Abstract
The translesion DNA synthesis (TLS) polymerase REV1 is implicated in the bypass of the irreparable DNA damage such as interstrand crosslinks (ICLs). However, the potential role of REV1 in DNA damage response (DDR) pathway has not been determined. In this research communication, we provide evidence to demonstrate that REV1 plays a previously unidentified but important role in the ATR-Chk1 checkpoint activation in response to mitomycin C (MMC)-induced ICLs in Xenopus egg extracts. We further pinpointed that REV1 plays a downstream role of a checkpoint protein complex assembly including ATR, ATRIP, TopBP1 and the Rad9-Rad1-Hus1 complex to MMC-induced ICLs on chromatin in the DDR pathway. Notably, domain dissection analysis demonstrates that a C-terminal domain, but not the individual ubiquitin binding motifs, of REV1 is important for the binding of REV1 to MMC-damaged chromatin and the MMC-induced Chk1 phosphorylation. Yet, the ATR-Chk1 DDR pathway appears to be dispensable for the preferential association of REV1 to MMC-damaged chromatin. Taken together, REV1 is important for the DDR pathway in Xenopus egg extracts.
Keywords: ATR, Chk1, DNA damage response, REV1, TLS
1. Introduction
The genome of organisms is exposed to a variety of insults from cellular metabolism byproducts and environmental toxins, leading to several different types of DNA damage, including interstrand crosslinks (ICLs), double-strand breaks (DSBs), and single-strand breaks (SSBs) [1,2]. Lesions generated from ICLs are extremely cytotoxic, as irreparable ICLs prevent DNA replication and transcription, thereby threatening genome stability. Although DNA crosslinking agents, such as mitomycin C (MMC), are widely used in chemotherapy, tumor cells can develop resistance to such agents, possibly through bypass of the ICLs [3]. The DNA damage response (DDR) pathway is a surveillance mechanism used by cells to coordinate cell cycle progression, transcription activation, and activation of apoptosis and senescence [1,4]. ATM-Chk2 and ATR-Chk1 pathways are the two major DDR pathways in response to DNA damage or replication stress from endogenous or exogenous sources [5]. ATM is activated with the help of other mediator proteins in response to DSBs [6–8]. ATR activation requires RPA-bound single-strand DNA (ssDNA) and 5-primed ssDNA/dsDNA (double-strand DNA) junctions, which are derived from the functional uncoupling of MCM helicase and DNA polymerase activities, DSB end resection in the 5′–3′ direction, or SSB end resection in the 3′–5′ direction [4,9–11]. ATR activation also requires several mediator proteins including ATRIP (ATR-interaction protein), TopBP1 and the Rad9-Rad1-Hus1 (9-1-1) complex [12–15]. A variety of substrates including Chk1 are phosphorylated by the activated ATR kinase [16]. Our understanding of ICL repair and signaling pathways has progressed with the use of a defined plasmid-based ICL in Xenopus egg extracts [17,18]. However, it is still not completely understood how exactly ICLs activate the ATR-Chk1-mediated DDR pathway.
When DNA lesions cannot be replicated by high-fidelity replicative DNA polymerases (Pol δ/ε), they can be bypassed by low-fidelity translesion DNA synthesis (TLS) polymerases, increasing the risk of mutagenesis as a tradeoff for survival [19,20]. TLS polymerases include the Y-family DNA polymerases (REV1, Pol η, Pol κ and Pol ι) and a B-family DNA polymerase Pol ζ [21,22]. Although the REV1 protein has deoxycytidyl transferase activity that transfers a dCMP to a damaged nucleotide in an error-free fashion, its non-catalytic function may play an essential role in mutagenesis and cell survival, possibly through its interaction with other TLS polymerases via a C-terminal fragment [23,24]. As REV1 lacks an obvious PCNA-interaction protein box (PIP box), this TLS protein may be recruited to damage sites through its unique N-terminal BRCT domain and ubiquitin-binding motifs (UBMs) [25,26]. Together with Pol ζ, REV1 facilitates various DNA repair programs including ICL repair and homologous recombination of DSBs, promoting or preventing genome instability [27,28]. However, it has not been determined whether REV1 plays role in the ICL-induced ATR-Chk1 DDR pathway.
It is pivotal to understand how exactly TLS and DDR pathways regulate each other, as the dependency and regulation between them is a long-standing question in the field of genome integrity. The 9-1-1 complex associates with DinB (yeast homologue of Pol κ) and may regulate the recruitment of DinB to damage sites in fission yeast; however, it has not been tested whether the ATR kinase itself regulates DinB [29]. REV1 phosphorylation by Mec1 (yeast homologue ATR) is important for the Pol ζ-mediated TLS of UV damage in nucleotide excision repair-deficient, but not wild type, budding yeast cells [30]. Mec1 also mediates the recruitment of REV1 to a DSB site in budding yeast [31]. However, the putative Mec1 phosphorylation sites of REV1 are lacking in higher eukaryotic organisms including humans and Xenopus [32]. Although the DDR pathway may regulate the TLS pathway under some circumstances, it was demonstrated in a recent report that Pol κ actually contributes to ATR-Chk1 DDR pathway activation induced by stalled replication forks, suggesting a complicated regulation between TLS and DDR pathways [29,30,33]. It remains unknown, however, whether or not REV1 and ATR-Chk1 DDR pathways regulate each other and how this regulation might occur in response to ICLs in higher eukaryotes. Xenopus egg extracts has been demonstrated as an excellent cell-free model system for studies of ICL repair and DDR pathways [17,18,34]. In this communication, our compelling evidence suggests that REV1 plays a previously unidentified, but important, role in the activation of MMC-induced ATR-Chk1 DDR pathway in Xenopus egg extracts. Furthermore, we pinpointed the step of REV1 in the ATR-Chk1 DDR pathway and dissected the necessary domain within REV1 for such an important function. In contrast, ATR-Chk1 plays a negligible role for the recruitment of REV1 to ICLs on chromatin in Xenopus egg extracts.
2. Materials and Methods
2.1 Xenopus egg extract and related procedures
The use and care of Xenopus laevis were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Charlotte. Low-speed supernatant egg extract was prepared according to our previously published approach [10,35,36]. Sperm chromatin was added to egg extracts at a concentration of approximately 4000 sperm/μL. Chromatin fractions were isolated and examined as previously described [14]. Caffeine, KU55933, VE-822, and NU6027 were added to egg extracts at final concentrations of 1 ng/μL, 120 μM, 10 μM, and 1 mM, respectively [10,37,38]. Aphidicolin (APH) and Mitomycin C (MMC) were added to egg extracts at final concentrations of 100 ng/μL and 0.5 mM as previously described [14,34]. Immunodepletion of REV1 was performed in a similar fashion as previously described [14].
2.2 Expression vectors, recombinant proteins, and antibodies
His-REV1-NT300, corresponding to Xenopus laevis REV1 (MGC: 83743) nucleotide 246-1145, was cloned into the pET28a expression vector. Recombinant His-REV1-NT300 was expressed and purified from DE3 bacteria cells. Anti-REV1 antibodies were raised in rabbit against His-REV1-NT300 (Cocalico Biologicals). Wild type full length (WT) Myc-tagged REV1 corresponding to REV1 nucleotide 246-3938 was cloned into the pCS2+MT vector. ΔUBM1, ΔUBM2, and ΔCTD Myc-tagged REV1 variants were derived from WT Myc-REV1, in which nucleotides 2991-3086, 3225-3327, and 3513-3834 were removed, respectively. Recombinant WT and deletion versions of Myc-tagged REV1 were expressed in SP6 TnT transcription/translation coupled quick master mix kit (Promega). Antibodies against Xenopus ATR, ATRIP, TopBP1, Rad9, RPA32, and Orc2 have been described previously [10,14]. Additionally, antibodies against Chk1 P-S344 (Cell Signaling), Chk1 (Santa Cruz), c-Myc (Santa Cruz), GST (Santa Cruz), Histone 3 (Abcam), and PCNA (Santa Cruz) were purchased from respective vendors. Peroxidase-conjugated monoclonal mouse anti-rabbit IgG light chain specific (Jackson ImmunoResearch), peroxidase-conjugated goat anti-rabbit IgG (Thermo), and peroxidase-conjugated sheep anti-mouse IgG (GE Healthcare) were used as secondary antibodies in immunoblotting analysis as appropriate.
3. Results
3.1 Chk1 phosphorylation was triggered in response to MMC-induced ICLs in an ATR-dependent but ATM-independent manner in Xenopus egg extracts
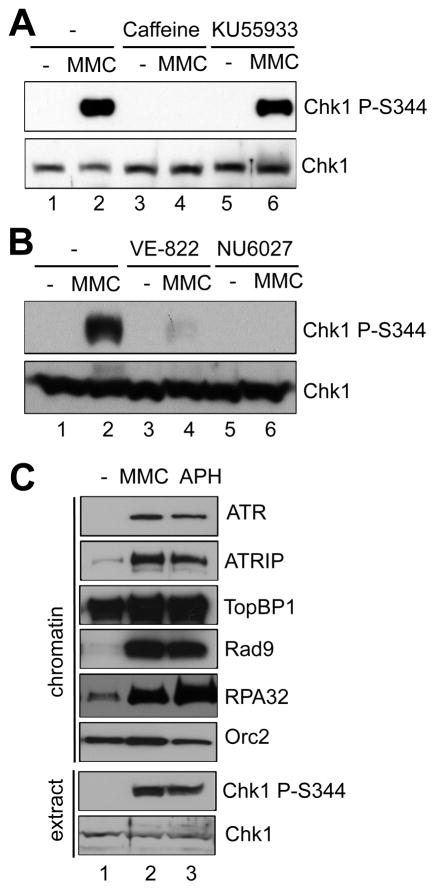
Chk1 phosphorylation at Serine 344 (Chk1 P-S344) is widely used as an indicator of ATR activation [18,39]. Chk1 phosphorylation was triggered by MMC treatment in Xenopus egg extracts (Fig. 1A), consistent with our recently published study [34]. The MMC-induced Chk1 phosphorylation was compromised by the ATR/ATM inhibitor, caffeine, but not the ATM specific inhibitor, KU55933 (Fig. 1A) [10]. Furthermore, MMC-induced Chk1 phosphorylation was compromised by the ATR specific inhibitors, VE-822 or NU6027 (Fig. 1B) [37,38]. These observations suggest that MMC-induced ICLs trigger Chk1 phosphorylation in an ATR-dependent, but ATM-independent fashion. To compare the MMC-induced ICLs and aphidicolin-induced stalled replication forks, we examined the recruitment of several key checkpoint proteins to chromatin in Xenopus egg extracts. Aphidicolin inhibits DNA polymerase activities, leading to stalled DNA replication forks, functional uncoupling of the helicase and polymerase, and the ATR-Chk1 DDR pathway activation [40]. As expected, aphidicolin triggered Chk1 phosphorylation and the assembly of a checkpoint complex containing ATR, ATRIP, and Rad9 in response to stalled replication forks (Fig. 1C) [14]. Orc2 was used as a loading control. Notably, the checkpoint protein complex (ATR, ATRIP, TopBP1, and Rad9) was also assembled onto MMC-damaged chromatin, suggesting DNA replication forks are stalled by MMC-induced ICLs (Fig. 1C). In addition, RPA32 was hyperloaded onto MMC-damaged chromatin, suggesting that long stretches of ssDNA are generated around ICL sites (Fig. 1C). These observations suggest that when DNA replication forks stall at ICLs, ssDNA is generated and bound by RPA, and that a checkpoint protein complex is recruited to ICL sites to trigger the ATR-Chk1 DDR pathway.
Fig. 1.
Chk1 phosphorylation and a checkpoint protein complex assembly in response to MMC-induced ICLs in Xenopus egg extract. (A) Caffeine or KU55933 was incubated in egg extracts supplemented with sperm chromatin and mitomycin C (MMC). After 1-hr incubation, Chk1 phosphorylation (Chk1 P-S344) and total Chk1 in extracts were examined via immunoblotting analysis. (B) VE-822 or NU6027 was incubated in egg extracts supplemented with sperm chromatin and MMC. After 1-hr incubation, Chk1 phosphorylation and total Chk1 in extracts were examined via immunoblotting analysis. (C) MMC or aphidicolin (APH) was incubated in egg extracts supplemented with sperm chromatin. After 1-hr incubation, chromatin fractions (“chromatin”) and total extract (“extract”) were isolated and examined via immunoblotting analysis as indicated. Orc2 was used as a loading control.
3.2 REV1 was preferentially recruited to MMC-induced ICLs in an ATR-Chk1-independent fashion
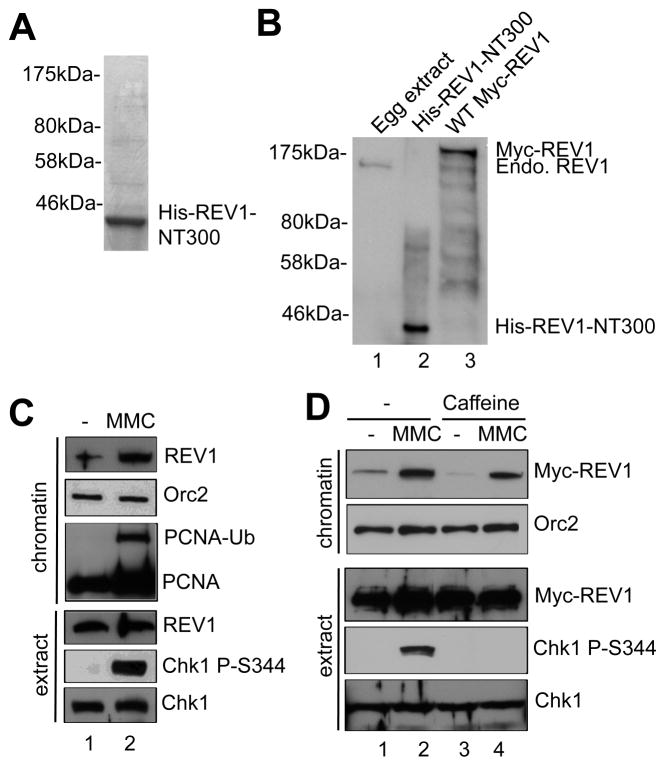
To study the role of REV1 in the MMC-induced DDR pathway, we expressed and purified His-tagged recombinant N-terminal 300-amino-acid fragment of REV1 (designated as His-REV1-NT300) from bacteria, as shown on SDS-PAGE gel (Fig. 2A). His-REV1-NT300 protein was then used to generate customized antibodies against REV1 in rabbits, which recognized endogenous REV1 in Xenopus egg extracts, recombinant His-REV1-NT300, and Myc-tagged wild type REV1 (WT Myc-REV1) (Figs. 2B and 4A). Notably, endogenous REV1 and recombinant WT Myc-REV1 were preferentially recruited to MMC-damaged chromatin in Xenopus egg extracts (Figs. 2C and 2D). PCNA was monoubiquitinated after MMC-treatment (Fig. 2C). These observations suggest that REV1 preferentially associates with MMC-induced ICL sites. Although caffeine compromised MMC-induced Chk1 phosphorylation, this ATR/ATM inhibitor had no noticeable effect on the recruitment of Myc-REV1 to MMC-damaged chromatin, suggesting that the ATR-Chk1 DDR pathway is not required for the recruitment of REV1 to MMC-induced ICLs in Xenopus at least under our experimental conditions (Fig. 2D).
Fig. 2.
REV1 preferentially associates with MMC-damaged chromatin. (A) Purified His-REV1-NT300 was examined via SDS-PAGE and Coomassie blue staining. Molecular weight markers were also labeled. (B) Xenopus egg extract, recombinant His-REV1-NT300 and SP6 TnT expressed WT Myc-REV1 were examined via immunoblotting analysis using anti-REV1 antibodies. “Endo. REV1” represents endogenous REV1. (C) MMC was incubated in egg extracts supplemented with sperm chromatin. After 1-hr incubation, chromatin fractions and total extract were isolated and examined via immunoblotting analysis. “PCNA-Ub” denotes monoubiquitinated PCNA. (D) WT Myc-REV1 was incubated in egg extracts at a similar concentration as the endogenous REV1, which was followed by addition of MMC and Caffeine. After 1-hr incubation, chromatin fractions and total extract were analyzed via immunoblotting analysis as indicated. Orc2 was used as a loading control.
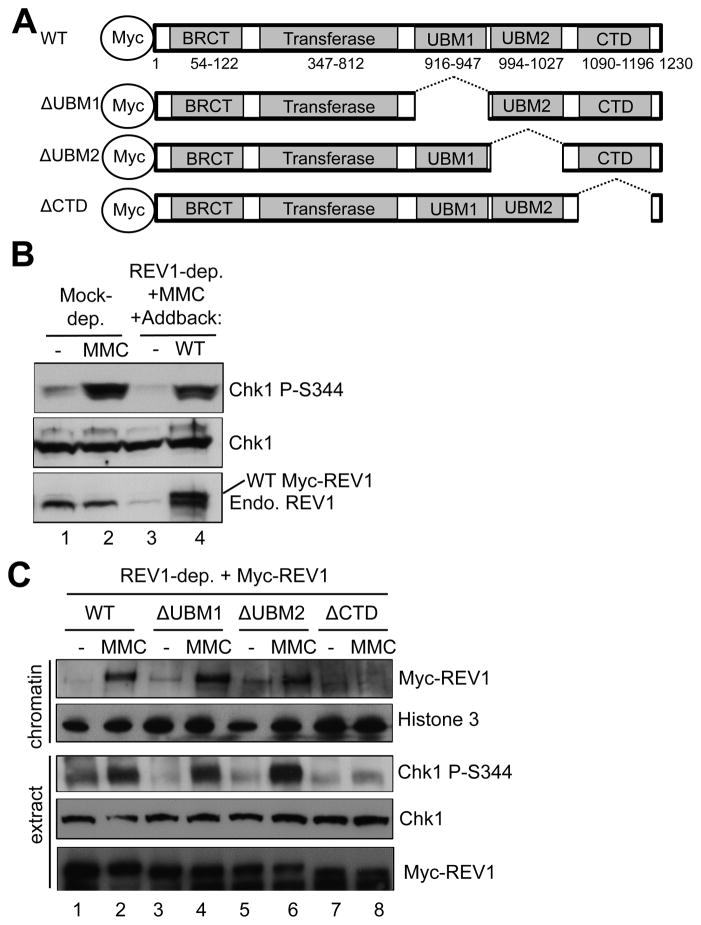
Fig. 4.
Domain dissection of REV1 for association with MMC-damaged chromatin and MMC-induced Chk1 phosphorylation. (A) Schematic demonstration of WT REV1 and domain deletion versions of REV1. (B) WT Myc-REV1 (“WT”) or an egg lysis buffer (“−”) was added to REV1-depleted egg extract, which was supplemented with sperm chromatin and MMC. Total extracts were examined for indicated proteins via immunoblotting analysis. “Endo. REV1” represents endogenous REV1. (C) WT REV1 or depletion mutant versions of REV1 were individually added back to REV1-depleted egg extract supplemented with sperm chromatin and MMC. Chromatin fractions and total extracts were analyzed for indicated proteins via immunoblotting analysis. Histone 3 was a loading control.
3.3 REV1 was important for the MMC-induced ATR-Chk1 DDR pathway
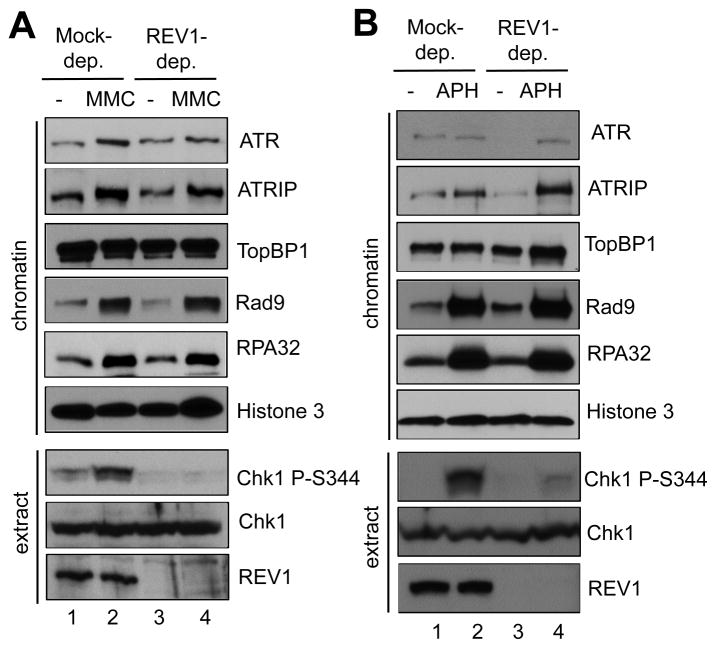
To test the potential role of REV1 in the MMC-induced Chk1 phosphorylation, we successfully immunodepleted endogenous REV1 from egg extract with customized anti-REV1 antibodies (Fig. 3A). Notably, the MMC-induced Chk1 phosphorylation was compromised significantly in the REV1-depleted egg extract, suggesting that REV1 plays an important role in the activation of the ATR-Chk1 DDR pathway (Fig. 3A). Interestingly, Chk1 phosphorylation induced by stalled replication forks was also compromised in the REV1-depleted egg extract, suggesting the novel role of REV1 in the ATR-Chk1 DDR pathway in response to replication stress (Fig. 3B). These observations suggest that REV1 is important for the ATR-Chk1 DDR pathway activation in response to both ICLs and stalled DNA replication forks.
Fig. 3.
REV1 is important for Chk1 phosphorylation in response to MMC-induced ICLs and stalled DNA replication forks. (A) MMC was incubated in mock-depleted or REV1-depleted egg extracts supplemented with sperm chromatin. After 1-hr incubation, chromatin fractions and total extracts were examined via immunoblotting analysis as indicated. (B) APH was incubated in mock-depleted or REV1-depleted egg extracts supplemented with sperm chromatin. After 1-hr incubation, chromatin fractions and total extracts were analyzed via immunoblotting analysis. Histone 3 was used as a loading control.
To determine the precise role of REV1 in the ATR-Chk1 DDR pathway, we examined the assembly of the checkpoint protein complex onto MMC-damaged chromatin. The recruitment of ATR, ATRIP, TopBP1, and Rad9 to MMC-damaged chromatin had no noticeable change in REV1-depleted egg extract in comparison with mock-depleted egg extract (Fig. 3A). RPA32 was hyperloaded to MMC-damaged chromatin in REV1-depleted egg extract, suggesting that REV1 has no effect on ssDNA generation. The recruitment of ATR, ATRIP, TopBP1, and Rad9 to aphidicolin-induced stalled replication forks demonstrated no noticeable change in REV1-depleted egg extract when compared to mock-depleted egg extract (Fig. 3B). Histone 3 was used as a loading control in these experiments. Similarly, REV1 depletion had no noticeable effect on RPA32 hyperloading to stalled replication forks (Fig. 3B). To confirm that the Chk1 phosphorylation defect by REV1 depletion is because of the absence of REV1, we added WT Myc-REV1 to REV1-depleted egg extract at a similar concentration as the endogenous REV1 and found that MMC-induced Chk1 phosphorylation was partially rescued (Figs. 4A and 4B). These observations suggest that REV1 may play a role downstream of the checkpoint protein complex assembly, but before Chk1 is phosphorylated by ATR in the DDR pathway and that the role of REV1 in the DDR pathway is not limited to MMC-induced ICLs.
3.4 A C-terminal domain (CTD) of REV1 was required for the recruitment of REV1 to MMC-damaged chromatin and the MMC-induced Chk1 phosphorylation
REV1 contains several distinct domains: BRCT (aa 54-122), dCMP transferase (aa 347-812), UBM1 (aa 916-947), UBM2 (aa 994-1027), and CTD (aa 1090-1193) (Fig. 4A). Through its dCMP transferase domain, REV1 protein transfers a dCMP opposite a damaged nucleotide [23,41]. The UBM1 and UBM2 are important for the binding of REV1 to the ubiquitin moiety of monoubiquitinated PCNA [42]. The CTD fragment of REV1 associates with other TLS proteins, such as Pol η, Pol ζ, and Pol κ [43,44]. To directly test the role of the UBMs and CTD region of REV1 for the DDR pathway, several deletion mutant versions of REV1 were added to REV1-depleted egg extract (Fig. 4A). Surprisingly, ΔUBM1 REV1 and ΔUBM2 REV1 retained binding capacity to MMC-damaged chromatin similar to that of WT REV1. Furthermore, WT REV1, ΔUBM1 REV1 and ΔUBM2 REV1 rescued the MMC-induced Chk1 phosphorylation in the REV1-depleted egg extract (Fig. 4C). However, ΔCTD REV1 failed to rescue MMC-induced Chk1 phosphorylation in the REV1-depleted egg extract (Fig. 4C). Notably, the binding of ΔCTD REV1 to MMC-damaged chromatin was also compromised (Fig. 4C). These observations suggest that recruitment of REV1 to MMC-damaged chromatin requires its CTD domain, but not UBMs, and that CTD of REV1 is essential for the activation of the ATR-Chk1 pathway in response to MMC-induced ICLs in Xenopus egg extracts.
4. Discussion
It is important to investigate the regulatory mechanisms between the DDR and TLS pathways. In this research communication, we report that the ATR-Chk1 DDR pathway is activated by MMC-induced ICLs on chromatin and that REV1 is important for the MMC-induced DDR pathway in Xenopus egg extracts. In addition, ATR-Chk1 pathway is dispensable for the preferential binding of REV1 to MMC-damaged chromatin. We further demonstrate that REV1 may function downstream of the assembly of the checkpoint protein complex containing ATR, ATRIP, and the 9-1-1 complex onto MMC-induced ICLs. Domain dissection analysis reveals that the CTD of REV1 is important for its recruitment onto MMC-induced ICLs and for its role in the ATR-Chk1 DDR pathway.
It was recently reported that Pol κ, but not Pol η, is important for the ATR-Chk1 DDR pathway induced by stalled DNA replication forks in both humans and Xenopus [33]. Furthermore, it was shown that Pol κ is implicated in the synthesis of short DNA intermediates at stalled replication forks, facilitating the recruitment of the 9-1-1 complex [33]. Here we show that another TLS member REV1 is important for the ATR-Chk1 DDR pathway in response to MMC-induced ICLs and stalled DNA replication forks (Fig. 3). The association of the 9-1-1 complex to MMC-induced ICLs and stalled replication forks was not compromised in the absence of REV1, suggesting that REV1 plays a role downstream of the assembly of the checkpoint protein complex. More studies are needed to clarify the potential regulation between Pol κ and REV1 in the DDR pathway.
It is important to determine how exactly REV1 is recruited to damaged sites. Since REV1 lacks a PIP box motif for PCNA association, it has been suggested that REV1 may associate with a ubiquitin moiety in monoubiquitinated PCNA via its UBMs [42]. Our observation in Figure 4 does not rule out the possibility that REV1 may have a defect in MMC-induced Chk1 phosphorylation when both UBM1 and UBM2 are removed. The CTD fragment of REV1 associates with other TLS polymerases such as Pol η, Pol ζ, and Pol κ [43,44]. Thus, it is also possible that REV1 is recruited by other TLS polymerases via its CTD fragment. In addition, it was recently reported that REV1 CTD forms a quaternary complex consisting of REV1, Pol ζ, and Pol κ [45]. The requirement of CTD for REV1 binding to MMC-damaged chromatin further supports the significance of the CTD of REV1 in the DDR pathway.
More investigation is necessary to test whether the important role of REV1 in the DDR pathway is conserved in humans or other model organisms. COSMIC (Catalogue of Somatic Mutations in Cancer) analysis reveals 100 missense, 7 nonsense, and 44 synonymous substitutions, as well as 3 insertion/deletion frame shifts in REV1 in various cancer patients including breast, ovary, large intestine, endometrium, lung, and pancreas carcinoma patients. These findings further highlight the significance of REV1 in the DDR pathway. Our findings from this work may elucidate novel avenues for cancer treatment through manipulation of REV1’s distinct role in the DDR pathway. Taken together, our presented evidence suggests that REV1 is important for the ATR-Chk1 DDR pathway in response to MMC-induced ICLs in Xenopus egg extracts.
Supplementary Material
Highlights.
REV1 preferentially associates with MMC-damaged chromatin in Xenopus.
REV1 is important for the Chk1 phosphorylation in response to MMC-induced ICLs.
A C-terminal motif of REV1 is important for ICL binding and Chk1 phosphorylation.
Acknowledgments
The Yan lab is supported, in part, by funds provided by the University of North Carolina at Charlotte (FRG 111764) and a grant from the NIGMS/NIH (R15 GM101571). We thank Drs. Matthew Michael, Karlene Cimprich, and Howard Lindsay for reagents. We thank Drs. Chandra Williams and Yvette Huet and the UNC Charlotte Vivarium staff (Alvaro Perez, Bryan Porter, and Hernando Gordils) for the care of our frogs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long DT, Walter JC. A novel function for BRCA1 in crosslink repair. Mol Cell. 2012;46:111–112. doi: 10.1016/j.molcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 8.Masai H. ATM in prevention of genomic instability. Cell Cycle. 2014;13:882–883. doi: 10.4161/cc.28216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan S, Sorrell M, Berman Z. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol Life Sci. 2014;71:3951–3967. doi: 10.1007/s00018-014-1666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willis J, Patel Y, Lentz BL, Yan S. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc Natl Acad Sci USA. 2013;110:10592–10597. doi: 10.1073/pnas.1301445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 13.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 14.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Sanchez Y. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair (Amst) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol Cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang DJ, Cimprich KA. DNA damage tolerance: when it’s OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TV, Scharer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 22.You C, Swanson AL, Dai X, Yuan B, Wang J, Wang Y. Translesion synthesis of 8,5′-cyclopurine-2′-deoxynucleosides by DNA polymerases eta, iota, and zeta. J Biol Chem. 2013;288:28548–28556. doi: 10.1074/jbc.M113.480459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 24.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 25.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 26.Bomar MG, D’Souza S, Bienko M, Dikic I, Walker GC, Zhou P. Unconventional ubiquitin recognition by the ubiquitin-binding motif within the Y family DNA polymerases iota and Rev1. Mol Cell. 2010;37:408–417. doi: 10.1016/j.molcel.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res. 2012;40:682–691. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Scharer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pages V, Santa Maria SR, Prakash L, Prakash S. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 2009;23:1438–1449. doi: 10.1101/gad.1793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pustovalova Y, Maciejewski MW, Korzhnev DM. NMR mapping of PCNA interaction with translesion synthesis DNA polymerase Rev1 mediated by Rev1-BRCT domain. J Mol Biol. 2013;425:3091–3105. doi: 10.1016/j.jmb.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 33.Betous R, Pillaire MJ, Pierini L, van der Laan S, Recolin B, Ohl-Seguy E, Guo C, Niimi N, Gruz P, Nohmi T, Friedberg E, Cazaux C, Maiorano D, Hoffmann JS. DNA polymerase kappa-dependent DNA synthesis at stalled replication forks is important for CHK1 activation. EMBO J. 2013;32:2172–2185. doi: 10.1038/emboj.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai L, Michael WM, Yan S. Importin beta-dependent nuclear import of TopBP1 in ATR-Chk1 checkpoint in Xenopus egg extracts. Cell Signal. 2014;26:857–867. doi: 10.1016/j.cellsig.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willis J, Destephanis D, Patel Y, Gowda V, Yan S. Study of the DNA damage checkpoint using Xenopus egg extracts. J Vis Exp. 2012:e4449. doi: 10.3791/4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan S, Willis J. WD40-repeat protein WDR18 collaborates with TopBP1 to facilitate DNA damage checkpoint signaling. Biochem Biophys Res Commun. 2013;431:466–471. doi: 10.1016/j.bbrc.2012.12.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, Vallis KA, Hammond EM, Olcina MM, Gillies McKenna W, Muschel RJ, Brunner TB. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, Cliby WA, Sarkaria J, Beale G, Edmondson RJ, Curtin NJ. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105:372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Wang J, Zhang Y, Wang Z. The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis. Nucleic Acids Res. 2010;38:5036–5046. doi: 10.1093/nar/gkq225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 44.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojtaszek J, Lee CJ, D’Souza S, Minesinger B, Kim H, D’Andrea AD, Walker GC, Zhou P. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) zeta, and Pol kappa. J Biol Chem. 2012;287:33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.