Abstract
Nodamura virus (NoV) lethally infects suckling mice and contains a segmented positive-strand RNA genome that encodes a potent suppressor of RNA interference (RNAi). Recent studies have demonstrated immune detection and subsequent processing of NoV dsRNA replicative intermediates by the mouse RNAi machinery. However, diverse RNA viruses, including Encephalomyocarditis virus that also triggers Dicer-dependent biogenesis of viral siRNAs in mouse cells, are targeted in mammals by RIG-I-like receptors that initiate an IFN-dependent antiviral response. Using mouse embryonic fibroblasts (MEFs) for NoV infection, here we show that MEFs derived from mice knockout for RIG-I, but not those knockout for MDA5, LGP2, TLR3 or TLR7, exhibited an enhanced susceptibility to NoV. Further studies indicate that NoV infection induced an IFN-dependent antiviral response mediated by RIG-I. Our findings suggest that RIG-I directs a typical IFN-dependent antiviral response against an RNA virus capable of suppressing the RNAi response.
Keywords: NoV, RIG-I, Type I Interferons, RNAi
1. Introduction
Host innate immune system provides protection against virus attack by recognizing pathogen-associated molecular patterns (PAMPs) and generates both inflammatory and antiviral responses through pattern recognition receptors (PRRs)[1,2]. PRRs are classified into several families. The family of RIG-I-like receptors (RLRs) contains RIG-I, MDA5, LGP2. RIG-I, retinoic acid inducible gene 1 protein is required for the innate immune sensing of many RNA viruses including Influenza A, B virus, Paramyxoviruses, Vesicular Stomatitis virus (VSV), Respiratory syncytial virus (RSV), Japanese encephalitis virus [3,4,5]. Melanoma differentiation associated gene–5 (MDA5) preferentially recognizes Picornaviruses including Encephalomyocarditis virus (EMCV) [5]. Several viruses, such as West Nile virus, Sendai virus (SeV), Dengue virus, are detected by both RIG-I and MDA5 [4,6,7]. By contrast, the role of Laboratory of Genetics and Physiology 2 (LGP2) in virus sensing is yet to be clearly defined; some studies suggest that LGP2 is required for the virus-induced production of type-1 interferons (IFNs) whereas others indicate a negative regulatory role [8,9,10]. During the RLR signaling, mitochondrial anti-viral signaling protein (MAVS) [11], also known as IPS-1 [12], VISA [13], Cardif [14], functions downstream of RIG-I and MDA5 as an essential adapter protein to mediate IRF3 and IRF7 activation, leading to IFN production and subsequent transcriptional induction of IFN-stimulated genes (ISGs). The family of Toll-like receptors (TLRs) consists of more than 10 members. TLR3 is known to participate in the ligand recognition of viruses such as RSV [15], West Nile virus [15], IAV [16]. TLR7 is essential for the recognition of IAV [17], HIV [18], Dengue virus [19], SeV [20], whereas TLR8 shares phylogenetic and functional similarity to TLR7 and recognizes HIV [17,21].
Recent studies have provided evidence for an antiviral function of RNA interference RNAi) in mammals [22,23]. Antiviral RNAi, characterized extensively in plants and invertebrates, begins with the processing of virus-specific dsRNA by the Dicer nuclease into small interfering RNAs (siRNAs), which are subsequently assembled into RNA-induced silencing complex (RISC) to guide specific virus clearance by an Argonaute protein [24]. Production of abundant viral siRNAs was detected in both mouse embryonic stem cells (mESCs) and suckling mice infected by a mutant Nodamura virus (NoV) defective in the expression of its B2 protein [22,23], a known viral suppressor of RNAi (VSR) that acts by inhibiting Dicer processing of long dsRNA into siRNAs [25,26,27]. Although wildtype NoV is lethal to suckling mice, the VSR-deficient NoV mutant fails to establish infection in suckling mice and mESCs, but replicates to high levels in mESCs knockout of the four mouse Argonaute genes [22,23]. Dicer-dependent production of the viral siRNAs was also readily detectable in mESCs infected with EMCV [23], indicating dual recognition of EMCV dsRNA by both MDA5 and Dicer.
In this work, we investigated if NoV infection triggers innate immune recognition by RLRs and TLRs known to restrict RNA virus infection in mammals. NoV contains a positive sense single-stranded RNA genome and is the type species of the genus Alphanodavirus in the Nodaviridae. Unlike other nodaviruses that are pathogens of insects and fishes, NoV can lethally infect both insects and mammals [28,29,30]. The genome of NoV is divided into RNA1 and RNA2 that encode RNA-dependent RNA polymerase (RdRP) and the viral capsid precursor protein, respectively [31]. The VSR protein B2 is translated from RNA3, which is a subgenomic RNA of RNA1. Here, we first developed a model for NoV infection in cultured mouse embryonic fibroblasts MEFs). Use of MEFs derived from wildtype and mutant mouse strains knockout for individual RLRs and TLRs allowed us to examine the role of these innate immune receptors in the mouse response to NoV infection. Our findings indicate a key role for RIG-I in the induction of an IFN-dependent response against NoV infection.
2. Materials and methods
2.1 Cells and viruses
Stocks of Nodamura virus (NoV) were produced by intraperitoneal injection of BALB/c suckling mice as previously reported [22]. We followed the guidelines described under the federal Animal Welfare Regulations Act with the protocol approved by the Institutional Animal Care and Use Committee at the University of California, Riverside. Mouse embryonic fibroblasts (MEFs) cell lines were generated from the wild-type (WT) mice with a C57BL/6 background and RIG-I−/−, MDA5−/−, LGP2−/−, MAVS−/−, TLR3−/−, TLR7−/− knockout mice as previously described [5,10,32,33,34,35]. C57BL/6, MDA5−/−, MAVS−/−, TLR3−/−, and TLR7−/− mice were purchased from Jackson Laboratory whereas RIG-I−/− and LGP2−/− were kindly provided by Drs. Adolfo García-Sastre, Shizuo Akira and Michael Gale, Jr. Cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine solution, 37°C, 5% CO2.
2.2 Infection in MEFs
MEFs (1×106) were infected by NoV with the same amount of viral genome copies (5×106). MEFs were harvested at 12, 24, 48, 72-hour post infection (hpi). For IFN pre-treatment, mouse IFN-β at final concentration of 1 IU (International Units)/ml (PBL assay science) was added to MEFs 8 h prior to infection. Total RNA from MEFs was extracted using TRIzol reagent (Invitrogen). First strand cDNA synthesis was performed using iScript Reverse Transcription Supermix (Bio-Rad) according to the manufacturer’s instruction. The copy numbers of the viral genome RNA1 of NoV were analyzed by real-time PCR as previously described [22].
2.3 Real-time RT-PCR assay
Quantitative real-time PCR was used to determine the gene expression changes in MEFs. One µg of extracted total RNA was reverse-transcribed with iScript Reverse Transcription Supermix (Bio-Rad), and 1/10 of the cDNA products was mixed with iQ SYBR green Supermix (Bio-Rad). Mouse β-actin was used as an endogenous control. Primer sequences were as follows. Mouse β-actin forward primer 5’-ATT GGC AAC GAG CGG TTC C-3’ and reverse primer 5’-AGC ACT GTG TTG GCA TAG AGG-3’. Mouse RIG-I forward primer 5’- GAG AGT CAC GGG ACC CAC T -3’ and reverse primer 5’- CGG TCT TAG CAT CTC CAA CG -3’. Mouse MDA5 forward primer 5’- TGA TGC ACT ATT CCA AGA ACT AAC A -3’ and reverse primer 5’- TCT GTG AGA CGA GTT AGC CAA G -3’. Mouse LGP2 forward primer 5’- CAG CCT AGT CTG CTG CTA TTC -3’ and reverse primer 5’- CCA GAG CAG GTA AGA TCA CTT -3’. Mouse MAVS forward primer 5’- CTG GCT GAT CAA GTG ACT CG -3’ and reverse primer 5’- AAT GCA GAG GGT CCA GAA AC -3’. Mouse Dicer forward primer 5’- TGA ACC TTT TGA CAC CTC GG C-3’ and reverse primer 5’- TGA TGC TGG GAT TGG ATG TAT AG -3’. Mouse Argonaute 2 forward primer 5’- ATT CAG TTC TAC AAG TCC ACC C -3’ and reverse primer 5’- CTG ATA GTC CTT CTC CAG CTT G -3’. Mouse TRBP2 forward primer 5’- GGA GGG AAT GAG TGA AGA GG -3’ and reverse primer 5’- GGC GTC TTT CCT ATT CTG GTC -3’. Mouse PACT forward primer 5’- CCG AAC ACA GAC TAC ATC CAG -3’ and reverse primer 5’- CTC TGC GAG ACA CTG ATA CTG -3’. Changes in gene expression were expressed as a ratio of the level observed in mock-infected MEFs by Real-time PCR performed as previously described [22].
2.4 Statistical analysis
Data were expressed as mean ± S.E.M. from at least three independent experiments. Statistical analysis was done using student’s test where * =p<0.05, ** =p<0.01, and *** =p<0.001.
3. Results
3.1 RIG-I−/− MEFs are more susceptible to NoV infection than wildtype MEFs
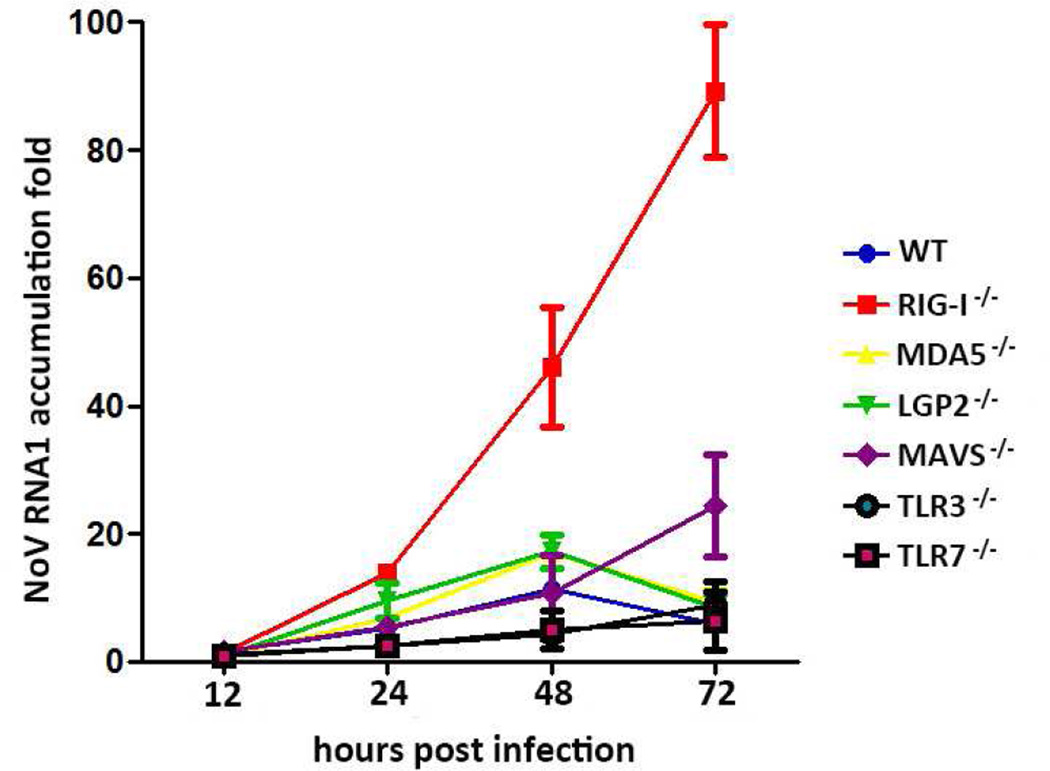
NoV has a limited cell and tissue tropism and was reported infectious in cultured BHK-21 and CHO cells that support high levels of replication [36]. Since fibroblasts from skeletal muscles of suckling mice were permissive to NoV [37], we isolated mouse embryonic fibroblasts (MEFs) from C57BL/6 mice for NoV infection. MEFs seeded in 6-well plate were infected with 5×106 genome copies of NoV as previously described [22]. Following infection with NoV, MEFs were collected without supernatants and total RNA was extracted at 12, 24, 48, 72 hours post infection (hpi) for real-time RT-PCR analysis of NoV genomic RNA accumulation. We detected an approximately 10-fold increase of NoV accumulation in the wildtype MEFs 48 hour after infection (Figure 1). This indicated that MEFs were susceptible to NoV although MEFs appeared more resistant to NoV than BHK-21 cells.
Figure 1. RIG-I deficiency leads to increased NoV replication in MEFs.
Mouse embryo fibroblasts from wild type (WT), RIG-I−/−, MDA5−/−, LGP2−/−, MAVS−/−, TLR3−/− and TLR7−/− were inoculated with NoV at amount of viral genome copies (5×106). RNA1 levels of NoV were determined at 12, 24, 48 and 72 hpi for each sample by real-time RT-PCR.
We next generated MEFs from RIG-I−/−, MDA5−/−, LGP2−/−, MAVS−/−, TLR3−/−, and TLR7−/− mice and determined whether any of these MEFs was more susceptible to NoV infection. We found that NoV RNA1 levels increased approximately 46 and 89 folds in RIG-I−/− MEFs at 48 and 72 hpi, respectively (Fig. 1). In contrast, no obvious differences in the accumulation of NoV were observed at 48 hpi in MEFs from WT, MDA5−/−, LGP2−/−, MAVS−/−, TLR3−/−, and TLR7−/− mice (Fig. 1). However, NoV accumulated to approximately 24-fold higher levels in MAVS−/− MEFs at 72 hpi when NoV accumulation remained low in MEFs from WT, MDA5−/−, LGP2−/−, TLR3−/−, and TLR7−/− mice (Fig. 1). These findings indicate that MEFs from RIG-I−/− and MAVS−/− mice were more susceptible to NoV infection than other MEFs. Therefore, NoV infection in MEFs may be detected by RIG-I, triggering a MAVS-dependent antiviral response.
3.2 RIG-I is necessary for the expression of IFN-β and ISGs induced by NoV
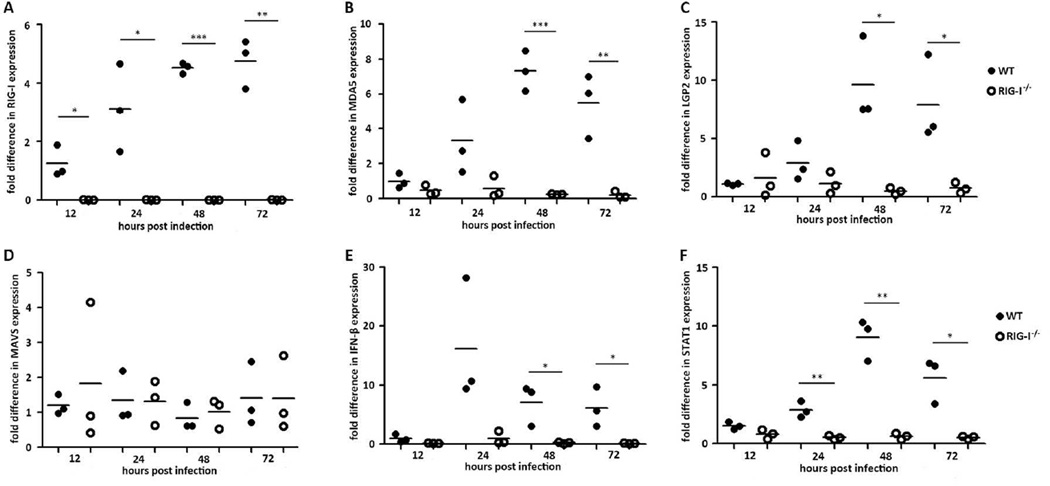
To investigate the role of RIG-I in the innate immune response to NoV, we next compared the expression of key innate immune genes in WT and RIG-I−/− MEFs following NoV infection. It is known that nonprofessional immune cells such as fibroblasts produce IFN-β in response to viral infection [7]. Therefore, we focused on IFN-β and four IFN-stimulated genes (ISGs), including RIG-I, MDA5, LGP2 and STAT1. As a control, we also analyzed the expression of MAVS, which is not induced by IFN. Our time course analyses by real-time RT-PCR at 12, 24, 48 and 72 hpi showed that RIG-I, MDA5, LGP2, and STAT1 as well as IFNβ expressed to significantly higher levels in WT MEFs after NoV infection (Fig. 2A, 2B, 2C, 2E, 2F). However, expression of MDA5, LGP2, STAT1 and IFNβ remained at the background levels in RIG-I−/− MEFs as found for RIG-I during NoV infection (Fig. 2A, 2B, 2C, 2E, 2F). In contrast, no significant difference in MAVS expression was detected between WT and RIG-I−/− MEFs in response to NoV infection (Fig. 2D). These findings together suggest that NoV infection induces the expression of IFNβ and ISGs in a manner dependent on RIG-I.
Figure 2. Expression profiles of innate immune-related genes in RIG-I−/− MEFs.
WT and RIG-I−/− MEFs were infected with NoV for 12, 24, 48 and 72hpi. Cells of each sample were harvested and total RNA extracted. RIG-I (A), MDA5 (B), LGP2 (C), MAVS (D), IFN-β (E) and STAT1 (F) mRNA levels were determined by real-time RT-PCR and normalized by mRNA levels expressed in mock-inoculated MEFs.
3.3 IFNβ pre-treatment rescues NoV resistance in RIG-I−/− MEFs
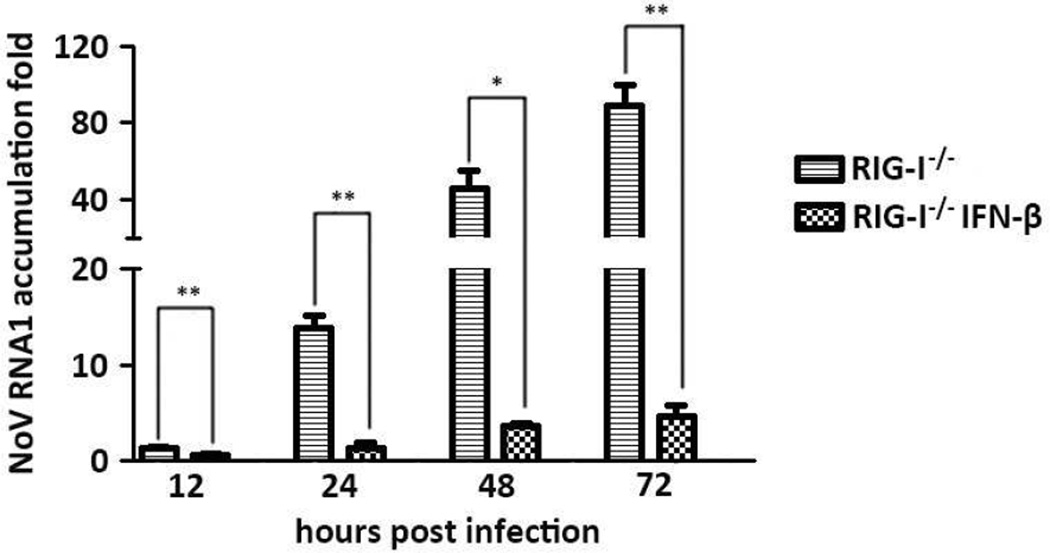
The above results suggest that RIG-I is necessary for NoV-triggered expression of IFNβ and the downstream signaling to the induction of ISGs and that NoV replicated to higher levels in MEFs derived from RIG-I−/− mice than in WT MEFs. Therefore, we investigated whether IFNβ pre-treatment could enhance the resistance of RIG-I−/− MEFs to NoV infection. We found that NoV replication was markedly inhibited in RIG-I−/− MEFs pre-treated with IFNβ at the final concentration of 1 IU (International Units)/ml (Fig. 3). The inhibitory effect of IFNβ on NoV infection was clearly detectable at all of the four time points examined after inoculation (Fig. 3). This result suggests that, in the absence of RIG-I, a potent antiviral state could be rescued in MEFs by IFNβ pre-treatment. Taken together, our findings indicate that NoV infection of MEFs induces an IFN-dependent antiviral response mediated by RIG-I.
Figure 3. Suppression of NoV replication in RIG-I−/− MEFs by IFN pre-treatment.
IFN-β (1unit/mL) was added in IFN pre-treated fibroblasts 8 hours prior to NoV infection. RNA1 levels of NoV were determined at 12, 24, 48 and 72 hpi for each sample by real-time RT-PCR. Data are shown as mean±S.E.M of triplicate samples from three independent experiments.
3.4 IFNβ pre-treatment does not alter expression of key RNAi pathway genes
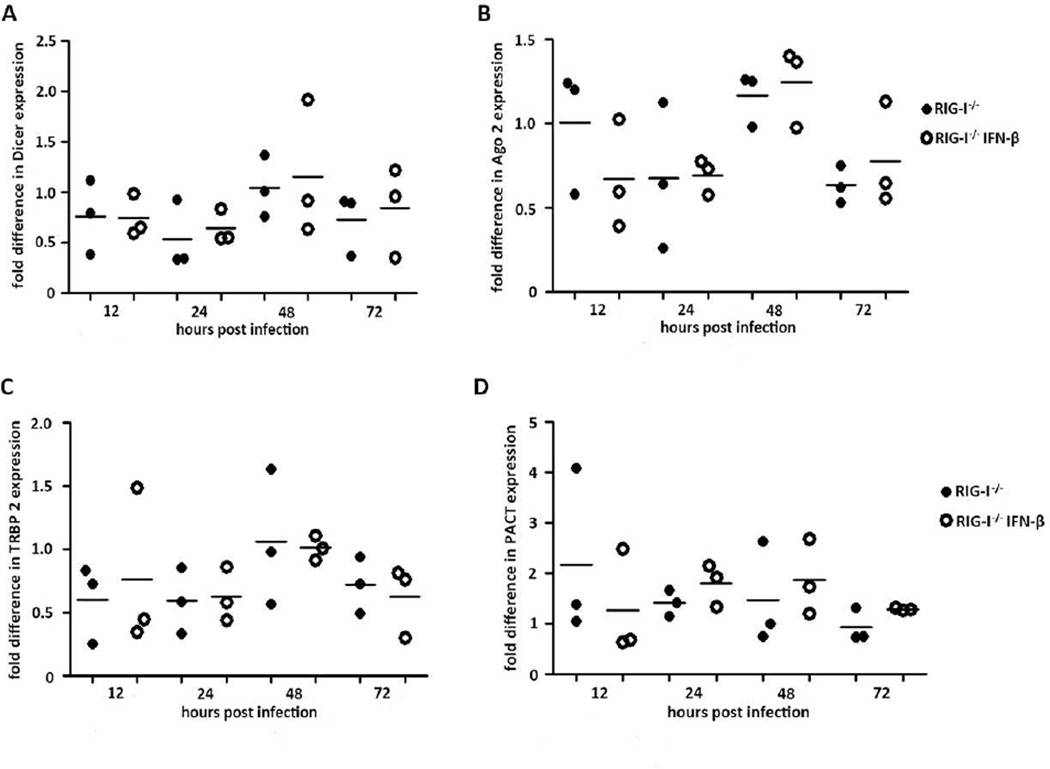
We next determined if IFNβ pre-treatment altered the expression of RNAi pathway genes in NoV-infected RIG-I−/− MEFs. To this end, we examined the expression level of four RNAi pathway genes by real-time RT-PCR in RIG-I−/− MEFs at four time points after NoV inoculation with and without IFNβ pre-treatment. Dicer and Argonaute 2 as well as the two dsRNA-binding proteins found in RISC, PACT and TRBP, were selected. The results showed that NoV infection does not alter the expression of these RNAi pathway genes in RIG-I−/− MEFs either with or without IFNβ pre-treatment (Fig. 4A–D). This suggests that the enhanced resistance of RIG-I−/− MEFs pre-treated with IFNβ is not associated with an altered expression of key RNAi pathway genes.
Figure 4. RNAi related gene expression profiles in IFN pre-treated RIG-I−/− fibroblasts.
Fibroblasts with and without IFN treatment were infected with NoV for 12, 24, 48 and 72hpi. Cells of each sample were harvested and total RNA was extracted. Dicer (A), Ago 2 (B), TRBP2 (C) and PACT (D) mRNA levels were determined by real-time RT-PCR and normalized by mRNA levels expressed in mock fibroblasts.
4. Discussion
In this manuscript we report the first investigation on the role of innate immune receptors RLRs and TLRs in the control of an RNA virus that encodes a VSR characterized in mouse infection. We showed previously that lethal infection of suckling mice with NoV is associated with the suppression of the biogenesis of viral siRNAs by the viral B2 protein [22]. Production of abundant viral siRNAs is readily detectable in mESCs infected with the B2-deficient mutant NoV, but not with wildtype NoV, indicating B2 suppression of viral siRNA biogenesis in the cultured mouse cells [23]. This study aimed to determine whether mouse RLRs and TLRs restrict the infection of NoV, an RNA virus that potently suppresses the processing of the viral dsRNA into siRNAs during infection. We found that NoV accumulation reached a peak at 48 hpi in WT MEFs and declined at 72 hpi, suggesting restriction of NoV infection in MEFs by an antiviral response. However, NoV replicated too much higher levels in RIG-I−/− MEFs than WT MEFs and the accumulation of NoV in RIG-I−/− MEFs continued to increase from 48 hpi to 72 hpi. In contrast, genetic knockout of other RLRs and the two TLRs had no apparent effect on the infection of MEFs by NoV. These observations suggest that RIG-I acts to suppress the infection of NoV, similarly to those reported previously on the infection of other RNA viruses [3,4,5,7,38]. Our quantitative RT-PCR analysis showed that NoV infection induced the expression of IFNβ and four ISGs in WT MEFs, but not in RIG-I−/− MEFs. We found that unlike WT MEFs, the resistance to NoV infection was not maintained beyond 48 hpi in MEFs from MAVS−/− mice, which are deficient in the production of type 1 IFNs triggered by RIG-I in response to virus infection [39,40]. Notably, IFNβ pre-treatment significantly enhanced the resistance of RIG-I−/− MEFs to NoV infection. These findings together suggest that NoV is targeted by an IFN-dependent antiviral immunity initiated by RIG-I.
Our results indicate that the IFN-dependent antiviral immunity mediated by RIG-I remains effective to target an RNA virus capable of strong suppression of antiviral RNAi during infection. One interpretation of this finding is that distinct mechanisms restrict NoV infection in IFN-dependent immunity and antiviral RNAi, but only the latter is targeted for suppression by the B2 protein. Future studies are necessary to investigate the possible functional interactions of these two antiviral responses. For example, recent studies have shown that two RLR proteins are essential for antiviral RNAi in Caenorhabditis elegans by controlling the biogenesis of viral siRNAs [41,42,43]. Interestingly, suppression of antiviral RNAi in plants by VSRs triggers a distinct counter-counter defense known as effector-triggered immunity [44,45,46]. Therefore, IFN-dependent response may have evolved in mammals to inhibit the infection of those viruses capable of escaping restriction by antiviral RNAi.
Supplementary Material
Highlights.
RIG-I−/− MEFs are more susceptible to NoV infection than wildtype MEFs.
RIG-I is necessary for the expression of IFN-β and ISGs induced by NoV.
IFNβ pre-treatment rescues NoV resistance in RIG-I−/− MEFs.
IFNβ pre-treatment does not alter expression of key RNAi pathway genes.
Acknowledgement
We thank Drs. Adolfo García-Sastre, Shizuo Akira and Michael Gale, Jr. for providing RIG-I−/− and LGP2−/− mice. This work was supported by National Natural Science Foundation of China (Grant no. 31272531 and 91129709), NIH grant (Grant no. AI52447) and in part by China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu SY, Sanchez DJ, Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol. 2011;23:57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Sousa CRE. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 4.Loo YM, Fornek J, Crochet N, Zeng H, Akira S, Gill MA, Tumpey TM, Garcia-Sastre A, Katze MG, Gale M. Distinct RIG-I and MDA5 signaling regulation by RNA viruses in innate immunity. Journal of Interferon and Cytokine Research. 2007;27:697–697. [Google Scholar]
- 5.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Sousa CRE, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Fredericksen BL, Fornek J, Keller BC, Katze MG, Gale M. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. Journal of Interferon and Cytokine Research. 2007;27:699–700. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. Journal of Immunology. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 9.Malur M, Gale M, Krug RM. LGP2 Downregulates Interferon Production during Infection with Seasonal Human Influenza A Viruses That Activate Interferon Regulatory Factor 3. Journal of Virology. 2012;86:10733–10738. doi: 10.1128/JVI.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seth RB, Sun LJ, Ea CK, Chen ZJJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Xu LG, Wang YY, Han KJ, Li LY, Zhai ZH, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Molecular Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp R. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 15.Kawai T, Akira S. Toll-like Receptor and RIG-1-like Receptor Signaling. Year in Immunology. 2008;143:1–20. doi: 10.1196/annals.1443.020. 2008. [DOI] [PubMed] [Google Scholar]
- 16.Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. Involvement of toll-like receptor 3 in the immune response of lung evithelial cells to double-stranded RNA and influenza A virus. Journal of Biological Chemistry. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 17.Diebold SS, Kaisho T, Hemmi H, Akira S, Sousa CRE. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 18.Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via toll-like receptor-viral RNA interactions. Journal of Clinical Investigation. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. Journal of Immunology. 2006;177:7114–7121. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- 20.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 21.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Lu JF, Han YH, Fan XX, Ding SW. RNA Interference Functions as an Antiviral Immunity Mechanism in Mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 25.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korber S, Shaik Syed Ali P, Chen JC. Structure of the RNA-binding domain of Nodamura virus protein B2, a suppressor of RNA interference. Biochemistry. 2009;48:2307–2309. doi: 10.1021/bi900126s. [DOI] [PubMed] [Google Scholar]
- 28.Scherer WF, Hurlbut HS. Nodamura Virus from Japan - a New and Unusual Arbovirus Resistant to Diethyl Ether and Chloroform. American Journal of Epidemiology. 1967;86 doi: 10.1093/oxfordjournals.aje.a120737. 271-&. [DOI] [PubMed] [Google Scholar]
- 29.Gant VU, Jr, Moreno S, Varela-Ramirez A, Johnson KL. Two membrane-associated regions within the Nodamura virus RNA-dependent RNA polymerase are critical for both mitochondrial localization and RNA replication. J Virol. 2014;88:5912–5926. doi: 10.1128/JVI.03032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrillo JE, Venter PA, Short JR, Gopal R, Deddouche S, Lamiable O, Imler JL, Schneemann A. Cytoplasmic granule formation and translational inhibition of nodaviral RNAs in the absence of the double-stranded RNA binding protein B2. J Virol. 2013;87:13409–13421. doi: 10.1128/JVI.02362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman JFE, Brown F. Further Physicochemical Characterization of Nodamura Virus - Evidence That Divided Genome Occurs in a Single Component. Journal of General Virology. 1978;38:83–95. doi: 10.1099/0022-1317-38-1-83. [DOI] [PubMed] [Google Scholar]
- 32.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 35.Kurt-Jones EA, Sandor F, Ortiz Y, Bowen GN, Counter SL, Wang TC, Finberg RW. Use of murine embryonic fibroblasts to define Toll-like receptor activation and specificity. J Endotoxin Res. 2004;10:419–424. doi: 10.1179/096805104225006516. [DOI] [PubMed] [Google Scholar]
- 36.Ball LA. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J Virol. 1992;66:2335–2345. doi: 10.1128/jvi.66.4.2335-2345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy FA, Scherer WF, Harrison AK, Dunne HW, Gary GW., Jr Characterization of Nodamura virus, an arthropod transmissible picornavirus. Virology. 1970;40:1008–1021. doi: 10.1016/0042-6822(70)90147-9. [DOI] [PubMed] [Google Scholar]
- 38.Ermler ME, Yerukhim E, Schriewer J, Schattgen S, Traylor Z, Wespiser AR, Caffrey DR, Chen ZJJ, King CH, Gale M, Colonna M, Fitzgerald KA, Buller RML, Hise AG. RNA Helicase Signaling Is Critical for Type I Interferon Production and Protection against Rift Valley Fever Virus during Mucosal Challenge. Journal of Virology. 2013;87:4846–4860. doi: 10.1128/JVI.01997-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, Finberg RW. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, Zhang R, Wang J, Ding SW, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci U S A. 2013;110:16085–16090. doi: 10.1073/pnas.1307453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashe A, Belicard T, Le Pen J, Sarkies P, Frezal L, Lehrbach NJ, Felix MA, Miska EA. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife. 2013;2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li HW, Lucy AP, Guo HS, Li WX, Ji LH, Wong SM, Ding SW. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 1999;18:2683–2691. doi: 10.1093/emboj/18.10.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sansregret R, Dufour V, Langlois M, Daayf F, Dunoyer P, Voinnet O, Bouarab K. Extreme resistance as a host counter-counter defense against viral suppression of RNA silencing. PLoS Pathog. 2013;9:e1003435. doi: 10.1371/journal.ppat.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.