Abstract
The survival of all living organisms is determined by their ability to reproduce, which in turn depends on accurate duplication of chromosomal DNA. In order to ensure the integrity of genome duplication, DNA polymerases are equipped with stringent mechanisms by which they select and insert correctly paired nucleotides with a deoxyribose sugar ring. However, this process is never 100% accurate. To fix occasional mistakes, cells have evolved highly sophisticated and often redundant mechanisms. A good example is mismatch repair (MMR), which corrects the majority of mispaired bases and which has been extensively studied for many years. On the contrary, pathways leading to the replacement of nucleotides with an incorrect sugar that is embedded in chromosomal DNA have only recently attracted significant attention. This review describes progress made during the last few years in understanding such pathways in both prokaryotes and eukaryotes. Genetic studies in Escherichia coli and Saccharomyces cerevisiae demonstrated that MMR has the capacity to replace errant ribonucleotides, but only when the base is mispaired. In contrast, the major evolutionarily conserved ribonucleotide repair pathway initiated by the ribonuclease activity of type 2 Rnase H has broad specificity. In yeast, this pathway also requires the concerted action of Fen1 and pol δ, while in bacteria it can be successfully completed by DNA polymerase I. Besides these main players, all organisms contain alternative enzymes able to accomplish the same tasks, although with differing efficiency and fidelity. Studies in bacteria have very recently demonstrated that isolated rNMPs can be removed from genomic DNA by error-free nucleotide excision repair (NER), while studies in yeast suggest the involvement of topoisomerase 1 in alternative mutagenic ribonucleotide processing. This review summarizes the most recent progress in understanding the ribonucleotide repair mechanisms in prokaryotes and eukaryotes.
Keywords: ribonucleotide excision repair, nucleotide excision repair, mismatch repair, ribonuclease H, flap endonuclease, DNA polymerase I
1. Introduction
The synthesis of vitally important macromolecules that encode and transmit genetic information in all living organisms relies on nucleic acid polymerases. The nucleotide substrate specificity separates these enzymes into two distinct groups: those that utilize ribonucleoside triphosphates (rNTPs) and those that utilize deoxyribonucleoside triphosphates (dNTPs). DNA polymerases, enzymes that are essential for genome duplication and repair synthesis, belong to the latter group. Deoxynucleotides, the building blocks of DNA, are synthesized by ribonucleotide reductases from ribonucleotides and are present throughout the cell cycle at much lower concentrations than their precursors. As a result, and since dNTPs and rNTPs are chemically and structurally very similar, it is imperative for DNA polymerases to exhibit a high degree of selectivity for deoxyribonucleotides over ribonucleotides.
Various structural, genetic and biochemical studies revealed that the major barrier for rNTPs is confined to a single residue in the active site of all DNA polymerases that is called the “steric gate” (for recent reviews see [1-3]). The side chain or backbone of this residue physically clashes with the 2'-OH group on the sugar ring of an incoming ribonucleotide and prevents its insertion. Moreover, most high-fidelity DNA replicases are equipped with 3' →5' exonucleolytic proofreading domains or subunits that are designed to improve enzymatic fidelity, and have the capacity to not only recognize and excise nucleotides with a wrong base, but also with a wrong sugar [4-6]. However, proofreading of errantly incorporated ribonucleotides is relatively poor and as with other enzymatic processes, the overall discrimination against ribonucleotide insertion is not 100% efficient, even for high-fidelity DNA polymerases [2, 3, 7]. As a result, a significant number of ribonucleotides are incorporated into nuclear DNA during the normal process of genome duplication. Estimations of this value for replicative and repair DNA polymerases from a variety of organisms [1, 3, 8-14] has led to the realization that among all non-canonical nucleotides embedded in chromosomal DNA, rNMPs are the most abundant. These findings hint at the possibility that incorporation of rNMPs during DNA replication or repair, is not simply a result of failed attempts to prevent it, but rather is an evolutionarily conserved property of DNA synthesis that may be of important biological significance (for recent review see [15, 16]). For example, this includes (i) marking the nascent DNA, thereby directing the mismatch repair (MMR) machinery to the correct strand [12, 17], (ii) improving the efficiency and fidelity of pol µ-dependent non-homologous end joining in the course of double-strand break repair [9], or (iii) directing the recombination important for mating type switching in Schizosaccharomyces pombe [18]. However, when accumulated at excessive levels, rNMPs scattered throughout the chromosome might pose serious danger for a living cell, mainly due to the reduced stability [19] and altered structure of the nucleic acid backbone ([20, 21] and references therein). This threat is imminent not only for dividing cells, but also for quiescent cells that have substantially lower dNTP:rNTP ratios. To prevent persistent ribonucleotide accumulation, cells rely on the help of repair systems with the capacity to monitor and excise rNMPs inadvertently incorporated by DNA polymerases into genomic DNA. Here, we review and summarize the most recent data that has led to the elucidation of ribonucleotide repair mechanisms with emphasis on our own in vivo and in vitro studies of prokaryotic pathways. We also present some previously unpublished data, which characterize specific features of excision/re-synthesis steps of the ribonucleotide repair pathway.
2. Approaches to study ribonucleotide repair
Ribonucleotide repair has been extensively investigated using Saccharomyces cerevisiae (S. cerevisiae) and Escherichia coli (E. coli) model systems. Studies in yeast were mainly performed using crude cell extracts from strains carrying a deletion of gene(s) encoding proteins implicated in RER and by reconstituting repair pathways in vitro using purified recombinant proteins [4, 6, 8, 22, 23]. We have elucidated RER in bacteria using biochemical and genetic approaches [24-27]. In particular, we have utilized low-fidelity E.coli pol V (UmuD'2C) and a steric gate mutant (umuC_Y11A) that avidly misincorporates ribonucleotides into genomic DNA to investigate the mechanisms of prokaryotic ribonucleotide repair.
E.coli pol V is best characterized by its ability to promote translesion DNA synthesis with a concomitant increase in damage-induced mutagenesis [28, 29]. However, in a recA730 lexA(Def) background, where pol V is maximally activated and the enzyme is able to compete with pol III for access to undamaged DNA, the low-fidelity pol V confers a significant spontaneous mutator phenotype in the absence of exogenous DNA damage. Biochemical characterization of wild-type pol V revealed that in addition to low base substitution fidelity, pol V readily incorporates ribonucleotides into DNA in vitro [24]. Furthermore, pol V is able to synthesize long RNA stretches in vitro when copying a DNA template in the presence of rNMPs. The umuC_Y11A steric gate mutant of pol V has an even greater propensity to incorporate ribonucleotides. This mutant is also characterized by reduced deoxyribonucleotide base specificity in vitro. We therefore expected that when expressed in vivo, the umuC_Y11A mutant would induce higher levels of spontaneous mutagenesis than wild-type pol V. However, the exact opposite phenotype was observed. Spontaneous mutagenesis in strain with the umuC_Y11A variant was only about 7% of spontaneous mutagenesis in strain with wild-type pol [24]. To explain this phenotype, we hypothesized that efficient and accurate repair specifically targeted to replace nucleotides with an incorrect sugar concomitantly replaces nucleotides with incorrect bases in the vicinity of the target ribonucleotide. In doing so, these repair pathways reduce the mutagenic consequences of DNA synthesis by the highly error-prone umuC_Y11A. Therefore by introducing the umuC_Y11A allele into a number of repair-deficient strains, we have been able to identify individual proteins and repair systems that make a significant contribution into the poorly mutable phenotype of umuC_Y11A-expressing cells, and thus have delineated both primary and back-up pathways of ribonucleotide repair in E. coli.
2.1 RNase H-dependent ribonucleotides excision repair
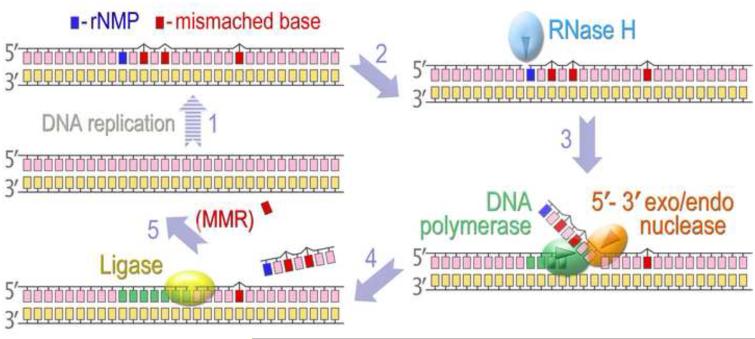
It should be noted that the accidental incorporation of ribonucleotides by DNA polymerases is not the major source of rNMPs embedded into chromosomal DNA. Initiation of DNA replication on both the leading and lagging strands in all organisms occurs through the synthesis of short RNA primers by primases, followed by primer elongation by replicative DNA polymerases. The RNA primers must then be replaced with deoxyribonucleotides before newly synthesized DNA can be ligated into an intact strand. Since replication of the lagging strand proceeds discontinuously, multiple Okazaki fragments form and the number of RNA primers that have to be removed during genome duplication is quite substantial, even in a bacterial chromosome, which has only a single origin of replication. Therefore, cells are equipped with an efficient system designed to detect and eliminate RNA patches from double-stranded DNA. Several distinct pathways have been implicated in RNA primer removal during Okazaki fragment maturation [30]. Naturally, at least one of these systems could be co-opted to remove errant ribonucleotides sporadically incorporated by DNA polymerases during replication and/or repair DNA synthesis. Indeed, it has been demonstrated in E. coli and S. cerevisiae, that the major pathway directed at the removal of isolated rNMPs from DNA is “RER” (Ribonucleotide Excision Repair), which is mechanistically very similar to the removal of ribonucleotides during Okazaki fragment maturation. In principal, this pathway in prokaryotes and eukaryotes consists of the following key steps (Fig. 1): (i) cleavage of the phosphodiester bond at the RNA-DNA junction 5' to the rNMP; (ii) DNA synthesis to replace excised nucleotides; (iii) a second cut is made 3' to the ribonucleotide; (iv) sealing of the nick by a DNA ligase that ultimately completes the repair pathway.
Fig. 1. A model for Ribonucleotide Excision Repair.
DNA replication catalyzed by an error-prone DNA polymerase might lead to occasional misinsertion of rNMPs along with erroneous bases (1). Type 2 RNase H recognizes a nucleotide with an extra 2'-OH group (2) and cleaves the phosphodiester bond at the RNA-DNA junction 5' to the rNMP (3). A DNA polymerase initiates strand-displacement DNA synthesis at the nick and the displaced single rNMP, or 5'-RNA-terminated DNA fragment is excised by the 5' nuclease activity either intrinsic to the polymerase or associated with a separate protein (4). When a DNA fragment in the vicinity of rNMP contains mispaired bases, its replacement by RER reduces the mutagenic consequences of DNA synthesis by an error-prone DNA polymerase. The DNA template left after completion of excision and re-synthesis steps contains a nick with 3'-hydroxyl and 5'-phosphate ends that is readily sealed by DNA ligase, thus completing the RER pathway (5). Mispaired bases outside the re-synthesis patch of RER can be replaced by the MMR. It has been proposed that in eukaryotes, a transient nick created during RER can serve as a signal that directs MMR to the mispaired base on the nascent strand.
As a general rule, DNA polymerases discriminate against rNMPs very efficiently and only rarely incorporate isolated ribonucleotides [2, 7]. In contrast to multiple consecutive rNMPs forming the RNA primer of an Okazaki fragment, these ribonucleotides are randomly scattered across the genome. Therefore, initiation of the RER process requires a ribonucleotide-specific endonuclease that can recognize a single rNMP embedded in double-stranded DNA. The enzymes that can hydrolyze the 3'-O-P bond on such substrates are well-conserved in all domains of life and are called type 2 ribonuclease H (RNase H) (Fig. 1). Even though RNases of this type, such as eukaryotic RNase H2 encoded by the rnh2 gene and prokaryotic RNase HII encoded by the rnhB, prefer to cleave the RNA moiety in DNA templates containing a single ribonucleotide, they are also able to incise templates containing multiple rNTPs [31-37].
In both bacterial and yeast strains expressing steric gate mutant DNA polymerases (pol V umuC_Y11A or pol ε M644G respectively), defects in type 2 RNase H have mutagenic consequences [8, 22, 26]. However, the pathway leading to an increase in mutagenesis and the origin and type of mutants themselves are very different. In agreement with our hypothesis, disabling the major ribonucleotide repair mechanism in E. coli ΔrnhB cells results in ~5-fold increase in pol V-dependent spontaneous mutagenesis because a significant number of misincorporated nucleotides errantly incorporated by umuC_Y11A remain unrepaired. In contrast, de novo mutations are generated in an S. cerevisiae strain expressing the M664G pol ε mutant as a result of triggering of an alternative rNMP processing when RNase H2 is inactivated. These mutations, among which is a signature 2 to 5-base-pair deletion, arise in rnh201Δlcells from the improper cleavage of the phosphodiester bond between ribo- and deoxyribonucleotides, creating unligatable nicks (discussed in more details below) [38].
It should be stressed that despite the mutagenic consequences described above, bacterial and yeast strains lacking type 2 RNase H are viable, while a defect in the catalytic activity of the mammalian enzyme is intolerable (recently reviewed in [16]). This is manifested by early embryonic lethality of RNase H2 knock-out mice [10] and by the fact that the human Aicardi– Goutières syndrome is caused by partial reduction in the enzyme’s catalytic activity [39], or by mutations in the non-catalytic RNase H2B subunit [40]. Nevertheless, similar to yeast, cells from RNase H2 mutant mice show signs of genome instability [10] emphasizing the importance of intact RNase H2 in the non-mutagenic processing of rNMPs in higher eukaryotes.
Type 1 RNases H (encoded by rnh1 in eukaryotes and by rnhA in prokaryotes), possess a similar mechanism of hydrolysis and are structurally related to type 2 RNase H. However, in contrast to type 2 enzymes, type 1 enzymes require a tract of at least four sequential ribonucleotides within the DNA strand for efficient nicking [35, 37]. It is therefore expected, that in general, type 1 RNase H enzymes are unable to substitute for type 2 RNase H enzymes in the repair of randomly incorporated ribonucleotides [31]. However, if a DNA polymerase manages to insert multiple consecutive rNMPs, the availability of RNase HI might become useful. For example, we have shown that in an E. coli ΔrnhB strain expressing umuC_Y11A, repair of a significant number of misincorporated ribonucleotides is dependent on RNase HI [26, 41].
3. Role of DNA polymerases and flap endonucleases in RER
Initiation of RER by RNase H-catalyzed hydrolysis of DNA at a ribonucleotide generates nicks with 3'-hydroxyl and 5'-phosphate termini that can be “nick-translated” and subsequently ligated. In eukaryotes this process occurs though the coordinated action of the replicative polymerase, pol δ, catalyzing strand displacement DNA synthesis, flap endonuclease 1 (FEN1), mediating cleavage of the displaced 5'-overhang, and DNA ligase I resealing the nick [31, 42](Fig. 1). By reconstituting RER in vitro using purified yeast enzymes, Sparks et al., have shown that, similar to Okazaki processing, strand displacement synthesis could be also accomplished by pol ε and excision of the resulting flap can be catalyzed by exonuclease 1 (Exo1), albeit at a reduced rate in both cases [31]. Another study demonstrating the significance of FEN1 in ensuring efficient RER was performed using crude cell extracts from human and yeast cells [36]. This study implied that after RNase H2 catalyzes the initial nick 5' to rNMP, a second cut is made by the exonuclease activity of FEN1, thereby releasing free ribonucleotide monophosphate and generating a single-nucleotide gapped DNA that could be filled in by an appropriate DNA polymerase, such as pol β. However, neither approach allowed one to definitively establish the sequence of the events. For example, it is possible that RNase H-initiated RER proceeds through the second cut made immediately 3' to the rNMP by the 5'→3' exonuclease activity of FEN1, followed by the gap-filling DNA synthesis. Alternatively, strand-displacement DNA synthesis initiated at the nick creates flap that is subsequently cleaved by the 5' endonuclease.
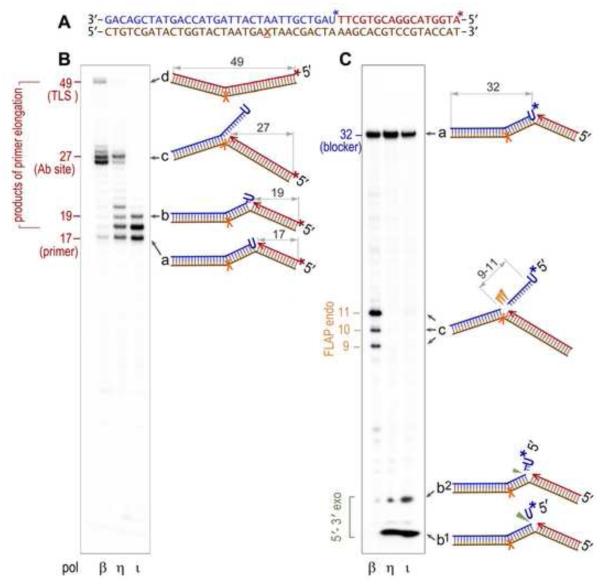
Accurate elucidation of the molecular mechanism of RER is essential, since the length of the repair patch is not only affected by the biochemical characteristics of the participating enzymes and the structure and composition of DNA, but also by the sequence of the events. As shown in Figure 2, the size of the re-synthesis patches and of the excised fragments clearly depends on the processivity of the DNA polymerase responsible for the strand-displacement DNA synthesis. Thus, in the presence of FEN1, displacement synthesis catalyzed by distributive DNA polymerases, such as pol η or pol ι (Fig. 2B) primarily favors the formation and cleavage of short 2-3 nucleotide flaps, or single nucleotides (Fig. 2C) (reminiscent of the short-patch repair pathway). In contrast, the relatively processive pol β (Fig. 2B) extends primers by efficiently displacing the downstream DNA strand until it encounters a kinetic block (a template abasic site in our model substrate, Fig. 2A), which obstructs further primer elongation. As a result, the branched structure containing a single-stranded 5' flap of ~10 nucleotides is created and cleaved by the 5' endonuclease activity of Fen 1 (Fig. 2C) (medium-patch RER). By analogy to the model proposed to elucidate removal of short RNA primer flaps [43, 44], short-patch RER can also be envisioned as resulting from strand-displacement synthesis catalyzed by a highly processive DNA polymerase when its forward progression is limited by the 3′→5′ exonuclease proofreading activity of the polymerase.
Fig. 2. Processing of a nicked DNA substrate with a single ribonucleotide (U) in the 5' terminus of the nick by the combined action of a DNA polymerase and nuclease.
A. Sequence of the oligonucleotides used to generate the nicked DNA substrate (also shown schematically to the right of each gel panel as “a”), consisting of the 49-mer DNA template (brown) with an abasic site (X) annealed with a 17-mer primer (red) and a 32-mer downstream blocker (blue). The template in which either primer, or blocker, is P32-labeled (indicated by the star, *), together with the purified recombinant human DNA polymerase (pol β, pol η, or pol ι) and Fen1 protein was used in in vitro reactions B. P32-labeling of the 17-mer primer (red *) allows for visualization of the primer extension products. The processivity of the strand displacement synthesis, decreasing in the order pol β> pol η> or pol τ□ can be judged by the intensity of the intermediate bands such as the 19-mer oligonucleotide, whose structure (“b”) is schematically shown to the right of the gel. The ~27 nt-long bands correspond to replication blockage by the abasic site on the DNA template (c), and the 49-mer band corresponds to the full-sized product generated by translesion replication to the end of the template (d). C. P32- labeling of the 32-mer blocker (blue *) allows one to visualize products of the 5'→3' exo-(green ▼), and flap endonucleolytic (orange ▼) cleavage.
In search of proteins facilitating RER in prokaryotes, we turned our attention to DNA polymerase I, an important component of the bacterial replication machinery and a central player in a number of excision repair pathways. While the recognition and initial processing of various modified nucleotides is initiated via different lesion-specific endonucleases, subsequent DNA handling requires the same group of proteins. Indeed, replacement of damage-containing DNA fragments in the framework of most repair pathways occurs by an identical mechanism and by default, relies on the polA gene product. Pol I possesses three distinct biochemical properties: DNA polymerase catalytic activity, 3'→5' proofreading activity, which excises misincorporated nucleotides from the 3' end of nascent DNA, and flap exo/endonuclease activity, which removes nucleotides from the 5' terminus. The combination of pol I’s enzymatic activities makes it a perfect RER player acting downstream of an endonuclease and via classical “nick translation” mechanism preparing cleaved substrates for ligation. Therefore, even though the overall pathway of ribonucleotide repair is evolutionary conserved, the generation and processing of the RER intermediates (i.e. substrates of the flap exo/endonuclease activity) can be accomplished by a single protein in prokaryotes, while this step normally requires the concerted action of two eukaryotic enzymes.
Historically, the biological role of pol I in various DNA repair pathways was elucidated to a large extent through genetic characterization of individual polA mutants. In a similar vein, based upon the quantitation of spontaneous mutagenesis in strains expressing different pol I and pol V variants, we can confidently conclude that pol I acts in RER to mediate replacement and excision of rNMP-containing DNA fragments [27]. Akin to its function in other DNA repair pathways and Okazaki fragments processing, pol I binds to the nicked DNA template, displaces the rNMP-containing fragment by strand-displacement DNA synthesis and excises the displaced nucleotides using 5'→3' exonuclease and/or 5' flap exo/endonuclease activity.
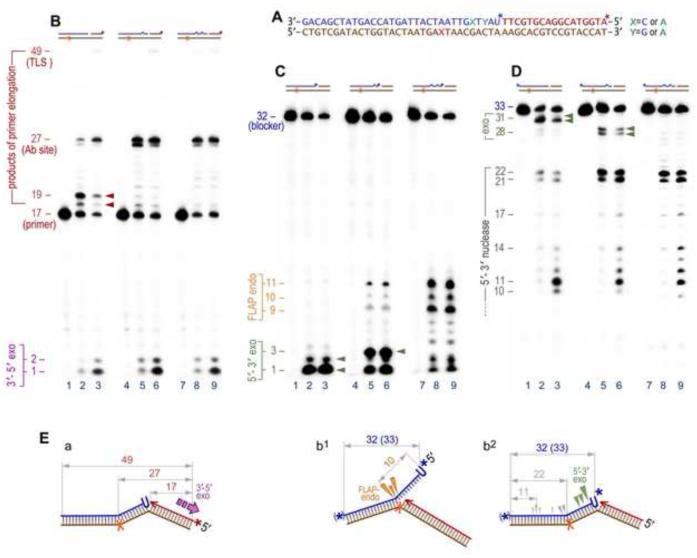
Biochemical characterization of pol I’s catalytic activities suggests that both enzymatic reactions take place within the same polypeptide chain. The polymerase acts first, producing an intermediate that is then passed to and cleaved by the 5'-nuclease [45]. When we reconstituted RER in vitro using oligonucleotide templates with nicks made 5' to a single ribonucleotide (rU) (Fig. 3A), pol I displayed poor processivity, displacing 1–2 nucleotides of the RNA/DNA hybrids (Fig. 3B, lanes 2, 3) and releasing mono- and short oligonucleotides through its 5'→3' exonuclease and/or 5' flap exo/endonuclease activity (Fig. 3C, lanes 2, 3). These findings are consistent with studies of Xu et al., [45], that were aimed at the characterization of the coupling of polymerase and 5'-nuclease activities of pol I. Their study demonstrated that the preferred substrate for the 5'-nuclease of pol I is formed by rearrangement of the displaced 5' end via branch migration generating a “double-flap” structure with both 3'- and 5'- single-stranded extensions. Cleavage of such a structure mainly occurs between the first two paired bases of the strand with the 5'-overhang. Cleavage of a nicked substrate is less efficient and predominantly occurs between the first two paired bases of the downstream strand [45]. When DNA synthesis is non-processive, such nuclease specificity would result in the release of very short excision products regardless of whether the cleavage occurs through the 5'- flap-endo or through the 5'- exonuclease activity. Ultimately, however, the size of the cleaved products depends on the extent to which synthesis proceeds into the downstream duplex. Indeed, pol I excised longer 5'-FLAPs in vitro when reactions were performed using substrates that allowed for more processive strand-displacement synthesis, i.e. when the downstream double-stranded DNA contained base mispairs (Figs. 3B-D, lanes 5, 6, 8, 9). In fact, pol I has been shown to be more processive (15-50 nucleotides per binding event) on long circular, nicked, or gapped DNA substrates [46] and its processivity further increases through an interaction with the β-sliding clamp when loaded onto DNA by the γ-complex. Although it is generally accepted that in vivo pol I is involved in single nucleotide, or relatively short (2-30 nt) patch repair, several lines of evidence suggest that in some sequence contexts, where it is capable of highly processive DNA synthesis [47], repair tracts generated by pol I and oligonucleotides released by its flap endonuclease activity could be very long (see for example [48]). It should be noted that the number of replaced nucleotides and the size of the excised fragments is not necessarily determined by the difference in the relative rates of synthesis and hydrolysis. Even when pol I is less processive, relatively long repair tracks could be generated in a cell through multiple rounds of synthesis and rebinding (Fig. 3D, compare lanes 2 & 3 with 5, 6, 8, & 9) [49], which is accompanied by the release of a series of short oligonucleotides (Fig. 3C).
Fig. 3. Enzymatic properties of pol I on a nicked DNA substrate containing an abasic site (X) located in the template strand and a 5'-terminal mono-ribonucleotide in the downstream blocking oligonucleotide.
A. The sequence of the oligonucleotides used to generate the various DNA substrates is essentially the same as indicated in Figure 1A. The structures of the DNA substrates are schematically presented on the top of each gel image (brown – template, red – primer, blue - blocker). B. The products of pol I-catalyzed strand displacement synthesis and 3'→5' exonucleolytic proofreading are detected using DNA substrates with the P32-labeled 17-mer primer (red star, *). C. 5'-end P32-labeling of the 32-mer blocker (blue star, *) is used to visualize the 5' nuclease activity of pol I. D. Labeling of the blocker at the 3' termini by addition of a single P32-ATP (and converting the blocker into the 33-mer) allows for the detection of repair track synthesis. E. Schematic representation of DNA templates with arrows pointing to the cleavage sites on the DNA made by the 3'→5' exo- (purple Ξ in a), 5'→3' exo- (green ▼ in b2), and FLAP-endonuclease (orange ▼in b1) activities of pol I. Lanes 1, 4, & 7 in each gel panel represent control reactions incubated in the absence of pol I. Pausing of strand-displacement DNA synthesis after incorporation of 1 or 2 nucleotides (indicated by the red arrows, lanes 2 & 3 on the panel B) is accompanied by the release of the mono- or di-nucleotides catalyzed by the 5'→3'exonucleolytic activity of pol I (indicated by the green arrows, lanes 2 & 3, panel C, and illustrated on the scheme b2, panel E). Blocking DNA synthesis with an abasic site (27-mer on the panel B) is accompanied by the emergence of the 3'- end labeled 21- and 22-mers (panel D). These bands are mainly formed as a result of 5'→3'exonucleolytic cleavage by pol I, although a low level contribution of FLAP-endonuclease cannot be excluded. The attempts of pol I to replicate past the lesion is largely overcome by the 3'→5' exonuclease proofreading (scheme a, panel E) as indicated by the emergence of the mono- or di-nucleotides (lanes 2 & 3, panel B), although a small amount of full-sized translesion replication products (49-mer) can also be seen. Futile cycling (bypass synthesis/3'→5' proofreading) causes release of a significant portion of the downstream blocker which is degraded by the 5' exo- and/or endonuclease activity of pol I, which is manifested by the 3'-end labeled 10-17-mer bands (panel D and scheme b2, gray arrows). Introduction of one (lanes 4-6, Y=A) or two (lanes 7-9, X & Y=A) mismatched base pairs facilitates strand displacement and as a consequence, increases the processivity of DNA synthesis (manifested by the significant reduction of band intensities at the +1 and +2 positions, lanes 5, 6, 8, 9, panel B). For the template with the single mismatch, this results in a shift in the size of the pol I cleavage products from a 2-mer to 3-mer (indicated by the green arrows, lanes 5 & 6, panel C). Correspondingly, length of the 3'-end labeled blocker reduces to 27-28 nucleotides (panel D). At the same time, products of the FLAP-endonuclease activity also become more visible as 9-11 nucleotide long oligonucleotides in panel C (also illustrated on the scheme b1, panel E). Products of the FLAP-endonuclease are much more prevalent with a DNA template containing two mispaired bases (lanes 8 & 9, panels C & D).
In eukaryotes, DNA replication is initiated through the generation of composite RNA-DNA primers catalyzed by a hetero-tetrameric complex consisting of primase and DNA polymerase α [50]. The 3'→5' exonuclease-deficient pol α is error-prone and its mistakes must be edited in order to prevent them from being fixed as mutations. One of the mechanisms utilized by cells to correct pol α errors involves their excision together with the removal of RNA primers [42]. This could be accomplished through FEN1-catalyzed cleavage of the displaced RNA/DNA flaps containing mispaired nucleotides, or by the 5' exonuclease activity of FEN1 that removes several ribo- and deoxyribonucleotides from the 5' end of the nick. In prokaryotes, primase hands off the DNA template directly to the high-fidelity replicative pol III without the need for an intermediate inaccurate DNA polymerase. Therefore, replacement of RNA primers does not require the correction of neighboring DNA bases. However, mechanisms similar to the eukaryotic correction of base mispairs in the vicinity of RNA primers do appear to operate in E. coli cells during RNase HII-initiated repair of ribonucleotides that are incorporated by the highly error-prone pol V. Akin to Okazaki fragment maturation in eukaryotes, replacement of mispaired nucleotides can be envisioned to occur in two ways in bacteria (Fig. 1). Together with the rNMP they could be displaced into the 5′ flap and excised through pol I-catalyzed nick-translation, or, alternatively, pol V errors are corrected by the 5'→3' exonuclease activity of pol I by excising 5' terminal nucleotides ahead of the polymerase domain catalyzing strand extension. Pol I is less accurate than replicative pol III, but even a 3'→5' proofreading deficient variant is significantly more accurate compared to pol V. Therefore, recruitment of pol I to sites of RER inevitably leads to a reduction in pol V-dependent mutation rates. As a consequence, the longer the pol I-dependent repair patch the greater number of pol V errors that can be corrected. By taking advantage of the differences in the fidelity of bacterial DNA polymerases and by using strains with different mutant polA alleles, we were able to define the role of pol I in RER [27].
Our studies were performed in a mismatch repair-deficient background, so that any transition mutations generated by pol I during RER would not be corrected. Indeed, in rnhB+ cells expressing umuC_Y11A, the absence of mismatch repair resulted in a three-fold increase in mutagenesis, which we ascribed to error-prone pol I-dependent RER. In support of this hypothesis, mutagenesis further increased when intrinsic 3'→5' exonucleolytic proofreading of pol I was inactivated [27]. In contrast, disruption of pol I’s polymerase activity in strains encoding a polA allele with a C-terminal truncation (polA_ΔC) resulted in a dramatic reduction in the number of mutations introduced during the course of the re-synthesis step of RER. The increased fidelity of RER in these cells can be easily explained by the substitution of pol I with the much higher fidelity replicase, pol III. Accordingly, the fidelity of RER in polA_ΔC cells dropped drastically when the DNA synthesis step was catalyzed by pol III lacking exonucleolytic proofreading. Overall, these findings provide compelling evidence that pol I is the polymerase of choice for RER.
4. Backup pathways of ribonucleotide removal
DNA lesions are often repaired by multiple pathways. It is therefore reasonable to expect that in addition to the major RNase H-dependent RER pathway (Fig. 4), alternative defense mechanisms are able to target nucleotides with a wrong sugar for repair, and other yet to be identified enzymes can cleave the phosphodiester bond at a newly incorporated rNMP. Indeed, even in the absence of both ribonucleases HI and HII, the levels of spontaneous mutagenesis detected in a umuC_Y11A-expessing strain is reduced compared to that of the same strain bearing wild-type pol V [41]. This is consistent with the presence of back-up enzymes ready to assume the responsibility of cleansing the bacterial genome from errant ribonucleotides, even though they may do it less effectively than RNase HII.
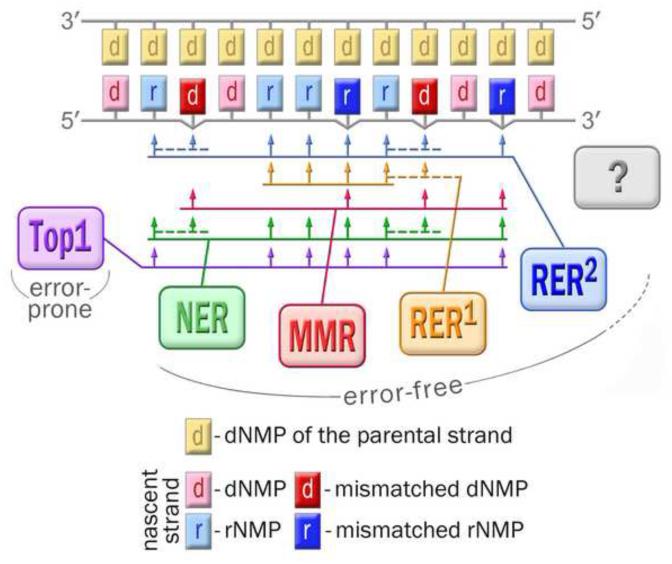
Fig. 4. Pathways for keeping genomic DNA free from errant ribonucleotides: from competition to cooperation.
While copying the parental DNA strand (shown in yellow), a low fidelity DNA polymerase, such as E. coli pol V, besides the correct nucleotides (shown in pink), often incorporates nucleotides with wrong base (shown in red) and/or wrong sugar moiety (correctly paired ribonucleotides are indicated in light blue, mispaired rNMPs are indicated in dark blue). These mistakes can be corrected by multiple repair pathways with distinct but overlapping specificities. The cartoon illustrates interplay between those pathways schematically presented as colored boxes connected with the corresponding target sites by a set of arrows. The first responder to the errantly incorporated rNMPs is type 2 RNase H. This enzyme initiates the primary and most efficient pathway of rNMP repair (RER2, blue). Type 2 RNase H preferentially cleaves DNA templates with a single ribonucleotide, although it is also able to incise templates containing multiple rNTPs. In contrast, type 1 RNase H initiates repair (RER1, orange) that operates only on a tract of at least four sequential ribonucleotides within the DNA strand. Ribonucleotides that for some reason escape RER, can be excised by means of NER (green), most likely activated later than the RNase H-dependent pathway, or when it is impaired. All three pathways, RER2, RER1, and NER, are specifically triggered by the rNMPs, either correctly paired or mispaired, but concomitantly they can remove mispaired dNMPs positioned within the repair patch (indicated by the dashed lines). In contrast, coupled to replication MMR (crimson) is able to replace nucleotides with wrong sugar only when the base is also wrong. In this sense MMR cannot be considered as truly rNMP-targeted repair. Similar to NER, Top1-initiated excision of rNMPs (violet) is activated later than RER and MMR. Moreover, it seems that this pathway operates only when RNase H is impaired. In contrast to other mechanisms of rNMP repair, the Top1-initiated pathway is highly mutagenic and to date has been detected only in yeast. It remains to be determined whether additional, as yet unidentified pathways of ribonucleotide repair (shown in grey and by question mark) exist in other species.
It seems that the simplest way for the cell to correct pol V errors would be to utilize the MMR apparatus drawn to the mispaired bases on the newly synthesized DNA strand. Indeed, as shown by several groups, the MMR machinery is able to bind and excise nucleotides with incorrect sugar, but only when the base is also incorrect [23, 41, 51]. These observations imply that a base mispair, not an incorrect sugar, is actually recognized by the MMR machinery (Fig. 4). We also were unable to find any evidence of a significant role of base excision repair (BER) as a back-up system for the RER pathway, at least in E coli [41], even though BER is a major pathway responsible for cleansing DNA of deoxyuracils misincorporated by DNA polymerases, or generated through cytosine deamination.
The breakthrough in our quest for alternative ribonucleotide repair pathways came from an unexpected quarter. We discovered that in the absence of RNase HII, the excision of rNMPs scattered throughout the E.coli genome is partly recovered through the concerted actions of the UvrA, UvrB, and UvrC proteins [41], that are normally required for damage recognition and excision steps of prokaryotic nucleotide excision repair (NER) (Fig. 4). A second E. coli endonuclease, Cho (UvrC homologue), capable of assisting the NER complex in the excision of damaged substrates that are poorly incised by UvrC, is apparently not involved in the removal of ribonucleotides. Interestingly, our studies also indicated that removal of rNMPs by the UvrABC excinuclease can be successfully completed in the absence of the UvrD helicase, a key player in both nucleotide and mismatch excision repair [41].
The ability of the UvrABC complex to remove rNMPs from double stranded DNA was confirmed by reconstituting the reaction in vitro [41]. Biochemical characterization suggests that similar to NER of various helix-distorting modifications, repair of rNMPs proceeds through dual incisions made at the 8th phosphodiester bond 5', and 4th–5th phosphodiester bonds 3' of the ribonucleotide that releases a DNA fragment of 12–13 bases. The NER complex has the capacity to recognize and excise not only an isolated ribonucleotide, but also multiple rNMPs within a short RNA fragment in double-stranded DNA. The efficiency of repair depends on the number of consecutive rNMPs, the presence of base mispairs in the vicinity of the target ribonucleotide, as well as the type and number of the mispaired bases. The biochemical data suggest that mismatch-induced distortions in DNA structure in close proximity to the ribonucleotide help to target it for NER. Nevertheless, the NER complex is able to recognize and excise a single ribonucleotide embedded in dsDNA even in the absence of additional errors. Furthermore, despite its small impact on the global helical properties of the DNA duplex, an isolated rNMP can be replaced as efficiently as various bulky DNA-distorting lesions. Based on the results of molecular dynamics simulations, Cai et al., [52] have proposed that ribonucleotide recognition by the UvrAB relies on hydrogen bonding and electrostatic interactions of the ribose 2'-OH group with several aromatic residues in UvrB that are responsible for damage recognition.
Even though in vitro NER is able to relatively efficiently excise a single as well as multiple consecutive rNMPs with different base identity and placed within different surrounding sequence contexts, in vivo it became apparent only in ΔrnhB cells [41]. These findings are consistent with the model in which the first repair enzyme encountering a newly incorporated ribonucleotide in bacterial cell is RNase HII. The primary repair pathway initiated by RNase HII with possible assistance of RNase HI is efficient enough to alleviate need for the additional mechanisms (Fig. 4). Our data are also consistent with the hypothesis that RNase HII-initiated RER is coupled to DNA replication [4]. Therefore, post-replicative NER, which is generally assumed to act in the G1 and G2 phases of the cell cycle, functions mainly as an effective substitute for the disabled RNase HII-dependent RER, although limited competition between RER and NER for the subset of errant nucleotides cannot be excluded.
It is clear that NER targeted at rNMP removal is capable of making a significant contribution to E. coli genome stability by helping reduce the number of errantly incorporated nucleotides. Several lines of indirect evidence suggest similar role for this mechanism in eukaryotic cells. Thus, association between defect in RNase H2 and changes in the expression of NER genes has been reported for yeast cells [53], and an increase in the rates of transcription-associated mutagenesis has been found in the rnh201Δ (RNase H2-defective) yeast strains upon loss of the NER pathway [54].
The mutagenic outcome of ribonucleotide removal by the NER complex depends on the differences in fidelity of the polymerase responsible for the rNMP insertion and enzymes involved in excision repair. Thus, in E. coli, replacement of rNMPs through both primary (RER) and secondary (NER) mechanisms drastically reduces the number of spontaneous mutations arising from umuC_Y11A errors. In contrast, an alternative pathway of rNMP processing that has been recently identified in yeast is inherently error-prone and its activation poses a serious threat to the integrity of the genome. The threat comes from the action of topoisomerase 1 (Top1) (Fig. 4), whose primary function in the cell is to regulate DNA supercoiling through the creation of transient single-strand breaks. Interestingly, while substitution of RNase H2 by Top1 in rNMP processing compromises S. cerevisiae genome integrity (reviewed in [4, 55]) the availability of at least one RNases H in top1Δ cells has an opposite effect. Thus, it has been shown that both RNaseH1 and RNase H2 can rescue genome integrity of yeast cells by resolving transcriptional R-loops (anomalous RNA–DNA hybrids), formation of which would otherwise be suppressed by Top1 [56].
In the absence of a proper substitute for RNase H2, Top1 gains access to the rNMP-containing DNA and hydrolyzes the phosphodiester bond between ribo- and deoxyribonucleotides [38] (Fig. 4). Such cleavage is irreversible because it creates 2',3'-cyclic phosphate ends that are refractory to re-ligation. The events initiated by Top1-catalyzed incision have a distinct mutagenic consequence, i.e. generation of 2- to 5-bp deletions within tandem repeat sequences [57]. These mutations, when accumulated at significant levels, result in replicative stress and genome instability. Such an effect was observed in rnh201Δ S. cerevisiae cells expressing a steric gate variant of the replicative polymerase, pol ε [8]. When RNase H2 is intact, excessive rNMP incorporation does not pose a threat to the yeast genome stability. The lack of 2 to 5 bp deletions from the mutagenic spectra of cells expressing the mutant pol ε, but with a wild type rnh2 gene, indicates that similar to the bacterial RNase HII protein, yeast RNase H2 has priority access to ribonucleotides randomly distributed within chromosomal DNA (Fig. 4). When this barrier is removed in rnh201Δ yeast strains, aberrant processing of abundantly accumulated ribonucleotides has an adverse mutagenic effect, even if Top1 is able to target only a subset of rNMPs [8]. It is plausible to assume that in RNase H2-defective mammalian cells Top1-dependent flawed attempts to excise ribonucleotides accompanied by an increased frequency of strand breaks, contribute to mouse embryonic lethality and to the pathogenesis of Aicardi-Goutières syndrome in humans [58]. A similar scenario of mutagenic rNMPs processing has been outlined by Jean-Sebastien Hoffmann and co-workers [59] in order to explain the enhanced genetic instability of mammalian cells over-expressing pol β, which is an enzyme characterized by relatively low sugar selectivity. Bergoglio et al., hypothesized that overly frequent ribonucleotide incorporation by up-regulated pol β saturates normal RER repair pathways, thereby allowing Top1 to initiate rNMP processing. Cleavage of rNMP-containing DNA by Top1 results in the formation of unligatable nicks with 2', 3'-cyclic phosphate and 5'- OH ends and ultimately leads to increased spontaneous mutagenesis [60], chromosome instability, and tumorigenesis [61]. This hypothesized pathway is virtually identical to the one later demonstrated in S. cerevisiae [8]. However, in yeast cells, the involvement of Top1 in rNMP excision only becomes evident when RNase H2 is inactivated, while in mammalian cells with elevated rNMPs incorporation, Τop1 appears to be able to gain an access to the rNMP-containing DNA even in presence of functional RNase H2.
5. Concluding remarks
In her 1997 commentary “Choosing the right sugar: How polymerases select a nucleotide substrate” [7], Catherine Joyce stated: “A quick glance at the recent scientific literature might give the impression that nucleic acid polymerases are suffering from an identity crisis” because of “the blurring of the distinction” between different polymerase classes. One of three studies Catherine Joyce cited to highlight her remark, was a paper entitled “Conferring RNA polymerase activity to a DNA polymerase” [62], which demonstrated that the identity of a DNA polymerase is virtually single-handedly defined by an evolutionarily conserved amino acid. This amino acid, which Gao et al., called a “steric gate”, serves as a guard blocking access to DNA for nucleotide substrates with the wrong sugar. Indeed, mutation of the steric gate residue essentially converted the Moloney murine leukemia virus reverse transcriptase into an RNA polymerase [62]. Several studies that followed this initial discovery seemed to indicate that the steric gate mechanism is a very reliable way to protect the integrity of DNA structure. However, the subsequent in-depth research revealed a surprising phenomenon. Errant ribonucleotides embedded in chromosomal DNA, which originally appeared to represent rare, potentially damaging mistakes of DNA polymerases, are now believed to be the most frequent non-canonical DNA inserts [9, 63]. It has been demonstrated that at physiological nucleoside concentrations, even high-fidelity replicative polymerases with an intact steric gate incorporate significant number of ribonucleotides during genome duplication (recently reviewed in [3, 4, 16]). Furthermore, besides being dangerous when accumulated at excessive amounts, rNMPs transiently present in DNA have important biological functions that are beneficial for the cell.
As mentioned above, living organisms are well prepared to limit the impact of mistakes made by DNA polymerases and to free their genomes from erroneous ribonucleotides. Some of the aberrant replacement mechanisms can have devastating consequences, but reassuringly, most of the ribonucleotide repair pathways identified to date are beneficial for the cell. Considering the large number of rNMPs embedded in genomic DNA and the importance of keeping the DNA structure intact, it would not be surprising if other, as yet unidentified pathways of ribonucleotide repair exist in various species (Fig. 4). It is possible, as well, that in order to preserve the positive outcome of rNMPs in genomic DNA cells utilize yet unidentified mechanisms that actually limit ribonucleotide excision.
Highlights.
We review progress in understanding the mechanisms of ribonucleotide excision repair
The key enzymes that initiate ribonucleotide excision repair are type 2 RNases
Back-up ribonucleotide excision repair pathways can be error-free, or error prone
Similarities and differences in prokaryotic and eukaryotic RER are highlighted
Original research findings are presented for the re-synthesis step of RER
Acknowledgements
This study was made possible by funding from the NIH/NICHD Intramural Research Program.
Abbreviations
- nt
nucleotide
- dNTP
deoxyribonucleoside triphosphate
- rNTP
ribonucleoside triphosphate
- pol
DNA polymerase
- RNase H
ribonuclease H
- Top1
topoisomerase 1
- Fen1
flap endonuclease 1
- PCNA
proliferating cell nuclear antigen
- Exo1
exonuclease 1
- Cho
UvrC homologue
- RER
ribonucleotide excision repair
- NER
nucleotide excision repair
- MMR
mismatch repair
- BER
base excision repair
- NHEJ
non-homologous end joining repair
- E. coli
Escherichia coli
- S. cerevisiae
Saccharomyces cerevisiae
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest
References
- [1].Cavanaugh NA, Beard WA, Batra VK, Perera L, Pedersen LG, Wilson SH. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J. Biol. Chem. 2011;286:31650–31660. doi: 10.1074/jbc.M111.253401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brown JA, Suo Z. Unlocking the sugar "steric gate" of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vaisman A, Woodgate R. Ribonucleotide selectivity of translesion DNA synthesis polymerases. Research Signpost. 2014 [Google Scholar]
- [4].Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair. 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goksenin AY, Zahurancik W, LeCompte KG, Taggart DJ, Suo Z, Pursell ZF. Human DNA polymerase ε is able to efficiently extend from multiple consecutive ribonucleotides. J. Biol. Chem. 2012;287:42675–42684. doi: 10.1074/jbc.M112.422733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nick McElhinny SA, Ramsden DA. Polymerase μ is a DNA-directed DNA/RNA polymerase. Mol.Cell.Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yao NY, Schroeder JW, Yurieva O, Simmons LA, O'Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol.Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cavanaugh NA, Beard WA, Wilson SH. DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gosavi RA, Moon AF, Kunkel TA, Pedersen LC, Bebenek K. The catalytic cycle for ribonucleotide incorporation by human DNA pol λ. Nucleic Acids Res. 2012;40:7518–7527. doi: 10.1093/nar/gks413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28:592–597. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- [16].Potenski CJ, Klein HL. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014;42:10226–10234. doi: 10.1093/nar/gku773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol.Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sayrac S, Vengrova S, Godfrey EL, Dalgaard JZ. Identification of a novel type of spacer element required for imprinting in fission yeast. PLoS Genet. 2011;7:e1001328. doi: 10.1371/journal.pgen.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- [20].Wahl MC, Sundaralingam M. B-form to A-form conversion by a 3'-terminal ribose: crystal structure of the chimera d(CCACTAGTG)r(G) Nucleic Acids Res. 2000;28:4356–4363. doi: 10.1093/nar/28.21.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nature Chemical Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε. DNA Repair. 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. Critical amino acids in Escherichia coli responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 2012;40:6144–6157. doi: 10.1093/nar/gks233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kuban W, Vaisman A, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. Escherichia coli UmuC active site mutants: effects on translesion DNA synthesis, mutagenesis and cell survival. DNA Repair. 2012;11:726–732. doi: 10.1016/j.dnarep.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by pol V. PLoS Genet. 2012;8:e1003030. doi: 10.1371/journal.pgen.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vaisman A, McDonald JP, Noll S, Huston D, Loeb G, Goodman MF, Woodgate R. Investigating the mecahnisms of ribonucleotide excision repair in Escherichia coli. Mutat. Res. 2014;761:21–33. doi: 10.1016/j.mrfmmm.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- [29].Fuchs RP, Fujii S. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect Biol. 2013;5:a012682. doi: 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb. Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-Initiated Ribonucleotide Excision Repair. Mol.Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Haruki M, Tsunaka Y, Morikawa M, Kanaya S. Cleavage of a DNA-RNA-DNA/DNA chimeric substrate containing a single ribonucleotide at the DNA-RNA junction with prokaryotic RNases HII. FEBS Lett. 2002;531:204–208. doi: 10.1016/s0014-5793(02)03503-2. [DOI] [PubMed] [Google Scholar]
- [33].Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol. Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects. J. Biol. Chem. 2011;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. Febs J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. Febs J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- [38].Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol.Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- [40].Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Baxter P, Benko WS, Bergmann C, Bertini E, Biancheri R, Blair EM, Blau N, Bonthron DT, Briggs T, Brueton LA, Brunner HG, Burke CJ, Carr IM, Carvalho DR, Chandler KE, Christen HJ, Corry PC, Cowan FM, Cox H, D'Arrigo S, Dean J, De Laet C, De Praeter C, Dery C, Ferrie CD, Flintoff K, Frints SG, Garcia-Cazorla A, Gener B, Goizet C, Goutieres F, Green AJ, Guet A, Hamel BC, Hayward BE, Heiberg A, Hennekam RC, Husson M, Jackson AP, Jayatunga R, Jiang YH, Kant SG, Kao A, King MD, Kingston HM, Klepper J, van der Knaap MS, Kornberg AJ, Kotzot D, Kratzer W, Lacombe D, Lagae L, Landrieu PG, Lanzi G, Leitch A, Lim MJ, Livingston JH, Lourenco CM, Lyall EG, Lynch SA, Lyons MJ, Marom D, McClure JP, McWilliam R, Melancon SB, Mewasingh LD, Moutard ML, Nischal KK, Ostergaard JR, Prendiville J, Rasmussen M, Rogers RC, Roland D, Rosser EM, Rostasy K, Roubertie A, Sanchis A, Schiffmann R, Scholl-Burgi S, Seal S, Shalev SA, Corcoles CS, Sinha GP, Soler D, Spiegel R, Stephenson JB, Tacke U, Tan TY, Till M, Tolmie JL, Tomlin P, Vagnarelli F, Valente EM, Van Coster RN, Van der Aa N, Vanderver A, Vles JS, Voit T, Wassmer E, Weschke B, Whiteford ML, Willemsen MA, Zankl A, Zuberi SM, Orcesi S, Fazzi E, Lebon P, Crow YJ. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. American Journal of Human Genetics. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vaisman A, McDonald JP, Huston D, Kuban W, Liu L, Van Houten B, Woodgate R. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet. 2013:e1003878. doi: 10.1371/journal.pgen.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 2008;283:34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Xu Y, Grindley ND, Joyce CM. Coordination between the polymerase and 5'-nuclease components of DNA polymerase I of Escherichia coli. J. Biol. Chem. 2000;275:20949–20955. doi: 10.1074/jbc.M909135199. [DOI] [PubMed] [Google Scholar]
- [46].Matson SW, Capaldo-Kimball FN, Bambara RA. On the processive mechanism of Escherichia coli DNA Polymerase I. The polA5 mutation. J. Biol. Chem. 1978;253:7851–7856. [PubMed] [Google Scholar]
- [47].Camps M, Loeb LA. When pol I goes into high gear: processive DNA synthesis by pol I in the cell. Cell Cycle. 2004;3:116–118. [PubMed] [Google Scholar]
- [48].Sung JS, Mosbaugh DW. Escherichia coli uracil- and ethenocytosine-initiated base excision DNA repair: rate-limiting step and patch size distribution. Biochemistry. 2003;42:4613–4625. doi: 10.1021/bi027115v. [DOI] [PubMed] [Google Scholar]
- [49].Robertson AB, Matson SW. Reconstitution of the very short patch repair pathway from Escherichia coli. J. Biol. Chem. 2012;287:32953–32966. doi: 10.1074/jbc.M112.384321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Walsh E, Eckert KA. Eukaryotic replicative DNA polymerases. In: Murakami KS, Trakselis MA, editors. Nucleic Acid Polymerases, Nucleic Acids and Molecular Biology. Springer-Verlag; Heidelberg: 2014. pp. 17–41. [Google Scholar]
- [51].Shen Y, Koh KD, Weiss B, Storici F. Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat. Struct. Mol. Biol. 2012;19:98–104. doi: 10.1038/nsmb.2176. [DOI] [PubMed] [Google Scholar]
- [52].Cai Y, Geacintov NE, Broyde S. Ribonucleotides as nucleotide excision repair substrates. DNA Repair. 2014;13:55–60. doi: 10.1016/j.dnarep.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Arana ME, Kerns RT, Wharey L, Gerrish KE, Bushel PR, Kunkel TA. Transcriptional responses to loss of RNase H2 in Saccharomyces cerevisiae. DNA Repair. 2012;11:933–941. doi: 10.1016/j.dnarep.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim N, Cho JE, Li YC, Jinks-Robertson S. RNA:DNA hybrids initiate quasi-palindrome-associated mutations in highly transcribed yeast DNA. PLoS Genet. 2013;9:e1003924. doi: 10.1371/journal.pgen.1003924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cerritelli SM, Chon H, Crouch RJ. Molecular biology. A new twist for topoisomerase. Science. 2011;332:1510–1511. doi: 10.1126/science.1208450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J.Exp. Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bergoglio V, Ferrari E, Hubscher U, Cazaux C, Hoffmann JS. DNA polymerase β can incorporate ribonucleotides during DNA synthesis of undamaged and CPD-damaged DNA. J. Mol. Biol. 2003;331:1017–1023. doi: 10.1016/s0022-2836(03)00837-4. [DOI] [PubMed] [Google Scholar]
- [60].Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffmann JS. Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12586–12590. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bergoglio V, Pillaire MJ, Lacroix-Triki M, Raynaud-Messina B, Canitrot Y, Bieth A, Gares M, Wright M, Delsol G, Loeb LA, Cazaux C, Hoffmann JS. Deregulated DNA polymerase β induces chromosome instability and tumorigenesis. Cancer Res. 2002;62:3511–3514. [PubMed] [Google Scholar]
- [62].Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. U.S.A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases α and δ. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]