Summary
Noncoding RNAs (ncRNAs) function with associated proteins to effect complex structural and regulatory outcomes. To reveal the composition and dynamics of specific noncoding RNA- protein complexes (RNPs) in vivo, we developed comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS). ChIRP-MS analysis of four ncRNAs captures key protein interactors, including a U1-specific link to the 3′ RNA processing machinery. Xist, an essential lncRNA for X-chromosome inactivation (XCI), interacts with 81 proteins from chromatin modification, nuclear matrix, and RNA remodeling pathways. The Xist RNA-protein particle assembles in two steps coupled with the transition from pluripotency to differentiation. Specific interactors include HnrnpK that participates in Xist-mediated gene silencing and histone modifications, but not Xist localization and Drosophila Split ends homolog Spen that interacts via the A-repeat domain of Xist and is required for gene silencing. Thus, Xist lncRNA engages with proteins in a modular and developmentally controlled manner to coordinate chromatin spreading and silencing.
Introduction
Many lncRNAs are recently recognized as functional regulators of gene expression (Rinn and Chang, 2012), but their mechanisms of action are largely unknown. RNA-binding proteins (RBPs) play key roles in lncRNA-mediated gene regulation, and obtaining the full interaction map of proteins bound to a lncRNA of interest is critical to our understanding of its function. Many tools have been developed to describe RNA-protein interaction from a protein-centric view, typically by immunoprecipitating a protein and analyzing the associated RNAs with a microarray or high-throughput sequencing (reviewed by Riley and Steitz, 2013). In contrast, fewer methods are available from the perspective of a particular RNA. This is usually achieved by 1) tagging the RNA with affinity-aptamers, which involves complicated genetic engineering; 2) using in-vitro transcribed RNA to retrieve proteins from native cell lysates (RNA chromatography), which is prone to the formation of non-physiological RNA-protein interactions; 3) using immobilized oligonucleotides to capture RNA:protein complex under native conditions, which suffers from both post-lysis re-associations and unpredictable specificity of target RNA retrieval (reviewed by Chu et al., 2015). The ideal strategy should capture in vivo lncRNA-protein interactions, achieve high yield and specificity without genetic tagging, and provide comprehensive portraits of lncRNP in diverse biological states.
Xist is a lncRNA (17Kb long in the mouse) required for X-chromosome inactivation (XCI) of one of the two X-chromosomes in female cells, thus enabling dosage compensation between XX females and XY males (Gendrel and Heard, 2011). XCI takes place early in embryonic development, and is thought to occur in multiple steps: counting and choosing the X chromosome to silence, spreading of Xist over the target X chromosome, and silencing of most of its active genes (Payer and Lee, 2008). The latter two steps are believed to be mediated by specific Xist-associated protein factors, which remain largely mysterious. Xist expression marks the future inactive X chromosome (Xi) and is sufficient to recruit silencing chromatin modifications complex such as the Polycomb proteins (Gendrel and Heard, 2011). It has been debated whether Xist RNA physically recruits one or more silencing factors, or whether Xist indirectly promotes transcriptional silencing via reinforcement of repressive chromatin. XCI is also developmentally regulated in several important ways. In the mouse, XCI can proceed by random inactivation of either paternal or maternal chromosome in somatic cells, or by always inactivating the paternally derived X in extra-embryonic cells, a process called imprinted XCI (Takagi and Sasaki, 1975). During random XCI, Xist is not expressed in pluripotent embryonic stem cells (ESCs), and is up-regulated during differentiation (Wutz and Jaenisch, 2000). Ectopic Xist RNA–coating can induce gene silencing in ESCs, although this is reversible during an early differentiation time window, becoming irreversible at later stages (Wutz and Jaenisch, 2000). Knowledge of the Xist lncRNP in these diverse states may provide insights into this classic and intricate epigenetic system.
Here we introduce comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS), an optimized method for the identification of lncRNA-bound proteome. Applying ChIRP-MS to three noncoding RNAs, we found known and validated novel functional interactors. By performing Xist ChIRP-MS in different cell states, lineages and cell types, and with mutant Xist alleles, we uncover mechanisms of dynamic and coordinate assembly of Xist binding partners, suggesting an organizing principle for lncRNPs.
Results
ChIRP-MS method
Extending on ChIRP-seq, a method using DNA oligonucleotides to capture lncRNAs and their genomic DNA binding sites (Chu et al., 2011), we optimized ChIRP-MS to identify lncRNA-associated proteins (Figure 1A). We crosslink cells extensively with formaldehyde, retrieve target RNA with oligonucleotide hybridization, and use a gentle biotin-elution to liberate associated proteins. The enriched proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). We conducted negative controls by use of non-interacting control probes, RNase treatment of chromatin prior to ChIRP, or genetic removal of the target RNA.
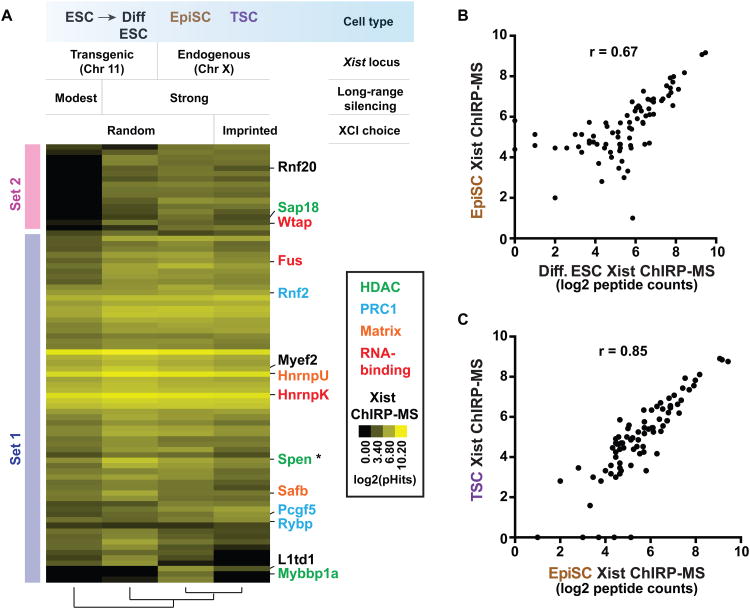
Figure 1. ChIRP-MS method and validation.
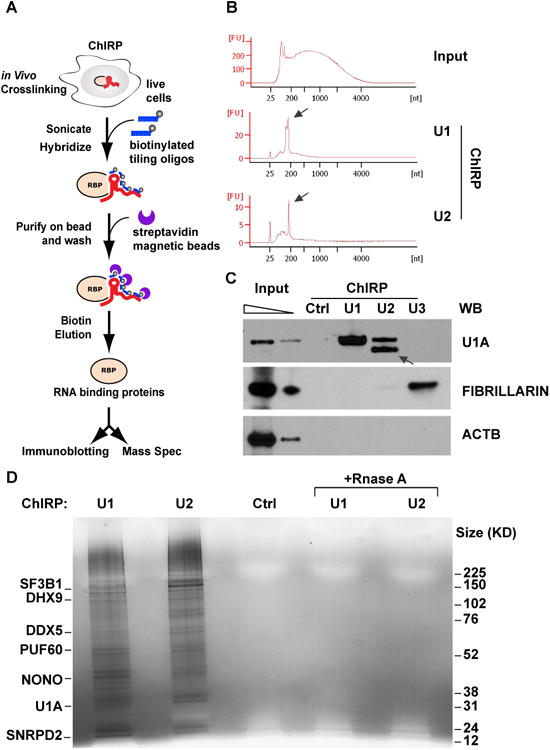
(A) outline of the ChIRP-MS workflow. Briefly, RNP complexes are crosslinked in vivo by 3% formaldehyde for 30min, and solubilized by sonication. Target ncRNA are pulled out by biotinylated anti-sense oligos, and associated proteins are eluted with free biotin, separated by electrophoresis, and each size fraction is subjected to LC/MS-MS identification. (B) Distribution of input, and U1 and U2-enriched RNA sizes, as determined by Bioanalyzer (Agilent). (C) Proteins retrieved by U1, U2, U3 and control probes, analyzed by immunoblotting. Arrow indicates the U1A close homolog, U2B, cross-identified by U1A antibody. (D) Proteins retrieved by U1, U2, Rnase-treated controls and non-targeting probe control, visualized by silver staining. Major proteins enriched are indicated on the left.
As a proof-of-principle, we performed ChIRP-MS of human U1 and U2 snRNAs in HeLa S3 cells. The snRNAs are ideal for validating ChIRP-MS because they are abundant (∼1 million copies of U1 per cell)(Gesteland and Atkins, 1993) and the spliceosome composition is well known (Pomeranz Krummel et al., 2009; Stark et al., 2001; Zhou et al., 2002). Furthermore, non-canonical roles of U1 in preventing premature mRNA cleavage and polyadenylation have been recently reported (Almada et al., 2013; Berg et al., 2012; Kaida et al., 2010), implying potential novel interactors that ChIRP-MS may discover. We designed antisense DNA oligonucleotides targeting U1 and U2 snRNAs respectively in regions previously found to be accessible for morpholino binding, and as a negative control, we chose a non-targeting probe that does not bind any human RNA (Berg et al., 2012; Kaida et al., 2010). While the input RNA spread over a large size range (due to shearing by sonication) with distinct tRNA peaks, after ChIRP-enrichment the two snRNAs predominated (Figure 1B). U1 probe retrieved the known direct binding protein U1A, while the control probe did not. U2 probe also enriched for U1A, although the indirect interaction resulted in reduced enrichment. U2 probe also retrieved known U2-binding protein U2B, which crossreacts with U1A antibody due to their close homology (arrow, Figure 1C). ChIRP of U3, an abundant small nucleolar RNA not involved in splicing, specifically retrieved the nucleolar protein fibrillarin but not U1A (Figure 1C). Beta-Actin (ACTB) was not enriched by any probe, serving as another negative control. These results indicate that ChIRP is specific even for very abundant RBPs.
U1 and U2 ChIRP-MS reveal known and novel interactors
We next scaled up experiments for MS-level analysis, including both RNase and non-targeting probe controls. Silver staining of ChIRP samples showed that U1 and U2 probes pulled down rich proteins from HeLa lysates, while all control samples are clean (Figure 1D), indicating that ChIRP-MS is highly specific on the proteome level. U1 and U2 ChIRP-MS enriched (by >log23.5, or >10-fold, see Methods) more than 400 proteins over respective negative controls (Figure 2A, full peptide count list in Table S1). The results were highly reproducible regardless of control strategies: for U1, 98% overlap between RNase and non-targeting probe controls; 99% for U2. The near-identical results from using two orthogonal methods for background removal highlights the robustness of the protocol.
Figure 2. U1/U2 ChIRP-MS.
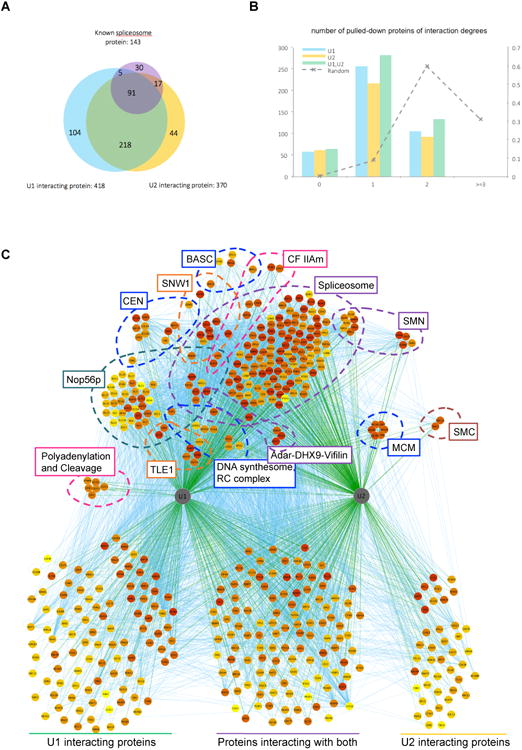
(A) Venn diagram of known spliceosome proteins, and proteins pulled-down by U1 or U2. The number of interactions in each set is given after the set label. (B) Numbers of U1/U2 pulled-down proteins by their degrees of separations from known spliceosome proteins. The dashed line represents the distribution of a randomly simulated set of the same number of proteins pulled-down by U1 and U2 (right axis). (C) Protein-protein and protein-RNA interaction network of U1/U2 pulled-down proteins. Proteins belonging to known complexes are organized and annotated in groups in top half of the plot, and proteins of unknown affiliation are presented at the bottom. Complexes and proteins more strongly enriched by U1 (left in graph) (e.g. Polyadenylation and cleavage, Nop56p) are positioned accordingly.
U1 and U2 snRNAs shared their RBPs extensively (309 in common, or 74% of U1 and 84% of U2-RBPs), as predicted from their common cellular function (Figure 2A). Both U1 and U2 strongly enriched for proteins involved in splicing and pre-mRNA biogenesis, as anticipated (Figure S1A). Together the two snRNAs retrieved 79% of the human spliceosome components (Figure 2A) and 8 of 9 direct U1 binding proteins verified by crystal structure (Pomeranz Krummel et al., 2009; Stark et al., 2001; Zhang et al., 2012). Analysis of known protein-protein interaction networks showed that the vast majority (96%) of all proteins identified were within two degrees of separations from the core spliceosome (Figure 2B) or the direct binding proteins of U1 (Figure S2A) (Pomeranz Krummel et al., 2009; Ruepp et al., 2008; Stark et al., 2001), suggesting that ChIRP-MS yields the immediate and most relevant protein network. Organization of U1/U2 interactomes into complexes based on curated protein interaction data confirmed extensive coverage of the spliceosome, SMN, and cap binding complexes (Figure 2C).
U1 selectively enriched for the CSTF complex involved in pre-mRNA cleavage and polyadenylation, a recently described non-canonical function of U1 (Figure 2C, S2B Gene Ontology (GO)-term “RNA 3′-end processing” in Figure S1A)(Berg et al., 2012; Kaida et al., 2010). Immunoblots validated U1-selective pulldown of CSTF2 over other snRNAs, which potentially explains this U1-exclusive function (Figure S1B) (Berg et al., 2012), and shows that ChIRP is specific for proximal interactions even within the same complex (e.g. the spliceosome). These and other protein complexes discovered represent a wealth of information for the snRNP community (Figure 2C, Figure S1A).
Xist ribonucleoprotein complex purification
We next turned to discover the protein partners of Xist. ChIRP-MS of Xist represents a substantial challenge in several ways: 1) Xist is far less abundant than U1 (<2000 copies per cell vs. 1million) (Buzin et al., 1994), making it more relevant to other regulatory lncRNAs; 2) Xist transcript is long and will be sheared into fragments, requiring a tiling-probe strategy not necessary for the study of U1/U2; 3) Xist is chromatin- and nuclear matrix-associated and therefore insoluble even by detergent and nuclease extraction (Clemson et al., 1996). Based on these considerations we designed 43 probes against the mouse Xist RNA (Table S2). In a female mouse cell line (Neuro2a) we confirmed that Xist RNA was completely solubilized by sonication (data not shown) and over 60% of Xist RNA was selectively retrieved without enrichment of housekeeping Gapdh mRNA (Figure 3A).
Figure 3. Xist ChIRP-MS.
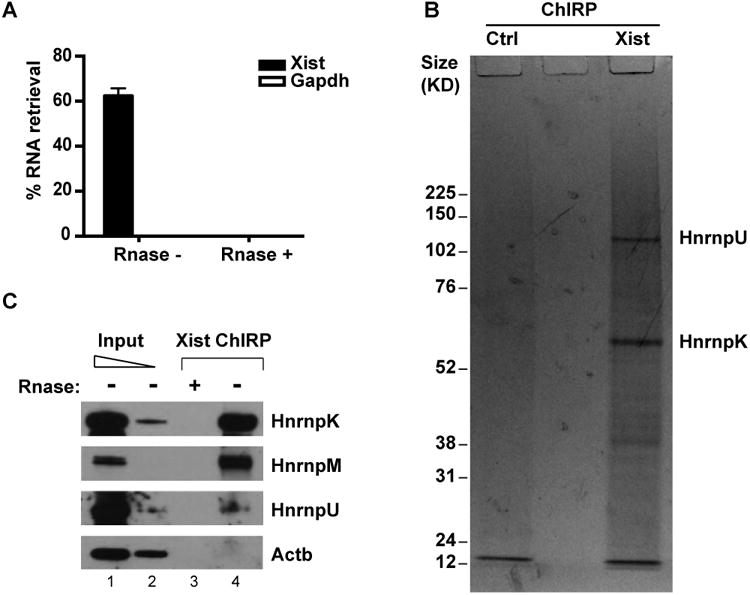
(A) >60% of Xist RNA was retrieved from the cell by ChIRP, while no Gapdh was detected. RNase treatment eliminates Xist transcripts prior to pulldown. (B) Proteins retrieved by Xist and isogenic control (no Xist), visualized by Coomassie blue staining. (C) Validation of ChIRP-enriched proteins by immunoblotting.
Xist probes retrieved rich protein analytes compared to the RNase control (Figure 3B). The most abundant proteins retrieved are HnrnpK and U, and M, the first two readily visualizable by Coomassie blue (Figure 3B). HnrnpU is required for the spread of Xist RNA across the chromosome in cis (Hasegawa et al., 2010), thus a positive control. Xist-dependent retrieval of all three proteins was validated by ChIRP-western, proving that they are not retrieved by virtue of their sheer abundance; the control protein beta-Actin was not enriched (Figure 3C).
Stepwise and developmentally regulated assembly of Xist RNP
We carefully selected biological systems to performed Xist ChIRP-MS that represent different stages of Xist-mediated silencing (Figure 4A). Although Xist is expressed in most differentiated female cells, it is largely dispensable for the maintenance of XCI (Brown and Willard, 1994; Csankovszki et al., 1999). To ensure that we catch Xist “in action,” we chose a male mouse embryonic stem cell (ESC) line that has been genetically engineered to harbor a Xist cDNA knocked into chromosome 11 (chr11) that is inducible by doxycycline (dox) (Wutz and Jaenisch, 2000). The exogenous Xist localizes to chr11, and silences chr11 genes at a long distance after 4 days of sustained expression and retinoic acid-induced differentiation (Wutz and Jaenisch, 2000) (Figure 4A, lanes 1 and 2). Turning on or off Xist transcription with dox creates an isogenically controlled experiment. Furthermore, the relatively rapid initiation of Xist silencing ensures synchronicity among cells, suppressing noise arising from population heterogeneity. To study the endogenous Xist lncRNP, we performed parallel ChIRP-MS in an epiblast stem cell line (EpiSC) (Gillich et al., 2012). EpiSCs are derived from E5.5-E6.5 epiblasts and represent cells that have just undergone random XCI (occurring ∼E5.5) (Hayashi and Surani, 2009; Rastan, 1982; Takagi et al., 1982) (Figure 4A, lane 3). Finally, we performed Xist ChIRP-MS in trophoblast stem cells (TSCs), where the paternal X-chromosome is always silenced (Calabrese et al., 2012), a phenomenon termed imprinted XCI that contrasts with the random XCI in somatic cells (Figure 4A, lane 4). RNase controls were performed side-by-side in the EpiSC and TSC experiments.
Figure 4. Xist partner proteins are developmentally regulated.
(A) Heatmap of Xist-RBPs pulled down in the four experiments. Color bars indicate abundance of peptides detected. Protein annotations were color-designated based on their class. (B) Similar proteins are enriched between differentiating ES cells vs. EpiSCs and (C) between EpiSCs and TSCs.
We also compared Xist ChIRP-MS to ChIRP-MS of three abundant nuclear RNAs-- U1, U2, and 7SK--to evaluate Xist-specific interactions (Methods). 7SK is a snRNA present at ∼200,000 copies per cell and is involved in transcriptional elongation control. We ranked peptides enriched by each ncRNA, and prioritized proteins that had Xist ChIRP-MS enrichment ranking at least two-fold better than rankings in any of the three comparator ncRNAs.
In total we identified 81 Xist binding proteins from the four experiments (Figure 4A and full list of enriched proteins with peptide counts reported in Table S3). When compared to U1/U2/7SK, only a minority of Xist hits (30/81) was also highly enriched by another ncRNA (rank ratio < 2, Table S4). These non-specific proteins are mainly involved in RNA processing (GO enrichment p=8.4E-28), and may be involved in nuclear ncRNA splicing, nuclear retention, or stability. They are likely bona-fide Xist binding proteins since they pass RNase and genetic controls, but they may not contribute to the specific gene regulatory function of Xist. We provide the list of nonspecific proteins retrieved by all four nuclear ncRNAs as a resource for the field (in red, Table S4). In contrast, the Xist-specific proteins selectively enriched for gene repressors (GO enrichment p=9.6E-8), which are highlighted in Table S4 and discussed below. We also overlapped the set of proteins retrieved by Xist with those retrieved by two other abundant nuclear lncRNAs NEAT1 and MALAT1, and found limited overlap (14 out of 81 shared by all three, Figure S2C) (West et al., 2014). As expected, the majority of overlapping proteins (8/14) are “nonspecific ChIRP hits” as defined above.
Xist ChIRP-MS in all four cell-types retrieved a common set of proteins (62/81, 77%), termed Set 1. An additional 19 proteins interacted with Xist only in differentiated ESC, EpiSC, and TSCs; these proteins are termed Set 2. We describe the identity of proteins in these two sets, and then discuss the dynamics of the interactions. Some of the binding proteins were known factors involved in XCI. We identified Rnf2 (also known as Ring2 or Ring1b), the catalytic subunit of Polycomb repressive complex 1 (PRC1) that deposits the repressive lysine119 monoubiquitination on histone H2A (H2AK119ub) over the inactive X chromosome (de Napoles et al., 2004; Fang et al., 2004). Other PRC1 components identified included Pcgf5 and Rybp (both in set 1); Rybp is a stoichiometric component of PRC1 that has been shown to accumulate on the Xi independently of PRC2 (Tavares et al., 2012). We also found the Sin3-HDAC1 components Spen, Sap18, and Mybbp1a, which are repressive transcriptional factors that recruit histone deacetylase (HDAC) complexes. Histone deacetylation correlates with reduced gene expression and is another hallmark of the inactive X chromosome (Keohane et al., 1996). The co-purification of these proteins may bridge the biochemical gap between Xist and HDAC that remains little explored in the field. Xist ChIRP-MS also recovered nuclear matrix proteins HnrnpU, Matrin 3, and Safb, consistent with the observation that Xist is probably anchored by nuclear matrix (Clemson et al., 1996). Notably, HnrnpU is required for Xist localization (Hasegawa et al., 2010). Finally, RBPs such as HnrnpK strongly and specifically interacted with Xist; HnrnpK was not retrieved by U1 or U2. Collectively, the two sets of proteins represent candidate factors that could play roles in Xist localization or function.
Comparison of Xist interactors in the four cell types revealed a potential step-wise assembly of Xist binding proteins from the pluripotent state to differentiation. Unsupervised hierarchical analysis showed that the Xist interactors are distinct in ESCs while the differentiated ESC, EpiSC and TSC shared a significant degree of overlap (Figure 4A). While Set 1 proteins remain associated with Xist from pluripotency to differentiation, Xist interaction with Set 2 proteins are observed only upon differentiation. Xist interacted with both Set 1 and Set 2 proteins in the latter three cell types; 77 of 81 Xist interactors (95%) were independently retrieved in these differentiated cells. The HDAC complex subunit Spen straddles these categories because it interacts with Xist in ESC but the interaction intensifies with differentiation (asterisk in Figure 4A). The distinction between Set 1 and 2 is unlikely due to lower efficiency of Xist ChIRP-MS in ESCs because the quantitative signal for Set1 proteins in ESC is on par with that in differentiated cells. While the Set 1 proteins may represent the ground state of Xist-interactome that prepares the lncRNA for action, the differentiation-coupled Xist interactors include intriguing chromatin-modifying proteins such as Spen, Rnf20, Mybbp1a, and Sap18. These may represent additional silencing factors recruited to Xist RNA when XCI is in full action. Quantitative comparison between Xist ChIRP-MS in differentiated ESC versus EpiSC or versus TSC showed that they are largely similar, especially for the strong interactors (r=0.67 and 0.85, respectively, Figure 4B, 4C). These results suggest that 1) transgenic Xist indeed phenocopies the endogenous RNA and shares similar binding proteins; 2) ChIRP-MS is robust and gives consistent results in multiple systems; 3) random XCI and imprinted XCI appear to employ nearly identical Xist-associated proteins and therefore extraembryonic trophoectoderm likely executes silencing in ways that are highly similar to that of the embryo proper.
HrnpK participates in Xist-mediated gene silencing
To assess the functional importance of Xist interacting proteins in gene silencing, we tested their dispensability in Xist-mediated silencing of the imprinted Grb10/Meg1 gene, previously shown to be silenced by Xist upon differentiation of ESC (Wutz and Jaenisch, 2000). The imprinted Grb10 gene is located 41 megabases away from the Xist transgene on chr11 and thought to be monoallelically expressed from the chr11 harboring the transgene (Figure 5A) (Wutz and Jaenisch, 2000). We showed it indeed was silenced by transgenic Xist (Figure 5B). We chose to first target HnrnpK, M, and U because they represent some of the most enriched Set 1 proteins (especially K), and because this simple heuristic identifies HnrnpU, a known key mediator of Xist function. Upon siRNA-mediated depletion (Figure 5C), only HnrnpU and HnrnpK had significant effects on Grb10 silencing (Figure 5D). We ruled out off-target effects by showing that all four individual siRNAs against HnrnpK produced the same de-repression effect (Figure S4A). We directly visualized transcription from the Xist-silenced allele using two color RNA-FISH (Figure 5E, F). We used a genomic (BAC) probe, allowing us to detect the Grb10 nascent transcript rather than its mature mRNA. In this way we scored for the presence or absence of Grb10 transcription adjacent to the Xist-coated chr11. HnrnpK depletion significantly increased the frequency of active Grb10 allele found close to within the Xist RNA coated chromosome 11, indicating that indeed it is less sensitive to Xist-mediated silencing in the absence of HnrnpK (Figure 5E, F).
Figure 5. Functional characterization of Xist RBPs.
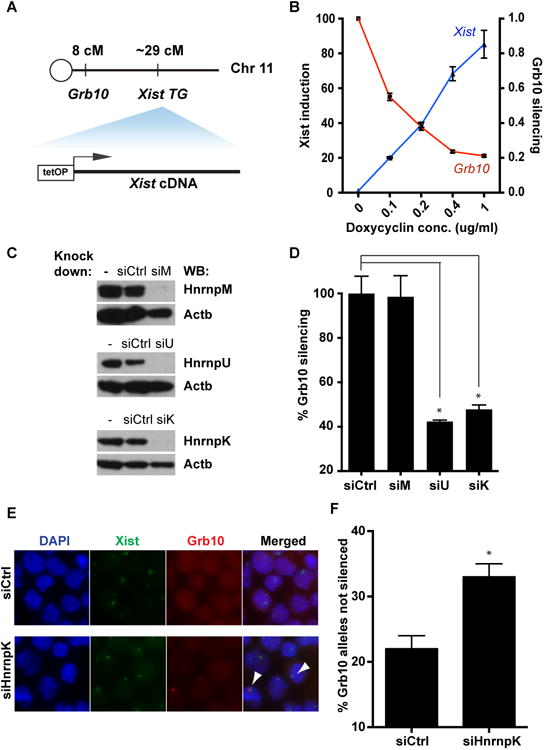
(A) Relative positions of Grb10 and Xist transgene (TG) on chr11. (B) Induction of Xist and repression of Grb10 by different doses of dox in e36 cells that have undergone RA-induced differentiation for 4 days. (C) Western validation of HnrnpU, K and M knockdown by siRNAs. (D) De-repression of Grb10 upon depletion of HnrnpU, K and M. (E) Dual-color FISH of Grb10 and Xist in e36 cells that are depleted of HnrnpK. Arrowheads indicate Grb10 allele escaping Xist silencing. (F) Quantification of cells with Grb10 expression on the Xist-coated chromosome by counting >150 cells from 3 replicates.
We also tested the requirement for HnrnpK in endogenous XCI in EpiSC. We converted ESC into EpiSC in the presence of Fgf2 and Activin (Guo et al., 2009). EpiSC conversion was confirmed by morphologic changes, marker expression, and induction of Xist expression, and Xist localization to the Xi (Figure S3A-D). We performed single molecule fluorescent in situ hybridization (sm-FISH) on Usp9x, an X-linked gene that is subject to random X-inactivation. We used FISH probes against the introns of Usp9x gene to exclusively detect its pre-mRNA that indicates active transcription. While only 10% of the cells show two Usp9x pinpoints in control cells, HnrnpK- or HnrnpU-depleted cells showed a two- to three-fold increase in cells with two Usp9x FISH signals (Figure S3E, S3F). The reduction in successful XCI for HnrnpU depletion matched observations from a prior study (Figure S3F) (Hasegawa et al., 2010). We conclude that HnrnpK is an important factor for Xist-mediated silencing.
HnrnpK contributes to Xist-mediated chromatin modifications but not Xist biogenesis or localization
We tested potential roles of HnrnpK early in the sequence of repressive events, including Xist biogenesis, localization and spreading, or chromatin silencing. Northern blots analysis showed that Xist abundance or splicing were not impacted by depletion of HnrnpK, U or M (the two minor isoforms upon HnrnpU depletion are consistent with previous report) (Hasegawa et al., 2010)(Figure 6A) although we cannot exclude that minor changes occurred given that Xist is present in multiple isoforms. Next, sm-FISH confirmed that HnrnpU depletion indeed delocalized Xist, but HnrnpK depletion did not (Figure 6B). Combined immunofluorescence and RNA FISH (IF-coFISH) showed that while Xist RNA colocalized with H2AK119ub and H3K27me3, HnrnpK depletion significantly reduced the accumulation of H2AK119ub and H3K27me3 on the Xi, without affecting Xist RNA localization (Figure 6C, D). HnrnpK depletion did not affect the global level of H3K27me3, showing that HnrnpK has a specific impact on Xist-mediated recruitment of repressive chromatin marks (Figure S4B). Given that both H3K27me3 and H2AK119ub modifications are among the earliest epigenetic changes occurring to the Xi, the results are consistent with our hypothesis that HnrnpK is a novel regulator of the initiation of X-inactivation. Xist ChIRP retrieved multiple PRC1 subunits, and PRC1 or PRC2 action can mutually recruit each other (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). Indeed, HnrnpK depletion also spatially dissociated Xist from the PRC2 subunit Eed (Figure S5). HnrnpK contains three RNA-binding KH domains that may directly bind Xist. UV-crosslinking RNP immunoprecipitation followed by reverse transcription PCR (CLIP-qRT-PCR) showed that HnrnpK directly bound Xist RNA, with the strongest interaction mapping downstream of repeat F in exon 1 (Figure S4C). HnrnpK retrieved Xist more efficiently in CLIP than HnrnpU, a known direct interaction that we reproduced (Hasegawa et al., 2010).
Figure 6. HnrnpK is required for repressive chromatin modifications of inactive X.
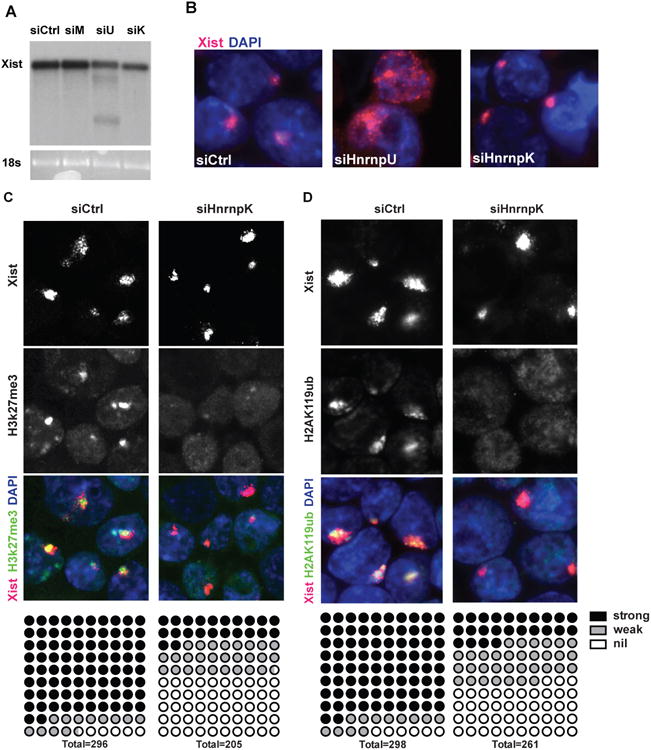
(A) Northern blot against Xist in e36 cells depleted of HnrnpM, U or K. (B) Xist sm-FISH in HnrnpU and K knockdown cells. (C-D) IF co-FISH of Xist and H3K27Me3 (C)/H2AK119ub (D) in HnrnpK knockdown cells. Number of cells with strong, weak and undetectable repressive marks overlapping with Xist foci were tallied and represented below.
The A-repeat of Xist interacts with Spen to mediate gene silencing
We next explored the use of ChIRP-MS to dissect domain-specific interactions of Xist with its partner proteins. A small 0.9 kb deletion of the very 5′ end of Xist that harbors the conserved A-repeat element of Xist is required for transcriptional silencing but not for chromatin interaction or spreading across the X chromosome (da Rocha et al., 2014; Wutz et al., 2002). In principle, deletion of A-repeat may alter RNA folding or modification to abrogate interaction of most of the silencing proteins; alternatively, the A-repeat may be selectively required for the interaction of a small number of key silencing factors. ChIRP-MS appears to be an ideal approach to distinguish between these models. Xist ChIRP-MS of ES cells harboring wild type Xist or A-repeat mutant knocked into the X chromosome (Wutz et al., 2002) revealed that most protein interactions were not affected by the deletion, but 3 proteins—Spen, Rnf20, and Wtap—were completely unable to bind the mutant (Figure 7A). Notably, Spen interaction with Xist is increased upon ESC differentiation, and Rnf20 and Wtap both belong to Set 2 proteins that interact with Xist only upon differentiation (Figure 4A). Thus the A-repeat appears to be a focus of the differentiation-coupled assembly of Xist RNP. The exclusive binding of these three proteins to full length Xist but not the A-repeat mutant was confirmed by ChIRP-western (Figure S6A). This result also implied that HnrnpK binding does not require A-repeat, which we confirmed by ChIRP-western (Figure S6A). Thus, two sets of silencing proteins bind to different domains of Xist.
Figure 7. Xist A-repeat binds Spen, a silencing factor that contributes to XCI.
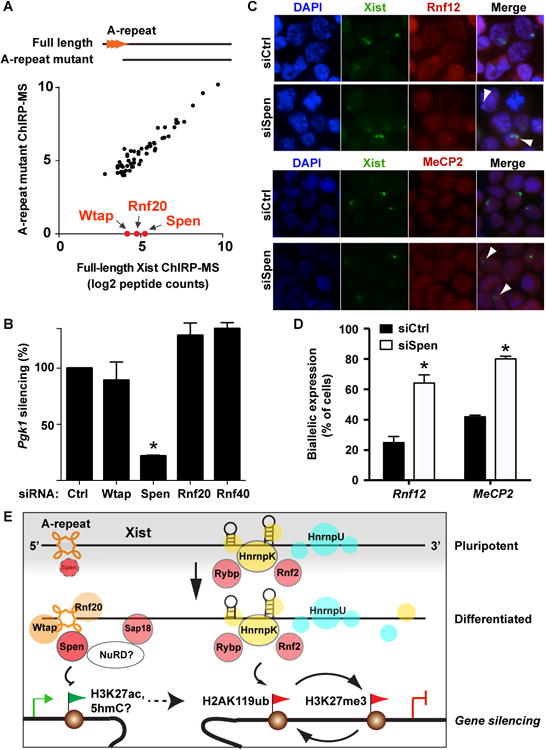
(A) Similar proteins are enriched by ChIRP-MS of full-length Xist and A-repeat mutant, except three highlighted proteins: Wtap, Rnf20, and Spen. (B) siRNA depletion of the indicated factors show only Spen is required for X-linked silencing of Pgk1. (C) siRNA depletion of Spen interferes with XCI in cells, as indicated by co-localization of Xist “cloud” and active transcription of X-linked genes Rnf12 and MeCP2 (arrowheads) on the same chromosome. (D) Quantification of cells with expression of Mecp2 and Rnf12 on the Xist-coated chromosome by counting > 100 cells from 3 replicates. A proportion of cells do not upregulate Xist and do not coat (around 40%); we counted only the cells with Xist domains. (E) Model of the cell-state- and scaffold-specific loading of Xist-RBPs, and their chromatin modifying functions.
We reasoned that one or more of the A-repeat binding factors may be required for XCI. RNAi depletion in ES cells harboring wild type Xist of each of these proteins as well as Rnf40, a functional partner of Rnf20, showed that only depletion of Spen, but not the other proteins, dramatically reduced Xist-mediated silencing of the X-linked gene Pgk1 (Figure 7B). Rnf20 and Rnf40 depletion actually slightly increased Pgk1 silencing, consistent with their known roles in enhancing transcription (Figure 7B) (Zhu et al., 2005). In addition, by two color RNA-FISH, we found that Spen depletion results in reduced silencing of the X-linked genes Mecp2 and Rnf12, with more frequent detection of nascent transcription from the Xist-coated inactive X chromosome in Spen-depleted cells, compared to control cells, or to cells depleted for Rnf20, Rnf40, or Wtap (Figure 7C, D). This experiment further illustrates that Spen is not apparently required for Xist RNA accumulation or spreading across the Xi, but is specifically needed for transcriptional silencing, consistent with its specific association with the A-repeat region of Xist. Furthermore, we validated the requirement of Spen for silencing of Grb10 in ESCs where an Xist transgene is ectopically expressed on chr11 (Figure S6B). Collectively, these results suggest that Spen could be a functional mediator of Xist-RNA driven gene silencing.
Spen is the mouse homolog of Drosophila homeotic mutant Split ends and encodes a transcriptional repressor (Arieti et al., 2014; Shi et al., 2001). Spen contains at least three RNA recognition motifs (RRMs) that can bind the lncRNA SRA to mediate RNA-directed transcriptional regulation (Arieti et al., 2014; Shi et al., 2001). Several existing Spen antibodies tested were not suitable for UV CLIP. Instead, we generated recombinant Spen RRM domains by in vitro translation, and found that two or three of the Spen RRMs preferentially retrieved with Xist A-repeat over GFP mRNA in vitro (Figure S6C). These results suggest that Spen RRM domains may interact directly with the Xist A-repeat region.
Discussion
ChIRP-MS: an RNA-centric interactome technology
ChIRP-MS provides a potentially universal interactome discovery strategy that can be readily applied to any RNA of interest. We found comparable results from RNase-treated samples or isogenic cells that lack the target RNA, suggesting application in non-genetic systems. The use of different crosslinking reagents allows the investigator to potentially tune the degree of interactions captured from the target RNA. The thorough re-discovery of the spliceosome complex proteins by ChIRP-MS of U1 and U2 snRNPs validates the robustness of ChIRP-MS. In addition, the novel factors found in U1, U2 and Xist RBPs (functionally validated in the latter), demonstrates the added sensitivity of ChIRP-MS over traditional methods of RBP identification, and provide a rich resource for future investigations. For example, U1-specific interaction with the cleavage stimulation and polyadenylation proteins has direct implications for “telescripting”, a critical process of U1-mediated protection from premature mRNA shortening (Berg et al., 2012; Kaida et al., 2010).
Dynamic plug-and-play of Xist binding proteins
Our analysis revealed two sets of proteins that interact with Xist in a developmentally regulated manner. As Xist expression and reversibility of Xist-mediated gene silencing are tightly coupled to ESC differentiation, Xist may gain new silencing functions, perhaps through newly acquired or strengthened protein interactions, upon exit from pluripotency. Consistent with this idea, “Set 2” proteins bind Xist exclusively in differentiating ESCs (and EpiSCs and TSCs); this developmentally controlled assembly of Xist RNP provides a fail-safe backup for premature Xist expression during pluripotency. The expression of most factors in Set 2 remains stable throughout the differentiation of mESC into mEpiSC (<2 fold change), as measured by whole-nucleus proteomic analysis (Song et al., 2012)(Figure S6D). Thus, the vast majority of Set 2 interactions are most parsimoniously explained by a change in Xist RNA that now allows interaction with a pre-existing set of proteins. In contrast, the compositions of Xist-RBPs are strikingly similar in differentiating ES cells, EpiSCs and TSCs. TSCs are derived from extra-embryonic trophectoderm cells, where the inactive X is always paternal (Takagi and Sasaki, 1975). It remains a standing debate in the field whether imprinted XCI differs from random XCI merely by a simple choice mechanism while sharing the same silencing machinery, or whether the imprinted vs. random XCI are fundamentally different. Our observations support the former hypothesis, and suggest that the difference between random vs. imprinted XCI is focused on the choice mechanism of the future Xi.
HnrnpU and HnrnpK emerged as the most enriched Xist-associated factors, and both functionally contribute to XCI. While HnrnpU is required for Xist spreading across X chromosome, HnrnpK knockdown affects Xist-directed deposition of silencing histone modifications H2AK119ub and H3K27me3, the products of PRC1 and PRC2 complexes respectively. Xist appears to directly bind PRC1 but not PRC2. This is consistent with recent reports demonstrating the PRC1-dependent recruitment of PRC2 complex (Blackledge et al., 2014; Cooper et al., 2014; Kalb et al., 2014). It has been reported that PRC2 binds specifically to the repeat A (repA) transcript of Xist, which is produced as a separate and shorter RNA (1.6kb, including the A-repeat region) (Zhao et al., 2008), although the exact function of this shorter transcript remains unclear. One explanation for our findings could be the existence of different RNA isoforms with different functions. Further tests will be required to dissect the events by which Polycomb proteins associate with Xi.
Modular Xist RNA domains link Spen- and HnrnpK-mediated silencing
Although the A-repeat was proposed to recruit PRC2 complex (Zhao et al., 2008), PRC2 itself is dispensable for the initiation of gene silencing during XCI (Kalantry and Magnuson, 2006). Furthermore, in the A-repeat deletion Xist mutant, PRC2 and H3K27me3 are still recruited to the Xist-coated chromosome (da Rocha et al., 2014; Plath et al., 2003). Imaging studies suggested that Xist RNA create a transcriptionally inactive nuclear compartment, independent of the A-repeat, but that the A-repeat is required for the movement of genes into this compartment as they become silenced (Chaumeil et al., 2006). These observations suggest that factors beyond PRC2 are at play.
Our results revealed the A-repeat—essential for Xist-mediated gene silencing (Wutz et al., 2002)-- as a key element for the developmentally regulated binding of several proteins. The selective abrogation of three protein interactions but full preservation of all others by the A-repeat deletion highlights the modular organization of Xist. We found Spen, a potent transcriptional repressor, to be important for Xist-mediated silencing. Spen interaction with Xist is increased upon differentiation, suggesting a gain of Spen-associated silencing activity to the Xist RNP. The Spen knockout is embryonic lethal at E12.5 (Kuroda et al., 2003), which is later than expected if XCI is fully defective. However, the knock out was not performed with a maternal germ line depletion of the protein; so an earlier phenotype masked by the maternal pool cannot be ruled out. On the other hand, Spen may well collaborate with other Xist-recruited silencing activities, and there may also be potential redundancy with two other mammalian Spit ends homologs.
The reported association between Spen and MBD3-NuRD complex nominates several gene silencing pathways, including ATP-dependent nucleosome remodeling, histone deacetylation via HDAC1/2, and modulation of DNA methylation (Shi et al., 2001; Zhang et al., 1999). NuRD complex decommissions ESC enhancers to enable differentiation and lineage commitment—the same developmental window where XCI takes place (Reynolds et al., 2012; Whyte et al., 2012). It is conceptually appealing that the same silencing mechanism that turns off pluripotency regulators may both enable Xist expression (by removing repression of Xist) and endow Xist with the silencing power to achieve XCI. Intriguingly, Spen interacts with Mbd3 (Shi et al., 2001); NuRD recruitment to active enhancers is believed to occur through Mbd3 recognition of 5-hydroxymethylcytosine (Yildirim et al., 2011). NuRD-mediated deacetylation of H3K27ac also permits PRC2-mediated H3K27me3 and gene silencing (Reynolds et al., 2012). Thus, the combination of NuRD and Polycomb activity can turn an active gene into an inactive one. We propose that Xist, may serve as a physical scaffold for organizing at least two chromatin modification activities— a writer to deposit silencing marks via PRC1 and an eraser to remove active marks via Spen-Mbd3-NuRD —that together coordinately enforce permanent epigenetic silencing (Figure 7E).
Although the other two A-repeat associating factors do not directly impact XCI in our limited analysis, they could conceptually still contribute to XCI. Rnf20 is the E3 ubiquitin ligase for H2BK120ub1, a histone modification that marks the gene bodies of transcriptionally active genes (Zhu et al., 2005). Xist has been proposed to preferentially target actively transcribed genes on X chromosome, exploiting the spatial proximity of actively transcribed loci to efficiently target Xist-associated silencing factors (Engreitz et al., 2013; Simon et al., 2013). Furthermore, the A-repeat mutant of Xist shows reduced binding to such active regions (Engreitz et al., 2013), which may be explained by the inability of the Xist A-repeat mutant to seek out Rnf20 complex loaded on active loci. Finally, Wtap is involved in the installation of the N6-methyladenosin (m6A) on RNAs. Wtap binding to the A-repeat of Xist is consistent with the presence of m6A in the same region of the RNA (data not shown). The functional impact of Wtap binding or m6A modification remains to be understood, but represents an exciting perspective given the strategic importance of the domain in question. Our results set the stage for future structure-function analysis of Xist and its interacting proteins as a paradigm to understand functional motifs in lncRNAs.
Materials and Methods
ChIRP-MS
10-20 15cm dishes of cells were used per ChIRP-MS experiment (100million - 500million cells depending on the cell type). Cell harvesting, lysis, disruption, and ChIRP were essentially performed as previously described (Chu et al., 2012), with the following modifications: 1) Cells are crosslinked in 3% formaldehyde for 30min, followed by 0.125M glycine quenching for 5min; 2) hybridization can be started late in the day and left running overnight to reduce hands-on time; 3) For mass spec experiments, lysates were pre-cleared by incubating with 30ul washed beads per ml of lysate at 37C for 30min with shaking. Prior to hybridization beads were removed twice from lysate using a magnetic stand; 4) for RNase control, lysates are pooled first and aliquoted into two equal amounts. 1/1000 volume of 10mg/ml Rnase A (Sigma) is added to the RNase control sample and both control and non-treated samples are incubated at 37C for 30min with mixing prior to hybridization steps. This can be done concurrently with pre-clearing. RNA extraction can be performed from a small aliquot of post-ChIRP beads as described (Chu et al., 2012). For protein elution, beads were collected on magnetic stand, resuspended in biotin elution buffer (12.5mM biotin (Invitrogen), 7.5mM HEPES pH 7.5, 75mM NaCl, 1.5mM EDTA, 0.15% SDS, 0.075% sarkosyl, and 0.02% Na-Deoxycholate), mixed at r.t. for 20min, and at 65C for 10min. Eluent was transferred to a fresh tube and beads were eluted again. The two eluents were pooled, and residual beads were removed again using the magnetic stand. ¼ total volume TCA was added to the clean eluent, and after thorough mixing proteins were precipitated at 4C overnight. Next day, proteins were pelleted at 16000rcf at 4C for 30min. Supernatant was carefully removed from the belly side of tubes and protein pellets on the spine of tubes (sometimes invisible at this step) were washed once with cold acetone, and pelleted again at 16000rcf at 4C for 5min and acetone was removed. Pellets (much more visible now) were briefly centrifuged again and after removal of residual acetone, left to air-dry for 1 min on bench-top. Proteins are then immediately solubilized in desired volumes of 1× laemmli sample buffer (Invitrogen), boiled at 95C for 30min with occasional mixing for reverse-crosslinking. Final protein samples were size-separated in bis-tris SDS-PAGE gels (Invitrogen) for western blots or MS. See Supplemental Methods and Table S2 for ChIRP probe design.
Defining proteins identified by ChIRP-MS
Potential MS artifacts were first filtered by removing low-confidence protein hits with fewer than 9 peptides from a single gel-C slice and fewer than 16 total peptides (a simpler cut-off of >10 peptides from any single gel-C slice was used for U1/U2). Thereafter a stringent cut-off of log2 >= 3.5 between experiment and control (>=11.3 fold enrichment) is applied to eliminate RNA-independent background interactions. Specific hits of 7SK ChIRP-MS will be reported elsewhere. To define specific vs. non-specific components of the Xist lncRNP, ChIRP-MS hits from Xist (Diff. ESC), U1, U2 and 7SK were first ranked based on peptide abundance. Xist-specific interactors are defined as proteins with Xist ChIRP-MS rank at least twice better than in ChIRP-MS of U1, U2, and 7SK. Non-specific interactors are proteins that show rank ratio <2 in Xist ChIRP vs. U1, U2, or 7SK. For the purpose of comparison, mouse protein names of 7SK and Xist hits were replaced with their human counterparts (no ambiguity).
Defining Xist-specific RBPs vs. promiscuous RBPs
The most enriched protein (most peptide counts in experiment) is ranked 1, the second most enriched is ranked 2, and so forth. “Specific interactors” for Xist are defined as proteins that have a rank that is at least two-fold better than in all three other ChIRP-MS of U1, U2, or 7SK.
Knockdown studies
siRNAs and shRNAs are purchased from Dharmacon and Invitrogen. Transfection was performed with nucleofector or RNAiMAX. See Supplemental Methods and Table S5 for full details.
Microscopy
Xist-FISH, Usp9x-FISH and co-IF are performed with sm-FISH probes with standard protocol. All other dual-color FISH were essentially performed as previously described (Chaumeil et al., 2008). See Supplemental Methods for full protocols and the list of reagents used.
RNA Crosslinking IP and interaction studies
Clip-qRTPCR was essentially performed as described (Flynn et al., 2015), and triple flag-tagged codon-optimized 2× RRM and 3× RRM Spen fragments were used in in-vitro interaction studies. See Supplemental Methods for full details.
Supplementary Material
Acknowledgments
We thank A. Wutz (ETH), A. Surani and J. Zylicz (U. Cambridge), A. Smith (U. Cambridge), A. Gillich (Stanford) for reagents and advice; W. Lane and R. Robinson (Harvard) for MS analysis; A. Olson for assistance with confocal microscopy (Stanford Neuroscience Microscopy Service supported by NIH NS069375); and P. Walker (Stanford PAN facility) for ChIRP oligo synthesis. Supported by NIH P50-HG007735 and California Institute for Regenerative Medicine. H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute. EH is supported as an Equipe labellisée “La Ligue Contre Le Cancer” (Equipe Labéllisé to E.H) and by Labex DEEP (ANR-11-LBX-0044) part of the IDEX Idex PSL (ANR-10-IDEX-000102 PSL) as well as the EpiGeneSys FP7 257082 Network of Excellence and ERC Advanced Investigator award 250367.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almada AE, Wu XB, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–U141. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieti F, Gabus C, Tambalo M, Huet T, Round A, Thore S. The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs. Nucleic Acids Res. 2014;42:6742–6752. doi: 10.1093/nar/gku277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang ZX, Cho SC, Sherrill-Mix S, Wan LL, et al. U1 snRNP Determines mRNA Length and Regulates Isoform Expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Buzin CH, Mann JR, Singer-Sam J. Quantitative RT-PCR assays show Xist RNA levels are low in mouse female adult tissue, embryos and embryoid bodies. Development. 1994;120:3529–3536. doi: 10.1242/dev.120.12.3529. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell. 2012;151:951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods in molecular biology. 2008;463:297–308. doi: 10.1007/978-1-59745-406-3_18. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes & development. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) Journal of visualized experiments : JoVE. 2012 doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nature structural & molecular biology. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. The Journal of cell biology. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell reports. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nature genetics. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. The Journal of biological chemistry. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Flynn RA, Martin L, Spitale RC, Do BT, Sagan SM, Zarnegar B, Qu K, Khavari PA, Quake SR, Sarnow P, et al. Dissecting noncoding and pathogen RNA-protein interactomes. Rna. 2015;21:135–143. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Fifty years of X-inactivation research. Development. 2011;138:5049–5055. doi: 10.1242/dev.068320. [DOI] [PubMed] [Google Scholar]
- Gesteland RF, Atkins JF. The RNA world : the nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Gillich A, Bao S, Grabole N, Hayashi K, Trotter MW, Pasque V, Magnusdottir E, Surani MA. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell stem cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Developmental cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Surani MA. Resetting the Epigenome beyond Pluripotency in the Germline. Cell stem cell. 2009;4:493–498. doi: 10.1016/j.stem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–U681. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS genetics. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nature structural & molecular biology. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill L P, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Developmental biology. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, et al. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annual review of genetics. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastan S. Timing of X-Chromosome Inactivation in Postimplantation Mouse Embryos. J Embryol Exp Morph. 1982;71:11–24. [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell stem cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Steitz JA. The “Observer Effect” in genome-wide surveys of protein-RNA interactions. Mol Cell. 2013;49:601–604. doi: 10.1016/j.molcel.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes & development. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Saha S, Gokulrangan G, Tesar PJ, Ewing RM. DNA and chromatin modification networks distinguish stem cell pluripotent ground states. Molecular & cellular proteomics : MCP. 2012;11:1036–1047. doi: 10.1074/mcp.M111.011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sugawara O, Sasaki M. Regional and Temporal Changes in the Pattern of X-Chromosome Replication during the Early Post-Implantation Development of the Female Mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature genetics. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & development. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZL, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.