Abstract
One of the great strengths of the mouse model is the wide array of genetic tools that have been developed. Striking examples include methods for directed modification of the genome, and for regulated expression or inactivation of genes. Within neuroscience, it is now routine to express reporter genes, neuronal activity indicators and opsins in specific neuronal types in the mouse. However, there are considerable anatomical, physiological, cognitive and behavioral differences between the mouse and the human that, in some areas of inquiry, limit the degree to which insights derived from the mouse can be applied to understanding human neurobiology. Several recent advances have now brought into reach the goal of applying these tools to understanding the primate brain. Here we describe these advances, consider their potential to advance our understanding of the human brain and brain disorders, discuss bioethical considerations, and describe what will be needed to move forward.
Introduction
Science lacks a full understanding of how the brain works in health and how it fails in disease. As a consequence, medical researchers do not have a well-defined long-term strategy for the development of new and effective treatments for mental disorders. The size of the problem cannot be overstated. The cost of neurological diseases to society is enormous. Dementias alone, for example, cost more than heart disease and cancer, exceeding $160 billion in the United States alone (Hurd et al., 2013), equivalent to $500 per US citizen per year. The toll in human suffering is immense, both to the patients and to their families. Progress on treatment for psychiatric conditions, such as schizophrenia, is comparably stalled. Schizophrenia is a life sentence and at best current drug therapy is palliative, with severe side effects. The etiology of autism, though intensively explored, remains frustratingly baffling, and neither amelioration of symptoms nor a cure seems imminent. For autism too, human misery takes a truly staggering toll. We now know of 600 diseases of the nervous system, with a high likelihood that each of us will suffer from one of them in our lifetime. At this stage, there is no effective treatment, and little if anything to assist with prevention. With increases in the size of the aging population, the human and economic costs will certainly increase in step, possibly to crushing proportions.
One of the major obstacles to progress in understanding and developing treatments for these diseases is the relatively limited set of genetic tools currently available to systematically study and test relevant neural circuits in primates, the mammalian order of which we are members. Rodent models play an essential role in neurobiology, where a powerful array of modern genetic tools has been successfully applied. Striking examples include methods for targeted inactivation of endogenous genes and for regulated expression of transgenes, yielding cell-type-specific expression of opsins, fluorescent markers, and neuronal activity indicators. These tools have enabled major advances in neurobiology, and they will continue to be used to great effect in rodents. There are, however, considerable anatomical, physiological and behavioral differences between the rodent and the human. This means that for many disorders, especially those involving high-level cognitive functions, studies of rodents may not reveal the mechanisms at work in the human brain. The development of primate models for human diseases also addresses a major concern articulated in 2011 by a British independent panel chaired by Sir Patrick Bateson (“the Bateson report”), which is that while much non-human primate work is of high quality, its impact on our understanding of human disease and its treatment has been limited (Bateson et al., 2011, http://www.wellcome.ac.uk/stellent/groups/corporatesite/@policy_communications/documents/web_document/wtvm052279.pdf). Arguably this limitation arises in part because the lack of genetic tools for cell-type specific targeting of protein expression has limited our understanding of neural circuits in the primate brain. Without these tools, primate models of genetically based diseases cannot be created and studied. Equally important, the lack of tools to cause cell-type-specific expression of proteins such as opsins and genetically encoded neuronal activity indicators severely limits basic scientific understanding of the primate brain.
Concern over these critical limitations led to a recent symposium at the Salk Institute for Biological Studies, in which world leaders in multiple disciplines met to consider how to bring modern genetic tools to bear directly on understanding the primate brain. The purpose of this Perspective is to describe the findings of this symposium and to motivate its conclusion that the goal of developing genetically modified primates for use in studying the primate brain is both necessary and within reach. Advances in methods of gene editing and stem cell technology, coupled with successes in germline transmission of transgenes in the common marmoset (Callithrix jacchus), position researchers to make critical advances in our fundamental scientific understanding of the primate brain. At the same time, we acknowledge and discuss the ethical considerations of engaging in work with transgenic non-human primates.
This new line of research promises to significantly accelerate progress in understanding the fundamental organizing principles of the primate brain and the etiology of human neurological and psychiatric disorders, progress on which so many victims and their families have pinned their hopes.
1. The need for non-human primates as a model for studying the human brain
Rodents serve as important animal models in many domains of biomedical research. Within neuroscience, powerful genetic tools are being used to probe the functions of different components of the murine brain. This work is highly relevant to understanding the workings of the human brain because mouse and human brains share many of the same circuit components and there are important similarities in the ways these components are wired together. Further, the social, cognitive, and perceptual abilities of rodents are more impressive than at first assumed, which has enabled researchers to study neural mechanisms underlying some of these functions in the behaving mouse. As we come to understand these mechanisms in mice, it is likely that this will enhance our understanding of the human brain, shedding light on its disorders.
These advantages notwithstanding, rodents do differ in important ways from humans. Brain circuitry, cognitive capacities and behavioral repertoires have evolved over the 83 million years that have passed since the rodent and primate lineages separated (Meredith et al., 2011). Over this time, natural selection has endowed primates with specialized brain structures that give rise to our particular motor, perceptual and cognitive capacities (Kaas, 2013). These specializations include prominent expansion of the frontal cortex, parts of which are implicated in psychiatric disorders and have no homolog in other mammals (Wise, 2008).
To take a simple concrete example, humans and non-human primates differ from rodents in how they explore the visual environment. The primate oculomotor system serves to move the eyes to align the high-resolution fovea with objects of interest in a scene. The fovea has a huge impact on the way visual information is processed, not simply because it yields higher acuity, but because it changes in a fundamental way how primates use their eyes to acquire information about their world. Evolution has endowed primates with efficient strategies for moving their eyes so the fovea is rapidly positioned over targets of interest. Rapid eye movements (saccades), are made 2 to 3 times every second as the brain samples the visual scene, and in a remarkable computational feat, these signals are smoothly integrated across time so that it looks to the observer as though a wide visual field is seen crisply during a period of viewing. Primates also have stereoscopic vision across the majority of the visual field, and the computational capacity to construct a three-dimensional representation of the visual world. They possess the ability to smoothly track objects moving through that world, a capacity that is associated with special cortical circuits for motion analysis (MT and MST) and oculomotor control (FPA) that appear to be unique to primates. Although rodents can and do move their eyes, so far as is known, they do not track objects or integrate information across eye movements in the way primates do. Rather, they seem mostly to use eye movements as part of the optokinetic (OKN) and vestibulo-ocular reflexes (Faulstich et al., 2004) and to maximize the overhead binocular visual field (Wallace et al., 2013).
Moving toward more cognitive domains, consider attention, the cognitive process by which we concentrate on one aspect of the environment while ignoring others. This process is severely impaired in multiple brain disorders including autism, schizophrenia and Alzheimer’s’ Disease. Many animals, including chickens (Sridharan et al., 2014; Ben-Tov et al., 2014) exhibit forms of attentional selection, and some of the mechanisms that play a role in attention in primates are shared with rodents. For example, neurophysiological studies in non-human primates have found that when attention is directed toward a visual stimulus, this increases the gain of neurons responsive to the attended stimulus (McAdams and Maunsell, 1999) while suppressing the activity of neurons selective for nearby unattended stimuli (Desimore and Duncan, 1995), via activation of inhibitory interneurons (Mitchell et al., 2007; Sundberg et al., 2009). Mouse studies have contributed to our understanding of the mechanisms underlying these forms of attentional modulation. Mouse neocortical neurons exhibit increases in gain when mice are actively engaged in locomotion (Niell and Stryker, 2010), and studies have begun to elucidate the cellular mechanisms underlying gain control in mouse visual cortex (Polack et al., 2013), including the roles of different classes of interneurons in gain modulation and sensory discrimination (Otte et al., 2010; Atallah et al., 2011; Fu et al., 2014; Lee et al., 2012). Thus, many of the neural substrates of attention may be shared between rodents and primates.
Critical differences, however, do exist. In humans and non-human primates the allocation of attention is determined by a fronto-parietal control network (Kastner and Ungerleider, 2000). This network includes parts of the oculomotor system, which not only aligns the fovea with objects of interest, but also provides spatially-selective feedback signals to extrastriate visual cortex that cause attention-dependent changes in gain (Moore et al., 2003; Moore and Fallah, 2004). In this respect, rodents are strikingly different. Although rodents can and do move their eyes, they lack a fovea. Consequently, they do not have the oculomotor infrastructure that serves to deploy spatial attention in primates.
Another example of primate-specific brain specializations is drawn from the domain of social cognition. Primates form intricate social systems involving hierarchies, kin attachments, friendships and other social relations (Chang et al., 2013; Brent et al., 2014). Many monkey species have rich vocal repertoires for communication, which depend on specialized higher-order cortical auditory processing regions (Eliades and Wang, 2013; Romanski and Averbeck, 2009). Non-human primates also have a highly refined capacity to recognize faces and to interpret facial gestures, mediated by a specialized network of brain areas devoted to face processing (Bruce et al., 1981; Tsao et al., 2006). At the core of primate social cognition is a conceptual understanding of our social and familial relationships, and the ability to use that information to form alliances, to strategically manipulate conspecifics, and to conform to a system of social norms (Cheney and Seyfarth, 2007; Rosati et al., 2010; Seyfarth and Cheney, 2014). Primates have demonstrated a sense of fairness in social behavior (Lakshiminarayanan and Santo, 2008; Hare et al., 2007, 2010). For example, only those chimpanzees helpful in hunting monkeys, will receive a share of the spoils (Boesch, 1994). While other species, such as vampire bats also exhibit a sense of fairness (Wilkinson, 1984) primate social cognition reflects a degree of sophistication not known to occur in other taxonomic groups. Primates also appear able to attribute mental states such as goals, perceptions, and feelings, to others. This capacity for mental state attribution, also referred to as ‘Theory of Mind’, is a pervasive feature in human social cognition. Our closest living relative, the chimpanzee, also appears to possess this ability (Hare et al., 2001, 2006), as do some other primate species (Flombaum and Santos, 2005). Rodents, so far as anyone can tell, do not demonstrate any capacity for attributing mental states, limiting their use as model organisms for understanding the failure of social cognition in neural disorders such as autism spectrum disorder.
Monkeys, like humans, also have enhanced tactile specializations that enable recognition and discrimination of objects based on shape and texture (Johnson and Hsaio, 1992). Monkeys have multiple representations of the body in the somatosensory cortex (Kaas, 1993). Cortical area 3b of the primary somatosensory cortex (S1) displays a disproportionally large representation of the hand and the face Jain et al., 2001). In the hand representation, multiple sub-regions encode the fingers, and discrete areas interspaced by septal regions respond to stimulation of individual digits (Krubitzer and Kaas, 1990). Monkeys also have a secondary somatosensory cortex (S2), organized in parallel to S1 (Zhang et al., 2001). These somatosensory areas interact with the visual system to enhance object recognition (Macaluso and Maravita, 2010).
Primates use their hands extensively to manipulate objects in their environments (Gaziano, 2006). To guide these more controlled movements, they have extensive direct connections from motor cortex to spinal motor neurons that can augment more primitive motor programs mediated by spinal interneurons and spinal reflex pathways. Indeed, the cortical neurons with direct input to motor neurons (cortico-motoneuronal (CM) cells) are located in a spatially separate part of the primary motor cortex that is not found in rodents or cats (Rathelot and Strick, 2006, 2009). This cortico-motor circuitry is used for dexterous manipulation, such as allowing relatively independent control of the digits. A simple example is precision grip, a unique primate behavior that has been used extensively to study the functional role of cortical-motorneuronal circuitry (Davare et al., 2011). Rodents have only a basic grasp, nothing like a precision grip using the thumb and index finger.
Finally, there is tool use. Though primates are not unique in using tools (Hunt, 1996), they are by far the most adept. In addition to the numerous examples of tool use for procuring food (Whiten et al., 1999), examples of tools used as weapons and for hunting have been reported (Pruetz and Bertolani, 2007). Even primate species that do not use tools in the wild readily use tools in captivity with little to no training (Yamazaki et al., 2011). This domain of physical cognition does not occur in isolation: it is affected by related cognitive processes, such as social learning (Hobaiter et al., 2014). Differences in tool use across populations of chimpanzees throughout Africa illustrate cultural traditions (Gruber et al., 2009). One key mechanism for social learning is imitation. Imitation in the use of objects has only been described in primates, ranging from humans and chimpanzees (Whiten, 1998) to common marmosets (Yamazaki et al., 2011). This complex form of social learning is a critical feature of human culture, but its underlying mechanisms may have existed in some form in the last common ancestor with extant primates. Its absence amongst other taxonomic groups suggests an important difference to primates that is likely the result of critical differences in brain architecture.
In each of these cases, key parts of our perceptual, cognitive, and behavioral repertoires depend on primate-specific brain specializations. We do not know where the key discoveries that will transform our understanding of the human brain will occur. Some may come from greater understanding of mechanisms we share with mice. Others may derive from insights into the primate brain itself. The non-human primate thus has a unique role to play as a model for studying the human brain. Thus, even as we continue to extend our understanding of the mouse brain, it is essential to proceed, in parallel, to develop the means to apply the best available tools to studying the non-human primate brain, as well.
2. Primate gene editing is now within reach
The ability to manipulate the mouse genome has transformed modern biology. Gene editing allows reliable spatial and cell type specific inactivation of endogenous genes and expression of exogenous genes, including fluorophores, calcium indicators and opsins. As a result, it is possible to trace connections, manipulate and monitor neural activity, or modulate gene expression, with cell-type specificity. These techniques are only now becoming available in primates. Localized gene editing has been routinely performed in primates via local injection of viral vectors. For example, promoter-based strategies, coupled with viral tropism, have enabled selective expression of opsins in pyramidal neurons (Han et al., 2009), and viral approaches have also been used to achieve pathway selective blocking of neural transmission (Kinoshita et al., 2012). Notwithstanding these successes, these procedures have not provided a generally applicable means of achieving cell-type-specific transgene expression (Luo et al., 2008).
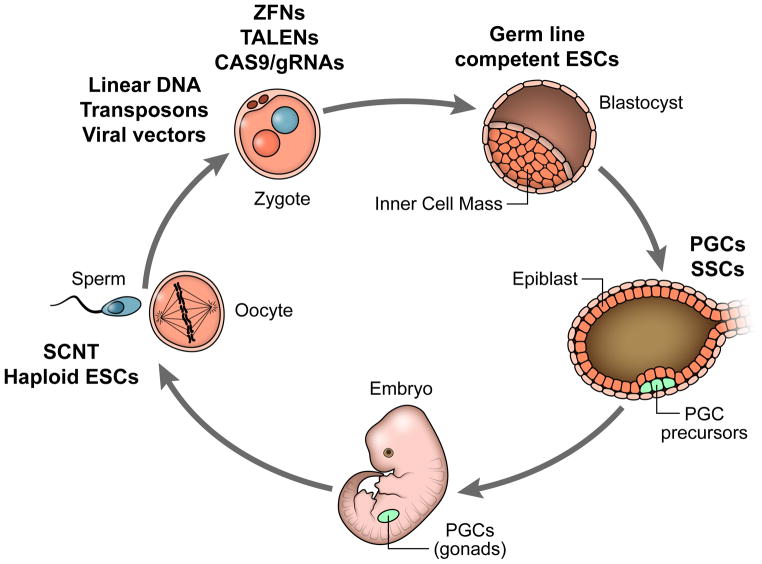
However, several key advances have occurred, including the development of new genome editing tools (ZFNs, TALENs, the CRISPR/Cas9 system), demonstration of germline transmission of a transgene in the common marmoset (Sasaki et al., 2009), and the complete sequencing of the marmoset genome (Marmoset Genome Sequencing and Analysis Consortium, 2014). As a result, the goal of adapting mouse transgenic techniques to non-human primates, though long an unreachable aspiration, has now become feasible. Here we review the current status of technologies for producing genetically modified non-human primate models. We begin with approaches that have already been proven to work, including direct genome editing of early pre-implantation embryos, which has successfully been used to yield both germline transmission of transgenes and the creation of several gene knockout lines. We next describe techniques that are on the near horizon, such as the development of primate embryonic stem cell approaches, which hold the promise of cell-type specific expression of transgenes. Finally, we describe techniques that, while further off, hold the potential to accelerate the pace of development of lines in future (See Figure 2).
Figure 2.
Approaches to primate transgenesis. Different methods now in use or having the potential to be used in the creation of genetically modified non-human primates, indicated in bold, with each method placed within the reproductive cycle. See text for details.
Direct genome editing in pre-implantation embryos
In the mouse, several transgenic technologies can be directly applied to early embryos, with the aim of creating lines of mice in which an endogenous gene is knocked out or a transgene is expressed. These include direct delivery of linear DNA (Palmiter et al., 1982), DNA transposon vectors such as Sleeping Beauty and PiggyBac (Ding et al., 2005; Ivics et al., 1997) or viral based vectors into the cytoplasm of unfertilized oocytes and zygotes or targeted to the pronuclei of zygotes (Niu et al., 2010). As virus-mediated gene transfer in rodent germlines has long been possible, it is natural that the first efforts toward primate transgenesis used this technology. The first transgenic macaques were produced more than a decade ago, with the founder animals showing the expression of transgenes that had been inserted into the embryo (Chan et al., 2001; Wolfgang et al., 2001; Yang et al., 2008). However, germline transmission of these transgenes – a necessary step in the creation of a transgenic line – was not demonstrated in a primate until 2009, when Sasaki and colleagues injected lentiviruses expressing EGFP into early stage pre-implantation marmoset embryos produced either by in vitro fertilization (IVF) or natural mating (Sasaki et al., 2009). Demonstration of germline transmission was an essential step forward in the aim of achieving genetic non-human primate models of human neurological disorders such as schizophrenia, Alzheimer’s disease, and Parkinson’s disease (Okano et al., 2012). Some inherent limitations to virally mediated gene transfer need to be overcome. Most notably, the integration of transgenes is random, so there is no control over the site of integration in the genome. As well, the size of the gene insert is limited to the size of the native HIV genome (< 9 kb), with a large penalty in virus titer and efficiency of infection of the embryonic cells and integration of the transgene for larger gene inserts.
In recent years, gene editing has become much more precise through use of programmable nucleases (Doudna and Charpentier, 2014). These include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats (CRISPR) with RNA-guided nucleases, such as Cas9. Each of these systems uses principles of site-specific DNA-protein or DNA-RNA recognition to target nucleases to specific genomic loci for generating double strand breaks in the DNA. Based on sequences of unique base pairs associated with specific genes or control regions, these systems exploit the cell’s natural capacity to repair double strand breaks (DSBs) and are able to achieve genome modification through non-homologous end joining (NHEJ) and homology-directed repair (HDR) (Critchlow and Jackson, 1998). Error-prone NHEJ has been used to successfully produce gene knockouts in the mouse, rat, pig, macaques and marmosets (Geurts et al., 2009; Hai et al., 2014; Hauschild et al., 2011; Sung et al., 2013; Wang et al., 2013; Ferguson et al., 2013; Niu et al., 2014; Liu et al., 2014; Kishi et al., 2014). While HDR offers an attractive way to introduce targeted gene mutations or whole gene insertions the efficiency of which is low, since HDR is dependent on exogenous repair templates with sequence homology to a specific site. Thus, while the CRISPR/Cas9 system is able to create knockins and conditional mutant rodents, its efficacy varies depending on the constructs (Yang et al., 2013).
There are two significant challenges associated with using programmable nucleases for creating transgenic lines. The first is with regard to fidelity: while programmable nucleases generally cleave target sites reliably, they can also cause off-target mutations and unwanted chromosomal translocations resulting from off-target DNA cleavage. This has been found to be the case with the CRISPR/Cas9 system (Cradick et al., 2013; Fu et al., 2013; Hsu et al., 2013). Off-target effects are less common with TALENs, though their on-target efficiency is generally lower. Several strategies have been developed to restrict editing to the target site (Hsu et al., 2013). Another issue associated with nuclease-based gene editing in early embryos is mosaicism, in which two or more populations of cells with different genomes are present in an individual animal. This can be caused by delayed and/or multiple DNA cleavage by injected nucleases during embryogenesis (Sung et al., 2013; Niu et al., 2014; Li et al., 2013; Tesson et al., 2011; Yen et al., 2014).
Creation of germline competent non-human primate embryonic stem (ES) cells
Primate embryonic stem cell technologies hold the promise of overcoming limitations of direct genome editing in pre-implantation embryos, because one can use low efficiency gene insertion techniques, followed by selection of genetically modified ES cells via co-insertion of an antibiotic resistance gene, to isolate ES cells that have been appropriately modified. In mouse, sustainable ES lines provide researchers with the capacity to systematically investigate and characterize genomic loci following recombination. For example, it is possible to examine the number of off-target genome integrations and the trade-offs associated with inserting larger gene cassettes. In addition, it is possible to directly investigate the effects of gene editing on cellular physiology and behavior, both before and after differentiation of the cells into the desired cell type. Furthermore, mouse ES cells can be maintained in their naïve state, capable of fully integrating into mouse embryos that develop into chimeric transgenic offspring. Gene targeting in mouse ES cells has thus become a mainstay to establish genetically modified mouse strains.
In primates, ES cells have been derived from several non-human primate species, including rhesus macaques (Mitalipov et al., 2006; Thomson et al., 1995), cynomolgus macaque (Suemori et al., 2001), baboons (Simerly et al., 2009) and the common marmoset (Thomson et al., 1996; Sasaki et al., 2005). Some of these ES cell lines have been modified using conventional gene targeting methods (Shiozawa et al., 2011) and more recent genome editing technologies such as ZFNs and CRISPR/Cas9 (Okano et al, unpublished results). However, for reasons that are yet fully understood, non-human primate ES cells appear incapable of incorporating into developing embryos to generate chimeric offspring (Tachibana et al., 2013). This difference may be rooted, in part, in molecular signals: the growth factors and other molecules used to sustain primate ES cells are distinct from those required for mouse ES cells (Kim et al., 2013; Tesar et al., 2007). In fact, the primate ES lines more closely resemble the mouse epiblast stem cells (EpiSCs) than the mouse ES cells. Despite many years of research, the derivation of true naïve ES cells from non-human primate species has not yet been successful, though naïve induced pluripotent stem cells (iPSCs) have recently been generated from apes (Marchetto et al., 2013), rhesus monkey (Fang et al., 2014) and common marmoset (Okano et al, unpublished results).
Several recent studies give reason to expect advances in this direction. These studies have reported culture conditions with the capacity to revert human ES cells into a more naïve state, and thus more similar to mouse ES lines (Chan et al., 2013; Gafni et al., 2013; Ware et al., 2014; Theunissen et al., 2014; Takashima et al., 2014; Wang et al., 2014). In coming years, it will be of great interest to apply species optimized culture conditions, either for converting existing “primed” non-human primate ES cells to a chimeric-competent naïve state or for direct derivation of naïve ES cells from pre-implantation non-human primate embryos. These naïve ES cells can then be tested for generating chimeric non-human primates. If chimeric-competent non-human primate ES cells with germline competency can be derived using these novel culture conditions, this will represent a significant step forward in the development of genetically modified non-human primate models for neuroscience. Importantly, it will for the first time allow for targeted gene insertion followed by selection of desired cells prior to implantation, greatly increasing the efficiency of producing transgenic animal lines.
A potential limitation of using non-human primate ES cells as means of generating transgenic animals is longer gestational and sexual maturity lengths as compared to rodents. Macaques have a gestation period of approximately 165 days and though they are sexually mature at 3–4 years, they typically do not become sexually active until they reach adult size at age 8 (Bercovitch et al., 2003; Dixon and Nevison, 1997). It will thus likely take a decade before germline chimeras are identified and sufficient numbers of transgenic founder animals are produced by natural breeding. Fortunately, in the common marmoset, the challenges are reduced, due to their shorter gestational period (145–148 days), an early onset of puberty (sexual maturity at 15–18 months), and relatively large litter sizes (2 to 3).
Another potential limitation for gene editing in primates, based on experience in the mouse, is that factors affecting germline transmission of ES cells are complex. Examples include clone-to-clone variation, number of passages, genetic backgrounds, culture conditions and genomic instability. Efforts need to be invested to minimize these factors’ negative impact on germline competency. One approach is to directly differentiate ES cells into functional gametes. In mice, primordial germ cells (PGCs) can be induced directly from ES cells and it has been found that they can mature into functional oocytes and sperm after ovary and testis transplantation (Hayashi and Saitou, 2013; Hayashi et al., 2011). To do so, Hayashi and colleagues induced a transient epiblast-like cell population (EpiLCs) from naïve ES cells that bear high efficiency for PGC induction. Similar strategies could be developed for non-human primates if naïve non-human primate ES cells can be stabilized in culture, as has recently been demonstrated in humans (Irie et al., 2014). Also an in vivo maturation platform similar to what has been developed in the mice is possible in the rhesus macaque (Hermann et al., 2012). Alternatively, strategies of culturing germline progenitors in vitro (discussed below) offer a more direct means of editing the germline for the production of transgenic non-human primates. The limitation that may prove to be the most difficult to overcome is the diverse genetic backgrounds found in outbred non-human primates. Unlike inbred colonies of mice with a common genetic background, the insertion of a gene into non-human primates with different genetic backgrounds will have more diverse effects. Ultimately inbred strains may be needed.
Cloning by somatic cell nuclear transfer
Somatic cell nuclear transfer (SCNT) relies on the capacity of an enucleated oocyte to reprogram a somatic nucleus into a state equivalent to that of a fertilized oocyte. To date, this remains the sole technique to reinstate totipotency in the somatic genome (Ogura et al., 2013). SCNT promises to be a preferred method for generating knockin and/or knockout animals using donor nuclei derived from gene-edited cells. Cloned rhesus monkeys have been generated through nuclear transfer using blastomere nuclei from early cleaving embryos (Meng et al., 1997). However, reproductive cloning of non-human primates using somatic or ES cell nuclei has not yet been achieved. Both rhesus monkey ES cells (Byrne et al., 2007) and human ES cells (Tachibana et al., 2013) have been successfully generated from embryos produced by SCNT. However, rhesus SCNT embryos failed to produce viable pregnancies, suggesting that reprogramming to totipotency is not as complete in primates as compared to other species.
Creation of germline progenitors
The ability to generate stem cells of germ lineages is especially attractive for the production of transgenic animals because the gene editing can be transmitted to the germline with high efficiency. This is particularly germane to the derivation of genetically modified non-human primates, which have considerably longer reproductive cycles than mice. Primordial germ cells (PGC) are precursors of both oocytes and spermatozoa. In principle, manipulation of the genome in PGCs, accompanied by successful induction of gametogenesis, holds the potential to yield both genome-edited oocytes and sperm. This would substantially shorten the time required for developing homozygous knockout and knockin animal models by eliminating the time required for breeding and screening of germline transmission in chimera. However, it is not currently possible to maintain cultures of PGC lines. The only way, at present, to culture PGCs in vitro is to reprogram them back to pluripotent embryonic germ cells (EGCs) (Matsui et al., 1992; Resnick et al., 1992). EGCs, however, are pluripotent and share many features of ESCs and have lost the ability to exclusively commit to germline development. Recent progress (Juan Carlos Izpisua Belmonte, Personal communication) suggests that murine PGCs can be stabilized in culture and are amenable in that form to genetic manipulation. If this method proves successful, a similar approach in non-human primates will provide a means of compensating for their relatively long reproductive cycles.
In addition to PGCs, there are other stem cells in the germ lineage. Spermatogonial stem cells (SSCs) are a small population in the testis that have the unique ability to self-renew as well as undergo meiosis and produce daughter spermatids throughout adult life (Kanatsu-Shinohara and Shinohara, 2013). SSCs arise from gonocytes in the postnatal testis, which originate from PGCs. Rodent SSCs have been successfully derived and could be cultured long term in vitro while still retaining their capability of differentiation into functional sperm after testis transplantation (Hamra et al., 2005; Kanatsu-Shinohara et al., 2003; Ryu et al., 2005). Genetically modified animals have been generated via gene-targeting in rodent SSCs (Hamra et al., 2005; Kanatsu-Shinohara et al., 2003, 2006). Although non-human primate SSC culture conditions have not been established, freshly isolated macaque SSCs could generate functional sperm after autologous and allogeneic testis transplantation into recipient macaques that had previously been rendered infertile with alkylating chemotherapy (Hermann et al., 2012). This opens the possibility that once long-term culture systems are established, non-human primate SSCs could offer an attractive solution for shortening the generation period of transgenic non-human primates
Analogous to the SSCs, oogonial stem cells (OSCs) are present in small numbers in the postnatal mammalian ovary. They have been shown by several groups to be capable of expansion in vitro. They could potentially be genetically manipulated for the production of transgenic offspring (Johnson et al., 2004; Zou et al., 2009). OSCs have also recently been reported in the human (White et al., 2012). However, the existence of such cells is still under debate (Lei and Spradling, 2013; Zhang et al., 2012). At present, the issue of whether female mammals posses such a population of renewable OSCs remains unresolved.
Cloning of Haploid ES cells
Most animals are diploid and natural haploid cells are typically limited to mature germ cells. Generation of homozygous transgenic animals has been complicated by the diploid genome. Recently both androgenetic (male) and parthenogenetic (female) haploid ES cells (haESCs) have been derived in mice and rats (Leeb and Wutz, 2011; Li et al., 2014, 2012; yang et al., 2012). HaESCs contain only one copy of allelic genes of diploid cells and thus provide an effective platform for studying gene function. HaESCs are amenable to genetic modification with traditional gene targeting approaches, and with new nuclease-based gene editing strategies (Li et al., 2014, 2012). More interestingly, androgenetic haESCs can produce viable and fertile offspring after intracytoplasmic injection into mature oocytes (Li et al., 2014, 2012). Most recently, haploid parthenogenetic mouse haESCs were also shown to be able to produce fertile mice when injected into oocytes in place of the maternal genome (Wan et al., 2013). Both strategies provide advantages for introduction of genetic modifications to progeny.
Parthenogenetic haESCs have also been established in Macaca fascicularis and are readily genetically-manipulatable (Yang et al., 2013). Androgenetic haploid monkey ES cells have not been reported. There are several limitations for applying haESCs strategies to generate transgenic non-human primates. First, the haploid phenotype has been found to be unstable in culture. haESCs undergo spontaneous auto-diploidization and need several rounds of haploid purification by flow activated cell sorting (FACS) before becoming stable in culture. Also, there is a lack of androgenic haESCs containing the Y chromosome 9 Li et al., 2012). This is due to the poor developmental potential of androgenetic embryos of YY chromosomes (Latham et al., 2000; Tarkowski et al., 1977). Therefore only female animals can currently be created. With further breeding, males can then be obtained. Another major drawback is that the efficiency for androgenic haESCs to fertilize an egg is very low (less than 5% in mice and less than 2% in rat). This poses a challenge for the derivation of transgenic primates, as large numbers of eggs would be needed. Despite these limitations, this is a promising direction and warrants further investigation.
3. Factors to consider in selecting among potential primate models for genetic editing
Given the possibility of creating targeted gene knock-in primates, the scientific community faces a question of which primate model or models are the most likely to be useful. Factors include phylogenetic proximity and genetic similarity to humans, similarity of cognitive and behavioral functions, similarity neuroanatomical organization, applicability as a model of human brain disorders, existing knowledge of brain organization, generation time and reproductive rate, as well as cost and availability.
In past decades, many species of non-human primates have been used for neuroscience research, including prosimian primates (e.g. lemurs), New World monkeys (e.g. squirrel monkeys, marmosets) and Old World monkeys (e.g. rhesus and cynomolgus macaques). The primate of choice for studying mechanisms in the human brain has traditionally been the macaque, due to several factors, including phylogenetic proximity of the Old World monkeys to humans, their intelligence and capacity to be trained to perform complex tasks, close similarities between the brains of Old World monkeys and humans, and in part, due to their availability. As a result, a large amount of information has been accumulated regarding their brain structure, circuit assembly, neurophysiology and behavioral repertoire. As a routine genetic model, an important factor to consider is the long generation time and slow reproductive cycle of the macaques. Rhesus and cynomolgus macaques live up to 30 and 40 years in captivity, respectively. They reach sexual maturity at the age of 3–4 years and give birth once a year to a single offspring. If techniques can be developed to enable genome modifications to be accomplished in a single generation without cross breeding, it may become feasible to develop knock-in macaques. However, until such major technical advances occur, the creation of transgenic macaque lines is likely to be time consuming.
Two smaller primate species, the common marmoset (Callithrix jacchus) and the mouse lemur (Microcebus murinus), have several advantages as candidates for creating transgenic lines. The marmoset is a small (300–400g) New World primate that reaches sexual maturity around 15–18 months of age and thus establishes germline transmission with each generation is 2–3 times faster than in macaques (Sasaki et al., 2009). Mature females give birth twice a year, usually to non-identical twins. Compared to macaques, they are born developmentally immature, thus they are good models to study primate brain development (Bourne and Rosa, 2006; Hikishima et al., 2013; Sawada et al., 2014). They are also the shortest-lived of the anthropoid primates (New World monkeys, Old World Monkeys, apes and humans). They exhibit age-related changes that are similar in many respects to those of humans, including declines in lean muscle mass, circulating albumin, hemoglobin, and hematocrit, as well as increasing prevalence of cancer, amyloidosis, diabetes, and chronic renal disease as they age. These factors strongly suggest that marmosets could be a revealing model of neuro-degeneration, since they display reduced neurogenesis, beta amyloid deposition in cerebral cortex, loss of calbindin binding, and age-related hearing loss (Tardif et al., 2011). In addition, marmosets are highly social with a tight family structure and they are highly communicative (Takahashi et al., 2013). They may therefore be particularly suitable for studying brain disorders with social communication defects, such as autism spectrum disorders.
Mouse lemurs, native only to the island of Madagascar, have also been proposed as a possible transgenic model system (Bons et al., 2006; Languille et al., 2012). Mouse lemurs are even smaller than marmosets (80–100g), with a somewhat longer life span (8–18 years in captivity, 5 years in the wild), are nearly as fecund (2–3 offspring per year), and reach sexual maturity even more rapidly, at the age of 10 months. Mouse lemurs are particularly suitable for aging and Alzheimer’s disease research because in aged mouse lemurs (5–6 years of age), about 5–15% develop behavior indicative of “pathologic aging” (such as aggressiveness, loss of social contact, loss of biorhythm and cognitive deficits). Aging mouse lemur brains also show pathological alterations similar to those associated with Alzheimer’s disease. Unlike marmosets, mouse lemurs are prosimians and occupy a relatively specialized, nocturnal niche, suggesting that their brain is evolutionarily adapted in ways that differ from the human brain. The availability of mouse lemurs for neuroscience research is more restrictive and somewhat more uncertain compared to marmosets.
4. Bioethical considerations in genetic modification of non-human primates
The use of animals in research must be justified in terms of the value of the research to understanding fundamental biological processes and ameliorating devastating human diseases. Where experimental alternatives exist, those alternatives are preferred and used. Where no alternatives exist, established regulations allow for the use of animals in biomedical research while demanding that scientists justify the species appropriate to the specific problem to be solved, and to use only as many animals as is necessary (http://grants.nih.gov/grants/olaw/Guide-for-the-care-and-use-of-laboratory-animals.pdf). Other practical considerations such as availability, cost, husbandry arrangements and collaboration opportunities are also relevant constraints. High quality care stands out as paramount, certainly because the animals are in our care, but also because healthy animals are experimentally favored. In following these principles the scientists, veterinarians, animal technicians, institutional officials, medical charities and others involved and supportive of the research aim to position themselves at the intersection of animal welfare, science quality, and public confidence (Blakemore et al., 2012; Hyman, 2014).
The detailed case for augmenting mouse models of nervous system diseases with non-human primate models was presented in Section 1 above. It hinges on the fact that important features of the primate central nervous system are distinct from that of other mammals. Where these differences exist, the genetic and mechanistic determination of certain human neurological and psychiatric diseases may be better approximated by primate models. These diseases include but are not limited to autism spectrum disorder, Alzheimer’s disease, Parkinson’s disease, and psychiatric disorders such as schizophrenia, depression, and anxiety disorders. In recent decades, primate models have figured centrally into the investigation of the brain circuits affected in these diseases, particularly in the realm of basic science investigation. In fact, much of our understanding of the human brain can be traced to a range of experiments in primates, and in particular the macaque monkey. With the development of Cre lines, it will become possible to understand these circuits at the level of cell types, as is now routine in the mouse.
The genetic component of these diseases, which is critical for understanding their etiology and progression, has not been studied intensively in primates because of a lag in the availability of the genetic methods, including those described in Section 3. The creation of genetic models for complex disorders with incomplete penetrance, such as autism and schizophrenia, is a current challenge in any animal model (Silverman et al., 2010; Nestler and Hyman, 2010). Mouse models will continue to be important in this regard, as multiple lines can be cheaply derived to study the effects of different genetic variants. However, it is important to work in parallel to understand the genetic basis of these disorders in the primate brain. The natural genetically heterogeneous background in non-human primates may be useful in revealing the risk impact of individual genetic variants to multi-genetic diseases. Moreover, the development of Cre lines will enable basic research directed at understanding the neural circuits of the primate brain, including circuits and cell types that are misexpressed in these disorders. Basic research on primate cortical circuits thus promises to provide insight, even as challenges remain in our understanding of the many genetic factors that contribute to circuit dysfunctions characteristic of these brain disorders.
Now that non-human primates are candidates for genetic modification, they are likely to become an essential factor for making progress in understanding, diagnosing, and treating human diseases that were previously out of reach. One non-human primate research success story that is likely to become even more successful with transgenic models is research that has led to successful treatment of Parkinson’s disease (PD). This research that was recently recognized with a Lasker-DeBakey award to Mahlon DeLong and Alim Louis Benabid. In the US alone there are between 500,000 and 1 million people living with PD, with about 50 to 60 thousand new diagnoses every year. The National Institutes of Neurological Disorders and Stroke (NINDS) estimates the cost to our society is at least $5.6 billion, including both direct medical expenses and indirect costs from lost income and disability payments. Currently successful therapies developed for PD stem from a long history of investigation into a mechanistic understanding of the disease and the normal functioning of the relevant circuits in nonhuman primates. In one milestone, Parkinson’s symptoms were replicated pharmacologically in monkeys, and could then be readily relieved by the administration of L-dopa. With this animal model, it was possible to test a number of hypotheses related to the development and modification of pharmacological treatments for human patients. In another milestone, the electrical stimulation of certain structures within the human brain, so called deep brain stimulation (DBS) (Bronstein et al., 2011), has provided another effective treatment. This therapy benefited from both from the slow and meticulous charting out of the primate basal ganglia over decades, as well as targeted experiments in non-human primates that tested specific hypotheses related to the potential relief of PD symptoms (Emborg, 2007). The result of this research is that millions of patients have benefited from pharmacological management of PD symptoms. More recently over 80,000 patients have benefited from deep brain stimulation to alleviate their suffering. Given these important achievements in ameliorating suffering, even noted animal rights philosopher Peter Singer has considered such research as being morally justifiable [ http://www.bbc.co.uk/blogs/legacy/ni/2006/11/peter_singer_defends_animal_ex.html]. This ongoing story is an important one because it shows that complementary work in human and nonhuman primates, initially through anatomical, pharmacological, and electrophysiological investigation, and now through genetic perturbation, can lead steadily to medical breakthroughs that improve the quality of life for millions of people.
It is also worth emphasizing that the most efficient attack on these diseases may not be a head-on, direct search for treatments. Clearly, a basic understanding of brain function would put us in a better position to develop treatments for mental disorders. In this regard, understanding basic functions of the brain, such as how we allocate attention, how we store and retrieve memories, how we produce speech, how we recognize faces, and so on, are important scientific questions about brain function that are relevant to many neurological disorders. In other words, outcomes of science pursued merely for its own sake, usually with only the faintest inkling of possible practical implications, has taught us that basic, fundamental science sometimes yields the most monumental of unexpected dividends. One need only reflect on the discovery of the structure of DNA to see the lesson writ clear. Francis Crick said on numerous occasions that in the first several decades of molecular biology he did not have even the faintest idea that understanding this molecule would one day yield the stunning array of practical applications now in daily use in medical research. Nor, as is well known, was medical benefit a motivation for Watson and Crick in seeking the structure of DNA. They just passionately wanted to know how information passed from parent to offspring. A related point concerns the Human Genome Project. At its inception, a widely held view among molecular geneticists was that such a project was utterly misguided and probably useless. What the opposition could not foresee was the transformative impact of genomics on every aspect of biology and medicine once that the cost of sequencing reached nominal levels.
Any decision to bring transgenic nonhuman primates, such as marmosets, into the laboratory must be weighed in the context of relevant ethical considerations (Bateson and Ragan, 2014). Desperate human need must be balanced against the welfare and life quality of animal subjects. Regulations at the federal, state and local levels provide a matrix within which current animal research, including transgenic animal research, is conducted, and these regulations will continue to provide institutional protection for the animals. Research using transgenic models is already thoroughly regulated, and these regulations are readily extended to research in transgenic nonhuman primates.
As with many new developments in biology, a project proposing to apply gene-editing technology to nonhuman primates for disease research deserves careful examination from many angles. Moral problems in real life typically involve balancing many competing interests and taking the wisdom of diverse points of view into account. We are obligated to weigh the consequences to lives -- both if the proposal moves forward and if it does not. Assessing alternatives, not fancifully but realistically, is also part of our moral duty. Those who are fortunate enough to be spared an agonizing confrontation with nervous system diseases will benefit from acquainting themselves with the stark reality of what such diseases mean for those who suffer them. Bearing these considerations in mind, we see the weight of the argument in favor of moving forward on transgenic nonhuman primate disease models with due care, responsibility and transparency.
A Way Forward
The development of transgenic nonhuman primate models holds great promise for improving human health worldwide and for increased scientific understanding of the human nervous system. To succeed, this effort will require a thoughtful and coordinated approach. The development of gene targeted lines for studying genes associated with brain disorders, and Cre lines to enable the study of neuronal and non-neuronal cell types will require concerted research efforts at universities and research institutions. The field will need the support of governments, private foundations, and research institutions, as well as the development of critical infrastructure.
Given the regulatory hurdles and the high cost to import and export primates, it is essential that we consider appropriate strategies tailored for different countries. Japan has taken a significant step forward with the Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) initiative (Okano et al., 2012) (http://brainminds.jp/en/). This is a large-scale national program that will support three groups, one of which will focus on structural and functional mapping of the marmoset brain. The initiative will also support development of innovative neurotechnologies for brain mapping and relating these findings to human brain disorders. While there is no project announced at the national level in China, a flurry of recent publications indicates that several active research programs have been devoted to developing genetically modified primates in that country.
For the United States, the NIH should take a leading role in assessing the importance of this endeavor and in establishing a suitable strategy that would include any private or charitable input and support. The UK and the EU have comparable research funding bodies that could convene and lead similar initiatives. Within the United States, research will depend critically on one or more national primate breeding centers with the expertise needed to apply the technologies, described above, that are now being used to develop genetically modified lines. They should also have the capacity to incorporate new technologies for targeted gene insertion, as these technologies mature. Existing National Primate Research Centers would be well suited to fulfill this function. A system of peer review should be established to prioritize the development of those lines most likely to lead to the discovery of new principles of primate brain organization and those most likely to lead to breakthroughs in understanding human brain disorders. Infrastructure will be needed to disseminate primate lines to individual research institutions. Funding, both public and private, can play a significant role in providing support for the infrastructure to enable individual laboratories to incorporate transgenic primates into their research efforts. These include veterinary support, space to house transgenic primates and, in some cases, local breeding facilities. As we embark on an era of multi-collaborative brain initiatives, we feel strongly that collaboration, both scientifically and fiscally, will ensure that this effort will add significant value to our efforts to improve human health and reduce suffering, as well as to invaluable scientific understanding of the human brain.
Conclusion
In summary, while mouse models will continue to be of great value to neuroscience, the complementary use of non-human primates in basic neuroscience research and in the study of brain disorders, continues to be of great value because non-human primates and humans share many anatomical, perceptual, cognitive and behavioral specializations. Recent advances in gene-editing techniques have made it possible to create genetically modified primates, opening up new and exciting ways to gain insight into neuronal types and neural circuits in the primate brain, as well as to study the genetic underpinnings of brain function and brain pathology, in ways that are highly likely to build on the information already being obtained in rodents. There is a pressing need for establishing a thoughtful and coordinated effort to efficiently and ethically develop genetically engineered primate lines, including lines with cell-type-specific expression of Cre, both to deepen our understanding of the fundamental principles of brain function in healthy brains and to empower the study of neural circuits in neuropsychiatric disorders. This effort will require the coordinated support of both governmental and private funding institutions, as well as the development of the needed technological and housing infrastructure.
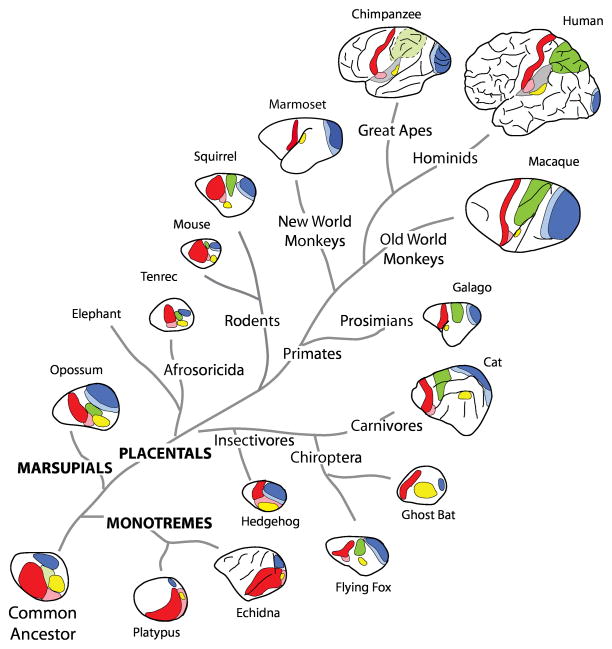
Figure 1.
Cladogram illustrating the phylogenetic relationships for the major subclasses of mammals and some of the orders within each subclass, with illustrations indicating some cortical fields that are shared across different mammals. Primary visual cortex, dark blue; Secondary visual cortex, light blue; Posterior parietal cortex, green; presumptive posterior parietal cortex, light green; Auditory cortex, yellow; Primary somatosensory cortex, red; Second somatosensory area, pink. Adapted from (Cooke et al., 2014), with permission.
Acknowledgments
We thank the following for thoughtful conversations that were helpful in preparing this manuscript: Michael C. Avery, Michele Basso, Hagai Bergman, Vince Ferrera, Fred H. Gage, Paul Glimcher, Josh Gold, Mickey Goldberg, Neng Gong, John D. Harding, Atsushi Iriki, Leah Krubitzer, Mathias Leblanc, Daeyol Lee, Steve Lisberger, Julio Martinez-Trujillo, John Maunsell, Samuel L. Pfaff, Michael Platt, Muming Poo, Nicholas Priebe, Louise Reichardt, Jeff Schall, Steve Scott, John Spiro, Stefan Treue, Bob Wurtz. Work in the laboratory of J.H.R. was supported, in part, by the Gatsby Charitable Foundation, the Crick Jacobs Center of the Salk Institute, a Salk Innovation Award, National Institutes of Health (R01 EY021827), Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS), Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and by the Intramural Research Program of the US National Institutes of Health, NINDS and NIMH. Work in the laboratory of J.C.I.B. was supported by G. Harlod and Leila Y. Mathers Charitable Foundation and The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002). Work in the laboratory of G.F. was supported by Poitras Center for Affective Disorders Research at McGovern Institute for Brain Research at MIT, Stanley Center for Psychiatric Research at Broad Institute of MIT and Harvard, and a Science Innovation Award from Brain Research Foundation. Work in the laboratory of K-F.L. was supported by a Salk Innovation Grant, the Clayton Foundation, the National Institute of Aging and the National Institute of Neurological disorders and Stokes. Work in the laboratory of D.A.L. was supported, in part, by the Intramural Research Program of the US National Institutes of Health, NINDS and NIMH. Work in the laboratory of C.T.M. was supported by NSF-IDBR, NIDCD and NIMH. Work in the laboratory of J.F.M was supported by NIH (R21 MH104756). Work in the laboratory of S.M. was supported by grants from National Institutes of Health R01-HD063276, R01-HD057121, R01-HD059946, R01-EY021214, P51-OD011092, a grant from the Leducq Foundation, and OHSU institutional funds. Work in the laboratory of A.R.M was supported by NIH Director’s New Innovator Award Program (1-DP2-OD006495-01) and a NARSAD Independent Investigator Grant. Work in the laboratory of D.R. was supported by NEI. Work in the laboratory of T.J.S. was supported by Howard Hughes Medical Institute and Office of Naval Research. Work in the laboratory of A.C.S. was supported by The Intramural Research Program of the NINDS, NIH. Work in the laboratory of F.Z. was supported by the National Institutes of Health through (NIMH: 5DP1-MH100706) and (NIDDK: 5R01-DK097768), a Waterman Award from the National Science Foundation, the Keck, New York Stem Cell, Damon Runyon, Searle Scholars, Merkin, and Vallee Foundations, and Bob Metcalfe. F.Z. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hurd MD, Martorell P, Delavande A, Mullen KJ, Langa KM. Monetary costs of dementia in the United States. The New England journal of medicine. 2013;368(14):1326–34. doi: 10.1056/NEJMsa1204629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–4. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 3.Kaas JH. The Evolution of Brains from Early Mammals to Humans. Wiley interdisciplinary reviews Cognitive science. 2013;4(1):33–45. doi: 10.1002/wcs.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise SP. Forward frontal fields: phylogeny and fundamental function. Trends in neurosciences. 2008;31(12):599–608. doi: 10.1016/j.tins.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faulstich BM, Onori KA, du Lac S. Comparison of plasticity and development of mouse optokinetic and vestibulo-ocular reflexes suggests differential gain control mechanisms. Vision research. 2004;44(28):3419–27. doi: 10.1016/j.visres.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DJ, Greenberg DS, Sawinski J, Rulla S, Notaro G, Kerr JN. Rats maintain an overhead binocular field at the expense of constant fusion. Nature. 2013;498(7452):65–9. doi: 10.1038/nature12153. [DOI] [PubMed] [Google Scholar]
- 7.Sridharan D, Ramamurthy DL, Schwarz JS, Knudsen EI. Visuospatial selective attention in chickens. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(19):E2056–65. doi: 10.1073/pnas.1316824111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAdams CJ, Maunsell JH. Effects of attention on orientation-tuning functions of single neurons in macaque cortical area V4. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19(1):431–41. doi: 10.1523/JNEUROSCI.19-01-00431.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desimone R, Duncan J. Neural mechanisms of selective visual attention. Annual review of neuroscience. 1995;18:193–222. doi: 10.1146/annurev.ne.18.030195.001205. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell JF, Sundberg KA, Reynolds JH. Differential attention-dependent response modulation across cell classes in macaque visual area V4. Neuron. 2007;55(1):131–41. doi: 10.1016/j.neuron.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 11.Sundberg KA, Mitchell JF, Reynolds JH. Spatial attention modulates center-surround interactions in macaque visual area v4. Neuron. 2009;61(6):952–63. doi: 10.1016/j.neuron.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niell CM, Stryker MP. Modulation of visual responses by behavioral state in mouse visual cortex. Neuron. 2010;65(4):472–9. doi: 10.1016/j.neuron.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polack PO, Friedman J, Golshani P. Cellular mechanisms of brain state-dependent gain modulation in visual cortex. Nature neuroscience. 2013;16(9):1331–9. doi: 10.1038/nn.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otte S, Hasenstaub A, Callaway EM. Cell type-specific control of neuronal responsiveness by gamma-band oscillatory inhibition. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(6):2150–9. doi: 10.1523/JNEUROSCI.4818-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atallah BV, Bruns W, Carandini M, Scanziani M. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron. 2012;73(1):159–70. doi: 10.1016/j.neuron.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Tucciarone JM, Espinosa JS, Sheng N, Darcy DP, Nicoll RA, Huang ZJ, Stryker MP. A cortical circuit for gain control by behavioral state. Cell. 2014;156(6):1139–52. doi: 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Kwan AC, Zhang S, Phoumthipphavong V, Flannery JG, Masmanidis SC, Taniguchi H, Huang ZJ, Zhang F, Boyden ES, Deisseroth K, Dan Y. Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature. 2012;488(7411):379–83. doi: 10.1038/nature11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kastner S, Ungerleider LG. Mechanisms of visual attention in the human cortex. Annual review of neuroscience. 2000;23:315–41. doi: 10.1146/annurev.neuro.23.1.315. [DOI] [PubMed] [Google Scholar]
- 19.Moore T, Armstrong KM, Fallah M. Visuomotor origins of covert spatial attention. Neuron. 2003;40(4):671–83. doi: 10.1016/s0896-6273(03)00716-5. [DOI] [PubMed] [Google Scholar]
- 20.Moore T, Fallah M. Microstimulation of the frontal eye field and its effects on covert spatial attention. Journal of neurophysiology. 2004;91(1):152–62. doi: 10.1152/jn.00741.2002. [DOI] [PubMed] [Google Scholar]
- 21.Chang SW, Brent LJ, Adams GK, Klein JT, Pearson JM, Watson KK, Platt ML. Neuroethology of primate social behavior. Proceedings of the National Academy of Sciences of the United States of America. 2013;110 (Suppl 2):10387–94. doi: 10.1073/pnas.1301213110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brent LJ, Chang SW, Gariepy JF, Platt ML. The neuroethology of friendship. Annals of the New York Academy of Sciences. 2014;1316:1–17. doi: 10.1111/nyas.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eliades SJ, Wang X. Comparison of auditory-vocal interactions across multiple types of vocalizations in marmoset auditory cortex. Journal of neurophysiology. 2013;109(6):1638–57. doi: 10.1152/jn.00698.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruce C, Desimone R, Gross CG. Visual properties of neurons in a polysensory area in superior temporal sulcus of the macaque. Journal of neurophysiology. 1981;46(2):369–84. doi: 10.1152/jn.1981.46.2.369. [DOI] [PubMed] [Google Scholar]
- 25.Tsao DY, Freiwald WA, Tootell RB, Livingstone MS. A cortical region consisting entirely of face-selective cells. Science. 2006;311(5761):670–4. doi: 10.1126/science.1119983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seyfarth RM, Cheney DL. The evolution of language from social cognition. Current opinion in neurobiology. 2014;28:5–9. doi: 10.1016/j.conb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Lakshminarayanan VR, Santos LR. Capuchin monkeys are sensitive to others’ welfare. Current biology : CB. 2008;18(21):R999–1000. doi: 10.1016/j.cub.2008.08.057. [DOI] [PubMed] [Google Scholar]
- 28.Hare B, Melis AP, Woods V, Hastings S, Wrangham R. Tolerance allows bonobos to outperform chimpanzees on a cooperative task. Current biology : CB. 2007;17(7):619–23. doi: 10.1016/j.cub.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 29.Hare B, Kwetuenda S. Bonobos voluntarily share their own food with others. Current biology : CB. 2010;20(5):R230–1. doi: 10.1016/j.cub.2009.12.038. [DOI] [PubMed] [Google Scholar]
- 30.Hare B, Call J, Tomasello M. Do chimpanzees know what conspecifics know? Animal behaviour. 2001;61(1):139–51. doi: 10.1006/anbe.2000.1518. [DOI] [PubMed] [Google Scholar]
- 31.Hare B, Call J, Tomasello M. Chimpanzees deceive a human competitor by hiding. Cognition. 2006;101(3):495–514. doi: 10.1016/j.cognition.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Flombaum JI, Santos LR. Rhesus monkeys attribute perceptions to others. Current biology : CB. 2005;15(5):447–52. doi: 10.1016/j.cub.2004.12.076. [DOI] [PubMed] [Google Scholar]
- 33.Johnson KO, Hsiao SS. Neural mechanisms of tactual form and texture perception. Annual review of neuroscience. 1992;15:227–50. doi: 10.1146/annurev.ne.15.030192.001303. [DOI] [PubMed] [Google Scholar]
- 34.Jain N, Qi HX, Catania KC, Kaas JH. Anatomic correlates of the face and oral cavity representations in the somatosensory cortical area 3b of monkeys. The Journal of comparative neurology. 2001;429(3):455–68. doi: 10.1002/1096-9861(20010115)429:3<455::aid-cne7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 35.Krubitzer LA, Kaas JH. The organization and connections of somatosensory cortex in marmosets. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1990;10(3):952–74. doi: 10.1523/JNEUROSCI.10-03-00952.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang HQ, Zachariah MK, Coleman GT, Rowe MJ. Hierarchical equivalence of somatosensory areas I and II for tactile processing in the cerebral cortex of the marmoset monkey. Journal of neurophysiology. 2001;85(5):1823–35. doi: 10.1152/jn.2001.85.5.1823. [DOI] [PubMed] [Google Scholar]
- 37.Macaluso E, Maravita A. The representation of space near the body through touch and vision. Neuropsychologia. 2010;48(3):782–95. doi: 10.1016/j.neuropsychologia.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Graziano M. The organization of behavioral repertoire in motor cortex. Annual review of neuroscience. 2006;29:105–34. doi: 10.1146/annurev.neuro.29.051605.112924. [DOI] [PubMed] [Google Scholar]
- 39.Rathelot JA, Strick PL. Muscle representation in the macaque motor cortex: an anatomical perspective. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(21):8257–62. doi: 10.1073/pnas.0602933103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathelot JA, Strick PL. Subdivisions of primary motor cortex based on cortico-motoneuronal cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(3):918–23. doi: 10.1073/pnas.0808362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davare M, Kraskov A, Rothwell JC, Lemon RN. Interactions between areas of the cortical grasping network. Current opinion in neurobiology. 2011;21(4):565–70. doi: 10.1016/j.conb.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiten A, Goodall J, McGrew WC, Nishida T, Reynolds V, Sugiyama Y, Tutin CE, Wrangham RW, Boesch C. Cultures in chimpanzees. Nature. 1999;399(6737):682–5. doi: 10.1038/21415. [DOI] [PubMed] [Google Scholar]
- 43.Pruetz JD, Bertolani P. Savanna chimpanzees, Pan troglodytes verus, hunt with tools. Current biology : CB. 2007;17(5):412–7. doi: 10.1016/j.cub.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 44.Yamazaki Y, Echigo C, Saiki M, Inada M, Watanabe S, Iriki A. Tool-use learning by common marmosets (Callithrix jacchus) Experimental brain research. 2011;213(1):63–71. doi: 10.1007/s00221-011-2778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hobaiter C, Poisot T, Zuberbuhler K, Hoppitt W, Gruber T. Social network analysis shows direct evidence for social transmission of tool use in wild chimpanzees. PLoS biology. 2014;12(9):e1001960. doi: 10.1371/journal.pbio.1001960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gruber T, Muller MN, Strimling P, Wrangham R, Zuberbuhler K. Wild chimpanzees rely on cultural knowledge to solve an experimental honey acquisition task. Current biology : CB. 2009;19(21):1806–10. doi: 10.1016/j.cub.2009.08.060. [DOI] [PubMed] [Google Scholar]
- 47.Whiten A. Imitation of the sequential structure of actions by chimpanzees (Pan troglodytes) Journal of comparative psychology. 1998;112(3):270–81. doi: 10.1037/0735-7036.112.3.270. [DOI] [PubMed] [Google Scholar]
- 48.Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62(2):191–8. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinoshita M, Matsui R, Kato S, Hasegawa T, Kasahara H, Isa K, Watakabe A, Yamamori T, Nishimura Y, Alstermark B, Watanabe D, Kobayashi K, Isa T. Genetic dissection of the circuit for hand dexterity in primates. Nature. 2012;487(7406):235–8. doi: 10.1038/nature11206. [DOI] [PubMed] [Google Scholar]
- 50.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57(5):634–60. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, Habu S, Okano H, Nomura T. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459(7246):523–7. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 52.Marmoset Genome S, Analysis C. The common marmoset genome provides insight into primate biology and evolution. Nature genetics. 2014;46(8):850–7. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, Birnberg NC, Evans RM. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300(5893):611–5. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–83. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91(4):501–10. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 56.Niu Y, Yu Y, Bernat A, Yang S, He X, Guo X, Chen D, Chen Y, Ji S, Si W, Lv Y, Tan T, Wei Q, Wang H, Shi L, Guan J, Zhu X, Afanassieff M, Savatier P, Zhang K, Zhou Q, Ji W. Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(41):17663–7. doi: 10.1073/pnas.1006563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan AW, Chong KY, Martinovich C, Simerly C, Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291(5502):309–12. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- 58.Wolfgang MJ, Eisele SG, Browne MA, Schotzko ML, Garthwaite MA, Durning M, Ramezani A, Hawley RG, Thomson JA, Golos TG. Rhesus monkey placental transgene expression after lentiviral gene transfer into preimplantation embryos. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10728–32. doi: 10.1073/pnas.181336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, Snyder B, Larkin K, Liu J, Orkin J, Fang ZH, Smith Y, Bachevalier J, Zola SM, Li SH, Li XJ, Chan AW. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453(7197):921–4. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okano H, Hikishima K, Iriki A, Sasaki E. The common marmoset as a novel animal model system for biomedical and neuroscience research applications. Seminars in fetal & neonatal medicine. 2012;17(6):336–40. doi: 10.1016/j.siny.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 61.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 62.Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends in biochemical sciences. 1998;23(10):394–8. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 63.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Menoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325(5939):433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell research. 2014;24(3):372–5. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):12013–7. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nature biotechnology. 2013;31(1):23–4. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 67.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Onestep generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferguson C, McKay M, Harris RA, Homanics GE. Toll-like receptor 4 (Tlr4) knockout rats produced by transcriptional activator-like effector nuclease (TALEN)-mediated gene inactivation. Alcohol. 2013;47(8):595–9. doi: 10.1016/j.alcohol.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–43. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 70.Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, Jing B, Si C, Lin Q, Chen X, Lin H, Pu X, Wang Y, Qin B, Wang F, Wang H, Si W, Zhou J, Tan T, Li T, Ji S, Xue Z, Luo Y, Cheng L, Zhou Q, Li S, Sun YE, Ji W. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell stem cell. 2014;14(3):323–8. doi: 10.1016/j.stem.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kishi N, Sato K, Sasaki E, Okano H. Common marmoset as a new model animal for neuroscience research and genome editing technology. Development, growth & differentiation. 2014;56(1):53–62. doi: 10.1111/dgd.12109. [DOI] [PubMed] [Google Scholar]
- 72.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic acids research. 2013;41(20):9584–92. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31(9):822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nature biotechnology. 2013;31(8):681–3. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 77.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nature biotechnology. 2011;29(8):695–6. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 78.Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Swinton PG, Martin JF, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Developmental biology. 2014;393(1):3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem cells. 2006;24(10):2177–86. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- 80.Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7844–8. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suemori H, Tada T, Torii R, Hosoi Y, Kobayashi K, Imahie H, Kondo Y, Iritani A, Nakatsuji N. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Developmental dynamics : an official publication of the American Association of Anatomists. 2001;222(2):273–9. doi: 10.1002/dvdy.1191. [DOI] [PubMed] [Google Scholar]
- 82.Simerly CR, Navara CS, Castro CA, Turpin JC, Redinger CJ, Mich-Basso JD, Jacoby ES, Grund KJ, McFarland DA, Oliver SL, Ben-Yehudah A, Carlisle DL, Frost P, Penedo C, Hewitson L, Schatten G. Establishment and characterization of baboon embryonic stem cell lines: an Old World Primate model for regeneration and transplantation research. Stem cell research. 2009;2(3):178–87. doi: 10.1016/j.scr.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Hearn JP. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biology of reproduction. 1996;55(2):254–9. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- 84.Sasaki E, Hanazawa K, Kurita R, Akatsuka A, Yoshizaki T, Ishii H, Tanioka Y, Ohnishi Y, Suemizu H, Sugawara A, Tamaoki N, Izawa K, Nakazaki Y, Hamada H, Suemori H, Asano S, Nakatsuji N, Okano H, Tani K. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus) Stem cells. 2005;23(9):1304–13. doi: 10.1634/stemcells.2004-0366. [DOI] [PubMed] [Google Scholar]
- 85.Shiozawa S, Kawai K, Okada Y, Tomioka I, Maeda T, Kanda A, Shinohara H, Suemizu H, James Okano H, Sotomaru Y, Sasaki E, Okano H. Gene targeting and subsequent site-specific transgenesis at the beta-actin (ACTB) locus in common marmoset embryonic stem cells. Stem cells and development. 2011;20(9):1587–99. doi: 10.1089/scd.2010.0351. [DOI] [PubMed] [Google Scholar]
- 86.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, Masterson K, Larson J, Eaton D, Sadler-Fredd K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer RL, Wolf D, Mitalipov S. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153(6):1228–38. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim H, Wu J, Ye S, Tai CI, Zhou X, Yan H, Li P, Pera M, Ying QL. Modulation of beta-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nature communications. 2013;4:2403. doi: 10.1038/ncomms3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 89.Fang R, Liu K, Zhao Y, Li H, Zhu D, Du Y, Xiang C, Li X, Liu H, Miao Z, Zhang X, Shi Y, Yang W, Xu J, Deng H. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell stem cell. 2014;15(4):488–96. doi: 10.1016/j.stem.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 90.Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell stem cell. 2013;13(6):663–75. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 91.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, Amit I, Tanay A, Massarwa R, Novershtern N, Hanna JH. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504(7479):282–6. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 92.Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, Tesar PJ, Okada J, Margaretha L, Sperber H, Choi M, Blau CA, Treuting PM, Hawkins RD, Cirulli V, Ruohola-Baker H. Derivation of naive human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(12):4484–9. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D’Alessio A, Zhang J, Gao Q, Dawlaty MM, Young RA, Gray NS, Jaenisch R. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell stem cell. 2014;15(4):471–87. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–69. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, Schumann GG, Chen W, Lorincz MC, Ivics Z, Hurst LD, Izsvak Z. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516(7531):405–9. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 96.Bercovitch FB, Widdig A, Trefilov A, Kessler MJ, Berard JD, Schmidtke J, Nurnberg P, Krawczak M. A longitudinal study of age-specific reproductive output and body condition among male rhesus macaques, Macaca mulatta. Die Naturwissenschaften. 2003;90(7):309–12. doi: 10.1007/s00114-003-0436-1. [DOI] [PubMed] [Google Scholar]
- 97.Dixson AF, Nevison CM. The socioendocrinology of adolescent development in male rhesus monkeys (Macaca mulatta) Hormones and behavior. 1997;31(2):126–35. doi: 10.1006/hbeh.1997.1374. [DOI] [PubMed] [Google Scholar]
- 98.Hayashi K, Saitou M. Generation of eggs from mouse embryonic stem cells and induced pluripotent stem cells. Nature protocols. 2013;8(8):1513–24. doi: 10.1038/nprot.2013.090. [DOI] [PubMed] [Google Scholar]
- 99.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146(4):519–32. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 100.Irie N, Tang WW, Azim Surani M. Germ cell specification and pluripotency in mammals: a perspective from early embryogenesis. Reproductive medicine and biology. 2014;13(4):203–15. doi: 10.1007/s12522-014-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G, Peterson K, Masterson K, Ramsey C, Ward T, Lienesch M, Volk A, Cooper DK, Thomson AW, Kiss JE, Penedo MC, Schatten GP, Mitalipov S, Orwig KE. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell stem cell. 2012;11(5):715–26. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368(1609):20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meng L, Ely JJ, Stouffer RL, Wolf DP. Rhesus monkeys produced by nuclear transfer. Biology of reproduction. 1997;57(2):454–9. doi: 10.1095/biolreprod57.2.454. [DOI] [PubMed] [Google Scholar]
- 104.Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450(7169):497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 105.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70(5):841–7. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 106.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359(6395):550–1. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 107.Kanatsu-Shinohara M, Shinohara T. Spermatogonial stem cell self-renewal and development. Annual review of cell and developmental biology. 2013;29:163–87. doi: 10.1146/annurev-cellbio-101512-122353. [DOI] [PubMed] [Google Scholar]
- 108.Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, Garbers DL. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(48):17430–5. doi: 10.1073/pnas.0508780102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biology of reproduction. 2003;69(2):612–6. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 110.Ryu BY, Kubota H, Avarbock MR, Brinster RL. Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(40):14302–7. doi: 10.1073/pnas.0506970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kanatsu-Shinohara M, Ikawa M, Takehashi M, Ogonuki N, Miki H, Inoue K, Kazuki Y, Lee J, Toyokuni S, Oshimura M, Ogura A, Shinohara T. Production of knockout mice by random or targeted mutagenesis in spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(21):8018–23. doi: 10.1073/pnas.0601139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson J, Canning J, Kaneko T, Pru JK, Tilly JL. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428(6979):145–50. doi: 10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 113.Zou K, Yuan Z, Yang Z, Luo H, Sun K, Zhou L, Xiang J, Shi L, Yu Q, Zhang Y, Hou R, Wu J. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nature cell biology. 2009;11(5):631–6. doi: 10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 114.White YA, Woods DC, Takai Y, Ishihara O, Seki H, Tilly JL. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nature medicine. 2012;18(3):413–21. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lei L, Spradling AC. Female mice lack adult germ-line stem cells but sustain oogenesis using stable primordial follicles. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(21):8585–90. doi: 10.1073/pnas.1306189110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang H, Zheng W, Shen Y, Adhikari D, Ueno H, Liu K. Experimental evidence showing that no mitotically active female germline progenitors exist in postnatal mouse ovaries. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):12580–5. doi: 10.1073/pnas.1206600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479(7371):131–4. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li W, Li X, Li T, Jiang MG, Wan H, Luo GZ, Feng C, Cui X, Teng F, Yuan Y, Zhou Q, Gu Q, Shuai L, Sha J, Xiao Y, Wang L, Liu Z, Wang XJ, Zhao XY, Zhou Q. Genetic modification and screening in rat using haploid embryonic stem cells. Cell stem cell. 2014;14(3):404–14. doi: 10.1016/j.stem.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 119.Li W, Shuai L, Wan H, Dong M, Wang M, Sang L, Feng C, Luo GZ, Li T, Li X, Wang L, Zheng QY, Sheng C, Wu HJ, Liu Z, Liu L, Wang L, Wang XJ, Zhao XY, Zhou Q. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature. 2012;490(7420):407–11. doi: 10.1038/nature11435. [DOI] [PubMed] [Google Scholar]
- 120.Yang H, Shi L, Wang BA, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X, Li D, Xu GL, Li J. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149(3):605–17. doi: 10.1016/j.cell.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 121.Wan H, He Z, Dong M, Gu T, Luo GZ, Teng F, Xia B, Li W, Feng C, Li X, Li T, Shuai L, Fu R, Wang L, Wang XJ, Zhao XY, Zhou Q. Parthenogenetic haploid embryonic stem cells produce fertile mice. Cell research. 2013;23(11):1330–3. doi: 10.1038/cr.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang H, Liu Z, Ma Y, Zhong C, Yin Q, Zhou C, Shi L, Cai Y, Zhao H, Wang H, Tang F, Wang Y, Zhang C, Liu XY, Lai D, Jin Y, Sun Q, Li J. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell research. 2013;23(10):1187–200. doi: 10.1038/cr.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Latham KE, Patel B, Bautista FD, Hawes SM. Effects of X chromosome number and parental origin on X-linked gene expression in preimplantation mouse embryos. Biology of reproduction. 2000;63(1):64–73. doi: 10.1095/biolreprod63.1.64. [DOI] [PubMed] [Google Scholar]
- 124.Tarkowski AK, Witkowska A, Opas J. Development of cytochalasin in B-induced tetraploid and diploid/tetraploid mosaic mouse embryos. Journal of embryology and experimental morphology. 1977;41:47–64. [PubMed] [Google Scholar]
- 125.Bourne JA, Rosa MG. Hierarchical development of the primate visual cortex, as revealed by neurofilament immunoreactivity: early maturation of the middle temporal area (MT) Cerebral cortex. 2006;16(3):405–14. doi: 10.1093/cercor/bhi119. [DOI] [PubMed] [Google Scholar]
- 126.Hikishima K, Sawada K, Murayama AY, Komaki Y, Kawai K, Sato N, Inoue T, Itoh T, Momoshima S, Iriki A, Okano HJ, Sasaki E, Okano H. Atlas of the developing brain of the marmoset monkey constructed using magnetic resonance histology. Neuroscience. 2013;230:102–13. doi: 10.1016/j.neuroscience.2012.09.053. [DOI] [PubMed] [Google Scholar]
- 127.Sawada K, Hikishima K, Murayama AY, Okano HJ, Sasaki E, Okano H. Fetal sulcation and gyrification in common marmosets (Callithrix jacchus) obtained by ex vivo magnetic resonance imaging. Neuroscience. 2014;257:158–74. doi: 10.1016/j.neuroscience.2013.10.067. [DOI] [PubMed] [Google Scholar]
- 128.Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The marmoset as a model of aging and age-related diseases. ILAR journal/National Research Council, Institute of Laboratory Animal Resources. 2011;52(1):54–65. doi: 10.1093/ilar.52.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takahashi DY, Narayanan DZ, Ghazanfar AA. Coupled oscillator dynamics of vocal turn-taking in monkeys. Current biology : CB. 2013;23(21):2162–8. doi: 10.1016/j.cub.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 130.Bons N, Rieger F, Prudhomme D, Fisher A, Krause KH. Microcebus murinus: a useful primate model for human cerebral aging and Alzheimer’s disease? Genes, brain, and behavior. 2006;5(2):120–30. doi: 10.1111/j.1601-183X.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- 131.Languille S, Blanc S, Blin O, Canale CI, Dal-Pan A, Devau G, Dhenain M, Dorieux O, Epelbaum J, Gomez D, Hardy I, Henry PY, Irving EA, Marchal J, Mestre-Frances N, Perret M, Picq JL, Pifferi F, Rahman A, Schenker E, Terrien J, Thery M, Verdier JM, Aujard F. The grey mouse lemur: a non-human primate model for ageing studies. Ageing research reviews. 2012;11(1):150–62. doi: 10.1016/j.arr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 132.Blakemore C, MaCarthur Clark J, Nevalainen T, Oberdorfer M, Sussman A. Implementing the 3Rs in neuroscience research: a reasoned approach. Neuron. 2012;75(6):948–50. doi: 10.1016/j.neuron.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 133.Hyman SE. How far can mice carry autism research? Cell. 2014;158(1):13–4. doi: 10.1016/j.cell.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 134.Bronstein JM, Tagliati M, Alterman RL, Lozano AM, Volkmann J, Stefani A, Horak FB, Okun MS, Foote KD, Krack P, Pahwa R, Henderson JM, Hariz MI, Bakay RA, Rezai A, Marks WJ, Jr, Moro E, Vitek JL, Weaver FM, Gross RE, DeLong MR. Deep brain stimulation for Parkinson disease: an expert consensus and review of key issues. Archives of neurology. 2011;68(2):165. doi: 10.1001/archneurol.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Emborg ME. Nonhuman primate models of Parkinson’s disease. ILAR journal/National Research Council, Institute of Laboratory Animal Resources. 2007;48(4):339–55. doi: 10.1093/ilar.48.4.339. [DOI] [PubMed] [Google Scholar]
- 136.Bateson P, Ragan CI. Lab animals: Can GM marmoset use be justified? Nature. 2014;514(7524):567. doi: 10.1038/514567a. [DOI] [PubMed] [Google Scholar]
- 137.Cooke DF, Goldring A, Recanzone GH, Krubitzer . The Evolution of Parietal Areas Associated with Visuomanual Behavior: From Grasping to Tool Use. In: Chalupa L, Werner J, editors. The New Visual Neurosciences. Cambridge, MA: MIT Press; 2014. pp. 1049–1063. [Google Scholar]
- 138.Ben-Tov M, Donchin O, Ben-Shahar O, et al. Pop-out visual search of moving targets in the archer fish. Journal of Molecular Neuroscience; Conference: 22nd Annual Meeting of the Israel-Society-for-Neuroscience (ISFN)/2nd Bi National Italy-Israel Neuroscience Meeting; Eilat, ISRAEL. DEC 14–17, 2013; pp. S16–S17. Sponsor(s): Israel Soc Neuroscience. Published: AUG 2014. [Google Scholar]
- 139.Romanski LM, Averbeck BB. The Primate Cortical Auditory System and Neural Representation of Conspecific Vocalizations. Annual Review of Neuroscience Book Series: Annual Review of Neuroscience. 32:315–346. doi: 10.1146/annurev.neuro.051508.135431. Published: 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cheney DL, Seyfarth RM. Baboon Metaphysics: The Evolution of a Social Mind. University of Chicago Press; Chicago: 2007. [Google Scholar]
- 141.Rosati AG, Santos LR, Hare B. Primate social cognition: thirty years after Premack and Woodruff. In: Ghazanfar A, Platt M, editors. Primate Neuroethology. Oxford: Oxford University Press; 2010. pp. 117–143. [Google Scholar]
- 142.Boesch C. Cooperative hunting in wild chimpanzees. Animal Behaviour. 1994;48:653–667. [Google Scholar]
- 143.Wilkinson GS. Reciprocal Food Sharing in the Vampire. Nature. 1984;308(5955):181–184. [Google Scholar]
- 144.Kaas JH. The Functional-Organization of Somatosensory Cortex in Primates. Annals of Anatomy-Anatomicscher Anzeiger. 1993;175(6):509–518. doi: 10.1016/s0940-9602(11)80212-8. [DOI] [PubMed] [Google Scholar]
- 145.Hunt GR. Manufacture and Use of Hook-Tools by New Caledonian Crows. Nature. 1996;379(6562):249–251. [Google Scholar]
- 146.Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, Yeo GW, Muotri AR, Gage FH. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;28:503(7477):525–9. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural Phenotyping assays for mouse models of autism. Nature Reviews Neuroscience. 2010;11(7):409–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nestler EJ, Hyman SE. Animal models of neurpsychiatric disorders. Nature Neuroscience. 2010;13(10):1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]