Abstract
Background
Heat stress is a critical threat to tall fescue in transitional and warm climate zones. Identification of association between molecular markers and heat tolerance-related functional traits would promote the efficient selection of heat tolerant tall fescue cultivars. Association analysis of heat tolerance-related traits was conducted in 100 diverse tall fescue accessions consisting of 93 natural genotypes originating from 33 countries and 7 turf-type commercial cultivars.
Results
The panel displayed significant genetic variations in growth rate (GR), turfgrass quality (TQ), survival rate (SR), chlorophyll content (CHL) and evapotranspiration rate (ET) in greenhouse and growth chamber trials. Two subpopulations were detected in the panel of accessions by 1010 SSR alleles with 90 SSR markers, but no obvious relative kinship was observed. 97 and 67 marker alleles associated with heat tolerance-related traits were identified in greenhouse trial and growth chamber trial (P < 0.01) using mix linear model, respectively. Due to different experimental conditions of the two trials, 2 SSR marker alleles associated with GR and ET were simultaneously identified at P < 0.01 level in two trials in response to heat stress.
Conclusion
High-temperature induced great variations of functional traits in tall fescue accessions. And the identified marker alleles associated with functional traits could provide important information about heat tolerance genetic pathways, and be used for molecular assisted breeding to enhance tall fescue performance under heat stress.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0494-5) contains supplementary material, which is available to authorized users.
Keywords: Association mapping, Tall fescue, Population structure, High-temperature stress, Functional traits
Background
Tall fescue (Festuca arundinacea Schreb.) is a major cool-season grass species from the family Poaceae. Native to Northern Europe, North Africa, Middle East, Central Asia, and Siberia, tall fescue is most widely utilized as forage and turfgrass attributed to its adaptability, yield, persistence, and other ecosystem services such as soil improvement, recreation, protection, and carbon sequestration. Tall fescue is a self-incompatible allohexaploid (2n = 6x = 42) out-crossing species containing three genomes (P, G1, and G2) with a genome size of approximately 5.27-5.83 × 106 kb [1].
Heat stress limits the growth and development of tall fescue in transitional and warm climatic regions. High summer temperature of 30 to 35°C could constrain growth, reduce turf quality, induce leaf withering, and inhibit photosynthesis [2], which would pose severe effects on global climate change. While effective agronomic measures, including heat acclimation, soil temperature reduction, and growth regulators application, could enhance heat tolerance of tall fescue. Heat tolerant cultivars would be key alternative in alleviation of the negative influences of abiotic stress on plant breeding programs [3]. However, plant heat tolerance is a complex quantitative trait, involving multiple regulatory mechanisms, signal transduction pathways, and metabolic pathways. Therefore, a study on genetic and molecular basis for heat tolerance in plants would be necessary. Detailed study in plant physiological responses to heat stress and identification of molecular markers linked to heat tolerance would enhance the efficiency of traditional breeding programs to developing heat tolerant cultivars.
The quantitative inheritances of heat tolerance and interaction between gene expression and environment make challenges to our knowledge of genetic basis of heat tolerant traits of plant. During last two decades, molecular marker has applied to insight into complex traits in plant. Many studies on quantitative trait locus (QTLs) mapping have been conducted to dissect numerous vital agronomical and morphological traits under abiotic stress. The results have improved the efficiency of conventional crop breeding via marker-assisted selection (MAS) in some crop species e.g. rice, maize, barley, soybean, and chickpea [4-8]. However, many linkage mapping based on QTLs studies presented modest and unreliable results due to several factors. First, mapping-based cloning of QTL is time-consuming and costly for construction of populations. Secondly, the restricted number of recombination events per chromosome during mapping population development limits the resolution of genetic map [9]. In addition, QTL mapping could not exploit the extensive genetic variation of natural germplasm resources. On the contrary, association mapping could exploit all recombination events and mutations including historical and evolutionary recombination in natural populations with unobserved ancestry [10]. Association mapping has been widely applied to explore the genetic basis of complex quantitative traits in plant species, and reported under favorable conditions like drought [11-14]. For example, a candidate gene, ZmDREB2.7 associated with drought stress, was identified to be effective in imparting plant tolerance to drought stress in maize [13]. In turfgrass species, a few studies on association mapping have been carried out involving flowering time, leaf length, submergence tolerance, salinity tolerance, and drought tolerance in perennial ryegrass [15-17]. Four single nucleotide polymorphisms from LpLEA3, LpFsSOD, and Cu-ZnSOD have been associated with drought tolerance traits in diverse perennial ryegrass accessions [14]. However, there was limited information on the association between marker genes and heat tolerance of plants [8].
Simple sequence repeats (SSRs) or microsatellites are widely distributed in all eukaryotic genomes. They are powerful tools for dissecting cultivar fingerprinting, genetic diversity assessment, evolutionary study, linkage map construction, and marker assisted breeding [18-20]. Alternatively, the SSR markers were developed for allohexaploid tall fescue, an out-crossing species with high intra-specific polymorphism, utilized for genomic mapping, identification of variety, population genetic analysis and diversity evaluation of germplasm [21-25]. Recently, SSR markers have been applied in trait and marker association of plants, such as kernel size and milling quality in wheat [26], oil, starch, and protein concentration in maize [27], submergence tolerance in perennial ryegrass [17]. However, the application of association mapping in detecting links between markers with functional traits such as heat tolerance in tall fescue is undocumented.
The objective of this study was to identify marker-trait associations for phenotypic and physiological traits under heat conditions. It was hypothesized that tall fescue accessions had high diversity in high temperature response and the population structure would influence individual functional traits associated with heat tolerance. A set of 100 diverse tall fescue accessions originating from different geographical regions was grown in two heat environmental conditions in the greenhouse and controlled growth chambers. The population structure, relative pairwise kinship, and marker-trait association (MTA) by mixed linear model were statistically analyzed based on SSR markers.
Results
Heat stress effects and functional traits variation
In tall fescue heat stress imposed leaf yellowing and wilting, limited plant growth, and even death. Turfgrass quality (TQ), survival rate (SR), chlorophyll content (CHL), and growth rate (GR) decreased with prolonged heat stress in both trials, but the severity of decline varied with accession and duration. Significant accession and treatment time effects under heat stress were observed on GR, TQ, CHL, and SR in both trials (Table 1). However, no significant time effect for evapotranspiration rate (ET) in growth chamber trial was detected. There was also no significant interaction effect for functional traits between grass accessions and treatment time.
Table 1.
Mean squares of variance for evapotranspiration rate (ET), growth rate (GR), turf quality (TQ), chlorophyll content (CHL), and survival rate (SR) of 100 tall fescue accessions on different times of heat treatment
| Sources | Greenhouse trial | Growth chamber trial | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ET | GR | TQ | CHL | SR | ET | GR | TQ | CHL | SR | |
| Accession (A) | 1725.3** | 1828.9** | 755.5** | 519.5** | 578.7** | 10693.3** | 17087.7** | 1313.1** | 2127.2** | 1135.2** |
| Time (T) | 2229801.4** | 189804.5** | 128139.4** | 30165.8** | 57682.3** | 7400.5ns | 299505.0** | 73169.6** | 29394.0** | 96450.8** |
| A*T | 483.5ns | 453.8ns | 111.3ns | 166.0 ns | 90.2ns | 2214.4ns | 3555.4ns | 168.3ns | 731.3ns | 161.2ns |
**mean significant at P < 0.01, and nsmean no significant.
With prolonged heat stress at 1-3 weeks, the mean, maximum, and minimum values decreased in two trials (Table 2). Under heat stress, the average growth rate decreased from 0.24 g d-1 at initial time to 0.05 g d-1 at 14 WOT, turfgrass quality reduced from 6.55 to 2.56, survival rate decreased from 99.65% to 46.66%, chlorophyll content decreased from 2.35 mg g-1 FW to 1.47 mg g-1 FW, and evapotranspiration rate decreased from 61.55 g d-1 to 10.64 g d-1, respectively in greenhouse trial. Most of the functional traits decreased except for ET, which increased after one week of stress treatment, and then drastically dropped. In growth chamber trial, all functional traits displayed similar trend, whereby the average GR dropped from 0.11 g d-1 at initial time to 0.03 g d-1 at two WOT, TQ from 7.50 to 3.01, SR from 99.75% to 52.50%, CHLT from 2.03 mg g-1 FW to 1.77 mg g-1 FW, and ET from 22.82 g d-1 to 18.93 g d-1, respectively. After two weeks, heat stress significantly reduced GR by 79.17% in greenhouse and 72.73% in growth chamber trials compared with their relative controls (the time before heat stress). The decline levels of TQ, SR, and ET were lower than that of GR.
Table 2.
Descriptive statistics for growth rate (GR), turfgrass quality (TQ), survival rate (SR), evapotranspiration rate (ET), chlorophyll content (CHL) under heat stress in two trials
| Trait | Greenhouse trial | Growth chamber trial | ||||||
|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | Std. | Minimum | Maximum | Mean | Std. | |
| GR-1a (g/d) | 0.04 | 15.29 | 0.24 | 0.8 | 0.03 | 0.25 | 0.11 | 0.04 |
| GR-2b (g/d) | 0.01 | 0.41 | 0.11 | 0.05 | 0.01 | 0.19 | 0.06 | 0.03 |
| GR-3c (g/d) | 0.00 | 0.24 | 0.05 | 0.03 | 0.00 | 0.07 | 0.03 | 0.02 |
| TQ-1a | 3.00 | 8.50 | 6.55 | 0.93 | 6.00 | 9.00 | 7.50 | 0.58 |
| TQ-2b | 2.00 | 7.00 | 4.72 | 1.14 | 2.33 | 7.67 | 5.54 | 1.15 |
| TQ-3c | 0.00 | 6.00 | 2.56 | 0.88 | 1.00 | 6.17 | 3.91 | 1.24 |
| CHL-1a ( mg.g-1.Fw ) | 1.39 | 6.22 | 2.35 | 0.55 | 1.32 | 3.20 | 2.03 | 0.41 |
| CHL-2b ( mg.g-1.Fw ) | 0.70 | 3.08 | 1.76 | 0.44 | 1.26 | 3.05 | 2.05 | 0.44 |
| CHL-3c (mg.g-1.Fw) | 0.09 | 3.11 | 1.47 | 0.46 | 0.28 | 3.43 | 1.77 | 0.55 |
| SR-1a (%) | 90.00 | 100.00 | 99.65 | 1.33 | 98.33 | 100.00 | 99.75 | 0.60 |
| SR-2b (%) | 35.00 | 95.00 | 71.92 | 9.27 | 36.67 | 95.00 | 77.78 | 13.07 |
| SR-3c (%) | 5.00 | 75.00 | 46.66 | 16.49 | 6.67 | 86.67 | 52.5 | 16.08 |
| ET-1a (g/d) | 23.50 | 97.00 | 61.55 | 11.38 | 8.37 | 47.12 | 22.82 | 6.93 |
| ET-2b (g/d) | 30.00 | 142.00 | 84.84 | 10.99 | 6.96 | 49.66 | 21.17 | 7.70 |
| ET-3c (g/d) | 1.00 | 48.00 | 10.64 | 5.80 | 4.13 | 33.17 | 18.93 | 6.62 |
1a sampling time before heat treatment.
2b sampling time after 7d of heat treatment.
3c sampling time after 14d of heat treatment.
Significant correlations between survival rate with evapotranspiration rate, turfgrass quality, and turfgrass quality with evapotranspiration rate were found at two time of heat stress that the values had been standard to relative control in greenhouse trial, and relative values of SR of two weeks under heat stress had significant relationship with chlorophyll content (Table 3). Meanwhile, there were significant correlations between turfgrass quality with GR, CHL, ET and SR in growth chamber trail. There was significant correlation between CHL and SR, ET and GR, however there was no relationship between ET and SR in growth chamber trail (Table 4). High correlations were identified for all functional traits between the two sample times under heat stress in both trials, with the highest correlation for ET (r = 0.86, P < 0.01) in greenhouse trial, and SR (r = 0.768, P < 0.01) in growth chamber trial.
Table 3.
Pearson correlations coefficients among functional traits of different time in greenhouse trial
| GR-1 a | GR-2 b | TQ-1 a | TQ-2 b | ET-1 a | ET-2 b | CHL-1 a | CHL-2 b | SR-1 a | SR-2 b | |
|---|---|---|---|---|---|---|---|---|---|---|
| GR-1 a | 1 | |||||||||
| GR-2 b | 0.684** | 1 | ||||||||
| TQ-1 a | -0.129 | -0.209* | 1 | |||||||
| TQ-2 b | -0.002 | -0.085 | 0.698** | 1 | ||||||
| ET-1 a | 0.060 | -0.025 | 0.231* | 0.276** | 1 | |||||
| ET-2 b | 0.059 | -0.006 | 0.280** | 0.283** | 0.850** | 1 | ||||
| CHL-1 a | -0.100 | -0.080 | 0.097 | 0.141 | -0.130 | -0.086 | 1 | |||
| CHL-2 b | -0.328** | -0.361** | 0.278** | 0.155 | -0.030 | 0.015 | 0.444** | 1 | ||
| SR-1 a | 0.045 | -0.011 | 0.731** | 0.568** | 0.315** | 0.322** | 0.146 | 0.104 | 1 | |
| SR-2 b | -0.031 | -0.197* | 0.602** | 0.699** | 0.333** | 0.313** | 0.197* | 0.265** | 0.644** | 1 |
*significant at P < 0.05, **significant at P < 0.01.
1a the value at 7 d of heat stress relative the initial value before heat stress.
2b the reduction value at 14d of heat stress relative the initial value.
Abbreviations: TQ-turf quality, ET- evapotranspiration rate, Chl-chlorophyll content. SR-survival rate, GR- Growth rate.
Table 4.
Pearson correlations coefficients among functional traits of different time in growth chambers trial
| GR-1 a | GR-2 b | TQ-1 a | TQ-2 b | ET-1 a | ET-2 b | CHL-1 a | CHL-2 b | SR-1 a | SR-2 b | |
|---|---|---|---|---|---|---|---|---|---|---|
| GR-1 a | 1 | |||||||||
| GR-2 b | 0.608** | 1 | ||||||||
| TQ-1 a | 0.316** | 0.327** | 1 | |||||||
| TQ-2 b | 0.243* | 0.444** | 0.686** | 1 | ||||||
| ET-1 a | 0.184 | 0.262** | 0.251* | 0.323** | 1 | |||||
| ET-2 b | 0.085 | 0.315** | 0.159 | 0.417** | 0.688** | 1 | ||||
| CHL-1 a | -0.036 | 0.109 | 0.119 | 0.181 | 0.036 | 0.054 | 1 | |||
| CHL-2 b | 0.031 | 0.230* | 0.361** | 0.403** | 0.015 | 0.121 | 0.525** | 1 | ||
| SR-1 a | 0.059 | 0.246* | 0.373** | 0.365** | -0.044 | -0.016 | 0.264** | 0.587** | 1 | |
| SR-2 b | -0.022 | 0.193 | 0.300** | 0.326** | -0.022 | 0.001 | 0.182 | 0.606** | 0.768** | 1 |
*significant at P < 0.05, **significant at P < 0.01.
1a the value at 7 d of heat stress relative the initial value before heat stress.
2b the reduction value at 14d of heat stress relative the initial value.
Abbreviations: TQ-turf quality, ET- evapotranspiration rate, Chl-chlorophyll content. SR-survival rate, GR- Growth rate
Population structure, relative kinship
A total of 1010 SSR alleles were amplified from 90 SSR markers by genotyping 100 tall fescue accessions (Additional file 1 Table S1). The allele numbers of SSR marker varied from 3 to 27 alleles per marker with an average of 11.22 alleles per locus. For the co-dominant SSR marker transit to dominant marker in this study, the genetic diversity of the 100 tall fescue accessions was at a relative lower level, in which average of Nei’s genetic diversity was 0.255, and average of polymorphism information content was 0.211.
According to STRUCTURE analysis results based on Bayesian clustering approach model, a significant population structure was detected among the 100 accessions. The results were consistent with those from the preliminary runs, in which the average probability of the data likelihoods for the population structure in the panel of accessions were increased following the increase of K (Figure 1A). Therefore, the likely number of subpopulations was identified using the Delta method. The optimal number of groups was determined by the maximum likelihood, and k was set at 2 implying two structural groups (G1 and G2) were identified in the panel (Figure 1B). The population structure matrix (Q) identified at k = 2 was applied to define the membership probability for assigning accessions to subpopulation when the value was >0.7 (Additional file 2 Table S2). However, a few of wild accessions were obscure, such as 4 (Q1 = 0.542), 55 (Q2 = 0.578), 62 (Q1 = 0.553), and 96 (Q2 = 0.583). Most of value of accessions (80%) were >0.8. G1 that was the most diverse group contained 74 accessions of mixed origins, including all commercial cultivars (8 accessions) and the majority of wild accessions form European (25/32), U.S. (28/30), Asia (8/20), and South America (3/3) (Figure 2). G2 contained 26 accessions that mainly collected majorly from North Africa (5/6, Tunisia, Algeria, and Morocco), Asia (12/20, China, Iran, Turkey, Israel), European (7/32).
Figure 1.

Calculation of true K of tall fescue accessions and (A) Evolution of the average logarithm probability of the data likelihoods (LnP(D)) for tall fescue genotypes; (B) Magnitude of Δk for each K value according to Evanno et al. [58].
Figure 2.

Population structure analysis of 100 tall fescue accessions. Numbers on the x-axis indicate the accession and those on the y-axis show the group membership.
There was no obvious kinship (K) that detected based on 90 SSR markers in the panel of populations (Figure 3). More than 55.3% of the pair-wise kinship estimates were zero while approximately 89% of estimates were between 0 and 0.05. Less than 5% of estimates were >0.1, indicating that the familial relationships minimum among samples, and would not cause further complexity in association analysis.
Figure 3.
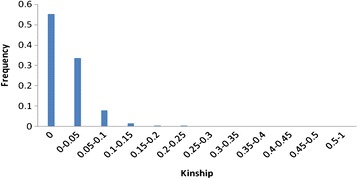
Distribution of pair-wise relative kinship estimates between 100 tall fescue accessions.
Association analysis and evaluation of association model
Combined with all SSR alleles and three traits including turfgrass quality, growth rate and leaf chlorophyll content in growth chamber trial, associations were performed to detect the effects of Q and K for controlling false associations. Owing to the complexity and population structure in out panel, the simple model that overlooked Q and K was not performed. For any trait, the P values from the three models were close to the expected P value (Figure 4). However, the model of Q showed a different distribution with the other models for turfgrass quality and chlorophyll content. On the other hand, the K and Q + K model displayed similar distribution of P values, and the identified associations (P < 0.01) showed the high similarity in both models (Additional file 3 Table S3). The more stringent model was performed, and the less spurious associations were identified. So the results from Q + K model by MLM would be showed and discussed.
Figure 4.

Quantile-quantile plots of estimated –log10 (P) from association analysis using three models in three traits: a turfgrass quality, b growth rate, c leaf chlorophyll content. The black line is the expected line under the null distribution. The blue line represents the observed P values using GLM with Q model; the red line represents the observed P values using MLM model with K; the yellow line represents the observed P values using MLM model with Q and K.
Marker allele-trait associations
In MLM model with Q and K, a total of 97 SSR alleles were associated with five heat-relative traits at two time points (P < 0.01) in greenhouse trial, while that in growth chamber trial resulted in 67 SSR loci that were strongly associated with the 5 traits (P < 0.01) (Table 5, and Additional file 4 Table S4). In greenhouse trial, 15 alleles of marker NAF057 that amplified 22 alleles showed the association with ET at two time points by using Q + K model. The similar results also occurred in marker NFA87 associated with GR-1, marker NFA155 related with TQ-1 in growth chamber trial. Moreover, many marker alleles could be associated with a functional trait, and one marker allele was associated with more than one trait. For example, SSR marker alleles (NAF036-194, NAF013-250, NAFG17-136, NAFG023-207, and NAF138-211) were associated with SR and TQ at two sampling times under heat stress in greenhouse trial. Comparing with the same association alleles in two trials, only 2 marker alleles showed similar associations with traits (Table 6). NFA87-418 that located the linkage group 3B was associated with GR-2, and NFA91-152 was associated with ET-2 in both trials by MLM analysis.
Table 5.
Significant marker-trait associations identified for different traits by Q + K model of MLM
| Trial | Trait | Growth chamber trial | Greenhouse trial | ||||
|---|---|---|---|---|---|---|---|
| Number of markers | P-value range | Phenotypic variation (%) | Number of markers | P-value range | Phenotypic variation (%) | ||
| Greenhouse | GR-1 | 22 | 3.93 × 10-4-0.0096 | 7.23-17.38 | 11 | 3.06 × 10-4-0.0093 | 7.09-13.93 |
| GR-2 | 7 | 6.31 × 10-5-0.0069 | 7.77-21.86 | 2 | 0.0038-0.0057 | 13.22-14.34 | |
| TQ-1 | 8 | 0.0030-0.0096 | 7.11-12.62 | 8 | 1.57 × 10-4-0.0093 | 7.03-14.09 | |
| TQ-2 | 2 | 7.51 × 10-4-0.0087 | 7.20-12.17 | 4 | 0.0030-0.0075 | 9.06-10.10 | |
| CHLT-1 | 5 | 8.59 × 10-4-0.0099 | 7.06-15.51 | 17 | 0.0025-0.0094 | 8.80-12.99 | |
| CHLT-2 | 6 | 0.0024-0.0078 | 8.14-13.94 | 4 | 0.0049-0.0073 | 10.38-11.27 | |
| SR-1 | 2 | 0.0011-0.0073 | 8.77-17.22 | 8 | 8.09 × 10-4-0.0090 | 7.15-15.46 | |
| SR-2 | 4 | 0.0028-0.0065 | 8.90-13.92 | 3 | 0.0089-0.0099 | 7.66-10.99 | |
| ET-1 | 5 | 0.0039-0.0057 | 10.49-13.52 | 20 | 7.29 × 10-4-0.0089 | 7.25-15.17 | |
| ET-2 | 6 | 0.0028-0.0085 | 7.36-12.48 | 20 | 2.86 × 10-4-0.0092 | 7.17-17.06 | |
Abbreviations: TQ-turf quality, ET- evapotranspiration rate, Chl-chlorophyll content. SR-survival rate, GR- Growth rate.
Table 6.
Same marker allele-trait associations with percentage of functional traits after 7 and 14 d of heat stress relative to initial condition of tall fescue accessions for both trials by Q + K model at P < 0.01
| Marker allels | LG a | Greenhouse trial | Growth chamber | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | marker_F | marker_p | markerR2 | Trait | marker_F | marker_p | markerR2 | ||
| NFA87-418 | 3B | GR-2 | 5.9196 | 0.0038 | 0.1434 | GR-2 | 5.9196 | 1.88 E-04 | 0.1916 |
| NFA91-152 | NA | ET-2 | 4.9308 | 0.0092 | 0.1001 | ET-2 | 5.1261 | 0.0077 | 0.1046 |
LGa mean the locus of linkage groups of genetic linkage map of tall fescue according to Sara et al. [21].
Abbreviations: ET- evapotranspiration rate, GR- Growth rate.
Discussion
Heat responses of tall fescue
Heat stress is a major factor that limits growth of cool-season turfgrass on a global scale. Turfgrass survive under high temperature through tolerance or escape mechanisms, which involve many phenotypic and physiological characteristics including growth-restricted, higher photosynthesis rate, stay-green, cell membrane thermal stability, and earliness [28]. High temperature decreased turf quality, caused leaf water deficiency and yellowing, constrained growth, and reduced photosynthesis. So, leaf wilting, turfgrass quality, growth rate, evorpotranspiration rate, and chlorophyll content provided convenient and more efficient measurements for studying turfgrass responding mechanism under unfavorable conditions, which have been intensively applied for screening heat-tolerant germplasm of turfgrass [29-31].
In our trials, tall fescue accessions under heat stress exhibited varying degree of negative effects based on ANOVA analysis. Relatively low TQ, high leaf wilting, reduced CHL and severe water loss characteristics presented the damage level of heat stress of tall fescue. Large variations in these functional traits of accessions from different geographic locations and significant correlations between functional traits would provide the potential for selecting heat tolerant accessions and evaluating reliable SSR marker by association analysis between marker and functional traits. However, heat tolerance mechanisms of tall fescue would be different in the two trials. In growth chamber trial, heat tolerant accessions maintained relatively high growth rate and good turfgrass quality. Simultaneously, heat-sensitive accessions presented lost water rapidly, curled leaves and even died. Meanwhile, in the greenhouse trial, heat tolerant accessions maintained good turfgrass quality and appearance by restraining growth. Similarly heat-sensitive accessions experienced yellowing of leaves and withering. The probable explanation for variations in tolerance mechanisms under heat stress were due to the different stress conditions including soil properties and temperature [32]. In the greenhouse trial the roots temperature was buffered because of properties of soil. But flasks with roots were directly exposed to heat stress in the chamber trial due to utilizing the nutrition solution, which made the turfgrass in flask to be more sensitive to high temperature than in the greenhouse trial. In addition, both trials displayed highly significant correlations in most of the functional traits. This indicated that heat tolerant traits had mutual influence, and these traits could provide adequate parameters for evaluating the heat tolerance in the field. Tall fescue accessions from different collection areas indicated diversity in phenotypic and physiological characteristics [33]. However the trend and level of heat damage of the accessions were roughly consistent in two time points. Therefore, heat tolerant tall fescue accessions would be effectively selected according the phenotypic traits when heat stress conducted early days.
Population structure
Tall fescue accessions native to Europe and North Africa, were introduced to North and South America in the late 1800s. They eventually became a prominent forage grass in 1940s in the United States where many commercial cultivars were produced through selective breeding [34]. Tall fescue samples were collected from more than 40 cities representing diverse geographical origins. So in view of the geographical origins, local adaptation, and breeding history of genotypes in association mapping panel, the non-independent samples would often encompass both population structure and familiar relatedness [35,36]. In our study, the Bayesian clustering approach model based analysis divided the panel of samples into two sub-populations. The most of the accessions from European, North America, and all commercial cultivars were separated into the main subpopulation. The wild accessions from North Africa and Asia were separated into the second subpopulation. The division rule cannot be simple explained geographically due to overlapping of several accessions from the same region (European and Asia) in two groups, which indicated regional breeding objectives, the probable different evolutionary paths and methods of ecological adaptation in morphology and agronomic characteristics of ecogeographic races would be considered [37,38].
Presence of population structure could make some allele frequencies significantly differ between subpopulations, which would lead to spurious association (false positives) of markers with traits [39]. Flint-Garcia et al. [40] presented that 33 to 35% of variation of phenotypic traits about flowering time in a diverse maize panel would be attributed to population structure. Therefore, if subpopulation structure is not taken into account, spurious associations may be identified at other loci that were differentially distributed among subpopulations. Moreover, spurious associations cannot be controlled entirely by GLM model. This is because the Q matrix can only carry a rough dissection of population differentiation. Therefore, a unified mixed-model approach for association mapping that incorporates the pairwise kinship (K matrix) and Q matrix to correct multiple levels of relatedness have been developed. This would be a powerful approach for improving accuracy of association in many cases [40,41]. Kang [42] demonstrated that the distribution of P values ideally should follow a uniform distribution with less deviation from the expected P value. In our panel, SSR marker-trait associations were performed for three traits using the Q, K, and Q + K models, and all the three showed a good fit for P values. However, the models showed the different effects of controlling the population structure for different traits. K model was more superior to the Q model, but similar to the Q + K model. This is consistent with some previous studies [41,43]. The K matrix could capture the relatedness between each possible pair of individual in panel. The Q matrix considers a few axes only [44]. Consequently, no vivid familiar relatedness (from the recent co-ancestry) has been detected in the panel. Therefore a model that would test for complex quantitative traits would be necessary for improving the accuracy of association.
Marker allelic effects on functional traits
Little is known about the association of SSR loci with heat tolerance related traits in plant species. In our study 97 marker-trait associations (MTAs) in greenhouse trial and 67 MTAs were identified for five heat tolerant traits at two time points (P < 0.01). A total of 13 MTAs in greenhouse trial and 29 MTAs in growth chamber trial were identified in present study for growth rate. High temperature would affect pollen viability, fertilization and seed development leading to yield losses. A large number of MTAs for yield and yield related traits under unfavorable conditions were reported in many crop species [7,8,13]. Furthermore, most of marker NAF057 amplifying 22 alleles were associated with ET in greenhouse trial, which implied the marker may be linked with a crucial gene that is necessary for regulating water loss and transpiration cooling under heat stress [45]. Similar results were observed between survival rate and turfgrass quality in greenhouse trial, suggesting survival rate, turfgrass quality and evapotranspiration rate are vital functional traits reflecting heat tolerance of tall fescue and might be regulated by genetically linked homologous genes. Therefore, these associated markers and identified genotypes with favorable alleles can be deployed after validation for molecular marker breeding to develop heat tolerant in tall fescue.
It is interesting to found that many marker alleles presented significant association with single trait, or associations with more than one trait. For instance, 5 associations were associated with SR and TQ in greenhouse trial, which would be considered to be pleiotropic or co-localized MTAs [8]. These co-localized or pleiotropic associations may be beneficial to detect some important genomic regions or genes for heat tolerance related traits. Furthermore, the markers associated with more than one trait may be made effectively use of improving more than one trait by marker assisted selection.
For screening heat tolerant accessions by phenotypic and physiological traits, our experimental population is relative small, which influence the power of association analysis. Yan et al. [46] showed that association study with a set of 500 individuals would supplyan 80% probability of detecting a gene that explains 3% or more of the phenotypic variation, and increasing the number of population could be more substantial effect on the power of MTAs than increasing the density of markers in genome-wide association (GWS). More reliable markers could be identified for developing elite heat tolerant tall fescue cultivars through marker assisted selection under various conditions: first if higher density DNA polymorphism databases would have been evenly distributed in all genome chromosomes. And secondly larger mapping populations and phenotypic traits under more sites of heat stress would have been used for association mapping.
A large challenge for association analysis for complex quantitative traits in plant is large number of loci identified with small effects in some plant species such as barley, maize, wheat and rice. In our study by MLM analysis, the explained variation (Marker R2) for the identified associations were low to modest, ranging from 7.03% (turfgrass quality)-19.21% (turfgrass quality) in greenhouse trial, and 7.06% (leaf chlorophyll content) -21.86% (growth rate) in growth chamber trial, respectively. The explained variation by marker-trait associations (MTAs) for abiotic stress related traits in association analysis of plant species is changeable. Thudi et al. (2014) used DArT, SNP, and SSR markers to study 300 accessions of chickpea for drought tolerance related root traits, heat tolerance, yield and yield component traits cross 6 environments, and showed that phenotypic variance explained of MTAs ranged from low (4.14%) to very high (96.55%). However, Varshney et al.[47] studying a diverse barley panel at a dry and wet location for drought tolerance related traits, found that explained variation for all of identified MTAs was rather low, ranging from 0.1% to 6.7%. Some other studies on GWA analysis in barley also showed that MTAs contributing large phenotypic variation are highly heritable, and MTAs of explained variation >10% seem hard to be identified for the complex quantitative traits like drought tolerance in association analysis [6,48]. Large effect QTL may be due to the inbreeding nature of some species, while out-crossing plants such as tall fescue and maize may have very large number of genes contributing a very small amount to a quantitative trait [46].
Associations identified in our study were not only small, but also little consistent across environments like barley or wheat. In the two trials of our study, there were only two associated SSR alleles were identified in two trials at low threshold, -Log (P-value) ≥2.0 by MLM analysis, which showed influence of environment on heat tolerance related traits that would be low heritability. The observed differences of marker-trait associations in both trials may be due to the different experimental conditions of heat stress, including temperature, heat intensity, duration, and matrix cultivated, which lead to the variation of phenotypic, physiologic and biochemical characteristics in response to heat stress, and even trigger different genetic pathways and mechanisms of heat tolerance. In greenhouse trial, the temperature of greenhouse often exceeded 45°C in summer, and reached 50°C at the noon, which restrained growth of tall fescue and caused severe thermal damage. Most tall fescue accessions halted growth, withered rapidly, and the leaves yellowed after 2 or 3 day of heat stress. However, the growth chambers controlled the temperature at 35°C moderate high temperature. The extreme high temperature would induce specific membrane damage, expression of HSP [49], and alteration of activity of enzymes, which was not prevalent at moderate heat stress [50]. Simultaneously, immersing grasses into nutrient solution in growth chamber trial made the roots that are more sensitive to heat stress than leaves to be directly exposed to high temperature and severe damage [51,52]. Therefore, many heat sensitive tall fescue accessions presented dehydration wilting and even death in growth chamber trials. Specifically, many factors resulted in the differences of phenotypic and physical traits when tall fescue accession responded to heat stress, which made a few SSR alleles associated with functional traits to be simultaneously identified. The observation that the majority of the SSR alleles associated with heat tolerant-related traits could only be identified in a specific condition of heat stress indicated that tall fescue is very sensitive to variation of high temperature. The similar conditions had also reported in association analysis for drought tolerance related traits of barley, wheat, maize, and chickpea [7,8,13,46]. So identified markers may be not suitable for direct application in marker-assisted selection (MAS) programme for developing more stable heat tolerant tall fescue varieties or cultivars. Vast studies including complex crosses and QTLs mapping with well chosen parents on the basis of results obtained in our study for verifying effectiveness of marker alleles are necessary for breeding heat tolerance cultivars by marker assisted selection.
Conclusion
In summary, we initial focus on association mapping analysis of heat tolerance-related functional traits in tall fescue. Five quantitative traits GR, TQ, SR, CHL and ET showed high diversity and significant mutual correlations in response to heat stress in tall fescue. Two subpopulations were detected in the panel of accessions, but no obvious relative kinship was observed. But for any trait, the K model controlling relative kinship showed the similar distribution of P value and associations with Q + K model that controlling both population structure and relative kinship in our study. So model testing is necessary to reduce the spurious associations. By mixed linear model (Q + K) as the best model for association analysis, 97 associations in greenhouse trial and 67 associations in chamber trial were identified for five heat tolerant traits at two time points (P < 0.01). It is necessary for tall fescue selection breeding because these markers would enhance efficiency of identifying heat tolerant accessions bringing desirable alleles. However, only two SSR alleles associated with GR and ET were identified due to the different environments between two trials. And inadequate samples and limited markers were utilized in our study which might have weakened the reliability and effectiveness of associated SSR markers. Hence, it was necessary to confirm the associated marker locus by genotypes F2 grasses and phenotype F3 progeny, or QTL mapping with a high resolution linkage mapping in the next step. Simultaneously, for identification of more effective markers or genes by association analysis, further research need to focus on selecting candidate genes regulating heat tolerance of tall fescue or developing a large amount of single nucleotide polymorphism for genotyping larger association population.
Methods
Plant materials and growth conditions
100 diverse accessions of tall fescue were employed in this study, including 93 accessions obtained from the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) and 7 turf-type commercial cultivars obtained from the seed industry (Table 7). The collection of accessions was based on geographical locations for maximizing genotypic diversity. All accessions were confirmed to be hexaploid by flow cytometry (data not shown). This study was conducted at Wuhan Botanical Garden, Chinese Academy of Science, beginning in 2012. A single seed from each accession was sown in petri dishes with a layer of filter paper soaked in water and kept in dark at 22°C for germination. After one week, the accessions were transplanted into plastic pots (15 cm deep, 11 cm wide) containing a mixture of sand and soil (1:1, v/v) in a greenhouse with temperature ranging from 20°C to 26°C, 1000-1500 μmol photons m-2 s-1, 14 h photoperiod of natural sunlight, and 76% average relative humidity. Plants were irrigated daily to maintain sufficient water supply conditions, fertilized weekly with half-strength Hoagland’s solution [53], and mowed to 7 cm canopy height once a week. Each accession was propagated through tillers multiple times for genetic uniformity.
Table 7.
Origin and grouping information of tall fescue accessions used in this research
| ID a | PI number | Origin | Q b | ID a | PI number | Origin | Q b |
|---|---|---|---|---|---|---|---|
| 1 | Justice | Cultivar | 1 | 51 | PI 438521 | Japan | 1 |
| 2 | PI 527504 | France | 1 | 52 | PI 442490 | Belgium | 1 |
| 3 | PI 531230 | USA | 1 | 53 | PI 469244 | USA | 1 |
| 4 | PI 595072 | Romania | 1 | 54 | PI 499494 | China | 1 |
| 5 | PI 596701 | USA | 1 | 55 | PI 499495 | China | 2 |
| 6 | PI 598491 | Netherlands | 1 | 56 | PI 174210 | Turkey | 2 |
| 7 | PI 598493 | Romania | 1 | 57 | PI 200339 | Israel | 2 |
| 8 | PI 636532 | Tunisia | 2 | 58 | PI 203728 | Uruguay | 1 |
| 9 | PI 636597 | USA | 1 | 59 | PI 208679 | Algeria | 2 |
| 10 | PI 636601 | France | 1 | 60 | PI 208681 | Algeria | 2 |
| 11 | PI 538006 | USA | 1 | 61 | PI 211032 | Afghanistan | 1 |
| 12 | PI 538330 | USA | 1 | 62 | PI 224975 | South Africa | 1 |
| 13 | PI 577082 | Yugoslavia | 1 | 63 | PI 231563 | Portugal | 2 |
| 14 | PI 578717 | USA | 1 | 64 | PI 234881 | Switzerland | 2 |
| 15 | PI 578718 | USA | 1 | 65 | PI 234883 | Switzerland | 1 |
| 16 | PI 578724 | USA | 1 | 66 | PI 235036 | Sweden | 1 |
| 17 | PI 583747 | USA | 1 | 67 | PI 235125 | Netherlands | 1 |
| 18 | PI 583822 | USA | 1 | 68 | PI 249738 | Greece | 1 |
| 19 | PI 655104 | USA | 2 | 69 | PI 257742 | Sweden | 1 |
| 20 | PI 655112 | USA | 1 | 70 | PI 269894 | Pakistan | 1 |
| 21 | PI 655113 | USA | 2 | 71 | PI 274617 | Poland | 2 |
| 22 | PI 423090 | Spain | 2 | 72 | PI 283281 | UK | 1 |
| 23 | PI 422638 | France | 1 | 73 | PI 283304 | Denmark | 1 |
| 24 | PI 502373 | Russian | 1 | 74 | PI 311044 | Romania | 2 |
| 25 | PI 504538 | Greece | 1 | 75 | PI 314685 | Russian | 2 |
| 26 | PI 577094 | Switzerland | 1 | 76 | PI 380844 | Iran | 2 |
| 27 | PI 505833 | Kazakhstan | 2 | 77 | PI 388897 | Japan | 1 |
| 28 | PI 508603 | Argentina | 1 | 78 | PI 388898 | Japan | 1 |
| 29 | PI 578719 | USA | 1 | 79 | PI 578714 | USA | 1 |
| 30 | PI 512305 | Portugal | 1 | 80 | PI 601106 | USA | 1 |
| 31 | PI 512315 | Spain | 1 | 81 | PI 601227 | USA | 1 |
| 32 | PI 547396 | Iran | 1 | 82 | PI 601447 | USA | 1 |
| 33 | PI 559374 | USA | 1 | 83 | PI 608024 | USA | 1 |
| 34 | PI 561430 | USA | 1 | 84 | PI 608025 | USA | 1 |
| 35 | PI 574522 | USA | 1 | 85 | PI 619025 | China | 2 |
| 36 | PI 577081 | Yugoslavia | 1 | 86 | PI 632516 | USA | 1 |
| 37 | PI 598496 | Hungary | 1 | 87 | PI 608787 | USA | 1 |
| 38 | PI 598574 | Kazakhstan | 2 | 88 | PI 600739 | USA | 1 |
| 39 | PI 598930 | Italy | 1 | 89 | PI 600801 | USA | 1 |
| 40 | PI 598860 | Morocco | 2 | 90 | 3rd Millennium | Cultivar | 1 |
| 41 | PI 610909 | Morocco | 1 | 91 | Stone wall | Cultivar | 1 |
| 42 | PI 610933 | Italy | 1 | 92 | Davinci | Cultivar | 1 |
| 43 | PI 610951 | Morocco | 2 | 93 | Pixie | Cultivar | 1 |
| 44 | Pure Gold | Cultivar | 1 | 94 | PI 184041 | Yugoslavia | 1 |
| 45 | PI 618971 | China | 2 | 95 | PI 255874 | Poland | 1 |
| 46 | PI 618973 | China | 2 | 96 | PI 283287 | Czechoslovakia | 2 |
| 47 | PI 619005 | China | 2 | 97 | PI 578713 | USA | 1 |
| 48 | PI 440345 | Russian | 2 | 98 | PI 608808 | USA | 1 |
| 49 | PI 423045 | Spain | 2 | 99 | Grand II | Cultivar | 1 |
| 50 | PI 427127 | Chile | 1 | 100 | Smirna | Cultivar | 1 |
aID number representing accessions used in this research.
bQ identified population structure groups in this research.
Heat treatment and experimental design
Two trails were conducted. One was processed in the greenhouse in June, 2012, the other in growth chambers repeated in August, September, and October 2012, respectively.
Greenhouse trail:
All 100 accessions were transferred into a natural greenhouse in June 8th to July 14th, 2012 after growing in the controlled greenhouse for 30 d. The maximum temperatures varied from 39°C to 51°C during 21 d of heat treatment. Each accession had three replications with same genotypes, and all plots were arranged in a completely randomized block design. The greenhouse had a photosynthetically active radiation (PAR) of 1000-2000 μmol s-1 m-2 of natural sunlight. Grasses were irrigated daily until water could freely drain from the holes under the plots.
-
2.
Growth chamber Experiment
The trial was repeated three times in growth chamber in August, September and October 2012. 100 accessions were transformed into 250 mL Erlenmeyer flask wrapping with aluminum foil, containing half-strength Hoagland’s solution and 0.1 μmol magnesium oxide to provide additional oxygen after 30 d growing in controlled greenhouse. Grasses with 7-10 tillers were sealed with parafilm to prevent water escaping from gaps. Before heat treatment, all flasks of grasses were pre-incubated 10 d. Two growth chambers during experimental period were controlled in 14 photoperiod, 70% ± 10% relative humidity, and approximately average 450 μmol photons m-2 s-1. Every other day all flasks were exchanged layers and half-strength Hoagland solution added. This trail included an unheated control (25/16°C, day/night) and heat stress (38/30°C, day/ night) treatment sustaining 15 d. The heat treatment was subjected in different chambers for each replication.
Growth and physiological measurements
Many growth and physiological traits were measured before and after heat stress interval 7 d in two trails, including turf grass quality (TQ), survival rate (SR), leaf chlorophyll content (CHL), evapotranspiration rate (ET) and growth rate (GR) . Turf quality was evaluated visually using a scale of 0 (yellow, brown or dead) to 9 (optimum greenness, uniformity, cover) based on density, texture, turf color, and smoothness. Survival rate was also assessed by visual rate using a ratio between survival canopy and total plant. Every 7 d leaves of pots were cut at 7 cm canopy height, were collected, immediately killed at 105°C 30 min, dried at 70°C in an oven for 72 h. Growth rate was calculated as dry weight per growth day. Evapotranspiration rate was measured by weight loss of the plant plot every 24 h and the relative transproation was normalized according to a method described by Hu et al. [54]. Leaf chlorophyll content was measured using the method described by Hiscox and Israelstam [55].
Data was collected from the non-heat and heat treatment across all accessions of tall fescue from two trails to examine the efficiency and consistency. The percentage of reduction of all traits, calculated as [(control value or initial value -heat value)/ control or initial value] × 100, was used to indicated the grass heat tolerance. The main treatment effect, variance analysis (ANOVA) and correlation between growth and physiological traits were performed using SPSS18.0 (IBM Corporation, New York, USA).
DNA isolation and SSR analysis
Young leaves of each accession were collected for DNA isolation using a cetyltrimethyl ammonium bromide (CTAB) method [56]. A set of 90 published genome-wide SSR markers [21,57] mapped in 22 linkage groups in tall fescue were analyzed in all accessions (Additional file 1 Table S1). All forward primer sequence of markers were labeled with four fluorescent dyes of different colors [FAM (blue), HEX (green), TAMRA (yellow), and ROX (red)]. Each 10 μL PCR reaction in 96 microplates consisted of 1 × supplied Taq-buffer, 2.5 mM MgCl2, 200 μM dNTPs, 0.2 mM of each primer pair, 0.5 U of Taq DNA polymerase, and 30 ng of template DNA. PCR reaction was started at 95°C for 10 min; followed by 25 cycles of 50 s at 95°C, 50 s at 68°C with a decrease of 0.6°C in each consequent cycle, 60 s at 72°C; then ran for 15 cycles at 95°C for 50 s, 54°C for 50 s, 72°C for 60 s; and a final extension step 72°C for 10 min. All PCR reactions were used a touch-down program in a 96-well My Cycler thermal cycler (Bio-Rad Inc., Hercules, CA, USA). The PCR amplified fragments were separated by an ABI 3730 DNA Sequence (Applied Biosystems Inc., Foster City, CA, USA). Alleles were scored by GeneMarker 1.5 software (Soft Genetics, LLC, State College, PA, USA) and checked twice manually for accuracy. If more than one fragment were amplified by a primer in accession and appeared differently in other accessions, they were scored as different loci. For allohexaploid genome of F. arundinacea, band scores of SSR loci were entered into a binary matrix as presence (1) or absence (0) following Sara et al. [21]. All confirmed polymorphic alleles were applied for population structure and kinship analysis.
Population structure and relative kinship
As a result of labeling all SSR markers as dominant in each genotype, no information on marker linkage could be obtained in population structure model. A Bayesian model-based clustering method carried out in STRUCTURE 2.0.1 software [58] was employed to determine population structure (Q) and division accessions into subpopulation. The basis of the clustering method is that it prevented admixture of correlated allele frequencies, therefore the allocation of individual genotype to K subpopulations is in such a way that Hardy-Weinberg and linkage equilibrium is valid within populations. The structure was run ten times by setting pre-defined k (the number of population groups) ranging from 1 to 15 using admixture models with 10,000 MCMC (Markov Chain Monte Carlo) replications and 10,000 burn-in time for each run. Population based on the maximum likelihood was determined by the probability of data likelihood LnP(D) in the output and an ad hoc statistic △K based on the second-order rate of change in LnP(D) between successive K values [58]. 15 independent runs were operated 100 000 iterations of each run after burn-in of 100 000 for a value of K setting from one to five. Then SPAGeDi software [59] was applied to evaluating relative pairwise kinship (K) by 90 SSR markers, and then the pairwise kinship matrix (100 × 100) was produced by the Loiselle coefficient [60]. All negative kinship values between the individuals were assigned to zero, according to Yu et al. [41].
Model testing and association mapping
Based on the differences in the regime of heat treatment (density, duration time and matrix cultivated plant), the functional traits of heat tolerant in two trials were used for identifying association with SSR loci, respectively. Turfgrass quality, growth rate and leaf chlorophyll content in growth chamber trial were selected to perform marker-trait associations. Three models were used to access the effects of relative kinship (K) and population structure (Q) for marker-trait associations. The Q model was performed using general linear model (GLM). The K and K + Q models were performed using MLM in TASSEL 2.0.1 software [58]. The quantile- quantile plots of estimated –log10 (P) were drawn using the observed P values from SSR alleles-trait associations and the expected P values assuming that there was no associations identified between marker and trait. The significant threshold for marker-trait associations was set at P < 0.01.
Acknowledgement
This research was supported by the General Program (Grant #: 31071822; 31470363) from the National Natural Science Foundation of China, the National High Technology Research and Development Program (No.2011AA100209-2) from “863” plan of China, and the Special Fund of Industrial (Agriculture) Research (No.200903001) for Public Welfare of China. We thank the United States Department of Agriculture––Agricultural Research Service (USDA-ARS) for contributing germplasm from their collection.
Abbreviations
- CHL
Chlorophyll content
- ET
Evapotranspiration rate
- FW
Fresh leave weight
- GLM
General linear model
- GR
Growth rate
- GWS
Genome-wide association
- MAS
Marker-assisted selection
- MLM
Mixed linear model
- MTA
Marker-trait association
- SR
Survival rate
- SSR
Simple sequence repeat
- TQ
Turfgrass quality
Additional files
Supplementary Table S1. List of the amplified information of polymorphism SSR markers.
Supplementary Table S2. The values of membership probability for assigning tall fescue accessions to subpopulation when the k = 2.
Supplementary Table S3. The associated marker alleles with five traits at two time points by Q model, K model and Q + K model in growth chambers trial (P < 0.01).
Supplementary Table S4. The associated marker alleles with five functional traits at two sampling times by Q + K model in greenhouse trial and growth chambers trial, respectively (P < 0.01).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XYS performed the experiments and wrote the manuscript. JR and ZMD analyzed the data. JMF and TH conceived and designed the experiments. EA and TH helped to draft the manuscript and revise the manuscript. All authors read and approved the final manuscript.
Contributor Information
Xiaoyan Sun, Email: 544842269@qq.com.
Zhimin Du, Email: 283452193@qq.com.
Jin Ren, Email: renjing879@163.com.
Erick Amombo, Email: amomboeric@gmail.com.
Tao Hu, Email: hut420@wbgcas.cn.
Jinmin Fu, Email: jfu@wbgcas.cn.
References
- 1.Seal AG. DNA variation in Festuca. Heredity. 1983;50:225–36. doi: 10.1038/hdy.1983.26. [DOI] [Google Scholar]
- 2.Wang JZ, Cui LJ, Wang Y, Li JL. Growth, lipid peroxidation and photosynthesis in two tall fescue cultivars differing in heat tolerance. Biologic Plantarum. 2009;53(2):237–42. doi: 10.1007/s10535-009-0045-8. [DOI] [Google Scholar]
- 3.Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327:818–22. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- 4.Hattori Y, Nagai K, Furukawa S. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–30. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 5.Konishi S, Izawa T, Lin SY. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1306. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 6.Korff V, Wang MH, Léon J, Pillen K. AB-QTL analysis in spring barley: II Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum) Theor Appl Genet. 2006;112:1221–31. doi: 10.1007/s00122-006-0223-4. [DOI] [PubMed] [Google Scholar]
- 7.Rahman H, Pekic S, Lazic-Jancic V, Quarrie SA, Shah SM, Pervez A, et al. Molecular mapping of quantitative trait loci for drought tolerance in maize plant. Genet Mol Res. 2011;10(2):889–901. doi: 10.4238/vol10-2gmr1139. [DOI] [PubMed] [Google Scholar]
- 8.Mahendar T, Hari DU, Abhishek R. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. Plos one 2014, doi:10.1371/journal.pone. 0096758 [DOI] [PMC free article] [PubMed]
- 9.Gupta PK, Rustgi S, Kulwal PL. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol. 2005;57(4):461–85. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- 10.Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES. Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetic. 2001;28:286–9. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 11.Aranzana MJ, Kim S, Zhao K. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 2005;1(5) doi: 10.1371/journal.pgen.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Wei X, Sang T, Zhao Q, Feng Q. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–7. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 13.Liu SX, Wang XL, Wang HW, Xin HB. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance ant seedling stage of Zea mays L. Plos Genetics. 2013;9(9) doi: 10.1371/journal.pgen.1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X, Bai G, Liu S. Association of candidate genes with drought tolerance traits in diverse perennial ryegrass accessions. J Exp Bot. 2013;64(6):1537–51. doi: 10.1093/jxb/ert018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auzanneau J, Huyghe C, Escobar-Gutiérrez AJ, Julier B, Gastal F, Barre P. Association study between the gibberellic acid insensitive gene and leaf length in a Lolium perenne L. synthetic variety. BMC Plant Biol. 2011;11:183. doi: 10.1186/1471-2229-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang JC, Camberato JJ, Yu XQ, Luo N, Bian SM, Jiang YW. Growth response, carbohydrate and ion accumulation of diverse perennial ryegrass accessions to increasing salinity. Sci Hortic. 2013;154:73–81. doi: 10.1016/j.scienta.2013.02.021. [DOI] [Google Scholar]
- 17.Yu X, Bai G, Luo N. Association of simple sequence repeat (SSR) markers with submergence tolerance in diverse populations of perennial ryegrass. Plant Sci. 2011;180(2):391–8. doi: 10.1016/j.plantsci.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Cipriani G, Spadotto A, Jurman I, Di Gaspero G, Crespan M, Meneghetti S, et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor Appl Genet. 2010;121:1569–85. doi: 10.1007/s00122-010-1411-9. [DOI] [PubMed] [Google Scholar]
- 19.Garris A, Tai T, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–8. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L, et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet. 2004;109(7):1448–58. doi: 10.1007/s00122-004-1760-3. [DOI] [PubMed] [Google Scholar]
- 21.Saha MC, Mian R, Zwonitzer JC. An SSR-and AFLP-based genetic linkage map of tall fescue (Festuca arundinacea Schreb.) Theor Appl Genet. 2005;110(2):323–36. doi: 10.1007/s00122-004-1843-1. [DOI] [PubMed] [Google Scholar]
- 22.Xu WW, Sleper DA, Chao S. Genome mapping of polyploid tall fescue ( Festuca arundinacea Schreb) with RFLP markers. Theor Appl Genet. 1995;91:947–55. doi: 10.1007/BF00223905. [DOI] [PubMed] [Google Scholar]
- 23.Hand ML, Cogan NO, Forster JW. Molecular characterisation and interpretation of genetic diversity within globally distributed germplasm collections of tall fescue (Festuca arundinacea Schreb.) and meadow fescue (F. pratensis Huds.) Theor Appl Genet. 2012;124:1127–37. doi: 10.1007/s00122-011-1774-6. [DOI] [PubMed] [Google Scholar]
- 24.Romina C, Beatriz R, Elba P. Genetic diversity in a world germplasm collection of tall fescue. Genet Mol Biol. 2013;36(2):237–42. doi: 10.1590/S1415-47572013005000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tehrani MS, Mardi M, Sahebi J, Catalán P, Díaz-Pérez A. Genetic diversity and structure among Iranian tall fescue populations based on genomic-SSR and EST-SSR marker analysis. Plant Syst Evol. 2009;282:57–70. doi: 10.1007/s00606-009-0207-3. [DOI] [Google Scholar]
- 26.Breseghello F, Sorrells ME. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–77. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Lu XQ, Song XF, Yan JB, Song TM. Mapping quantitative trait loci for oil, starch, and protein concentrations in grain with high-oil maize by SSR markers. Euphytica. 2008;162:335–244. doi: 10.1007/s10681-007-9500-9. [DOI] [Google Scholar]
- 28.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: an overview. Environ Exp Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 29.Jansen M, Gilmer F, Biskup B. Simultaneous phenotyping of leaf growth and chlorophyll luorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol. 2009;36:902–14. doi: 10.1071/FP09095. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt JM, Sears RG, Paulsen GM. Wheat high temperature tolerance during reproductive growth. I. Evaluation by chlorophyll fluorescence. Crop Sci. 1990;30:881–5. doi: 10.2135/cropsci1990.0011183X003000040024x. [DOI] [Google Scholar]
- 31.Petkova V, Denev I, Cholakov D, Porjazov I. Field screening for heat tolerant common bean cultivars (Phaseolus vulgaris L.) by measuring of chlorophyll fluorescence induction parameters. Hortic Sci. 2007;111:101–6. doi: 10.1016/j.scienta.2006.10.005. [DOI] [Google Scholar]
- 32.Sun XY, Hu LX, Xie Y, Fu JM. Evaluation of genotypic variation in heat tolerance of tall fescue by functional traits. Euphytica. 2014;199(3):247–60. doi: 10.1007/s10681-014-1122-4. [DOI] [Google Scholar]
- 33.Majidi MM, Mirlohi A, Amini F. Genetic variation, heritability and correlations of agro-morphological traits in tall fescue (Festuca anndinacea Schreb.) Euphytica. 2009;167:323–31. doi: 10.1007/s10681-009-9887-6. [DOI] [Google Scholar]
- 34.Boller B, Posselt U, Veronesi F. Fodder Crops and Amenity Grasses [M]// Handbook of Plant Breeding. New York: Spring Science Business Media; 2010. [Google Scholar]
- 35.Yu J, Buckler ES. Genetic association mapping and genome organization of maize. Curr Opin Biotechnol. 2006;17:155–60. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 37.Burner DM, Balasko JA, O’Brien PM. Attributes of tall fescue germplasm of diverse geographic origin. Crop Sci. 1988;28(3):459–62. doi: 10.2135/cropsci1988.0011183X002800030005x. [DOI] [Google Scholar]
- 38.Robson MJ, Jewiss OR. A comparison of British and North African varieties of tall fescue (Festuca arundinacea). III. Effects of light, temperature and day length on relative growth rate and its components. J Appl Ecol. 1968;5:191–204. doi: 10.2307/2401283. [DOI] [Google Scholar]
- 39.Ersoz ES, Yu J, Buckler ES. Applications of linkage disequilibrium and association mapping in maize. In: Kriz AL, Larkins BA, editors. Molecular Genetic Approaches to Maize Improvement. New York, NY: Springe-Verlag; 2009. pp. 173–95. [Google Scholar]
- 40.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Yu J, Pressoir G, Briggs W, Bi I, Yamasaki M, Doebley J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetic. 2006;38:203–8. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 42.Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178(3):1709–23. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Yan JB, Trushar S, Marilyn L, Zheng Y. Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor Appl Genet. 2010;121:417–31. doi: 10.1007/s00122-010-1320-y. [DOI] [PubMed] [Google Scholar]
- 44.Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang ZW, Costich DE, et al. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonos SA, Murphy JA. Growth response and performance of Kentucky Bluegrass under summer stress. Crop Sci. 1999;39:770–4. doi: 10.2135/cropsci1999.0011183X003900030026x. [DOI] [Google Scholar]
- 46.Yan J, Warburton M, Crouch J. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 2011;51:433–49. doi: 10.2135/cropsci2010.04.0233. [DOI] [Google Scholar]
- 47.Varshneya RK, Paulo MJ, Grando S, Graner A. Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.) Field Crop Res. 2012;126:171–80. doi: 10.1016/j.fcr.2011.10.008. [DOI] [Google Scholar]
- 48.Wang M, Jiang N, Jia T, Leach L, Cockram J, Waugh R, et al. Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor Appl Genet. 2011 doi: 10.1007/s00122-011-1697-2. [DOI] [PubMed] [Google Scholar]
- 49.Vierling RA, Nguyen HT. Heat shock protein gene expression in diploid wheat genotypes differing in thermal tolerance. Crop Sci. 1992;32:370–7. doi: 10.2135/cropsci1992.0011183X003200020019x. [DOI] [Google Scholar]
- 50.Stone PJ, Nicolas ME. A survey of the effects of high temperature during grain filling on yield and quality of 75 wheat cultivars. Aust J Agr Res. 1995;46:475–92. doi: 10.1071/AR9950475. [DOI] [Google Scholar]
- 51.Huang BR, Liu XZ, Fry JD. Effects of high temperature and poor soil aeration on root growth and viability of creeping bentgrass. Crop Sci. 1998;38:1618–22. doi: 10.2135/cropsci1998.0011183X003800060034x. [DOI] [Google Scholar]
- 52.Xu QZ, Huang BR. Effects of differential air and soil temperature on carbohydrate metabolism in creeping bentgrass. Crop Sci. 2000;40:1368–74. doi: 10.2135/cropsci2000.4051368x. [DOI] [Google Scholar]
- 53.Hoagland DR, Arnon DI: The water-culture method for growing plants without soil. Circular. Circular. Calif Agric Exp Circ 1950, pp 247.
- 54.Hu T, Li HY, Zhang XZ, Luo HJ, Fu JM. Toxic effect of NaCl on ion metabolism, antioxidative enzymes and gene expression of perennial ryegrass. Ecotoxicol Environ Saf. 2011;77:2050–6. doi: 10.1016/j.ecoenv.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Hiscox JD, Israelstam GF. A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot. 1979;57:1332–4. doi: 10.1139/b79-163. [DOI] [Google Scholar]
- 56.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–5. [Google Scholar]
- 57.Saha MC, Cooper JD, Mian MAR. Tall fescue genomic SSR markers: development and transferability across multiple grass species. Theor Appl Genet. 2006;113(8):1449–58. doi: 10.1007/s00122-006-0391-2. [DOI] [PubMed] [Google Scholar]
- 58.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 59.Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecology Notes. 2002;2:618–20. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- 60.Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae) Am J Bot. 1995;82:1420–5. doi: 10.2307/2445869. [DOI] [Google Scholar]


