ion injection time (IT), mass accuracy was verified to be maintained within 2 ppm using two diisooctyl phthalate peaks (m/z 391.28428 [M+H+]+ and m/z 413.26623 [M+Na+]+) as lockmasses in the instrument control software (43, 44). The mass range for the orbitrap acquisition was set to m/z 150–600 for experiments involving incubated tissue, and otherwise set to m/z 250–1000. The mass resolving power was set to 140,000 at m/z 200.
For MS2 imaging (MS2I) acquisition, a targeted MS2I method file was created using an inclusion list for isolating the protonated ion of cocaine (m/z 304.1550) with a 4 m/z window and a 1.5 m/z offset followed by ion accumulation in the C-trap. The accumulated ion packet was then fragmented in the HCD cell at a normalized collision energy of 25%. Direct infusion of cocaine validated assignment of the unique transitions. The normalized collision energy was optimized from the analysis of a cocaine standard evenly applied onto a glass microscope slide using the pneumatic sprayer. The mass resolving power was set to 140,000 at m/z 200 for the MS2I acquisition in the orbitrap.
Data Analysis
To create ion heatmaps, the raw data (.raw) from the Thermo Q Exactive was converted to the mzXML format using the MSConvert software from Proteowizard (45). For concatenated ion images, the raw files were converted to mzML files using the MSConvert software from Proteowizard and were then converted to individual imzML files using imzMLConverter(46). The imzML Converter was then used to stack the individual imzML files into one master imzML file. The mzXML or imzML files were then loaded into the standalone version of MSiReader which is freely available software developed in our lab for processing MSI data (47). In order to demonstrate the quality of the raw data, ion images were neither interpolated nor normalized (unless otherwise specified). Dimension scales associated with all presented ion heatmaps reflect length in millimeters. Supervised analysis of the MSI data was undertaken with MSiReader for untargeted discovery of ions associated with tissue. In this approach, the software averages voxel spectra over a region of interest specified by the user and identifies unique peaks associated with this region relative to a user-specified reference region, chosen to be off-tissue. Putative identification of ions selected by MSiReader based on exact mass has been performed where possible using the LIPID MAPS (48) and METLIN structure databases (49). No further targeted SRM was undertaken to confirm these assignments, and as a result no attempt is made to distinguish structural isomers. These ions are identified by their molecular formula and compound class.
RESULTS and DISCUSSION
Optimization of Desorption Conditions using an Ice Matrix
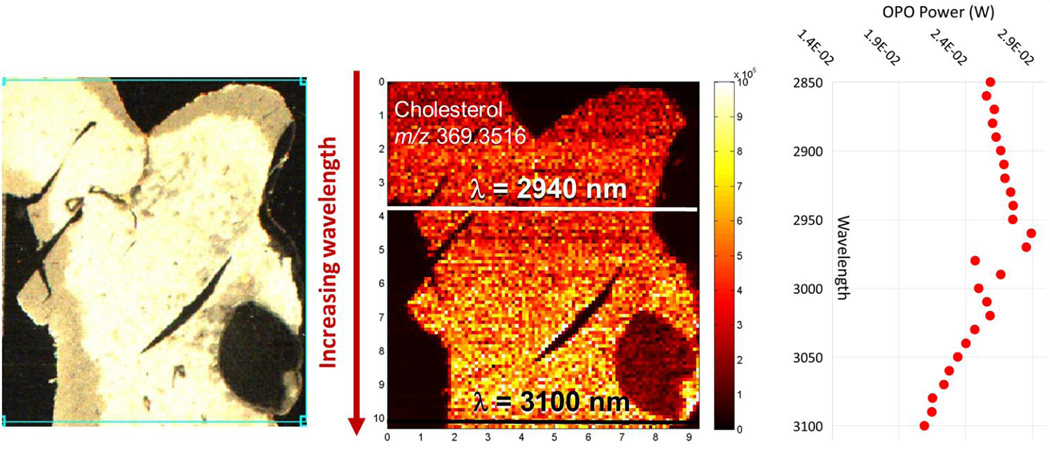
Initial efforts to evaluate the influence of excitation wavelength on IR-MALDESI response were performed by scanning the OPO emission wavelength while imaging a homogeneous 10 µm thick tissue slice of mouse liver. Imaging of the tissue was conducted in a standard scan pattern proceeding from the top left of the prescribed region of interest to the bottom right, with the incident wavelength ranging from 2850 nm to 3100 nm and increasing in 5 nm increments every 2 scan lines across the tissue. Ion abundance of many endogenous lipids within mouse liver such as cholesterol increased with wavelength, as shown in Figure 1. Unattenuated OPO pulse energy is wavelength-dependent over this spectral region, peaking at 2940 nm as shown in Figure 1, and while the trend in lipid response does not match that of OPO pulse energy, the influence of fluence and wavelength could not be entirely decoupled when interpreting these results. An optical attenuator was added to the beam path in order to match fluence while scanning emission wavelength, and a fluence scan (data not shown) was conducted using wavelengths of 2940 nm and 3100 nm on a serial tissue slice in a similar manner to Figure 1 with pulse energies varied from maximum OPO output to the desorption threshold. The fluence scan suggested that optimal ion abundance was achieved at each incident wavelength for laser fluences below the maximum output of the OPO.
Figure 1.
IR-MALDESI ion map of cholesterol response in mouse liver while scanning the incident wavelength from 2850 nm to 3100 nm. OPO wavelength was incremented 5 nm after the completion of every two scan lines. The position of key wavelengths, 2940 nm and 3100 nm, are highlighted on the sample. Unattenuated OPO power is also shown as a function of emission wavelength.
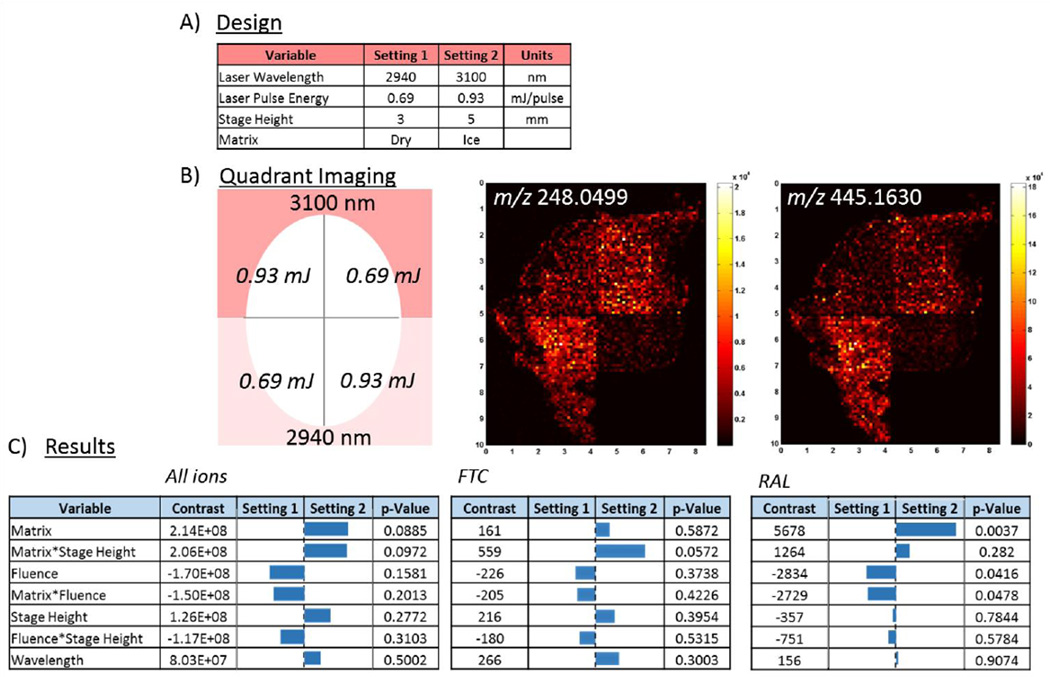
To systematically explore variables hypothesized to influence desorption plume dynamics, a full factorial screening design of experiments was conducted. This two-level statistical design investigated four parameters (laser fluence, excitation wavelength, absorption matrix, sample height) resulting in 16 total experiments. Selection of relevant parameter values for the screening experiment was informed by the wavelength and fluence scans described above, and these settings are summarized in Figure 2A. Experiments were conducted on ARV-incubated cervical tissue to investigate the influence of desorption conditions on IR-MALDESI response to both endogenous and xenobiotic compounds from tissue samples. Each of four quadrants of a morphologically homogeneous tissue cryosection was imaged with a unique combination of wavelength and matched fluence, as depicted in Figure 2B. This procedure was repeated four times with and without addition of an ice matrix for each sample height using serial slices, accounting for all combinations of variables in the 16 screening experiments. Also shown in Figure 2B are concatenated ion maps from each quadrant of FTC and RAL distributions within the incubated cervical tissue, analyzed with an ice matrix and with the sample stage 5 mm below the plane of the ESI emitter and the MS inlet. In addition to the ARV drugs, ions unique to the cervical tissue were identified from the IR-MALDESI imaging results using the MSiReader software. Representative ion maps of these species and, where available, their putative identification based on the METLIN database (49) can be seen in the Supporting Material (Figure S1). The average ion abundance in each of the 16 investigated quadrants was used as the figure of merit to evaluate the effect, or contrast, of each variable on the resulting response for all ions or each ion individually through statistical analysis using JMP software. Results of the full factorial screening experiment can be seen in Figure 2C. While the aggregate response for all detected ions does not indicate universal statistical significance between variable settings, the DOE screening results show common trends in variable dependence. Ion abundance is cumulatively enhanced when ice is present as an absorption matrix, when OPO laser pulse energy is reduced, and, to a lesser extent, at greater sample height and incident laser wavelength. The significance of each variable is analyte dependent, as shown for the ARV drugs in Figure 2C and for the other representative components in Figure S1, and the significance of the cross-terms suggests coupled effects.
Figure 2.
A) Parameters tested in screening design of experiments. B) Pictogram of approach for quadrant imaging and representative IR-MALDESI ion maps with ice matrix for selected ARV therapies. C) DOE screening results for the average response of all identified ions unique to tissue, as well as targeted results for individual ARV drugs indicating some specificity of parameter importance to individual analytes.
The importance of the ice matrix is consistent with previous work using IR-MALDESI. Shadowgraphy of the IR-MALDESI laser desorption process using an ice matrix has indicated complex desorption plume dynamics that involve absorption of energy by, and interaction between, tissue, ice, and liquid water (21). The presence of an ice matrix reduces the ejection of large tissue fragments that are not likely to contribute to mass spectral response, while also allowing the accumulation of ion signal over multiple ablation events(21).
This study is the first demonstration of the role of laser fluence on the IR-MALDESI mechanism and sensitivity. While signal abundance of IR-MALDI using an ice matrix has been observed to increase exponentially with laser fluence at low fluence values (< 0.2 J/cm2) (31), higher fluences may lead to a reduction in the effective absorption cross-section of the matrix as has been observed and modeled for liquid water (50). Based on measurements of incident OPO energy and beam diameter measured on thermal paper, the laser fluence over the on-tissue ablation threshold diameter of 150 µm can be estimated for this work assuming a Gaussian beam profile. Pulse energies utilized in this work correspond to fluences of approximately 2.8 – 3.8 J/cm2, which are above the fluence level at which Shori et al predict a “blue shift” in the absorption cross-section of water due to thermal excitation. Though the OPO pulse duration of 7 ns limits the thermal diffusion of incident energy, a reduction in the fluence likely results in more efficient absorption of energy by the matrix or simply better overlap between the desorbed plume and the orthogonal electrospray.
Analyte specificity toward desorption variables may arise from multiple sources. Infrared photosensitivity of compounds within the biological sample may contribute to wavelength-dependent differences in energy deposition, which have been demonstrated with IR-MALDI for matrices possessing different functionalities (25). Pirkl et al have suggested that the preferential IR-MALDI response at 2940 nm to a single analyte, Substance P, using an ice matrix is due to transient melting of the ice during ablation and predominant absorption of the laser light by liquid water. This certainly may be true, particularly for spectra acquired over 100s of laser pulses (31), but may also have been further influenced by analyte absorption. The excitation wavelengths investigated here correspond to absorption of free O-H and N-H stretching modes around 3000 nm present in many endogenous compounds. Any changes to the chemical composition or dynamics of the neutral plume due to energy absorption by the ice matrix and sample will alter the complexion of analytes ionized by electrospray, particularly for a complex biological sample.
Whole Body Imaging
To extend the results of the DOE experiments and to consider differences in response across different tissue types, a lipidomic MSI analysis of a neonatal mouse pup was performed. To preserve the integrity of all organs during cryosectioning, sagittal cross-sections of the mouse (roughly 1.5 cm × 2.5 cm) were cut to 25 µm thickness. Serial sections were imaged by IR-MALDESI using a full MS scan for untargeted lipidome analysis at incident wavelengths of both 2940 nm and 3100 nm while using the other optimized MSI conditions from the DOE experiments (ice matrix, reduced fluence, 5 mm stage height). Images of the whole body sections typically comprised 35,000 – 40,000 voxels, and were collected over 6 – 7 hours of imaging time at a scan rate of 1.6 scans/s.
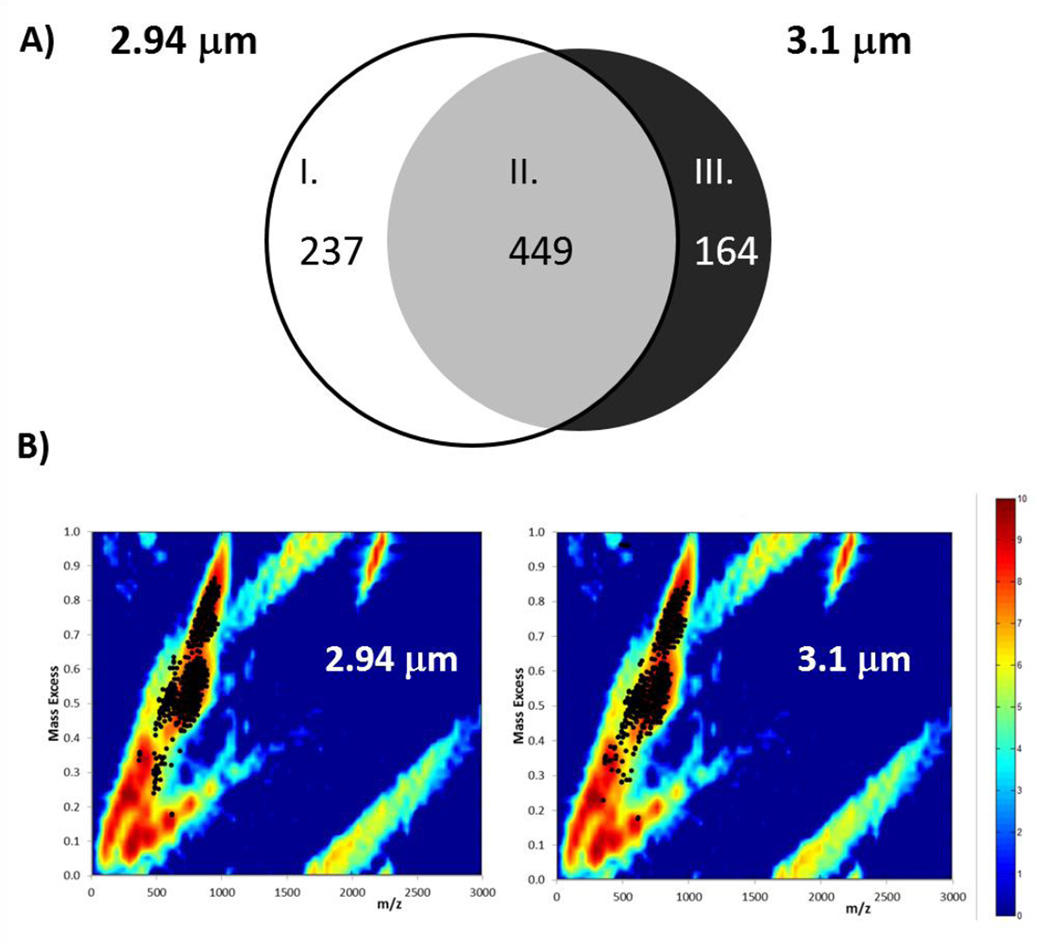
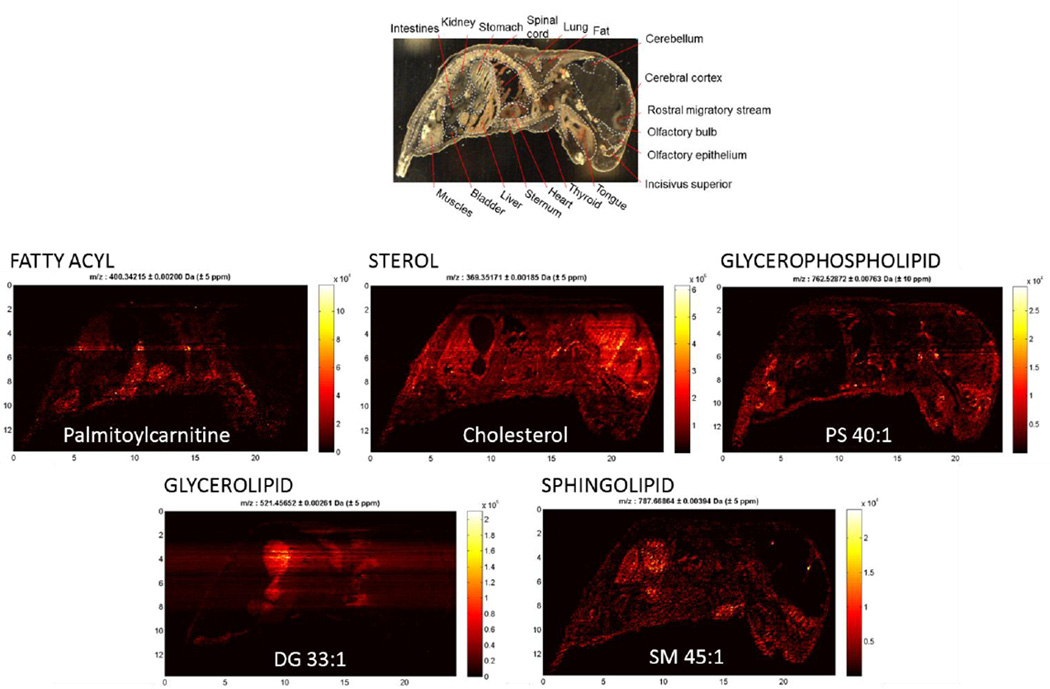
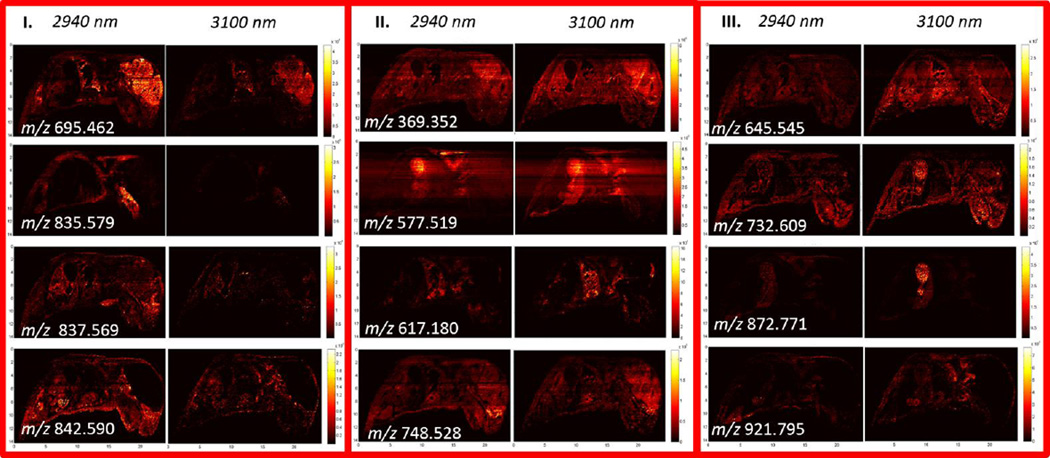
IR-MALDESI whole body imaging data was analyzed using MSiReader, which yielded 686 and 613 ions associated with tissue for imaging conducted with incident wavelengths of 2940 nm and 3100 nm, respectively. Of these detected compounds, 237 were detected preferentially at 2940 nm, 449 compounds were detected with similar relative ion abundance at both wavelengths, and 176 compounds were detected preferentially at 3100 nm, corresponding to classes I–III in Figure 3A. Mass excesses of all tissue related peaks, which represent the difference between the nominal and monoisotopic ion mass, indicate a high degree of overlap with the mass excess distribution for lipid classes from the LIPID MAPS structural database (48) (Figure 3B). While full MS acquisition does not eliminate the detection of isobaric species, the high resolution exact mass of ions measured by the Q Exactive significantly reduces the number of species to be considered during peak assignment when searching a lipid database for anticipated cations ([M+H+]+, [M+Na+]+, [M+H+-H2O]+) with 5 ppm mass accuracy. As such, lipid subclasses associated with many of the detected ions were identified unambiguously. IR-MALDESI is sensitive to a range of lipid classes (Table 1) which include fatty acyls, sterol lipids, glycerophospholipids, glycerolipids, and sphingolipids. Representative ion maps from each of these lipid classes can be seen in Figure 4, along with an anatomically annotated cryosection. These images demonstrate the capability of IR-MALDESI to evaluate the discrete biodistribution of ions across a whole body organism, exhibiting localization within individual or functionally related regions. Differences in ion response factors between tissue types will be addressed in the next section, and the ion distributions shown here do not account for such effects. Nonetheless, IR-MALDESI ion maps show similar patterns of lipid subclasses across tissues to a recent systematic lipid survey in 17 different mouse tissues by LC-MS (51): Fatty acylcarnitines appear to concentrate in the heart and liver; Sterols like cholesterol are concentrated in the brain; Glycerophospholipids like PS 40:1 are distributed across many tissues; Diradyl and triradylglycerolipids were observed to concentrate in the brown fat and intestines; and, Sphingomyelins like SM 45:1 are concentrated in kidney and stomach.
Figure 3.
A) Diagram of tissue-related ions identified by MSiReader during whole body imaging of a neonatal mouse using an excitation wavelength of λ = 2940 nm and 3100 nm, indicating: (I) ions detected preferentially at 2940 nm; (II) ions detected equally at both wavelengths; and, (III) ions detected preferentially at 3100 nm. B) Mass excess of all tissue related peaks overlapped with mass excess distribution for lipids from LIPID MAPS Structure Database. Location of the peaks found on tissue indicate that most of the 686 peaks and 613 peaks found for λ = 2940 nm and 3100 nm, respectively, correspond to lipids.
Table 1.
Summary of putative lipid class and subclass assignments for the measured exact masses of on-tissue ions during whole body imaging of a neonate mouse based on comparison to the METLIN database.
| Lipids | IR-MALDESI RESPONSE | ||||
|---|---|---|---|---|---|
| Class | Subclass | Annotation | Class I 2940 nm |
Class II Both |
Class III 3100 nm |
| Fatty Acyls | Fatty acyl carnitine | 0 | 0 | 2 | |
| Glycerophospholipids | Glycerophosphocholine | PC | 0 | 5 | 3 |
| Glycerophosphoethanolamine | PE | 4 | 4 | 2 | |
| PC/PE | 0 | 56 | 0 | ||
| Glycerophosphoserine | PS | 3 | 17 | 1 | |
| PC/PE/PS | 24 | 0 | |||
| Glycerophosphate | PA | 6 | 21 | 10 | |
| Glycerophosphoglycerols | PG | 0 | 9 | 2 | |
| Glycerophosphoinositols | PI | 0 | 1 | 0 | |
| Glycerolipids | Diradylglycerol | DG | 5 | 27 | 31 |
| Triradylglycerol | TG | 0 | 51 | 6 | |
| Sphingolipids | Sphingosine | 0 | 0 | 1 | |
| Ceramide | Cer | 0 | 6 | 3 | |
| Sphingomyelin | SM | 1 | 5 | 1 | |
| Sulfatide | 0 | 0 | 0 | ||
| Sterol Lipids | Steryl esters | CE | 2 | 3 | 1 |
| Total | 18 | 221 | 59 | ||
Where there is ambiguity in lipid class assignment due to the identification of isobaric species from multiple classes within the 5ppm database search tolerance, ions have been grouped accordingly (ie PC/PE or PC/PE/PS).
Figure 4.
Selected ion maps representing lipid classes identified by IR-MALDESI in whole body sections of neonate mouse, along with putative ion identification from Lipid Maps where available. An abbreviated mouse anatomy is included at top of the image to highlight regions where ion-specific localization of IR-MALDESI response has been observed.
A comparison of IR-MALDESI response to serial sections imaged at different wavelengths can be seen in Figure 5. Ion distributions exhibit localization within individual or functionally related regions including: the whole brain (m/z 695.462) or discrete brain regions such as the Rostral migratory system (m/z 837.569); the musculature (m/z 835.579); the heart and lungs (m/z 617.180); and, the stomach (m/z 732.609 and m/z 872.771). Class II ion maps show a similar magnitude of response at both 2940 and 3100 nm, but may exhibit differences in response to varying tissue types as seen in the liver (m/z 577.519) and the olfactory epithelium (m/z 748.528). Both class I and class III ion maps illustrate differences in the ion abundance between the experiments conducted at different wavelengths.
Figure 5.
Selected ion maps of serial 25 µm thick whole mouse corresponding to each of the three classes of ions defined in Figure 3. Pairs of ion maps corresponding to the matching MSI experiments are shown with matching intensity scales to illustrate differences in ion abundances between the sections imaged at λ = 2940 nm and 3100 nm.
Changes in the response to a given analyte observed between the cryosections imaged at 2940 and 3100 nm may be the result of differences in analyte concentration or differences in the desorption conditions. Each cryosection examined was separated by 25 µm, such that changes in tissue composition or morphology between the cryosections used for each experiment are expected to be minimal. The amount of incident laser energy coupled into an analyte via the ice matrix and through direct absorption is wavelength-dependent, which influences threshold fluence (25) and resulting plume height(52)of an analyte. Given a finite ionization capacity of the electrospray, alterations to the composition of the desorbed plume interacting with the orthogonal electrospray are likely to lead to differences in MS response to a complex sample. While the untargeted nature of the whole body MSI precludes distinguishing whether the observed differences arise more strongly from selective direct excitation of analytes or from differences to the plume behavior as a result of the tissue type and/or ice matrix, it is clear that imaging the whole body sections at these two discrete excitation wavelengths has led to enhanced coverage of lipid response.
While whole body lipidome studies are limited in number, this work represents a significantly more comprehensive positive ion lipid response across whole body mouse sections than previously reported with MALDI MSI (32), where the MS response between m/z 650 to 900 was characterized as being typical of phospholipids but no attempt was made to identify lipid subclasses. IR-MALDESI shows a strong response to phospholipids such as phosphatidylcholines (PC), which have been the focus of many mouse MSI studies(53), presumably due not only to their abundance and ionization efficiency but also due to their suppression of other ions(54). Additionally, IR-MALDESI is capable of response to a greater breadth of species like glycerolipids and sphingolipids than MALDI (3) and DESI (55)without requiring multiple matrices (56) or additional tissue preparation such as washing (57) or enzymatic digestion (58). Simultaneous acquisition of these species has great potential for future lipid imaging studies of disease pathologies, for example.
Response factors and Internal Energy Distribution across Whole body
Differences in the ionization efficiency across tissue types were evaluated by monitoring IR-MALDESI MS2I response to an analyte standard, cocaine, sprayed onto the glass mounting slide beneath a whole body cryosection. Homogeneity of analyte deposition onto the slide and variability of IR-MALDESI response to it beneath biological tissue has recently been characterized (19). While that study utilized the analyte to normalize ion response as part of a quantitative MSI method, here it is employed in an effort to evaluate the relative internal energy distribution of ions generated at the two different incident wavelengths, 2940 and 3100 nm across a plethora of tissue types. Cocaine’s simple fragmentation pattern(59) offers facile interpretation of its CID spectra for monitoring ion fragmentation ratios during MS2I analysis using the “survival yield” method (60), which has been used to evaluate internal energy deposition of DESI (61) and LAESI (62).
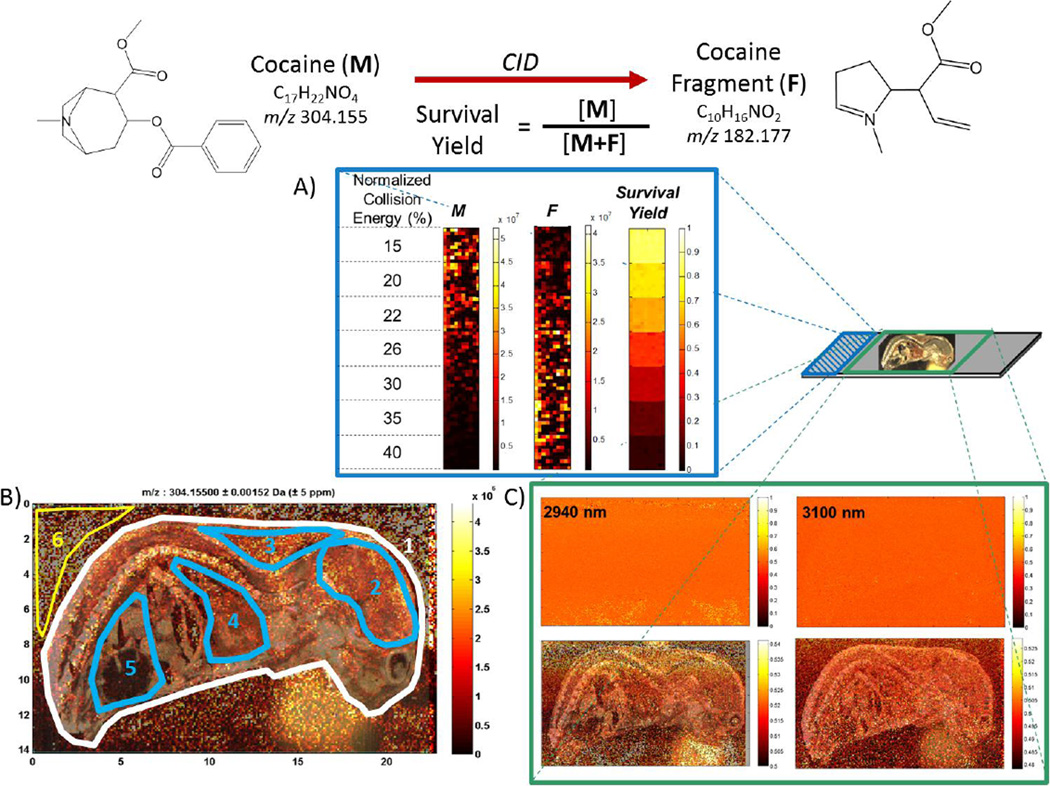
Discrete regions of a sprayed slide were imaged at varying normalized collision energies (NCE), shown in Figure 6A, to determine the IR-MALDESI breakdown diagram of cocaine in the QE HCD cell (Figure S2). The breakdown diagram indicated that ion internal energy in the HCD cell is similar for both +ESI direct infusion and IR-MALDESI. These experiments were repeated using the in-source CID capability of the QE to account for any collisional cooling within the HCD cell, yielding statistically the same results.
Figure 6.
IR-MALDESI MS2I monitoring of cocaine: A) Evaluation of cocaine survival yield breakdown diagram by varying the CID normalized collision energy over discrete regions on a glass slide; B) Cocaine [M+H+]+ response during imaging of a 25 µm thick whole mouse cryosection, with discrete anatomical regions highlighted that correspond to differences in response as summarized in text and in Table 2; C) Survival yield of cocaine during whole-body imaging (bottom panels show a narrower range of response and include overlayed optical images of analyzed sections).
Whole body sections of neonatal mouse were then thaw-mounted on top of glass slides sprayed with the cocaine analyte. MS2I of cocaine was performed across whole body cryosections using the incident wavelengths 2940 and 3100 nm with a fixed value of NCE, 25%, which corresponded to a cocaine survival yield of ~50% based on the breakdown diagram. IR-MALDESI response to cocaine was apparent across the entire cryosection and varied beneath different tissue types corresponding to anatomical features, as can be seen in Figure 6. Unlike previous MALDI MSI analysis of a homogeneous standard sprayed onto a whole body mouse section (35), there are no imaged regions exhibiting complete ion suppression. Differences in the cocaine [M+H+]+ ion abundance across discrete regions of the cryosection highlighted in Figure 6 are summarized in Table 2. Voxel-to-voxel variability of response below tissue is similar to, and in some cases better than, the response from off-tissue regions of the coated slide. Average voxel response varies by less than a factor of 5 between all virtual micro-dissected regions. Since MALDESI is not subject to tissue-specific issues regarding matrix application or analyte extraction efficiency (35, 36, 39), the observed differences in response factor are more likely to result from desorption conditions. While tissue response factors are expected to be analyte specific, this approach provides a pathway to quantitative whole body imaging with IR-MALDESI for targeted small molecules (63) or untargeted analysis of lipids with adroit selection of representative lipid standards (64).
Table 2.
Summary of MS2I response to Cocaine [M+H+]+, m/z 304.155, beneath different tissue types of a whole body mouse cryosection.
| Region | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Tissue Type |
Whole mouse |
Brain |
Adipose tissue |
Heart and Lung |
Gut | Off-tissue |
| Average | 9.0E+05 | 1.6E+06 | 1.4E+06 | 6.6E+05 | 3.4E+05 | 2.67E+06 |
| Max | 1.4E+07 | 8.7E+06 | 5.4E+06 | 6.6E+06 | 2.2E+06 | 2.15E+07 |
| Min | 1.7E+03 | 1.7E+04 | 2.5E+04 | 3.8E+03 | 2.3E+03 | 6.77E+03 |
| %RSD | 92 | 63 | 54 | 101 | 99 | 87 |
The survival yield of cocaine at each wavelength is shown in Figure 6C. These images were obtained by normalizing the cocaine response to the sum of the response for all molecular ion and fragment ions (m/z values of 304.1550, 182.177, and 150.092) on a voxel-by-voxel basis using MSiReader. Survival yields at each wavelength across the entire cryosections (SY2940nm = 46.5% ± 9.2% and SY3100nm = 51.3% ± 11.5%) are similar on average to the value of 50% derived from IR-MALDESI analysis of the sprayed slide alone. These results indicate complete collisional cooling of desorbed neutral molecules in the gas phase(15)or during uptake into ESI droplets, and show no evidence for direct ionization. As such, they support the mechanistic view of IR-MALDESI neutrals that are rapidly collisionally cooled during their partitioning into ESI droplets as has been posited previously (16, 65), but do not allow conclusions regarding initial differences in internal energy attributable to excitation wavelength. While an MS2I approach was selected to eliminate the possibility of endogenous isobaric species interfering with calculation of the survival yield, further information regarding wavelength dependence in the IR-MALDESI mechanism may be gained by full MS imaging of biological tissues using traditional benzylpyridinium thermometer ions with lower bond dissociation energies (61, 62) as a sprayed analyte.
CONCLUSIONS
This work has shown that the IR-MALDESI response to individual analytes within a complex biological system is sensitive to factors controlling analyte desorption, such as laser wavelength, fluence, stage height, and matrix. Overall trends of pharmaceutical drugs and endogenous lipids indicated enhanced response when reducing laser fluence and utilizing an ice matrix. While the influence of incident wavelength on the internal energy of desorbed neutrals could not be determined, it was shown that altering the excitation wavelength resulted in enhanced lipid response to whole body sections of neonate mouse. IR-MALDESI was shown to be responsive to a broad range of lipid classes present across an organism without any requirement for sample manipulation. Through the analysis of a standard deposited beneath the whole body sections, tissue-specific response factors were determined that will be a critical component for quantitative MSI on a multi-organ scale.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Prof. Angela Kashuba and Corbin Thompson of the Eshelman School of Pharmacy, UNC Chapel Hill for providing ARV-incubated tissue samples. The authors gratefully acknowledge the financial support received from the National Institutes of Health R01GM087964 and R01AI111891.
Footnotes
Special Issue Honoring Richard Caprioli for ASMS Award for Distinguished Contribution in Mass Spectrometry
REFERENCES
- 1.Prideaux B, Stoeckli M. Mass spectrometry imaging for drug distribution studies. Journal of Proteomics. 2012;75:4999–5013. doi: 10.1016/j.jprot.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 2.Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50(Suppl):S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gode D, Volmer DA. Lipid imaging by mass spectrometry - a review. Analyst. 2013;138:1289–1315. doi: 10.1039/c2an36337b. [DOI] [PubMed] [Google Scholar]
- 4.Burnum KE, Frappier SL, Caprioli RM. Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry for the Investigation of Proteins and Peptides. Annu Rev Anal Chem. 2008;1:689–705. doi: 10.1146/annurev.anchem.1.031207.112841. [DOI] [PubMed] [Google Scholar]
- 5.Greer T, Sturm R, Li LJ. Mass spectrometry imaging for drugs and metabolites. Journal of Proteomics. 2011;74:2617–2631. doi: 10.1016/j.jprot.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwamborn K. Imaging mass spectrometry in biomarker discovery and validation. Journal of Proteomics. 2012;75:4990–4998. doi: 10.1016/j.jprot.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 7.Calligaris D, Caragacianu D, Liu X, Norton I, Thompson CJ, Richardson AL, Golshan M, Easterling ML, Santagata S, Dillon DA, Jolesz FA, Agar NYR. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proceedings of the National Academy of Sciences. 2014;111:15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Gingras J, Ganley KP, Vismeh R, Teffera Y, Zhao Z. Whole-body tissue distribution study of drugs in neonate mice using desorption electrospray ionization mass spectrometry imaging. Rapid Communications in Mass Spectrometry. 2014;28:185–190. doi: 10.1002/rcm.6775. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin RJ. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. Journal of proteomics. 2012;75:4893–4911. doi: 10.1016/j.jprot.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 11.Eikel D, Vavrek M, Smith S, Bason C, Yeh S, Korfmacher WA, Henion JD. Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Communications in Mass Spectrometry. 2011;25:3587–3596. doi: 10.1002/rcm.5274. [DOI] [PubMed] [Google Scholar]
- 12.Sampson JS, Murray KK, Muddiman DC. Intact and top-down characterization of biomolecules and direct analysis using infrared matrix-assisted laser desorption electrospray ionization coupled to FT-ICR mass spectrometry. Journal of the American Society for Mass Spectrometry. 2009;20:667–673. doi: 10.1016/j.jasms.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkenkamp S, Kirpekar F, Hillenkamp F. Infrared MALDI mass spectrometry of large nucleic acids. Science. 1998;281:260–262. doi: 10.1126/science.281.5374.260. [DOI] [PubMed] [Google Scholar]
- 14.Dreisewerd K, Berkenkamp S, Leisner A, Rohlfing A, Menzel C. Fundamentals of matrix-assisted laser desorption/ionization mass spectrometry with pulsed infrared lasers. International Journal of Mass Spectrometry. 2003;226:189–209. [Google Scholar]
- 15.Levis RJ. Laser desorption and ejection of biomolecules from the condensed phase into the gas phase. Annual Review of Physical Chemistry. 1994;45:483–518. doi: 10.1146/annurev.pc.45.100194.002411. [DOI] [PubMed] [Google Scholar]
- 16.Sampson JS, Hawkridge AM, Muddiman DC. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. Journal of the American Society for Mass Spectrometry. 2006;17:1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Analytical Chemistry. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 18.Rezenom YH, Dong J, Murray KK. Infrared laser-assisted desorption electrospray ionization mass spectrometry. Analyst. 2008;133:226–232. doi: 10.1039/b715146b. [DOI] [PubMed] [Google Scholar]
- 19.Bokhart M, Rosen E, Thompson C, Sykes C, Kashuba AM, Muddiman D. Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Analytical and Bioanalytical Chemistry. 2014:1–12. doi: 10.1007/s00216-014-8220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X, Murray KK. Wavelength and Time-Resolved Imaging of Material Ejection in Infrared Matrix-Assisted Laser Desorption. Journal of Physical Chemistry A. 2010;114:1492–1497. doi: 10.1021/jp9077163. [DOI] [PubMed] [Google Scholar]
- 21.Robichaud G, Barry J, Muddiman D. IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix. Journal of the American Society for Mass Spectrometry. 2014:1–10. doi: 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barry J, Robichaud G, Bokhart M, Thompson C, Sykes C, Kashuba AM, Muddiman D. Mapping Antiretroviral Drugs in Tissue by IR-MALDESI MSI Coupled to the Q Exactive and Comparison with LC-MS/MS SRM Assay. Journal of The American Society for Mass Spectrometry. 2014:1–10. doi: 10.1007/s13361-014-0884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin RJA, Iverson SL, Andren PE. The significance of ambient-temperature on pharmaceutical and endogenous compound abundance and distribution in tissues sections when analyzed by matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Communications in Mass Spectrometry. 2012;26:494–498. doi: 10.1002/rcm.6125. [DOI] [PubMed] [Google Scholar]
- 24.Berkenkamp S, Karas M, Hillenkamp F. Ice as a matrix for IR-matrix-assisted laser desorption/ionization: Mass spectra from a protein single crystal. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7003–7007. doi: 10.1073/pnas.93.14.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menzel C, Dreisewerd K, Berkenkamp S, Hillenkamp F. Mechanisms of energy deposition in infrared matrix-assisted laser desorption/ionization mass spectrometry. International Journal of Mass Spectrometry. 2001;207:73–96. [Google Scholar]
- 26.Fisher RA. Statistical Methods for Research Workers. New York, New York: Hafner Publishing Company; 1973. [Google Scholar]
- 27.Riter LS, Vitek O, Gooding KM, Hodge BD, Julian RK. Statistical design of experiments as a tool in mass spectrometry. Journal of Mass Spectrometry. 2005;40:565–579. doi: 10.1002/jms.871. [DOI] [PubMed] [Google Scholar]
- 28.Schriver-Mazzuoli L, Schriver A, Hallou A. IR reflection–absorption spectra of thin water ice films between 10 and 160 K at low pressure. Journal of Molecular Structure. 2000;554:289–300. [Google Scholar]
- 29.Williams P. Time of flight mass spectrometry of DNA laser-ablated from frozen aqueous solutions: applications to the Human Genome Project. International Journal of Mass Spectrometry and Ion Processes. 1994;131:335–344. [Google Scholar]
- 30.Focsa C, Mihesan C, Ziskind M, Chazallon B, Therssen E, Desgroux P, Destombes JL. Wavelength-selective vibrationally excited photodesorption with tunable IR sources. Journal of Physics-Condensed Matter. 2006;18:S1357–S1387. [Google Scholar]
- 31.Pirkl A, Soltwisch J, Draude F, Dreisewerd K. Infrared Matrix-Assisted Laser Desorption/Ionization Orthogonal-Time-of-Flight Mass Spectrometry Employing a Cooling Stage and Water Ice As a Matrix. Analytical chemistry. 2012;84:5669–5676. doi: 10.1021/ac300840b. [DOI] [PubMed] [Google Scholar]
- 32.Chaurand P, Cornett DS, Angel PM, Caprioli RM. From Whole-body Sections Down to Cellular Level, Multiscale Imaging of Phospholipids by MALDI Mass Spectrometry. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Analytical chemistry. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 34.Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, Marshall PS, West A, Princivalle AP, Clench MR. Matrix-Assisted Laser Desorption/Ionization-Ion Mobility Separation-Mass Spectrometry Imaging of Vinblastine in Whole Body Tissue Sections. Analytical Chemistry. 2008;80:8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 35.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. International Journal of Mass Spectrometry. 2007;260:195–202. [Google Scholar]
- 36.Crossman L, McHugh NA, Hsieh YS, Korfmacher WA, Chen JW. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Communications in Mass Spectrometry. 2006;20:284–290. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 37.Reyzer M, Chaurand P, Angel P, Caprioli R. In: Mass Spectrometry Imaging. chap. 17. Rubakhin SS, Sweedler JV, editors. Vol. 656. Humana Press; 2010. pp. 285–301. [Google Scholar]
- 38.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Analytical chemistry. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 39.Tomlinson L, Fuchser J, Fuetterer A, Baumert M, Hassall DG, West A, Marshall PS. Using a single, high mass resolution mass spectrometry platform to investigate ion suppression effects observed during tissue imaging. Rapid Communications in Mass Spectrometry. 2014;28:995–1003. doi: 10.1002/rcm.6869. [DOI] [PubMed] [Google Scholar]
- 40.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer. Journal of the American Society for Mass Spectrometry. 2013;24:92–100. doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nazari M, Muddiman D. Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling. Analytical and Bioanalytical Chemistry. 2014:1–7. doi: 10.1007/s00216-014-8376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. Journal of the American Society for Mass Spectrometry. 2005;16:1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Barry JA, Robichaud G, Muddiman DC. Mass Recalibration of FT-ICR Mass Spectrometry Imaging Data Using the Average Frequency Shift of Ambient Ions. Journal of the American Society for Mass Spectrometry. 2013;24:1137–1145. doi: 10.1007/s13361-013-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen JV, de Godoy LMF, Li GQ, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Molecular & Cellular Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Race AM, Styles IB, Bunch J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. Journal of proteomics. 2012;75:5111–5112. doi: 10.1016/j.jprot.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 47.Robichaud G, Garrard K, Barry J, Muddiman D. MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. Journal of the American Society for Mass Spectrometry. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Therapeutic Drug Monitoring. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 50.Shori RK, Walston AA, Stafsudd OM, Fried D, Walsh JT. Quantification and modeling of the dynamic changes in the absorption coefficient of water at lambda=2.94 mu m. Ieee J Sel Top Quant. 2001;7:959–970. [Google Scholar]
- 51.Jain M, Ngoy S, Sheth SA, Swanson RA, Rhee EP, Liao R, Clish CB, Mootha VK, Nilsson R. A systematic survey of lipids across mouse tissues. 2014;306:E854–E868. doi: 10.1152/ajpendo.00371.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan X, Little MW, Murray KK. Infrared laser wavelength dependence of particles ablated from glycerol. Appl. Surf. Sci. 2008;255:1699–1704. [Google Scholar]
- 53.Hankin JA, Murphy RC. Relationship between MALDI IMS Intensity and Measured Quantity of Selected Phospholipids in Rat Brain Sections. Analytical Chemistry. 2010;82:8476–8484. doi: 10.1021/ac101079v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petković M, Schiller J, Müller M, Benard S, Reichl S, Arnold K, Arnhold J. Detection of Individual Phospholipids in Lipid Mixtures by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry: Phosphatidylcholine Prevents the Detection of Further Species. Analytical Biochemistry. 2001;289:202–216. doi: 10.1006/abio.2000.4926. [DOI] [PubMed] [Google Scholar]
- 55.Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2011;1811:946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellis SR, Brown SH, Blanksby SJ, Mitchell TW. Surface Analysis of Lipids by Mass Spectrometry: More than Just Imaging. Progress in lipid research. 2013 doi: 10.1016/j.plipres.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Angel PM, Spraggins JM, Baldwin HS, Caprioli R. Enhanced Sensitivity for High Spatial Resolution Lipid Analysis by Negative Ion Mode Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry. Analytical Chemistry. 2012;84:1557–1564. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones EE, Dworski S, Canals D, Casas J, Fabrias G, Schoenling D, Levade T, Denlinger C, Hannun YA, Medin JA, Drake RR. On-Tissue Localization of Ceramides and Other Sphingolipids by MALDI Mass Spectrometry Imaging. Analytical Chemistry. 2014;86:8303–8311. doi: 10.1021/ac501937d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dixon SJ, Brereton RG, Carter JF, Sleeman R. Determination of cocaine contamination on banknotes using tandem mass spectrometry and pattern recognition. Analytica Chimica Acta. 2006;559:54–63. [Google Scholar]
- 60.Kenttamaa HI, Cooks RG. Internal energy distributions acquired through collisional activation at low and high energies. International Journal of Mass Spectrometry and Ion Processes. 1985;64:79–83. [Google Scholar]
- 61.Nefliu M, Smith JN, Venter A, Cooks RG. Internal energy distributions in desorption electrospray ionization (DESI) Journal of the American Society for Mass Spectrometry. 2008;19:420–427. doi: 10.1016/j.jasms.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 62.Nemes P, Huang H, Vertes A. Internal energy deposition and ion fragmentation in atmospheric-pressure mid-infrared laser ablation electrospray ionization. Phys Chem Chem Phys. 2012;14:2501–2507. doi: 10.1039/c2cp23411d. [DOI] [PubMed] [Google Scholar]
- 63.Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. Journal of Proteomics. 2012;75:4952–4961. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 64.Lanekoff I, Thomas M, Laskin J. Shotgun Approach for Quantitative Imaging of Phospholipids Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Analytical chemistry. 2014 doi: 10.1021/ac403931r. [DOI] [PubMed] [Google Scholar]
- 65.Dixon RB, Muddiman DC. Study of the ionization mechanism in hybrid laser based desorption techniques. Analyst. 2010;135:880–882. doi: 10.1039/b926422a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.