Abstract
Background. Leprosy morbidity is increased by 2 pathologic immune reactions, reversal reaction (RR) and erythema nodosum leprosum (ENL).
Methods. To discover host factors related to immune reactions, global transcriptional profiles of peripheral blood mononuclear cells were compared between 11 RR, 11 ENL, and 19 matched control patients, with confirmation by quantitative polymerase chain reaction. Encoded proteins were investigated in skin biopsy specimens by means of immunohistochemistry.
Results. There were 275 genes differentially expressed in RR and 517 differentially expressed in ENL on the microarray. Pathway analysis showed immunity-related pathways represented in RR and ENL transcriptional profiles, with the “complement and coagulation” pathway common to both. Interferon γ was identified as a significant upstream regulator of the expression changes for RR and ENL. Immunohistochemical staining of skin lesions showed increased C1q in both RR and ENL.
Conclusions. These data suggest a previously underrecognized role for complement in the pathogenesis of both RR and ENL, and we propose new hypotheses for reaction pathogenesis.
Keywords: leprosy, reversal reaction, erythema nodosum leprosum, complement
Leprosy, or Hansen's disease, remains a significant challenge to global health despite the availability of antibiotic therapy. A total of 232 857 cases were reported in 2012, including 33 303 new cases in Brazil [1]. Infection with Mycobacterium leprae, the causative agent of leprosy, manifests as a spectrum of clinical presentations. These range from the tuberculoid form, with few skin lesions and strong cell-mediated immune response, to the lepromatous form (LL), with disseminated disease and predominant humoral response [2]. Individuals with intermediate immune response to M. leprae develop “borderline” clinical forms: borderline tuberculoid (BT), borderline-borderline, and borderline lepromatous (BL) [2]. About 30% of persons with leprosy will develop a pathologic immune reaction of leprosy, either reversal reaction (RR) or erythema nodosum leprosum (ENL) [3]. These reactions are common during the first 3 months of antileprosy therapy but can occur before, during, or after treatment of leprosy.
The first common reaction, RR, results in swelling and inflammation of existing skin lesions with increasing neuritis and nerve damage. It is thought to be related to an augmented cell-mediated immune response to M. leprae antigens in the skin and nerves in persons with borderline forms of leprosy [4, 5]. Clinical risk factors for RR include increased age, >5 skin lesions, and high M. leprae bacterial load [6]. A pronounced T-helper (Th) 1–type response to M. leprae antigens has been documented in the skin of patients with RR [7]. Persons with history of RR can maintain an altered response to M. leprae antigen that differs from patients with nonreaction leprosy for years after resolution of RR [8]. The other common reaction, ENL, occurs in lepromatous (BL or LL) leprosy with painful subcutaneous nodules and systemic findings, such as fever, arthritis, nephritis, and panniculitis [9]. Its pathogenesis seems to be related to formation and deposition of immune complexes in skin and organs [9]. Risk factors for ENL include LL clinical form and high bacterial load [10].
The immune reactions of leprosy are typically chronic and recurrent and can require years of treatment with corticosteroids or other immunomodulatory medications [10]. Neurologic sequelae from leprosy reactions can be irreversible even with appropriate therapy, significantly increasing the morbidity and disability due to leprosy [11]. Potential biomarkers have been proposed for RR and ENL but have not been validated for predicting reactions [12, 13]. Transcriptional profiling of skin lesions has shown differences between tuberculoid and lepromatous skin lesions [14], although these were not studied in relation to systemic immune profiles in blood leukocytes. Increased interferon (IFN) α pathway transcripts in RR lesions suggest its involvement in pathogenesis [15], and a recent study showed that persons with history of RR had a unique gene expression profile in response to M. leprae antigen in vitro [8]. Studies of the transcriptional profiles of immune cells from subjects with active RR and ENL are needed to understand the systemic immune response during reactions. We hypothesized that RR and ENL are caused by distinctly different immune responses. To test this hypothesis, we generated transcriptional profiles of peripheral blood mononuclear cells (PBMCs) of carefully characterized subjects with symptomatic leprosy immune reactions and matched controls with validation studies including quantitative polymerase chain reaction (qPCR), flow cytometry, and immunohistochemistry.
MATERIALS AND METHODS
Human Subjects
Adult patients with leprosy were enrolled from Hansen's disease treatment centers in Rio Grande do Norte, Brazil. Leprosy clinical forms were assigned by a trained dermatologist (M. d. C. A. P. Q., L. L. M., or M. L. N.) based on Ridley-Jopling criteria [2]. Diagnoses of RR or ENL were based on clinical findings [16, 17]. Excluded from this study were persons with known immunodeficiency or who had received corticosteroids within 7 days or thalidomide within 28 days of enrollment.
Transcript microarrays were obtained from 22 patients with leprosy with immune reaction (11 RR and 11 ENL) and 19 controls without reactions matched to cases for age, sex, leprosy clinical form, and stage of treatment. PBMCs for reverse-transcription qPCR validation were derived from a subset of these participants and an additional 28 persons (11 RR, 6 ENL, and 11 controls). Monocyte populations were compared in the different leprosy clinical groups using flow cytometry of PBMCs. Skin biopsy immunohistochemical studies were completed for 16 patients with leprosy (3 RR, 3 ENL, 7 BT controls, and 3 BL/LL controls).
PBMC Isolation and RNA Extraction
PBMCs were isolated from heparinized peripheral blood using a Ficoll-Paque PLUS gradient (GE Healthcare Life Sciences). RNA was isolated using Trizol (Life Technologies) and purified with a RNeasy MinElute Cleanup Kit (Qiagen) with on-column DNase treatment.
Microarray and Analysis
Preparation of RNA for hybridization to Illumina BeadChips (Ambion) was performed at the University of Iowa DNA Core Facility using the manufacturer's protocol. After hybridization, the arrays were washed, blocked, and stained according to the Illumina Whole-Genome Gene Expression Direct Hybridization Assay protocol. BeadChips were scanned with the Illumina iScan System and data collected using GenomeStudio software (Illumina, version 2011). Data sets were compared using Rank Products Analysis (Bioconductor utilities; www.bioconductor.org, last accessed December 1, 2014) to optimize comparison of array groups of small sample size and with variability inherent in human clinical samples [18, 19]. Quantile normalized fluorescence results were analyzed with the RankProd library using RPadvance software in R package (v2.36.0, http://www.r-project.org, last accessed December 1, 2014), considering arrays run on separate days as different origins. The resultant orders were compared with topGene software, with a percentage false-positive (FPR) cutoff of 0.05. Reported fold changes for the array are given as log2 fold change.
Transcripts with a FPR cutoff ≤0.05 and fold change ≥1.5 were used for pathway analyses. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) Biological Process pathways were generated with Webgestalt (2014, Vanderbilt University) [20] with Bonferroni correction. Ingenuity Pathway Analysis (IPA) (Qiagen) was completed using experimentally observed direct and indirect relationships for humans.
qPCR With Taqman Low Density Arrays
Significantly changed microarray transcripts were selected for validation with Taqman Low Density Arrays. PBMC-derived complementary DNA was applied in duplicate to custom Taqman Low Density Arrays (Life Technologies). Fluorescence was normalized to the array baseline in Expression Suite software (version 1.0; Life Technologies) and mean cycle threshold (Ct) values were compared using 2-tailed t tests. Delta-delta cycle threshold (ΔΔCt) values were converted to fold changes using this calculation: fold change = 2(−ΔΔCt).
Immunohistochemistry of Skin Biopsy Specimens
Skin biopsy specimens were collected in formalin and fixed in paraffin. After sectioning, slides were deparaffinized and subjected to antigen retrieval. Sections were stained with rabbit anti-human C1q (clone 9A7; Abcam) and mouse anti-human CD21 (clone EP3093; Abcam) with secondary staining with goat anti-rabbit AlexaFluor488 and goat anti-mouse-AlexaFluor546 (Life Technologies). Stained tissue sections were imaged using a Zeiss Examiner.Z1 AX10 confocal (LSN 7100) microscope and recorded using Zen 2010BSP1 software (Zeiss). Intensity, laser range, and gain were constant. For each biopsy specimen, 2 adjacent 15-section micrographs were obtained 1 mm from the edge of the specimen at the dermal-epidermal border with an additional 2 dermal photographs just internal to the initial photographs. Mean intensities of fluorescence were normalized to the BT group for each day of staining and compared using analysis of variance with Tukey posttest. Imaging and analysis were completed by authors (F. M. A. and M. R. C.) blinded to the group assignment.
Flow Cytometry
A total of 250 000 PBMCs were stained with anti–CD14-allophycocyanin–cyanine 7 (Ebioscience), and 50 000 events were counted using a FACS-Canto II flow cytometer (Becton Dickinson) and recorded with FACSDiva software (Becton Dickinson). Data were analyzed using FlowJo software (v 9.5.3, TreeStar) with monocyte gate and CD14 positivity assigned as in Supplementary Figure 1A. Monocyte populations were compared using analysis of variance with Tukey multiple-comparison tests for normally distributed data, and Kruskal–Wallis test with Dunn multiple comparison for non-normally distributed data.
Ethical Considerations
The study was reviewed and approved by the institutional review boards of the Universidade Federal do Rio Grande do Norte, Brazil's National Ethical Review Board, and Weill Cornell Medical College. The Brazilian institutional review board is registered with the US National Institutes of Health. All participants provided written informed consent for participation in the study.
RESULTS
Study Population
The study population for the transcript microarray included 22 patients with leprosy with immune reaction (11 RR and 11 ENL) matched 1:1 for age, sex, leprosy clinical form, and stage of treatment with a control patient without reaction (Table 1). PBMCs used for qPCR were from a subset of these subjects and another 28 patients with leprosy. Additional samples included in the RR qPCR validation were RR (n = 11) and non-RR controls (n = 7). Age (mean, 50 vs 51 years; P = .89), sex (59.1% vs 55.6% male; P = .82), and clinical form of leprosy (P = .96) did not differ significantly between RR and non-RR groups. Additional samples included in the ENL qPCR validation were ENL (n = 6) and non-ENL controls (n = 4). Age (45 vs 51 years; P = .33), sex (82.4% vs 60% male; P = .24), and clinical form of leprosy (P = .47) did not differ significantly different between ENL and non-ENL groups.
Table 1.
Baseline Characteristics of the Patients With Leprosy Included in the Microarray Analysis
| Characteristic | RR (n = 11) | Controls Without RR (n = 11) | ENL (n = 11) | Controls Without ENL (n = 11) |
|---|---|---|---|---|
| Male sex, % | 54.5 | 54.5 | 81.8 | 81.8 |
| Age, mean (range), y | 47.6 (22–73) | 46.8 (28–65) | 43.9 (23–65) | 48.3 (27–69) |
| Leprosy treatment status, No. | ||||
| Pretreatment | 5 | 6 | 3 | 5 |
| On treatment | 5 | 5 | 2 | 6 |
| Posttreatment | 1 | 0 | 6 | 0 |
| Ridley-Jopling clinical form of leprosy | ||||
| BT | 3 | 2 | 0 | 0 |
| BB | 3 | 4 | 0 | 0 |
| BL | 5 | 5 | 6 | 6 |
| LL | 0 | 0 | 5 | 5 |
Abbreviations: BB, borderline-borderline; BL, borderline lepromatous; BT, borderline tuberculoid; ENL, erythema nodosum leprosum; LL, lepromatous leprosy; RR, reversal reaction.
Pathway Analysis of Transcriptomes
The first aim of this study was to characterize the transcriptome profiles associated with the transition of leprosy to either RR or ENL. Comparing RR with non-RR controls, there were 275 differentially expressed genes (n = 203 increased and n = 72 decreased in RR). Comparing ENL with non-ENL controls, there were 517 differentially expressed genes (n = 300 increased and n = 217 decreased in ENL). Differentially expressed genes are listed in Supplementary Table 1.
Considering all differentially expressed genes in RR, the top GO Biological Process pathways were related to the immune response: immune response, defense response, immune system process, response to stress, and innate immune response (Table 2). Consistently, the top 3 KEGG pathways were Staphylococcus aureus infection (adjusted P = 2.56 × 10−13), complement and coagulation cascades (adjusted P = 4.85 ×10−9), and systemic lupus erythematosus (adjusted P = 7.32 ×10−9) (Table 3). HLA genes (HLA-DRB1 and HLA-DOB), C1Q, C1 esterase inhibitor (SERPING1), FPR 1, Fc fragment of IgG receptor (FcγR), and histone (HIST2H2AA3 and HIST2H2AA4) transcripts were represented in both sets of pathways. The top canonical pathways for RR identified by IPA were granulocyte adhesion and diapedesis (P = 8 × 10−9), agranulocyte adhesion and diapedesis (P = 1.35 × 10−7), and B-cell development (P = 6.96 × 10−7). IPA identified IFN-γ as the most significant upstream regulator of the expression changes seen in the array (P = 1.44 × 10−13), followed by immunoglobulin (1.22 × 10−12). The top associated IPA-identified network for RR is shown in Figure 1A and includes pathogen receptors, chemokines and chemokine receptors, and molecules that interact with immunoglobulin.
Table 2.
Top 10 GO Biological Process Pathways Among the Set of Differentially Expressed Genes in RR
| GO Pathway | Genes in Pathway, No. | Genes Changed in RR, No. | R | Adjusted P Value |
|---|---|---|---|---|
| Immune response | 1071 | 52 | 3.92 | 3.96 × 10−15 |
| Defense response | 1107 | 52 | 3.80 | 1.64 × 10−14 |
| Immune system process | 1792 | 66 | 2.98 | 4.78 × 10−14 |
| Response to stress | 2952 | 82 | 2.25 | 2.32 × 10−11 |
| Innate immune response | 539 | 30 | 4.50 | 5.93 × 10−9 |
| Response to stimulus | 6636 | 125 | 1.52 | 1.21 × 10−7 |
| Response to wounding | 1109 | 40 | 2.92 | 8.09 × 10−7 |
| Humoral immune response | 124 | 13 | 8.47 | 6.51 × 10−6 |
| Response to external stimulus | 1323 | 42 | 2.57 | 1.26 × 10−5 |
| Fibrinolysis | 24 | 7 | 23.58 | 1.73 × 10−5 |
Abbreviations: GO, Gene Ontology; RR, reversal reaction.
Table 3.
Significant KEGG Pathways With Pathway Transcripts Differentially Transcribed in RR
| KEGG Pathway (Adjusted P Value) | Transcriptsa (Fold Change; P Value) |
|---|---|
| Staphylococcus aureus infection (2.56 × 10−13) | HLA-DRB1 (−1.59; <.0001), C1QB (2.21; <.0001), C1QA (1.64; .0004), HLA-DQA1 (1.61; .0001), FCGR1A (2.78; <.0001), FPR1 (1.80; .0001), C1QC (1.77; <.0001), C2 (2.31; <.0001), FCGR3B (2.12; <.0001), HLA-DOB (−1.50; .0001), FPR2 (2.28; <.0001) |
| Complement and coagulation cascades (4.85 × 10−9) | C1QB (2.21; <.0001), SERPING1 (2.17; <.0001), C1QA (1.64; .0004), PLAU (2.52; <.0001), PLAUR (1.84; .0002), C1QC (1.77; <.0001), PROS1 (1.50; .0003), C2 (2.31; <.0001), THBD (1.78; .0001) |
| Systemic lupus erythematosus (7.32 × 10−9) | HLA-DRB1 (−1.59; <.0001), C1QB (2.21; <.0001), C1QA (1.64; .0004), HIST2H2AA4 (1.72; <.0001), HLA-DQA1 (1.61; .0001), FCGR1A (2.78; <.0001), C1QC (1.77; <.0001), HIST2H2AA3 (1.77; <.0001), C2 (2.31; <.0001), FCGR3B (2.12; <.0001), HLA-DOB (−1.50; .0001) |
| Hematopoietic cell lineage (4.50 × 10−8) | HLA-DRB1 (−1.59; <.0001), FCER2 (−1.5; <.0001), FCGR1A (2.78; <.0001), CD19 (−1.76; <.0001), ITGB3 (1.63; .0001), ILI1R2 (2.12; <.0001), MME (1.59; .0003), ITGA2B (2.04; <.0001), ANPEP (1.82; <.0001) |
| Cytokine-cytokine receptor interaction (7.38 × 10−8) | CCL7 (3.03; <.0001), PF4V1 (1.81; <.0001), CXCR5 (−1.65; <.0001), CXCL10 (2.02; <.0001), CCR7 (−1.73; <.0001), CCL2 (2.90; <.0001), ILIR2 (2.12; <.0001), CXCL1 (1.64; .0001), TNFRSF12A (1.94; <.0001), CCL3 (1.67; .0002), CD27 (−1.60; <.0001), CXCR1 (1.61; <.0001), CCL3L1 (1.61; .0001) |
| Phagosome (4.13 × 10−7) | MARCO (1.77; <.0001), CTSL1 (2.11; <.0001), HLA-DRB1 (−1.59; <.0001), CLEC7A (1.71; .0002), TUBB2A (1.52; .0002), HLA-DQA1 (1.61; .0001), FCGR1A (2.78; <.0001), ITGB3 (1.63; .0001), FCGR3B (2.12; <.0001), HLA-DOB (−1.50; .0001) |
| Rheumatoid arthritis (1.35 × 10−6) | CTSL1 (2.11; <.0001), HLA-DRB1 (−1.59; <.0001), HLA-DQA1 (1.61; .0001), CCL2 (2.90; <.0001), CXCL1 (1.64; .0001), CCL3 (1.67; .0002), CCL3L1 (1.61; .0001), HLA-DOB (−1.5; .0001) |
| Chemokine signaling pathway (3.09 × 10−6) | CCL7 (3.03; <.0001), PF4V1 (1.81; <.0001), CXCR5 (−1.65; <.0001), CXCL10 (2.02; <.0001), CCR7 (−1.73; <.0001), CCL2 (2.90; <.0001), CXCL1 (1.64; .0001), CCL3 (1.67; .0002), CXCR1 (1.61; <.0001), CCL3L1 (1.61; .0001) |
| Chagas disease (.001) | C1QB (2.21; <.0001), CCL3 (1.67; .0002), C1QA (1.64; .0004), CCL2 (2.90; <.0001), CCL3L1 (1.61; .0001), C1QC (1.77; <.0001) |
| Leishmaniasis (.002) | HLA-DRB1 (−1.59; <.0001), HLA-DQA1 (1.61; .0001), FCGR3B (2.12; <.0001), FCGR1A (2.78; <.0001), HLA-DOB (−1.5; .0003) |
| ECM receptor interaction (.004) | SV2A (1.68; .0001), LAMA5 (−1.57; .0001), ITGA2B (2.04; <.0001), SDC4 (1.58; .0002), ITGB3 (1.63; .0001) |
| Osteoclast differentiation (.03) | FOSL1 (1.79; <.0001), FCGR3B (2.12; <.0001), FCGR1A (2.78; <.0001), FOSB (1.68; .0001), ITGB3 (1.63; .0001) |
| Asthma (.03) | HLA-DRB1 (−1.59; <.0001), HLA-DQA1 (1.61; .0001), HLA-DOB (−1.5; .0001) |
| Toxoplasmosis (.04) | HLA-DRB1 (−1.59; <.0001), BCL2L1 (1.61; <.0001), HLA-DQA1 (1.61; .0001), HLA-DOB (−1.5; .0001), LAMA5 (−1.57; .0001) |
| Cell adhesion molecules (.04) | HLA-DRB1 (−1.59; <.0001), CNTNAP2 (−2.03; <.0001), HLA-DQA1 (1.61; .0001), HLA-DOB (−1.5; .0001), SDC4 (1.58; .0002) |
| Antigen processing and presentation (.04) | CTSL1 (2.11; <.0001), HLA-DRB1 (−1.59; <.0001), HLA-DQA1 (1.61; .0001), HLA-DOB (−1.5; .0001) |
| Prion diseases (.04) | C1QB (2.21; <.0001), C1QA (1.64; .0004), C1QC (1.77; <.0001) |
Abbreviations: ECM, extracellular matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes; RR, reversal reaction.
a Downregulated transcripts with negative fold-changes are in italics.
Figure 1.
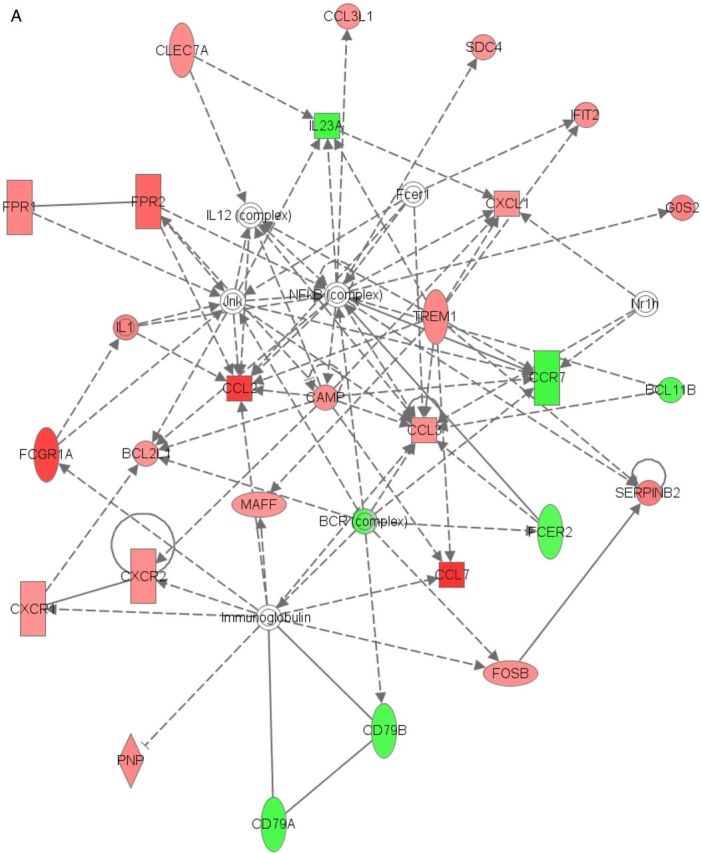
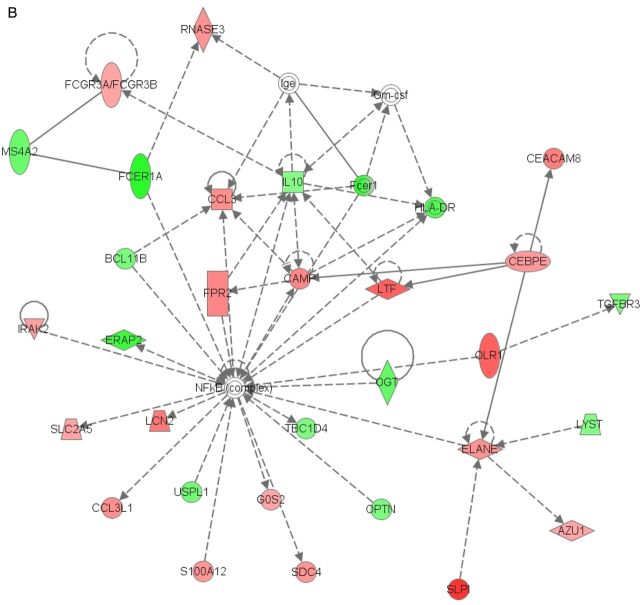
Networks generated by Ingenuity Pathway Analysis for reversal reaction (RR) (A) and erythema nodosum leprosum (ENL) (B). Red indicates genes upregulated in reaction; green, genes downregulated in reaction; darker shades, greater magnitude of fold change; broken lines, for indirect interactions; solid lines, direct interactions; arrows, activation; bars, inhibition.
Similar to RR, the top GO Biological Process pathways changed in ENL were related to the immune response: defense response, immune system process, response to bacterium, response to wounding, and immune response (Table 4). The top 3 KEGG pathways were S. aureus infection (adjusted P =1.9 × 10−16), systemic lupus erythematosus (adjusted P = 7.37 ×10−14), and cytokine-cytokine receptor interaction (adjusted P = 8.48 × 10−10) (Table 5). The complement and coagulation pathway (adjusted P = 3.78 × 10−9) was also associated with ENL. These pathways include components of the classic complement pathway, histones, cytokine receptors, and inflammatory response regulators. The top canonical pathways for ENL identified by IPA were granulocyte adhesion and diapedesis (P = 1.46 × 10−12), agranulocyte adhesion and diapedesis (P = 3.64 × 10−11), and interleukin 8 signaling (P = 2.68 ×10−6). IPA identified CCL5 as the most significant upstream regulator of the expression changes in the array (P = 9.78 ×10−16), followed by IFN-γ (P = 8.88 × 10−15). The top associated IPA-generated network for ENL is shown in Figure 1B and includes immunoglobulin receptors, pathogen recognition receptors, and chemokines.
Table 4.
Top 10 GO Biological Process Pathways Among the Set of Differentially Expressed Genes in ENL
| GO Pathway | Genes in Pathway, No. | Genes Changed in ENL, No. | R | Adjusted P Value |
|---|---|---|---|---|
| Defense response | 1107 | 83 | 3.39 | 1.55 × 10−20 |
| Immune system process | 1792 | 101 | 2.54 | 1.55 × 10−16 |
| Response to bacterium | 349 | 42 | 5.43 | 4.23 × 10−16 |
| Response to wounding | 1109 | 75 | 3.05 | 1.67 × 10−15 |
| Immune response | 1071 | 73 | 3.08 | 3.54 × 10−15 |
| Response to stress | 2952 | 133 | 2.03 | 7.38 × 10−15 |
| Inflammatory response | 484 | 45 | 4.20 | 4.86 × 10−13 |
| Response to biotic stimulus | 613 | 50 | 3.68 | 1.89 × 10−12 |
| Response to other organism | 583 | 48 | 3.72 | 5.33 × 10−12 |
Abbreviations: ENL, erythema nodosum leprosum; GO, Gene Ontology.
Table 5.
Significant KEGG Pathways With Pathway Transcripts Differentially Transcribed in ENL
| KEGG Pathway (Adjusted P Value) | Transcriptsa |
|---|---|
| Staphylococcus aureus infection (1.90 × 10−16) | C1QB (3.27; <.0001), PTAFR (1.89; .0001), C1QA (1.82; <.0001), HLA-DQA1 (1.50; .0003), FCGR1A (2.55; <.0001), SELP (1.62; <.0001), FPR1 (2.10 P < .0001), C1QC (2.61; <.0001), C5AR1 (1.83; <.0001), IL10 (−1.57; .0001), FCGR3B (1.61; .0002), HLA-DRB5 (−2.22; <.0001), FPR2 (2.15; <.0001), C3AR1 (1.66; <.0001), FCAR (1.68; .0003) |
| Systemic lupus erythematosus (7.37 × 10−14) | HIST1H3F (1.52; .0001), HIST2H3C (1.54; .0002), HIST2H2AA4 (2.09; <.0001), HLA-DQA1 (1.50; .0003), H2AFJ (1.65; <.0001), HIST2H2BE (1.77; <.0001), ELANE (1.92; <.0001), IL10 (−1.57; .0001), HIST1H3H (1.59; .0001), FCGR3B (1.61; .0002), HIST2H2AB (1.70; <.0001), C1QB (3.27; <.0001), C1QA (1.82; <.0001), FCGR1A (2.55; <.0001), C1QC (2.61; <.0001), HIST2H2AA3 (1.98; <.0001), HIST2H2AC (2.00; <.0001), HLA-DRB5 (−2.22; <.0001) |
| Cytokine-cytokine receptor interaction (8.48 × 10−10) | CCL7 (3.40; <.0001), IL11RA (−1.6; <.0001), CCL2 (2.70; <.0001), CXCL2 (2.81; <.0001), IL10 (−1.57; .0001), MPL (1.50; .0003), CCR1 (2.18; <.0001), CCL4L2 (1.72; <.0001), IL8 (1.84; <.0001), CCR3 (−1.63; <.0001), TNFRSF1A (1.69; .0005), IL1R2 (2.53; <.0001), CCL3L3 (1.72; <.0001), CXCL1 (2.63; <.0001), TNFRSF12A (1.94; <.0001), CCL3 (2.0; <.0001), CXCR1 (1.67; <.0001), CCL3L1 (2.0; <.0001), PPBP (1.63; <.0001) |
| Chagas disease (1.54 × 10−9) | C1QB (3.27; <.0001), JUN (1.84; <.0001), C1QA (1.82; <.0001), CCL2 (2.70; <.0001), GNA15 (1.82; <.0001), IL8 (1.84; <.0001), C1QC (2.61; <.0001), TNFRSF1A (1.69; .0005), CCL3L3 (1.72; <.0001), CCL3 (2.0; <.0001), IL10 (−1.57; .0001), FOS (1.68; <.0001), CCL3L1 (2.0; <.0001) |
| Complement and coagulation cascade (3.67 × 10−9) | C1QB (3.27; <.0001), C1QA (1.82; <.0001), PLAU (2.45; <.0001), PLAUR (1.71; <.0001), C1QC (2.61; <.0001), F13A1 (1.51; .0001), C5AR1 (1.83; <.0001), VWF (1.82; <.0001), PROS1 (1.57; <.0001), THBD (2.83; <.0001), C3AR1 (1.66; <.0001) |
| Rheumatoid arthritis (4.85 × 10−9) | CTSL1 (1.85; .0002), JUN (1.84; <.0001), CTLA4 (−1.77; .0004), HLA-DQA1 (1.50; .0003), CCL2 (2.70; <.0001), IL8 (1.84; <.0001), CCL3L3 (1.72; <.0001), CXCL1 (2.63; <.0001), CCL3 (2.0; <.0001), FOS (1.68; <.0001), HLA-DRB5 (−2.22; <.0001), CCL3L1 (2.0; <.0001) |
| Chemokine signaling pathway (2.93 × 10−7) | CCL7 (3.40; <.0001), CCR1 (2.18; <.0001), CCL4L2 (1.72; <.0001), CCL2 (2.70; <.0001), GNG10 (1.59; .0005), CCR3 (−1.63; <.0001), IL8 (1.84; <.0001), CCL3L3 (1.72; <.0001), CXCL1 (2.63; <.0001), CCL3 (2.0; <.0001), CXCL2 (2.81; <.0001), CXCR1 (1.67; <.0001), CCL3L1 (2.0; <.0001), PPBP (1.63; <.0001) |
| Cell adhesion molecules (4.15 × 10−7) | CD8A (−1.55; <.0001), ESAM (1.6; <.0001), CTLA4 (−1.77; .0004), HLA-DQA1 (1.50; .0003), PVRL2 (2.02; <.0001), SELP (1.62; <.0001), SDC4 (1.88; .0001), CD6 (−1.68; .0001), CNTNAP2 (−1.78; <.0001), HLA-C (1.85; <.0001), HLA-DRB5 (−2.22; <.0001), JAM3 (1.55; <.0001) |
| Hematopoietic cell lineage (8.40 × 10−7) | CD8A (−1.55; <.0001), CD24 (1.61; <.0001), IL11RA (−1.61; <.0001), FCGR1A (2.55; <.0001), ITGB3 (2.12; <.0001), IL1R2 (2.53; <.0001), GP9 (1.53; <.0001), HLA-DRB5 (−2.22; <.0001), ITGA2B (2.36; <.0001), ANPEP (2.14; <.0001) |
| Osteoclast differentiation (2.99 × 10−6) | JUN (1.84; <.0001), CYLD (−1.56; .0003), FCGR1A (2.55; <.0001), PPARG (2.27; <.0001), ITGB3 (2.12; <.0001), FOSB (1.96; <.0001), TNFRSF1A (1.69; .0005), FOS (1.68; <.0001), FCGR3B (1.61; .0002), FOSL1 (1.86; <.0001), SOCS3 (1.89; <.0001) |
| Phagosome (1.86 × 10−5) | CTSL1 (1.85; .0002), OLR1 (2.82; <.0001), HLA-DQA1 (1.50; .0003), FCGR1A (2.55; <.0001), ITGB3 (2.12; <.0001), HLA-C (1.85; <.0001), FCGR3B (1.61; .0002), ITGB5 (1.57; .0001), HLA-DRB5 (−2.22; <.0001), FCAR (1.68; .0003), THBS1 (2.28; <.0001) |
| Asthma (2.43 × 10−5) | FCER1A (−2.72; <.0001), IL10 (−1.57; .0001), RNASE3 (1.94; <.0001), HLA-DQA1 (1.50; .0003), HLA-DRB5 (−2.22; <.0001), MS4A2 (−1.90; <.0001) |
| Malaria (3.72 × 10−5) | IL10 (−1.57; .0001), SELP (1.62; <.0001), CCL2 (2.70; <.0001), GYPB (−2.45; <.0001), SDC4 (1.88; .0001), IL8 (1.84; <.0001), THBS1 (2.28; <.0001) |
| ECM receptor interaction (.0001) | GP9 (1.53; <.0001), VWF (1.82; <.0001), ITGB5 (1.57; .0001), SV2A (1.65; .0005), ITGA2B (2.36; <.0001), SDC4 (1.88; .0001), ITGB3 (2.12; <.0001), THBS1 (2.28; <.0001) |
| Leishmaniasis (.0004) | JUN (1.84; <.0001), IL10 (−1.57; .0001), FOS (1.68; <.0001), HLA-DQA1 (1.50; .0003), FCGR3B (1.61; .0002), FCGR1A (2.55; <.0001), HLA-DRB5 (−2.22; <.0001) |
| Toxoplasmosis (.003) | PPIF (1.67; .0005), BIRC3 (−1.59; .0002), HLA-DQA1 (1.50; .0003), LDLR (1.89; <.0001), TNFRSF1A (1.69; .0005), IL10 (−1.57; .0001), HLA-DRB5 (−2.22; <.0001), HSPA1B (1.68; .0001) |
| Amebiasis (.005) | CXCL1 (2.63; <.0001), IL10 (−1.57; .0001), ARG1 (2.10; <.0001), GNA15 (1.82; <.0001), SERPINB2 (4.82; <.0001), IL8 (1.84; <.0001), IL1R2 (2.53; <.0001) |
| T-cell receptor signaling pathway (.006) | JUN (1.84; <.0001), CD8A (−1.55; <.0001), IL10 (−1.57; .0001), CTLA4 (−1.77; .0004), NFKBIE (1.50; .0003), FOS (1.68; <.0001), RASGRP1 (−1.64; .0003) |
| Antigen processing and presentation (.006) | CTSL1 (1.85; .0002), CD8A (−1.55; <.0001), HLA-C (1.85; <.0001), HLA-DQA1 (1.50; .0003), HLA-DRB5 (−2.22; <.0001), HSPA1B (1.68; .0001) |
| Autoimmune thyroid disease (.008) | IL10 (−1.57; .0001), CTLA4 (−1.77; .0004), HLA-C (1.85; <.0001), HLA-DQA1 (1.50; .0003), HLA-DRB5 (−2.22; <.0001) |
| NOD-like receptor signaling pathway (.02) | CXCL1 (2.63; <.0001), CXCL2 (2.81; <.0001), BIRC3 (−1.59; .0002), CCL2 (2.70; <.0001), IL8 (1.84; <.0001) |
| Allograft rejection (.03) | IL10 (−1.57; .0001), HLA-C (1.85; <.0001), HLA-DQA1 (1.50; .0003), HLA-DRB5 (−2.22; <.0001) |
| P53 signaling pathway (.04) | ATM (−1.92; <.0001), SESN1 (−1.59; .0001), TP53I3 (1.67; .0004), GADD45G (1.59; .0006), THBS1 (2.28; <.0001) |
| Epithelial cell signaling in Helicobacter pylori infection (.04) | CXCL1 (2.63; <.0001), JUN (1.84; <.0001), CXCR1 (1.67; <.0001), JAM3 (1.56; <.0001), IL8 (1.84; <.0001) |
| PPAR signaling pathway (.04) | GK (2.0; <.0001), ACOX2 (2.10; <.0001), ACSL1 (1.66; <.0001), OLR1 (2.82; <.0001), PPARG (2.27; .0003) |
Abbreviations: ECM, extracellular matrix; ENL, erythema nodosum leprosum; KEGG, Kyoto Encyclopedia of Genes and Genomes; NOD, nucleotide-binding oligomerization domain; PPAR, peroxisome proliferator-activated receptor.
a Downregulated transcripts with negative fold changes are in italics.
Comparison of RR and ENL Transcriptomes
Another aim of this study was to describe the similarities and differences in gene expression in RR and ENL (compared with their respective controls). Considering all differentially expressed genes, there were 379 transcripts unique to ENL, 137 unique to RR, and 138 that had differential expression in both RR and ENL groups (Tables 6–8). Of those genes with increased expression, 104 were unique to RR, 201 unique to ENL, and 99 with increased expression in both RR and ENL groups. Among genes with decreased expression during an immune reaction, 57 were unique to RR, 202 unique to ENL, and 15 with decreased expression in both groups.
Table 7.
Differentially Expressed Transcripts Unique to ENL
| Transcript | Fold Changea | P Value |
|---|---|---|
| OLFM4 | 3.09 | <.0001 |
| LTF | 2.82 | <.0001 |
| OLR1 | 2.82 | <.0001 |
| CXCL2 | 2.81 | <.0001 |
| MMP8 | 2.46 | <.0001 |
| DEFA1B | 2.44 | <.0001 |
| FAM20A | 2.39 | <.0001 |
| LCN2 | 2.39 | <.0001 |
| SAMD14 | 2.33 | <.0001 |
| CEACAM8 | 2.30 | <.0001 |
| THBS1 | 2.28 | <.0001 |
| PPARG | 2.27 | <.0001 |
| CEACAM6 | 2.26 | <.0001 |
| PHACTR1 | 2.25 | <.0001 |
| CHST13 | 2.21 | <.0001 |
| CCR1 | 2.18 | <.0001 |
| DEFA3 | 2.17 | <.0001 |
| ACOX2 | 2.10 | <.0001 |
| ARG1 | 2.09 | <.0001 |
| HTRA1 | 2.08 | <.0001 |
| C19ORF59 | 2.07 | <.0001 |
| PTPN20 | 2.06 | <.0001 |
| RNU11 | 2.06 | <.0001 |
| RGL1 | 2.05 | <.0001 |
| PHLDA1 | 2.03 | <.0001 |
| HS.562219 | 2.02 | <.0001 |
| PVRL2 | 2.02 | <.0001 |
| HIST2H2AC | 2.01 | <.0001 |
| RNASE2 | 2.00 | <.0001 |
| CCRL2 | 1.99 | <.0001 |
| CEACAM1 | 1.98 | <.0001 |
| SLC22A18AS | 1.98 | <.0001 |
| COL17A1 | 1.96 | <.0001 |
| TNFAIP6 | 1.96 | <.0001 |
| ZDHHC19 | 1.96 | <.0001 |
| SPRY2 | 1.95 | <.0001 |
| RNASE3 | 1.94 | <.0001 |
| BPI | 1.93 | <.0001 |
| METTL7B | 1.93 | <.0001 |
| C5ORF32 | 1.92 | <.0001 |
| DDIT3 | 1.92 | <.0001 |
| ELANE | 1.92 | <.0001 |
| CYP1B1 | 1.89 | <.0001 |
| LDLR | 1.89 | <.0001 |
| LOC649210 | 1.89 | <.0001 |
| PTAFR | 1.89 | .0001 |
| RNASE1 | 1.89 | <.0001 |
| SOCS3 | 1.89 | <.0001 |
| PLP2 | 1.88 | <.0001 |
| TSPAN9 | 1.88 | <.0001 |
| CD300C | 1.87 | .0002 |
| ASGR2 | 1.86 | <.0001 |
| GSN | 1.86 | <.0001 |
| MAFB | 1.86 | <.0001 |
| S100A12 | 1.86 | <.0001 |
| CETP | 1.85 | <.0001 |
| HLA-C | 1.85 | <.0001 |
| RNU4-1 | 1.85 | <.0001 |
| IL8 | 1.84 | <.0001 |
| JUN | 1.84 | <.0001 |
| MGC29506 | 1.84 | <.0001 |
| SIGLEC9 | 1.84 | .0002 |
| AQP9 | 1.83 | <.0001 |
| C5AR1 | 1.83 | <.0001 |
| DOK3 | 1.83 | .0001 |
| LOC650263 | 1.83 | <.0001 |
| CEBPE | 1.82 | <.0001 |
| GNA15 | 1.82 | <.0001 |
| LOC100133477 | 1.82 | .0001 |
| VWF | 1.82 | <.0001 |
| DEFA1 | 1.81 | <.0001 |
| FBXL2 | 1.81 | .0001 |
| MIR1974 | 1.80 | <.0001 |
| RNU1-3 | 1.80 | <.0001 |
| RNU4-2 | 1.79 | <.0001 |
| SLC2A14 | 1.79 | <.0001 |
| C13ORF15 | 1.78 | <.0001 |
| LHFPL2 | 1.78 | .0001 |
| LOC100134331 | 1.78 | <.0001 |
| SERPINB8 | 1.78 | .0001 |
| ST14 | 1.78 | <.0001 |
| TMEM88 | 1.78 | <.0001 |
| HIST2H2BE | 1.77 | <.0001 |
| BASP1 | 1.76 | <.0001 |
| FLVCR2 | 1.76 | .0001 |
| PPP1R15A | 1.76 | <.0001 |
| TLE3 | 1.76 | .0001 |
| FAM129B | 1.75 | .0005 |
| RGL4 | 1.75 | <.0001 |
| KIFC3 | 1.74 | <.0001 |
| LOC653061 | 1.74 | <.0001 |
| NAB2 | 1.74 | <.0001 |
| NRIP3 | 1.74 | <.0001 |
| ABCA1 | 1.73 | <.0001 |
| IGFBP2 | 1.73 | <.0001 |
| LOC441481 | 1.73 | .0001 |
| ABLIM3 | 1.72 | <.0001 |
| CCL3L3 | 1.72 | <.0001 |
| CCL4L2 | 1.72 | <.0001 |
| LOC728744 | 1.72 | .0001 |
| PLIN2 | 1.72 | <.0001 |
| CTDSPL | 1.71 | <.0001 |
| MIR302C | 1.71 | .0004 |
| NOP10 | 1.71 | .0005 |
| PFKFB3 | 1.71 | <.0001 |
| HIST2H2AB | 1.70 | <.0001 |
| SLC24A3 | 1.70 | <.0001 |
| STAB1 | 1.70 | .0002 |
| BEX1 | 1.69 | .0001 |
| HLX | 1.69 | <.0001 |
| IRAK2 | 1.69 | .0001 |
| LIMK2 | 1.69 | .0001 |
| MIR223 | 1.69 | .0001 |
| RNU1-5 | 1.69 | <.0001 |
| SLC25A24 | 1.69 | .0003 |
| TNFRSF1A | 1.69 | .0005 |
| FCAR | 1.68 | .0003 |
| FOS | 1.68 | <.0001 |
| HSPA1B | 1.68 | .0001 |
| WIPI1 | 1.68 | <.0001 |
| ASPH | 1.67 | .0005 |
| PPIF | 1.67 | .0005 |
| RNU4ATAC | 1.67 | <.0001 |
| SPHK1 | 1.67 | .0004 |
| TP53I3 | 1.67 | .0004 |
| ACSL1 | 1.66 | <.0001 |
| C3AR1 | 1.66 | <.0001 |
| ECM1 | 1.66 | .0003 |
| FAH | 1.66 | .0002 |
| GAS6 | 1.66 | .0001 |
| HS.559602 | 1.66 | .0002 |
| IER3 | 1.66 | <.0001 |
| IGJ | 1.66 | <.0001 |
| STEAP4 | 1.66 | <.0001 |
| CEACAM4 | 1.65 | <.0001 |
| H2AFJ | 1.65 | <.0001 |
| LOC647506 | 1.65 | <.0001 |
| LOC650261 | 1.65 | .0006 |
| PSG9 | 1.65 | <.0001 |
| UBAP1 | 1.64 | .0001 |
| DCUN1D3 | 1.63 | <.0001 |
| LOC729040 | 1.63 | .0004 |
| PLSCR1 | 1.63 | .0005 |
| PPBP | 1.63 | <.0001 |
| BST1 | 1.62 | .0004 |
| LOC728835 | 1.62 | <.0001 |
| RNU1G2 | 1.62 | <.0001 |
| SBNO2 | 1.62 | .0004 |
| SELP | 1.62 | <.0001 |
| SGK1 | 1.62 | <.0001 |
| SLC2A5 | 1.62 | <.0001 |
| CD24 | 1.61 | <.0001 |
| DSE | 1.61 | <.0001 |
| HIST1H1C | 1.61 | <.0001 |
| LOC440731 | 1.61 | .0003 |
| ESAM | 1.60 | <.0001 |
| PDLIM7 | 1.60 | .0005 |
| CD63 | 1.59 | .0002 |
| CDA | 1.59 | <.0001 |
| GNG10 | 1.59 | .0005 |
| GPER | 1.59 | .0001 |
| HIST1H3H | 1.59 | .0001 |
| IGFBPL1 | 1.59 | .0004 |
| LOC100134728 | 1.59 | <.0001 |
| TRIB1 | 1.59 | <.0001 |
| AZU1 | 1.58 | <.0001 |
| H1F0 | 1.58 | .0005 |
| HS.521338 | 1.58 | .0006 |
| METRNL | 1.58 | <.0001 |
| TNNT1 | 1.58 | <.0001 |
| ITGB5 | 1.57 | .0001 |
| RNU5A | 1.57 | .0001 |
| UBTD1 | 1.57 | .0005 |
| 40425 | 1.56 | <.0001 |
| JAM3 | 1.56 | <.0001 |
| SH3BGRL2 | 1.56 | <.0001 |
| SLC6A6 | 1.56 | .0006 |
| HIST2H3C | 1.54 | .0002 |
| LOC100134379 | 1.54 | <.0001 |
| LOC649923 | 1.54 | <.0001 |
| NR1I2 | 1.54 | .0005 |
| TREML2 | 1.54 | .0006 |
| FLJ22662 | 1.53 | .0002 |
| GP9 | 1.53 | <.0001 |
| HOMER2 | 1.53 | .0002 |
| HS.557039 | 1.53 | <.0001 |
| IGLL1 | 1.53 | <.0001 |
| LOC554223 | 1.53 | .0001 |
| LOC647450 | 1.53 | <.0001 |
| NACC2 | 1.53 | .0003 |
| RAB13 | 1.53 | .0001 |
| SGK | 1.53 | <.0001 |
| C5ORF62 | 1.52 | .0003 |
| HIST1H3F | 1.52 | .0001 |
| HS.276854 | 1.52 | .0002 |
| LOC653506 | 1.52 | .0001 |
| ADORA2A | 1.51 | .0006 |
| F13A1 | 1.51 | .0001 |
| LOC652493 | 1.50 | <.0001 |
| MPL | 1.50 | .0003 |
| NFKBIE | 1.50 | .0003 |
| FCER1A | −2.72 | <.0001 |
| ERAP2 | −2.48 | <.0001 |
| HDC | −2.26 | <.0001 |
| HLA-DRB5 | −2.22 | <.0001 |
| NKTR | −2.16 | <.0001 |
| MCOLN2 | −2.05 | <.0001 |
| IFI44L | −2.01 | <.0001 |
| OGT | −1.98 | <.0001 |
| SAMD9L | −1.95 | <.0001 |
| FAM46C | −1.94 | <.0001 |
| HS.560343 | −1.93 | <.0001 |
| ATM | −1.92 | <.0001 |
| C7ORF54 | −1.91 | <.0001 |
| PNN | −1.91 | <.0001 |
| MS4A2 | −1.90 | <.0001 |
| ZBTB20 | −1.90 | <.0001 |
| HS.193767 | −1.85 | <.0001 |
| C14ORF106 | −1.84 | <.0001 |
| CD96 | −1.84 | <.0001 |
| SFRS18 | −1.84 | <.0001 |
| MTX3 | −1.83 | <.0001 |
| TMEM181 | −1.83 | <.0001 |
| HS.553301 | −1.82 | <.0001 |
| C6ORF111 | −1.81 | <.0001 |
| GVIN1 | −1.80 | <.0001 |
| HS.22689 | −1.80 | <.0001 |
| HS.556018 | −1.80 | <.0001 |
| PHIP | −1.80 | <.0001 |
| CCNJ | −1.79 | <.0001 |
| TMX3 | −1.79 | <.0001 |
| USPL1 | −1.78 | <.0001 |
| CTLA4 | −1.77 | .0004 |
| HS.356079 | −1.77 | <.0001 |
| MYBL1 | −1.77 | <.0001 |
| CEP350 | −1.76 | <.0001 |
| FAM190B | −1.76 | <.0001 |
| KIAA0528 | −1.76 | <.0001 |
| LOC642333 | −1.76 | .0001 |
| MGEA5 | −1.76 | <.0001 |
| PTAR1 | −1.76 | <.0001 |
| TTC3 | −1.76 | <.0001 |
| ZNF518A | −1.76 | <.0001 |
| C15ORF28 | −1.75 | <.0001 |
| C6ORF190 | −1.75 | .0002 |
| CROP | −1.75 | <.0001 |
| TC2N | −1.75 | <.0001 |
| C10ORF6 | −1.74 | <.0001 |
| HS.481659 | −1.74 | <.0001 |
| TTC14 | −1.74 | <.0001 |
| TTC37 | −1.74 | <.0001 |
| KNTC1 | −1.73 | .0002 |
| ANKRD12 | −1.72 | <.0001 |
| LOC100132247 | −1.72 | <.0001 |
| ZFC3H1 | −1.72 | <.0001 |
| DMTF1 | −1.71 | <.0001 |
| HS.143018 | −1.70 | <.0001 |
| MTM1 | −1.70 | .0002 |
| PSME4 | −1.70 | .0002 |
| RASGRP3 | −1.70 | <.0001 |
| SAMD9 | −1.70 | <.0001 |
| KIAA0907 | −1.69 | <.0001 |
| PDS5A | −1.69 | .0004 |
| CD6 | −1.68 | .0001 |
| HEMGN | −1.68 | .0002 |
| HS.202577 | −1.68 | <.0001 |
| LOC729645 | −1.68 | <.0001 |
| SAMD3 | −1.68 | .0003 |
| ZNF33B | −1.68 | <.0001 |
| EML4 | −1.67 | <.0001 |
| HS.549989 | −1.67 | <.0001 |
| HS.570988 | −1.67 | <.0001 |
| LOC729978 | −1.67 | <.0001 |
| OPTN | −1.67 | .0003 |
| RBM33 | −1.67 | <.0001 |
| ZNF512 | −1.67 | .0004 |
| KIAA1128 | −1.66 | .0003 |
| LOC613037 | −1.66 | .0001 |
| RAX2 | −1.66 | .0003 |
| SLC38A1 | −1.66 | .0004 |
| CEP135 | −1.65 | .0002 |
| HS.154336 | −1.65 | .0002 |
| HS.535028 | −1.65 | <.0001 |
| HS.571887 | −1.65 | .0004 |
| PTGDR | −1.65 | <.0001 |
| SLTM | −1.65 | .0002 |
| STAT4 | −1.65 | .0001 |
| TGFBR3 | −1.65 | .0001 |
| LOC100134241 | −1.64 | <.0001 |
| LOC23117 | −1.64 | <.0001 |
| MIR142 | −1.64 | <.0001 |
| RASGRP1 | −1.64 | .0003 |
| TBC1D4 | −1.64 | .0001 |
| CCDC66 | −1.63 | <.0001 |
| CCR3 | −1.63 | <.0001 |
| FLJ44342 | −1.63 | .0001 |
| KIAA1641 | −1.63 | .0005 |
| RBM25 | −1.63 | .0002 |
| ZRANB2 | −1.63 | <.0001 |
| ANAPC4 | −1.62 | .0004 |
| CCDC14 | −1.62 | .0001 |
| DKFZP586I1420 | −1.62 | <.0001 |
| DPYSL4 | −1.62 | <.0001 |
| FAM111A | −1.62 | .0004 |
| HS.574671 | −1.62 | <.0001 |
| MBNL1 | −1.62 | .0004 |
| PFAAP5 | −1.62 | .0001 |
| SLC30A7 | −1.62 | .0002 |
| YOD1 | −1.62 | .0002 |
| CRIPAK | −1.61 | .0003 |
| HS.374460 | −1.61 | <.0001 |
| HS.445274 | −1.61 | .0001 |
| HS.546375 | −1.61 | .0003 |
| IL11RA | −1.61 | <.0001 |
| ZNF721 | −1.61 | .0003 |
| HS.193784 | −1.60 | .0002 |
| HS.284464 | −1.60 | .0004 |
| LCOR | −1.60 | .0004 |
| LOC100131768 | −1.60 | .0001 |
| ACAD11 | −1.59 | .0001 |
| AHSA2 | −1.59 | <.0001 |
| BIRC3 | −1.59 | .0002 |
| HS.554324 | −1.59 | .0002 |
| LOC729120 | −1.59 | .0002 |
| SESN1 | −1.59 | .0001 |
| ANGEL2 | −1.58 | .0001 |
| C10ORF73 | −1.58 | .0004 |
| HS.444683 | −1.58 | <.0001 |
| KIAA1370 | −1.58 | .0003 |
| PDK4 | −1.58 | <.0001 |
| RASA4 | −1.58 | <.0001 |
| VPS36 | −1.58 | .0001 |
| C8ORF45 | −1.57 | <.0001 |
| CCDC84 | −1.57 | <.0001 |
| HS.473191 | −1.57 | .0002 |
| IL10 | −1.57 | .0001 |
| LOC728411 | −1.57 | .0002 |
| MKLN1 | −1.57 | .0003 |
| OLIG1 | −1.57 | <.0001 |
| TARBP1 | −1.57 | .0005 |
| ZNF529 | −1.57 | .0002 |
| ZNF786 | −1.57 | .0002 |
| CYLD | −1.56 | .0003 |
| FAM153B | −1.56 | .0004 |
| LOC441268 | −1.56 | <.0001 |
| LRFN3 | −1.56 | .0001 |
| ZNF224 | −1.56 | .0002 |
| C10ORF137 | −1.55 | .0002 |
| CD8A | −1.55 | <.0001 |
| CTGLF3 | −1.55 | .0002 |
| DMC1 | −1.55 | .0002 |
| HS.440088 | −1.55 | .0004 |
| HSD17B7 | −1.55 | .0005 |
| D2HGDH | −1.54 | .0004 |
| HS.371060 | −1.54 | .0002 |
| KRT72 | −1.54 | <.0001 |
| LYST | −1.54 | .0003 |
| TXK | −1.54 | <.0001 |
| C2ORF69 | −1.53 | .0002 |
| LOC202781 | −1.53 | .0002 |
| RAB12 | −1.53 | <.0001 |
| ZNF91 | −1.53 | .0001 |
| FLJ12078 | −1.52 | <.0001 |
| LOC100133950 | −1.52 | .0001 |
| LOC440353 | −1.52 | .0005 |
| LOC727908 | −1.52 | .0003 |
| TBL1XR1 | −1.52 | .0004 |
| BTAF1 | −1.51 | <.0001 |
| C2ORF89 | −1.51 | .0005 |
| CCDC45 | −1.51 | .0003 |
| DTWD2 | −1.51 | .0003 |
| LOC644297 | −1.51 | .0004 |
| RPS23 | −1.51 | <.0001 |
| SMCR5 | −1.51 | .0004 |
| ZNF700 | −1.51 | .0002 |
| INO80D | −1.50 | .0003 |
| KIAA1333 | −1.50 | .0002 |
| LOC645452 | −1.50 | .0002 |
| PARP15 | −1.50 | .0005 |
Abbreviation: ENL, erythema nodosum leprosum.
a Negative fold changes are in italic type.
Table 6.
Differentially Expressed Transcripts Unique to RR
| Transcript | Fold Changea | P Value |
|---|---|---|
| IFI27 | 3.04 | <.0001 |
| HBQ1 | 2.65 | <.0001 |
| TGM2 | 2.63 | <.0001 |
| HBE1 | 2.58 | <.0001 |
| GBP1 | 2.45 | <.0001 |
| RBPMS2 | 2.43 | <.0001 |
| SLC25A37 | 2.31 | <.0001 |
| C2 | 2.31 | <.0001 |
| LOC647307 | 2.22 | <.0001 |
| LOC654055 | 2.20 | <.0001 |
| SERPING1 | 2.17 | <.0001 |
| OSBP2 | 2.16 | <.0001 |
| LOC649143 | 2.15 | <.0001 |
| IFIT3 | 2.10 | <.0001 |
| BATF2 | 2.08 | <.0001 |
| LOC653778 | 2.08 | <.0001 |
| TRIM58 | 2.07 | <.0001 |
| RAP1GAP | 2.05 | .0001 |
| CXCL10 | 2.02 | <.0001 |
| LOC400759 | 1.99 | <.0001 |
| EPB49 | 1.95 | <.0001 |
| DPYSL5 | 1.92 | <.0001 |
| TACSTD2 | 1.91 | <.0001 |
| GMPR | 1.91 | <.0001 |
| LOC654103 | 1.89 | <.0001 |
| TMOD1 | 1.89 | <.0001 |
| SLC6A10P | 1.89 | <.0001 |
| E2F2 | 1.88 | <.0001 |
| IFITM3 | 1.87 | <.0001 |
| XK | 1.85 | <.0001 |
| GPR175 | 1.85 | <.0001 |
| GBP5 | 1.84 | <.0001 |
| TRIM16L | 1.83 | <.0001 |
| IL27 | 1.82 | <.0001 |
| FER1L3 | 1.81 | .0001 |
| PF4V1 | 1.81 | <.0001 |
| WARS | 1.81 | <.0001 |
| OR2W3 | 1.80 | <.0001 |
| HLA-DRB6 | 1.79 | <.0001 |
| KCNJ2 | 1.78 | <.0001 |
| SDSL | 1.78 | <.0001 |
| MARCO | 1.77 | <.0001 |
| BLVRB | 1.77 | <.0001 |
| MYOF | 1.77 | .0002 |
| VAMP5 | 1.76 | <.0001 |
| HBEGF | 1.76 | <.0001 |
| LOC642469 | 1.76 | <.0001 |
| LOC652616 | 1.73 | .0001 |
| LOC100133583 | 1.72 | .0003 |
| SERPINA13 | 1.71 | <.0001 |
| CLEC7A | 1.71 | .0002 |
| LOC642567 | 1.70 | .0003 |
| LOC729708 | 1.70 | <.0001 |
| HS.544245 | 1.69 | .0001 |
| IFIT2 | 1.67 | .0001 |
| SLC6A8 | 1.67 | <.0001 |
| TYMP | 1.65 | .0002 |
| LOC100133678 | 1.65 | .0001 |
| SLC6A12 | 1.65 | .0001 |
| PANX2 | 1.64 | .0002 |
| ODF3B | 1.63 | .0003 |
| MT2A | 1.63 | .0002 |
| LOC100131391 | 1.62 | .0001 |
| CDCA5 | 1.62 | .0002 |
| ALDH1A1 | 1.61 | .0001 |
| BCL2L1 | 1.61 | <.0001 |
| LGALS3BP | 1.61 | .0001 |
| PSG3 | 1.60 | .0001 |
| TK1 | 1.59 | .0002 |
| MME | 1.59 | .0003 |
| BATF3 | 1.59 | <.0001 |
| LOC100133875 | 1.59 | .0001 |
| LOC650557 | 1.59 | <.0001 |
| MAFF | 1.58 | .0003 |
| SAMD4A | 1.56 | .0003 |
| PLA2G4C | 1.56 | .0002 |
| CDC45L | 1.56 | .0003 |
| LOC388588 | 1.54 | <.0001 |
| HSPA7 | 1.53 | .0001 |
| TUBB2A | 1.52 | .0002 |
| VPREB3 | −1.98 | <.0001 |
| TCL1A | −1.98 | <.0001 |
| OSBPL10 | −1.97 | <.0001 |
| SNORD4A | −1.96 | <.0001 |
| ZNF256 | −1.87 | <.0001 |
| ZNF101 | −1.87 | <.0001 |
| ACACB | −1.77 | <.0001 |
| CD19 | −1.76 | <.0001 |
| SEL1L3 | −1.74 | <.0001 |
| CCR7 | −1.73 | <.0001 |
| FCGBP | −1.71 | <.0001 |
| MGC3020 | −1.71 | <.0001 |
| IRX3 | −1.71 | <.0001 |
| LOC791120 | −1.70 | <.0001 |
| FAIM3 | −1.69 | <.0001 |
| EOMES | −1.68 | <.0001 |
| LOC649841 | −1.67 | <.0001 |
| FAM84B | −1.67 | .0001 |
| SNORD104 | −1.65 | .0001 |
| LEF1 | −1.65 | .0001 |
| CXCR5 | −1.65 | <.0001 |
| BLR1 | −1.64 | <.0001 |
| CD79A | −1.64 | <.0001 |
| LOC283663 | −1.63 | <.0001 |
| CACNA1I | −1.63 | <.0001 |
| STRBP | −1.62 | .0002 |
| SNHG7 | −1.62 | <.0001 |
| MAL | −1.61 | <.0001 |
| LOC651751 | −1.61 | <.0001 |
| C21ORF2 | −1.61 | <.0001 |
| CRYBB2 | −1.60 | .0001 |
| LOC100132499 | −1.60 | <.0001 |
| CD27 | −1.60 | <.0001 |
| LRRN3 | −1.60 | <.0001 |
| CMTM8 | −1.60 | <.0001 |
| HLA-DRB1 | −1.60 | <.0001 |
| KLHL3 | −1.59 | .0001 |
| CYORF15A | −1.59 | <.0001 |
| CDR2 | −1.59 | .0001 |
| CD79B | −1.59 | <.0001 |
| MC1R | −1.57 | .0001 |
| LAMA5 | −1.57 | .0001 |
| KLRF1 | −1.57 | .0001 |
| LOC653316 | −1.56 | .0001 |
| LOC90925 | −1.56 | <.0001 |
| PLCH2 | −1.55 | .0001 |
| KIAA0114 | −1.55 | <.0001 |
| D4S234E | −1.55 | <.0001 |
| CCDC102A | −1.55 | <.0001 |
| HS.481464 | −1.54 | .0001 |
| POU2AF1 | −1.54 | .0001 |
| BLK | −1.53 | <.0001 |
| C16ORF74 | −1.53 | <.0001 |
| BEX2 | −1.53 | .0001 |
| FCER2 | −1.52 | <.0001 |
| HLA-DOB | −1.50 | .0001 |
| CENTG2 | −1.50 | .0001 |
Abbreviation: RR, reversal reaction.
a Negative fold changes are in italic type.
Table 8.
Differentially Expressed Transcripts in Both RR and ENL Groups
| Transcript | RR |
ENL |
||
|---|---|---|---|---|
| Fold Changea | P Value | Fold Changea | P Value | |
| ADM | 1.86 | .0001 | 2.15 | <.0001 |
| ADORA2B | 1.90 | <.0001 | 2.09 | <.0001 |
| AHSP | 2.74 | <.0001 | −2.52 | <.0001 |
| ALAS2 | 2.51 | <.0001 | −2.67 | <.0001 |
| ALPL | 2.53 | <.0001 | 3.65 | <.0001 |
| ANKRD22 | 3.15 | <.0001 | 1.75 | <.0001 |
| ANPEP | 1.82 | <.0001 | 2.14 | <.0001 |
| ANXA3 | 1.97 | <.0001 | 2.68 | <.0001 |
| AQP10 | 1.83 | .0001 | 1.54 | <.0001 |
| AXIN2 | −1.81 | <.0001 | −1.85 | <.0001 |
| BCL11B | −1.58 | .0002 | −1.71 | .0001 |
| BPGM | 1.53 | .0002 | −1.69 | .0004 |
| C15ORF48 | 2.60 | <.0001 | 2.38 | <.0001 |
| C1QA | 1.64 | .0004 | 1.82 | <.0001 |
| C1QB | 2.21 | <.0001 | 3.27 | <.0001 |
| C1QC | 1.77 | <.0001 | 2.61 | <.0001 |
| C9ORF45 | −1.76 | <.0001 | −1.63 | <.0001 |
| CA1 | 2.22 | <.0001 | −3.30 | <.0001 |
| CA4 | 2.26 | <.0001 | 1.91 | <.0001 |
| CAMP | 1.67 | <.0001 | 2.16 | <.0001 |
| CCL2 | 2.90 | <.0001 | 2.70 | <.0001 |
| CCL3 | 1.67 | .0002 | 2.00 | <.0001 |
| CCL3L1 | 1.61 | .0001 | 2.00 | <.0001 |
| CCL7 | 3.03 | <.0001 | 3.40 | <.0001 |
| CEACAM3 | 1.74 | <.0001 | 2.12 | <.0001 |
| CLEC5A | 2.12 | .0001 | 1.90 | <.0001 |
| CMTM2 | 1.73 | <.0001 | 1.93 | <.0001 |
| CMTM5 | 1.59 | .0001 | 1.97 | <.0001 |
| CNTNAP2 | −2.03 | <.0001 | −1.78 | <.0001 |
| CTSL1 | 2.11 | <.0001 | 1.85 | .0002 |
| CXCL1 | 1.64 | .0001 | 2.63 | <.0001 |
| CXCR1 | 1.61 | <.0001 | 1.67 | <.0001 |
| DEFA4 | 1.68 | <.0001 | 2.60 | <.0001 |
| DHRS9 | 2.34 | <.0001 | 1.83 | .0002 |
| DYSF | 2.18 | <.0001 | 1.86 | <.0001 |
| EDN1 | 1.62 | .0001 | 1.71 | .0003 |
| EGR2 | 1.76 | <.0001 | 1.58 | <.0001 |
| EMP1 | 1.92 | .0002 | 1.82 | <.0001 |
| EPB42 | 2.20 | <.0001 | −2.85 | <.0001 |
| FCGR1A | 2.78 | <.0001 | 2.55 | <.0001 |
| FCGR1B | 2.59 | <.0001 | 2.38 | <.0001 |
| FCGR1C | 2.87 | <.0001 | 2.49 | <.0001 |
| FCGR3B | 2.12 | <.0001 | 1.61 | .0002 |
| FCRL3 | −1.86 | <.0001 | −1.71 | <.0001 |
| FCRLA | −2.32 | <.0001 | −1.65 | <.0001 |
| FFAR2 | 2.45 | <.0001 | 2.02 | <.0001 |
| FOSB | 1.68 | .0001 | 1.96 | <.0001 |
| FOSL1 | 1.79 | <.0001 | 1.86 | <.0001 |
| FPR1 | 1.80 | .0001 | 2.10 | <.0001 |
| FPR2 | 2.28 | <.0001 | 2.15 | <.0001 |
| G0S2 | 1.86 | <.0001 | 1.60 | <.0001 |
| GADD45G | 1.63 | .0001 | 1.58 | .0006 |
| GAPDHL6 | 2.01 | <.0001 | 1.51 | <.0001 |
| GK | 2.05 | <.0001 | 2.00 | <.0001 |
| GOLGA8B | −1.52 | <.0001 | −2.13 | <.0001 |
| GPR109A | 2.11 | <.0001 | 2.93 | <.0001 |
| GPR109B | 1.74 | .0001 | 2.24 | <.0001 |
| GPR84 | 1.82 | .0001 | 2.47 | <.0001 |
| GPR97 | 1.85 | <.0001 | 1.90 | <.0001 |
| GYPB | 2.25 | <.0001 | −2.45 | <.0001 |
| GYPE | 1.62 | .0001 | −1.51 | .0003 |
| GZMK | −1.83 | <.0001 | −1.52 | <.0001 |
| HAMP | 1.74 | .0003 | 1.89 | .0004 |
| HBD | 2.53 | <.0001 | −2.48 | <.0001 |
| HBG1 | 1.76 | <.0001 | −2.14 | <.0001 |
| HBG2 | 1.67 | <.0001 | −2.06 | <.0001 |
| HBM | 2.84 | <.0001 | −2.85 | <.0001 |
| HIST2H2AA3 | 1.77 | <.0001 | 1.98 | <.0001 |
| HIST2H2AA4 | 1.72 | <.0001 | 2.09 | <.0001 |
| HLA-A29.1 | 16.26 | <.0001 | −3.05 | <.0001 |
| HLA-DQA1 | 1.61 | .0001 | 1.50 | .0003 |
| HP | 1.58 | .0001 | 2.55 | <.0001 |
| HS.572649 | −1.71 | <.0001 | −1.62 | <.0001 |
| IFIT1L | 1.92 | <.0001 | −3.03 | <.0001 |
| IL1R2 | 2.12 | <.0001 | 2.53 | <.0001 |
| IL1RN | 1.87 | <.0001 | 1.97 | <.0001 |
| IL8RB | 1.69 | <.0001 | 1.51 | .0004 |
| ITGA2B | 2.04 | <.0001 | 2.36 | <.0001 |
| ITGB3 | 1.63 | .0001 | 2.12 | <.0001 |
| KRT1 | 2.32 | <.0001 | −2.18 | <.0001 |
| LOC100131164 | 2.43 | <.0001 | −2.05 | <.0001 |
| LOC100133923 | −1.55 | <.0001 | −1.90 | <.0001 |
| LOC100190986 | −1.52 | .0001 | −2.01 | <.0001 |
| LOC389599 | 1.79 | <.0001 | −1.72 | .0001 |
| LOC440313 | 2.09 | <.0001 | −1.62 | .0001 |
| LOC642103 | 1.57 | .0002 | 1.58 | .0001 |
| LOC643332 | 1.68 | .0002 | 1.90 | <.0001 |
| LOC651309 | −1.65 | <.0001 | −1.56 | <.0001 |
| LOC651524 | 2.04 | <.0001 | 2.47 | <.0001 |
| LOC653600 | 1.61 | <.0001 | 3.17 | <.0001 |
| LOC653610 | 2.12 | <.0001 | 2.21 | <.0001 |
| LOC728499 | −1.66 | <.0001 | −2.43 | <.0001 |
| LOC728715 | 1.55 | .0002 | 1.71 | .0001 |
| LOC731682 | 1.93 | <.0001 | 1.78 | <.0001 |
| LRG1 | 1.54 | .0001 | 2.39 | <.0001 |
| LY6G6F | 1.66 | .0001 | 1.74 | <.0001 |
| MAP1A | 1.59 | .0002 | 1.83 | <.0001 |
| MIAT | −1.60 | <.0001 | −2.30 | <.0001 |
| MMP9 | 2.39 | <.0001 | 1.97 | <.0001 |
| MYL4 | 1.52 | .0001 | −1.74 | <.0001 |
| MYL9 | 2.14 | <.0001 | 2.60 | <.0001 |
| NAMPT | 1.87 | <.0001 | 1.68 | <.0001 |
| NFKBID | 1.66 | .0001 | 1.70 | <.0001 |
| NP | 1.80 | .0001 | 1.77 | <.0001 |
| ORM1 | 1.91 | .0001 | 1.95 | <.0001 |
| PGLYRP1 | 1.92 | <.0001 | 3.29 | <.0001 |
| PI3 | 2.30 | <.0001 | 2.41 | <.0001 |
| PLAU | 2.52 | <.0001 | 2.45 | <.0001 |
| PLAUR | 1.84 | .0002 | 1.71 | <.0001 |
| PROK2 | 1.98 | <.0001 | 1.98 | <.0001 |
| PROS1 | 1.50 | .0003 | 1.57 | <.0001 |
| PVALB | 1.91 | <.0001 | 2.46 | <.0001 |
| RAB20 | 1.90 | <.0001 | 1.93 | <.0001 |
| RETN | 1.72 | <.0001 | 2.97 | <.0001 |
| S100P | 1.90 | <.0001 | 2.59 | <.0001 |
| SDC4 | 1.58 | .0002 | 1.88 | .0001 |
| SELENBP1 | 1.97 | <.0001 | −2.19 | <.0001 |
| SERPINB2 | 1.96 | .0003 | 4.82 | <.0001 |
| SERTAD1 | 1.68 | .0001 | 1.68 | <.0001 |
| SIGLEC16 | 1.58 | .0003 | 1.64 | .0002 |
| SLC4A1 | 2.39 | <.0001 | −2.62 | <.0001 |
| SLPI | 2.04 | <.0001 | 3.72 | <.0001 |
| SNCA | 2.06 | <.0001 | −1.54 | .0003 |
| SOD2 | 1.79 | .0001 | 1.82 | <.0001 |
| STMN3 | −1.63 | <.0001 | −1.51 | .0003 |
| STRADB | 1.62 | .0001 | −1.92 | <.0001 |
| SV2A | 1.68 | .0001 | 1.65 | .0005 |
| TCN2 | 2.05 | <.0001 | 1.86 | <.0001 |
| TGM3 | 1.92 | <.0001 | 1.64 | .0005 |
| THBD | 1.78 | .0001 | 2.83 | <.0001 |
| TMEM158 | 1.57 | .0001 | 2.63 | <.0001 |
| TNFRSF12A | 1.94 | <.0001 | 1.94 | <.0001 |
| TNNI2 | 1.53 | .0001 | 1.72 | .0002 |
| TNS1 | 1.61 | <.0001 | −1.60 | .0001 |
| TP53INP2 | 1.70 | .0001 | 1.76 | <.0001 |
| TREM1 | 1.78 | .0002 | 1.88 | <.0001 |
| TREML1 | 1.64 | .0001 | 2.40 | <.0001 |
| WDR40A | 1.76 | <.0001 | −1.76 | <.0001 |
Abbreviations: ENL, erythema nodosum leprosum; RR, reversal reaction.
a Negative fold changes are in italic type.
A transcript uniquely increased in RR was CXCL10, which has previously been studied in association with RR [12]. Also increased in RR was transglutaminase 2 (TGM2), which is increased in autoimmune disease and may be related to antigen modification [21]. In the RR group, there were increased transcript levels of pattern recognition receptor C-type lectin domain family 7, member A (CLEC7A [dectin-1]), and a scavenger receptor, the macrophage receptor with collagenous structure (MARCO) [22]. There were decreased transcripts of B-cell-associated molecules CD79, CD19, and CD27 and T-cell signaling modulator CCR7. IPA biomarkers analysis identified chemokine (C-X-C motif) ligand 10 (CXCL10) and Fc fragment of IgE, low affinity II (FCER2) as transcripts with differential levels unique to RR.
Transcripts uniquely increased in ENL included the complement receptors C3AR1 and C5AR1 and 3 ribonucleases; RNASE1, RNASE2, and RNASE3. Uniquely decreased transcripts in ENL included interleukin 10 and cytotoxic t-lymphocyte associated protein 4 (CTLA4), modulators of T-cell responses. Decreased CTLA4 and interleukin 10 in ENL could either contribute to the inflammatory cascade observed clinically during ENL or reflect a relatively high level of the inhibitory protein in PBMCs of patients with lepromatous leprosy without ENL. IPA biomarkers analysis identified 20 genes as uniquely differentially expressed in ENL compared with RR including the chemokine ligands CCL3L3 and CXCL8.
Components of the innate immune response were increased in both RR and ENL, including C1q (C1QA, C1QB, and C1QC). Interestingly, both RR and ENL had increased expression of hepcidin (HAMP) and cathelicidin (CAMP) antimicrobial peptides. Defensins were increased in ENL (DEFA1, DEFA1B, DEFA3, and DEFA4) and in RR (DEFA4). Fc receptor-like 3 and Fc receptor-like A were both decreased in RR and ENL groups. IPA biomarkers analysis identified 12 transcripts which could be potential biomarkers for RR or ENL, including CCL2, CCL3, and SOD2. Transcripts increased in PBMCs from both RR and ENL also included FcγR1 (CD64), FPR1 and FPR2, and triggering receptor on myeloid cells 1 (TREM1) and the related molecule triggering receptor expressed on myeloid cells-like 1 (TREML1). FcγR1 recognizes immunoglobulin G; FPR1 and FPR2 recognize formylated peptides produced by bacteria and some mycobacteria [23].
Increased Monocyte-related Transcripts During RR and ENL
Changes in transcripts could reflect either a change in gene expression within PBMCs or a change in circulating cellular composition. To assess whether monocytosis was a potential contributor to differential expression, we quantified the monocyte population in PBMCs by gating and staining for CD14 (Supplementary Figure 1A) in patients with leprosy with RR (n = 7), ENL (n = 8), or leprosy without reaction (n = 16). Monocytes comprised 13.79%, 19.01%, or 15.77% of PBMCs in RR, ENL, or no reaction, respectively (P = .45) (Supplementary Figure 1B). Furthermore, the majority of gated monocytes were CD14+ in all groups (mean, 92.78%, 87.76%, or 92.28% in RR, ENL, or no reaction, respectively; P = .54) (Supplementary Figure 1C). Side scatter/forward scatter and CD14+ measures of monocytes do not confirm a significant difference in the proportion of circulating monocytes between reaction and nonreaction PBMCs.
Confirmation of Differential Gene Expression: Complement Components
Given the current theory of augmented Th1 response to antigen as the etiology of RR, findings of innate immune response as a top GO pathway and complement and coagulation cascade as a top KEGG pathway were interesting and suggestive of potential overlap in immunologic response during RR and ENL. The blue-pink o'gram representation of the array heat map for the complement pathway components of the array is shown in Supplementary Figure 2. Several components of the classic complement pathway had increased expression in PBMCs from persons with RR or ENL (Table 9). C1qA, B, and C, complement component 2 (C2); and C1 esterase inhibitor (SERPING1) were increased in RR. C1qA, B, and C and the complement receptors C3AR1 and C5AR1 were increased in ENL. Changes in C1QB, C2, C1 esterase inhibitor, and C5AR1 expression in PBMCs were validated with qPCR (Table 9).
Table 9.
Selected Complement Components and Monocyte Receptor Transcript Expression in RR and ENL, as Measured With Microarray and qPCR Validation of a Subset of Transcripts
| Complement Components and Monocyte Receptors | RR |
ENL |
||
|---|---|---|---|---|
| Fold Increase in Array (P Value) | Fold Increase in qPCR (P Value) | Fold Increase in Array (P Value) | Fold Increase in qPCR (P Value) | |
| Complement pathway component | ||||
| C1Q (subunit A) | 1.64 (.0004) | NA | 1.82 (<.0001) | NA |
| C1Q (subunit B) | 2.21 (<.0001) | 1.72 (.005) | 3.27 (<.0001) | 2.63 (.07) |
| C1Q (subunit C) | 1.77 (<.0001) | NA | 2.61 (<.0001) | 3.45 (.03) |
| C2 | 2.31 (<.0001) | 1.94 (.01) | Not significant | NA |
| C1 esterase inhibitor | 2.17 (<.0001) | 2.18 (.001) | Not significant | NA |
| C5AR1 | Not significant | NA | 1.83 (<.0001) | 1.60 (.005) |
| Monocyte receptor | ||||
| FcγRI (CD64) | 2.78 (<.0001) | 2.22 (.0002) | 2.55 (<.0001) | 2.39 (.005) |
| FPR-1 | 1.80 (.0001) | 1.81 (.0002) | 2.10 (<.0001) | 1.84 (.006) |
| FPR-2 | 2.3 (<.0001) | 2.05 (<.0001) | 2.15 (<.0001) | 1.87 (.01) |
| MARCO | 1.77 (<.0001) | 1.91 (.001) | Not significant | NA |
| CLEC7A or dectin-1 | 1.71 (.0002) | 1.67 (.004) | Not significant | NA |
Abbreviations: CLEC7A, C-type lectin domain family 7, member A; ENL, erythema nodosum leprosum; FcγRI, Fc fragment of IgG receptor-1; FPR, percentage false-positive; MARCO, macrophage receptor with collagenous structure; NA, not available; qPCR, quantitative polymerase chain reaction; RR, reversal reaction.
The skin is a primary site of RR and ENL signs and symptoms, so we hypothesized that increased expression of C1q in PBMCs during RR or ENL could reflect increased deposition of C1q in skin. Skin biopsy specimens from patients with leprosy obtained as part of diagnostic workup for RR (n = 3), ENL (n = 3), and borderline leprosy without reaction (n = 7 BT and n = 3 BL/LL) were studied with immunohistochemistry. The fluorescent intensity of C1q staining was significantly higher in both RR and ENL compared with nonreaction leprosy (P ≤ .01) (Figure 2), indicating increased deposition of C1q in reactional compared with nonreactional leprosy skin lesions.
Figure 2.
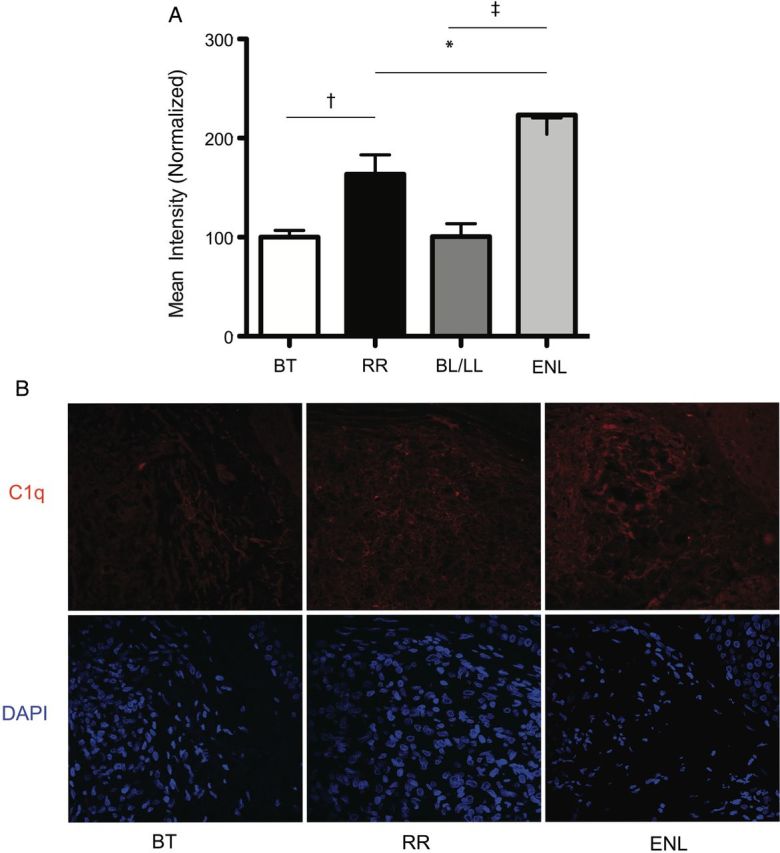
Intensity of fluorescent staining for C1q (A) with representative photographs (B) in skin lesions of reversal reaction (RR; n = 3), erythema nodosum leprosum (ENL; n = 3), borderline tuberculoid (BT) controls (n = 7), and borderline lepromatous (BL)/ lepromatous leprosy (LL) (n = 3) controls. Differences in groups were determined with analysis of variance and Tukey multiple-comparison test. *P ≤ .05; †P ≤ .01; ‡P ≤ .001. Error bars denote standard error of the mean.
DISCUSSION
Analysis of RR and ENL transcriptomes demonstrates distinct pathways associated with the immune response. No validated biomarker or effective prophylaxis is currently available for RR and ENL [7, 8, 12, 24–26], and gene expression studies are a crucial exploratory method to better understand reaction pathogenesis. Some findings support previously published results on leprosy immunology. Of particular interest was the increased expression of CXCL10 in the RR group, and its identification as a potential RR biomarker by IPA biomarkers analysis, as CXCL10 has previously been proposed as a biomarker for RR [12]. The nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway (KEGG) was associated with ENL, an interesting finding given the association of NOD2 with leprosy in a genome wide association study [27]. IFN-γ was identified as an upstream regulator of differential transcription in PBMCs of RR and ENL, which may be a stimulus for immune cascade of reactions.
Pathway analysis showed a significant involvement of the innate immune system with RR and ENL. HAMP and CAMP antimicrobial peptide transcripts were increased in RR and ENL. CAMP expression is increased by a Toll-like receptor 1/2–mediated process, and Toll-like receptor 2 polymorphisms have been associated with increased risk of RR [28]. The complement and coagulation pathway was an unexpected pathway to find enriched in the RR group, given the Th1 augmentation theory of RR pathology, but our results support a potential role in RR pathogenesis. We demonstrated increased deposition of C1q in skin lesions of patients with leprosy with RR or ENL, suggesting that complement is involved in both reaction types systemically and in skin lesions. Interestingly, we did not find a significantly different level of C1q deposition in tuberculoid and lepromatous leprosy lesions, despite the different bacterial burden and immune cell composition in the 2 leprosy poles. That BT and BL/LL controls without immune reaction have similar degree of C1q deposition in tissues supports association of complement deposition with reactions rather than leprosy per se. The increased complement deposition during reactions, compared with baseline pathology due to leprosy, suggests that complement deposition is part of the immune response in RR and ENL.
The frequent association of reactions with recent initiation of anti–M. leprae therapy parallels the pathologic immune activation that can occur when treatment is started for other infectious diseases, such as human immunodeficiency virus and Mycobacterium tuberculosis infection. Patients with tuberculosis who go on to develop immune reconstitution inflammatory syndrome (IRIS) have increased C1q expression at initiation and after 2 weeks of antiretroviral treatment [29, 30]. Patients with tuberculosis and IRIS have increased C1 inhibitor at baseline without further increase in C1 inhibitor 2 weeks after starting antiretroviral therapy, as was seen in controls. Tran et al [30] hypothesized that mycobacterial antigen load may drive the complement activation they observed during tuberculosis with IRIS. PBMCs from patients with leprosy with a history of RR have, in response to stimulation with M. leprae antigen, an increased expression of genes associated with monocyte recruitment and the innate immune response [8]. “Antigen processing and presentation” KEGG pathways were associated with RR and ENL in this microarray analysis, which supports contribution of antigen to RR and ENL pathogenesis. HLA associations have been made with leprosy [31, 32], although the contribution of HLA expression to antigen presentation and immune response during RR and ENL needs to be described. Our findings of transcriptional differences of HLA genes could inform future studies of HLA types and risk of immune reactions.
A major strength of this study is the description of PBMC gene expression from persons with active, untreated immune reactions compared with controls matched for age, sex, and clinical form of leprosy. Matching for stage of leprosy treatment in the array was a control for effects of antileprosy therapy and stage of leprosy disease on gene expression. Furthermore, samples were collected and processed before initiation of immunomodulatory therapies. We were also able to investigate correlation of gene expression differences of C1q in PBMCs to presence in skin during reactions. We minimized potential type 2 errors associated with large-scale array comparisons by applying stringent criteria (FPR, ≤0.05; P ≤ .05; fold change ≥1.5 or ≤−1.5) for transcripts used for functional and biologic pathway analyses. Study limitations include analysis of gene expression in PBMCs rather than whole blood, because conclusions are relevant for the isolated monocyte and lymphocyte populations. The PBMC and biopsy specimens used were not from the same patients, which needs to be considered when assessing experimental conclusions.
We hypothesize that both RR and ENL may have disordered recognition responses to M. leprae antigen with increased production of antibodies or heightened responsiveness to antibodies, with pathology mediated by complement and other components of innate immunity. Differences in the clinical presentation of RR and ENL may be related to several factors, including M. leprae burden and the balance between humoral and cell-mediated immune responses to M. leprae and other antigens. Genetic factors have also been proposed as risk factors for immune reactions [28]. Further investigations of the involvement of innate immunity, including complement and antimicrobial peptides, and IFN-γ–mediated processes are indicated to fully elucidate the pathophysiology of leprosy immune reactions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We acknowledge the patient volunteers and care teams at Giselda Trigueiro Hospital, Onofre Lopes Hospital, and Mossoró Hansen's disease clinics in Rio Grande do Norte, Brazil. We thank our colleagues from the Immunogenetics laboratory at Universidade Federal do Rio Grande do Norte, the Centro de Patologia Getúlio Sales, and the Confocal Microscopy core facility at the Brain Institute in Natal, Rio Grande do Norte, Brazil, and the DNA and Bioinformatics core facilities at the University of Iowa in Iowa City for their collaboration.
Financial support. This work was supported by the National Institutes of Health (grants T32-AI007613 and R25-TW009337-02 to K. M. D.), Burroughs Wellcome Fund and the American Society of Tropical Medicine and Hygiene (Postdoctoral Fellowship in Tropical Infectious Diseases to K. M. D.), and the Brazil National Institute of Science and Technology (tropical diseases grant CNPq 573839/2008-15).
Potential conflicts of interest. The authors have no conflicts of interest to declare. The funding agencies had no role in the study design or execution. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Global leprosy: update on the 2012 situation. Wkly Epidemiol Rec 2013; 88:365–79. [PubMed] [Google Scholar]
- 2.Ridley DS, Jopling WH. Classification of leprosy according to immunity: a five-group system. Int J Lepr Other Mycobact Dis 1966; 34:255–73. [PubMed] [Google Scholar]
- 3.Britton WJ, Lockwood DN. Leprosy. Lancet 2004; 363:1209–19. [DOI] [PubMed] [Google Scholar]
- 4.Walker SL, Lockwood DN. Leprosy type 1 (reversal) reactions and their management. Lepr Rev 2008; 79:372–86. [PubMed] [Google Scholar]
- 5.Lockwood D, Colston M, Khanolkar-Young S. The detection of Mycobacterium leprae protein and carbohydrate antigens in skin and nerve from leprosy patients with type 1 (reversal) reactions. Am J Trop Med Hyg 2002; 66:409–15. [DOI] [PubMed] [Google Scholar]
- 6.Ranque B, Nguyen VT, Vu HT, et al. Age is an important risk factor for onset and sequelae of reversal reactions in Vietnamese patients with leprosy. Clin Infect Dis 2007; 44:33–40. [DOI] [PubMed] [Google Scholar]
- 7.Lockwood DN, Suneetha L, Sagili KD, et al. Cytokine and protein markers of leprosy reactions in skin and nerves: baseline results for the North Indian INFIR cohort. PLoS Negl Trop Dis 2011; 5:e1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlova M, Cobat A, Huong NT, et al. Gene set signature of reversal reaction type I in leprosy patients. PLoS Genet 2013; 9:e1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahawita IP, Lockwood DN. Towards understanding the pathology of erythema nodosum leprosum. Trans R Soc Trop Med Hyg 2008; 102:329–37. [DOI] [PubMed] [Google Scholar]
- 10.Pocaterra L, Jain S, Reddy R, et al. Clinical course of erythema nodosum leprosum: an 11-year cohort study in Hyderabad, India. Am J Trop Med Hyg 2006; 74:868–79. [PubMed] [Google Scholar]
- 11.Lustosa AA, Nogueira LT, Pedrosa JI, Teles JB, Campelo V. The impact of leprosy on health-related quality of life. Rev Soc Bras Med Trop 2011; 44:621–6. [DOI] [PubMed] [Google Scholar]
- 12.Scollard DM, Chaduvula MV, Martinez A, et al. Increased CXC ligand 10 levels and gene expression in type 1 leprosy reactions. Clin Vaccine Immunol 2011; 18:947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa AL, Fava VM, Sampaio LH, et al. Genetic and immunological evidence implicates interleukin 6 as a susceptibility gene for leprosy type 2 reaction. J Infect Dis 2012; 205:1417–24. [DOI] [PubMed] [Google Scholar]
- 14.Bleharski JR, Li H, Meinken C, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 2003; 301:1527–30. [DOI] [PubMed] [Google Scholar]
- 15.Teles RM, Graeber TG, Krutzik SR, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 2013; 339:1448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ministério da Saúde. Define ações de controle da hanseníase. Vol. Portaria Conjunta No. 125. 26 March 2009. http://bvsms.saude.gov.br/bvs/saudelegis/svs/2009/poc0125_26_03_2009.html. Accessed 29 November 2014. [Google Scholar]
- 17.WHO Expert Committee on Leprosy: Eighth report . In: World Health Organization, ed. WHO Technical Report Series Geneva, Switzerland: World Health Organization, 2012. [PubMed] [Google Scholar]
- 18.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 2004; 573:83–92. [DOI] [PubMed] [Google Scholar]
- 19.Jeffery IB, Higgins DG, Culhane AC. Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinformatics 2006; 7:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013; 41:W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sollid LM, Jabri B. Celiac disease and transglutaminase 2: a model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Curr Opin Immunol 2011; 23:732–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraal G, van der Laan LJ, Elomaa O, Tryggvason K. The macrophage receptor MARCO. Microbes Infect 2000; 2:313–6. [DOI] [PubMed] [Google Scholar]
- 23.Chun T, Serbina NV, Nolt D, et al. Induction of M3-restricted cytotoxic T lymphocyte responses by N-formylated peptides derived from Mycobacterium tuberculosis. J Exp Med 2001; 193:1213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balagon M, Saunderson PR, Gelber RH. Does clofazimine prevent erythema nodosum leprosum (ENL) in leprosy? a retrospective study, comparing the experience of multibacillary patients receiving either 12 or 24 months WHO-MDT. Lepr Rev 2011; 82:213–21. [PubMed] [Google Scholar]
- 25.Iyer A, Hatta M, Usman R, et al. Serum levels of interferon-gamma, tumour necrosis factor-alpha, soluble interleukin-6R and soluble cell activation markers for monitoring response to treatment of leprosy reactions. Clin Exp Immunol 2007; 150:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith W, Anderson A, Withington S, et al. Steroid prophylaxis for prevention of nerve function impairment in leprosy: randomised placebo controlled trial (TRIPOD 1). BMJ 2004; 328:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med 2009; 361:2609–18. [DOI] [PubMed] [Google Scholar]
- 28.Bochud PY, Hawn TR, Siddiqui MR, et al. Toll-like receptor 2 (TLR2) polymorphisms are associated with reversal reaction in leprosy. J Infect Dis 2008; 197:253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran HT, Van den Bergh R, Loembe MM, et al. Modulation of the complement system in monocytes contributes to tuberculosis-associated immune reconstitution inflammatory syndrome. AIDS 2013; 27:1725–34. [DOI] [PubMed] [Google Scholar]
- 30.Tran HT, Van den Bergh R, Vu TN, et al. The role of monocytes in the development of Tuberculosis-associated Immune Reconstitution Inflammatory Syndrome. Immunobiology 2014; 219:37–44. [DOI] [PubMed] [Google Scholar]
- 31.da Silva SA, Mazini PS, Reis PG, et al. HLA-DR and HLA-DQ alleles in patients from the south of Brazil: markers for leprosy susceptibility and resistance. BMC Infect Dis 2009; 9:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw MA, Donaldson IJ, Collins A, et al. Association and linkage of leprosy phenotypes with HLA class II and tumour necrosis factor genes. Genes Immun 2001; 2:196–204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.